Abstract
ATP-binding-cassette (ABC) subunit MalK of the binding protein-dependent transport system for maltose of Salmonella typhimurium and Escherichia coli is crucial to the transport process but also exhibits a repressing activity on other genes of the maltose regulon. The latter function has been attributed to a carboxy-terminal extension by which MalK differs in length from a prototype ABC protein. In order to define the boundaries of putative functional domains of MalK, we have analyzed pairs of N- and C-terminally truncated MalK proteins of S. typhimurium. Coexpressed half molecules of about equal lengths (MalKN1: residues 1 to 179; MalKC1: residues 179 to 369) restored the transport activity of a malK strain and displayed substantial regulatory activity. The same regulatory activity was obtained when malKC1 was expressed separately. These results indicate that a covalent linkage is not absolutely essential for function and that the protein might be composed of two structurally distinct entities. To elucidate further the minimal structural requirements for the regulatory function of MalK, we have studied chimeric proteins that have C-terminal portions of MalK fused to the corresponding amino-terminal fragments of its close homolog LacK. Functional analyses revealed that a fusion containing only the C-terminal extension of MalK (Q263 to V369) is sufficient to display half-maximal regulatory activity. This activity increased with the lengths of the MalK portions present in the chimeras. Furthermore, the failure of two chimeras to support maltose transport suggests a structurally critical region between residues 243 and 264. In the absence of a crystal structure, this work contributes to the understanding of the multiple functions of MalK.
The rapidly growing family of ATP-binding-cassette (ABC) transport systems (14) (or traffic ATPases [1]) comprises an extremely diverse class of membrane transport proteins that couple the energy of ATP hydrolysis to the translocation of solutes across biological membranes. Members of this family not only accomplish the uptake of nutrients but also are involved in a large variety of processes, such as signal transduction, protein secretion, drug and antibiotic resistance, antigen presentation, bacterial pathogenesis, and sporulation (10). A prototype ABC transporter is composed of four parts: two membrane-integral domains, each of which spans the membrane six times, and two ATP-hydrolyzing domains (also referred to as ABC subunits or domains) (10, 28). While the membrane-spanning domains presumably constitute a translocation pore, the ABC domains are thought to provide the energy for the transport process. In eukaryotic systems, these modules are mostly fused to yield a single polypeptide chain, while bacterial ABC transporters are built up from individual subunits.
The binding protein-dependent transport system for maltose and maltodextrins of enterobacteria, such as Escherichia coli and Salmonella typhimurium, represents one of the best-studied (by genetic, molecular genetic, and biochemical means) members of the family (3). The membrane-bound complex is composed of one copy each of the integral membrane proteins MalF and MalG and two copies of ABC subunit MalK (5). Both the complete transport systems as well as the MalK protein have been purified and characterized (5, 6, 19, 32). Besides being indispensable for the transport process, the MalK protein displays additional regulatory functions. MalK, when overproduced, acts as a repressor of genes belonging to the maltose regulon (23), while deletion of the malK gene results in constitutive expression of these genes (4, 12). The mechanism of this activity is unknown, but interference of MalK with MalT, the positive regulator of the maltose regulon, has been suggested (8). Furthermore, MalK is the target of enzyme IIAGlc of the phosphoenolpyruvate phosphotransferase system for glucose in the process of inducer exclusion (7). Mutations that abolish these functions have been localized to the C-terminal extension of about 100 residues by which MalK differs in length from a prototype ABC domain (7, 15). These findings, together with the observation that an N-terminal fragment of MalK can be exchanged with a corresponding fragment of the homologous HisP protein without losing transport function (29), have led to the proposal of a domain structure of MalK (34), as presented in Fig. 1. In order to investigate, in the absence of a crystal structure, whether the separation of functional entities is reflected in structurally distinct domains, we have constructed and characterized N- and C-terminally truncated MalK proteins as well as chimeras of MalK with its close homolog LacK (Fig. 2). Our results demonstrate that (i) N- and C-terminal half molecules of MalK can support transport in the absence of a covalent linkage, indicating a two-domain structure of the protein, and (ii) a C-terminal fragment (Q263 to V369) is sufficient to allow half-maximal regulatory activity. The latter finding provides experimental evidence in support of the notion that the regulatory activity mainly resides in the C-terminal extension of MalK.
FIG. 1.
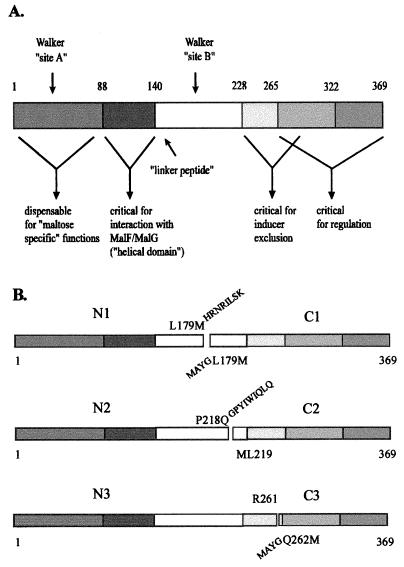
Structures of the truncated MalK proteins. (A) Linear representation of MalK with the assignment of putative functional domains (see text and reference 33 for details). (B) Structures of fragment pairs indicating the sites of separation relative to the functional domains. The last amino acid of each N-terminal fragment is shown above the respective bar, while the first residue of each C-terminal fragment is shown below. The numbers refer to the positions of the residues in mature MalK. Amino acid residues derived from the vector sequence are indicated.
FIG. 2.
Alignment of protein sequences of MalK of S. typhimurium and E. coli and LacK of A. radiobacter. Conserved sequence motifs of ABC proteins and the sites at which the MalK protein was split into fragment pairs are indicated above the alignment. LM1 to LM4 denote the joining points of the LacK′-′MalK chimeras described in the text. Sources of protein sequences are as follows: MalK of S. typhimurium (MALK STY), SwissProt accession no. P19566, corrected at position 141; MalK of E. coli (MALK ECO), SwissProt accession no. P02914; LacK of A. radiobacter (LACK ARA), SwissProt accession no. Q01937.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All cloning steps were performed with E. coli JM109 [recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] or TG1 [Δ(lac-proAB) supE thi hsdΔ5 F′ (traD36 proAB lacIq lacZΔM15)] (25). S. typhimurium ES25 (dhuA1 ΔhisF645 malK786 galE503 recA56) (29) was used for complementation studies and transport assays. The regulatory activity of MalK and its derivatives was monitored in E. coli SK1280 (MC4100 Θ[malK::lacZ] hyb1113 recA) (15). The plasmids used in this study are listed in Table 1. Bacteria were usually grown in Luria broth (18) or nutrient broth (24), supplemented with ampicillin (50 μg ml−1), if required. For complementation studies, minimal salt medium (omitting citrate) (24) supplemented with maltose (0.5%), l-histidine (0.4 mM), and ampicillin was used.
TABLE 1.
Plasmids used in this study
| Plasmida | Descriptionb | Reference or source |
|---|---|---|
| pSE380 | Apr ptrc | Invitrogen |
| pSW7 | malK+ | 33 |
| pSW48 | lacK+ | 34 |
| pGS96-22 | lacK11 (C234→G, ΔNcoI, silent) | This work |
| pGS94-11 | malKN1 (M1–L179Mhrnrilsk) | This work |
| pGS94-08 | malKC1 (maygL179M–V369) | This work |
| pGS94-13 | malKN2 (M1–P218Qgpyiwiqlq) | This work |
| pGS94-14 | malKC2 (mL219–V369) | This work |
| pGS94-20 | malKN3 (M1–R261) | This work |
| pGS95-06 | malKC3 (maygQ262M–V369) | This work |
| pGS95-19 | malKN1 malKC1 | This work |
| pGS95-11 | malKN2 malKC2 | This work |
| pGS95-09 | malKN3 malKC3 | This work |
| pGS97-30 | lacK′-′malK1 | This work |
| pGS97-14 | lacK′-′malK2 | This work |
| pGS97-24 | lacK′-′malK3 | This work |
| pGS97-31 | lacK′-′malK4 | This work |
Plasmids other than pSE380 are all derivatives of pSE380.
Small capitals indicate amino acid residues added on to the fragment encoded by the respective plasmid.
Construction of plasmids pGS94-11 (malKN1), pGS94-8 (malKC1), and pGS95-19 (malKN1 malKC1).
First, an SphI restriction site was created in the malK gene of S. typhimurium by changing codon 179 (CTG→ATG; resulting in amino acid change L179→M) as described previously (16), by using a malK derivative of M13mp18 as the template (33). For convenience, the mutagenized malK allele was subcloned as an NcoI-EcoRI fragment into expression vector pSE380, yielding pGS94-1. To create pGS94-11, an NcoI-SphI fragment of pGS94-1, encoding an N-terminal MalK peptide (M1 to L179M) was ligated with plasmid vector pJLA602 (26), thereby acquiring a translational stop codon. Due to this procedure, the encoded MalK fragment (M1 to L179M) is extended by peptide HRNRILSK. Subsequently, an NcoI-SalI fragment was inserted into plasmid vector pSE380, yielding plasmid pGS94-11.
For construction of pGS94-8, an SphI-EcoRI fragment of pGS94-1 (encoding the corresponding C-terminal MalK peptide) was first ligated with pJLA502, to add an NcoI cloning site at the 5′ end for convenience. By this procedure, the N terminus of the encoded MalK fragment was extended by peptide MAYG. Subsequently, an NcoI-SalI fragment was introduced into pSE380, yielding pGS94-8.
To allow expression of both gene fragments from one plasmid, a PvuII fragment of pGS94-8, which encodes C1 and includes the trc promoter sequence, was ligated with pGS94-11, previously linearized by double digestion with SmaI and StuI. The resulting plasmid was named pGS95-19.
Construction of plasmids pGS94-13 (malKN2), pGS94-14 (malKC2), and pGS95-11 (malKN2 malKC2).
A PvuII restriction site was introduced at codon 218 in malK (CCG→CAG; resulting in amino acid change P218→Q) by site-directed mutagenesis (11) as described above. A 5′ fragment encoding MalK fragment (M1 to P218Q) was ligated as an NcoI-PvuII fragment into pSE380, previously digested with NcoI and SmaI, to create pGS94-13. By this procedure, the C terminus of the resulting MalK fragment is extended by nine amino acid residues (GPYIWIQLQ).
Plasmid pGS94-14 (C2) was constructed by ligating a PvuII-EcoRI fragment of the mutagenized malK allele with pSE380, previously linearized with NcoI, blunt ended by treatment with Klenow enzyme, and subsequently digested with EcoRI. By this procedure a new start codon was placed 5′ of codon 219 (encoding Leu) in malK.
Plasmid pGS95-11 expressing both gene fragments was constructed in a manner similar to that for pGS95-19 by ligating a PvuII fragment of pGS94-14 with pGS94-13, previously double-digested with SmaI and StuI.
Construction of plasmids pGS94-20 (malKN3), pGS95-6 (malKC3), and pGS95-9 (malKN3 malKC3).
The DNA fragment encoding the N3 polypeptide was amplified by PCR from pSW7 in such a way that a new stop codon and an EcoRI restriction site were created 3′ of codon 261 (encoding Arg). Subsequently, the fragment was double digested with NcoI and EcoRI and ligated with pSE380, previously digested with the same enzymes, yielding plasmid pGS94-20.
To amplify by PCR a fragment encoding the corresponding C3 peptide, oligonucleotide primers, by which an SphI site was first placed 5′ to codon 263 (encoding Gln), were designed, thereby creating a new start codon (resulting in amino acid change Q262→M). Subsequently, the DNA fragment was digested with SphI and EcoRI, passaged through plasmid vector pJLA502 to add an NcoI restriction site at the 5′ end, and eventually introduced as an NcoI-EcoRI fragment into pSE380 to yield pGS95-6. As for MalKC1, the encoded fragment peptide was extended by the peptide MAYG at the N terminus. Construction of pGS95-9 carrying both fragments was achieved essentially as described for pGS95-11, with pGS95-6 as the donor and pGS94-20 as the recipient.
Construction of lacK′-′malK fusion genes.
As a prerequisite for subsequent cloning steps, we first mutagenized nucleotide 234 (C→G; silent) in the wild-type lacK gene in pSW48 by a PCR-based protocol (11), thereby eliminating an NcoI restriction site. The resulting plasmid was named pGS96-22. The same PCR protocol was then used to construct the lacK′-′malK hybrid genes. Briefly, the respective DNA fragments of the lacK and malK genes to be fused were separately amplified with Vent polymerase (New England Biolabs) by using pGS96-22 and pSW7, respectively, as the templates. The oligonucleotide primers were designed in such a way that both fragments overlap in the region of the desired joining point. Subsequently, the PCR products were purified by agarose gel electrophoresis and combined to be used as templates in a third PCR step to amplify the hybrid gene. The resulting DNA was then purified, double digested with NcoI and EcoRI, and subcloned into pSE380. The constructs were verified by nucleotide sequence analysis of the complete hybrid genes.
Assay for regulatory activity.
The effects of truncated MalK proteins and chimeras on the expression of other maltose-inducible genes were assayed by monitoring the β-galactosidase activity of E. coli SK1280, which carries a chromosomal malK-lacZ fusion under the control of the pmalK promoter (15). Enzyme activity was measured by the method of Miller (18), modified as described in reference 9.
Miscellaneous techniques.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotting, protein determination, transformation in S. typhimurium, and transport assays were carried out as described previously (29).
RESULTS
Construction of malK deletions.
The multiple activities of the MalK protein in transport and regulation can be separated by missense mutations (15). This prompted us to test whether these activities can be assigned to structurally distinct domains of the protein by studying the functions of truncated MalK variants. To this end, plasmids that carry malK alleles of S. typhimurium, with regions coding either for the N-terminal or C-terminal portion of MalK deleted, were constructed (see Materials and Methods for details) (Table 1; Fig. 1).
Limited proteolysis of the purified MalK protein has revealed an exposed tryptic cleavage site at R185; this site was largely unaffected by conformational changes upon the binding of MgATP (30). This observation led to the construction of the truncated genes encoding the N1/C1 fragment pair (Fig. 1).
The E. coli and S. typhimurium MalK proteins are functionally interchangeable, and their primary structures are 94% identical. Most of the mismatches cluster between residues 252 and 274, including a two-residue deletion in the S. typhimurium protein (Fig. 2). Thus, another pair of truncated MalK fragments (N3/C3) was obtained by splitting the malK gene at codon 262 (Fig. 1). The C3 fragment also comprises the C-terminal 100 amino acid residues by which MalK and other closely related proteins are extended compared to the prototype ABC protein (2, 3).
In addition, a third pair (N2/C2) was constructed in order to cover the region between codons 179 and 262.
Gene fragments encoding N-terminally truncated MalK proteins were provided with termination codons by the vector sequence (N1 and N2) or by the respective oligonucleotide primer used in the PCR (N3). Initiation codons of alleles coding for carboxy-terminal MalK fragments were derived from newly inserted restriction sites (C1 and C2) or from oligonucleotide primers as described above (C3). Finally, the gene fragments were placed under the control of the trc promoter in plasmid vector pSE380, either separately or in pairs. In the latter case, the fragments were oriented in tandem, each equipped with its own trc promoter sequence.
Immunochemical detection of truncated MalK proteins.
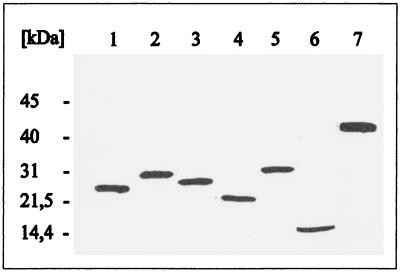
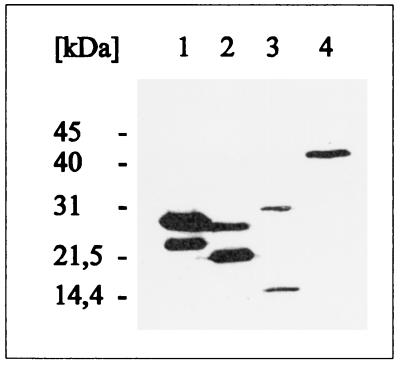
S. typhimurium ES25, which carries a defective malK gene, was subsequently transformed with the respective plasmids, and cells were then grown in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside). The total cellular proteins were separated by SDS-PAGE and subsequently analyzed by immunoblotting with a polyclonal antiserum raised against purified MalK. Individual transformants revealed the presence of unique protein bands with apparent molecular masses that corresponded favorably to those predicted from the nucleotide sequences for the truncated MalK proteins (Fig. 3). Moreover, cells harboring plasmid pGS95-19, pGS95-11, or pGS95-09 coproduced the encoded amino- and carboxy-terminal MalK fragment pairs, as shown in Fig. 4. Under these conditions, the amounts of N3 and C3 synthesized were reduced compared to those of the other pairs (Fig. 4, lane 3).
FIG. 3.
Immunoblot analysis of separately expressed malK fragments. Cells of host strain ES25 harboring the described plasmids were grown in the presence of IPTG (except pSW7; for details, see Materials and Methods). At early stationary phase, the cells were harvested and subjected to SDS-PAGE. Subsequently, the proteins were transferred to nitrocellulose and probed with a MalK-specific polyclonal antiserum. The blot was developed with the enhanced chemiluminescence kit from Amersham. Lanes: 1, MalKN1 (pGS94-11); 2, MalKC1 (pGS94-8); 3, MalKN2 (pGS94-13); 4, MalKC2 (pGS94-14); 5, MalKN3 (pGS94-20); 6, MalKC3 (pGS95-6); 7, MalK (wild type) (pSW7).
FIG. 4.
Immunoblot analysis of coproduced MalK fragments. Experimental details are as described in the legend to Fig. 3. Lanes: 1, N1 (lower band)/C1 (pGS95-19); 2, N2 (upper band)/C2 (pGS95-11); 3, N3 (upper band)/C3 (pGS95-9); 4, MalK (pSW7).
Coexpressed alleles malKN1 and malKC1 complement a malK mutation.
The transformants were then analyzed on minimal plates for their capability to grow on maltose as the sole source of carbon and energy. As anticipated, none of the plasmid-borne malK fragments encoding C-terminal portions of MalK restored the growth of ES25 (malK), since these truncated proteins lack the nucleotide binding motifs. The same phenotype was observed with plasmids carrying the corresponding fragments from the 5′ end of the gene, although all three proteins encompass the Walker A and B sites. Strikingly however, colonies comparable in size to those comprising control cells harboring the wild-type gene on the same plasmid vector (pSW7) appeared when malKN1 and malKC1 were coexpressed from plasmid pGS95-19 in the same cell. Again, none of the other fragment pairs, when coexpressed, allowed growth of the host strain on maltose. This result was confirmed by measuring the initial rate of uptake of [14C]maltose displayed by these cells (Table 2). Cells of strain ES25(pGS95-19) exhibited 81% of the transport rate measured for control cells harboring the plasmid-borne wild-type malK gene, while other combinations failed to allow uptake of the radiolabeled substrate.
TABLE 2.
Effects of separately and coexpressed malK fragments on maltose transport and mal gene expressiona
| Plasmid | Protein synthesized | Maltose uptake (% of control) | β-Galactosidase activity (% recovered) |
|---|---|---|---|
| pSE380 | None | ND | 100 |
| pSW7 | MalK | 100 | 6 ± 0.4 |
| pGS94-11 | MalKN1 | n.d. | 96 ± 3.7 |
| pGS94-8 | MalKC1 | n.d. | 46 ± 1.2 |
| pGS94-13 | MalKN2 | n.d. | 82 ± 2.7 |
| pGS94-14 | MalKC2 | n.d. | 102 ± 4.8 |
| pGS94-20 | MalKN3 | n.d. | 100 ± 4.3 |
| pGS95-6 | MalKC3 | n.d. | 95 ± 2.4 |
| pGS95-19 | MalKN1/C1 | 81 ± 1.7 | 49 ± 0.6 |
| pGS95-11 | MalKN2/C2 | ND | 102 ± 5.5 |
| pGS95-9 | MalKN3/C3 | ND | 104 ± 4.2 |
The effect of plasmid-encoded truncated MalK fragments on transport activity was measured for S. typhimurium ES25 (malK) (100% = 1.24 ± 0.1 nmol/min/109 cells). The effect on the expression of malK-lacZ was measured for E. coli SK1280 (100% = 148 ± 15 Miller units). Values are means ± standard deviations of three and four independent experiments, respectively. ND, not detectable; n.d., not determined.
These results suggested that coproduced N- and C-terminal half molecules of MalK, albeit nonfunctional as separate entities, spontaneously assemble in the absence of a covalent linkage in vivo and allow the formation of an active transport complex. The lack of activity by any of the N-terminal fragments as well as by other combinations might then be due to misfolded proteins. This conclusion is further supported by the finding that none of the fragments, even when overproduced relative to chromosomal levels, exerted a dominant-negative effect in a wild-type strain, thereby indicating a lack of interaction with MalF and MalG (data not shown). Moreover, attempts to assist folding by coexpression of the plasmid-borne groESL genes encoding E. coli chaperones were unsuccessful (data not shown). We also failed to purify the N1 fragment from inclusion bodies by a denaturation/renaturation protocol that has been established for the mature MalK protein (32). Since the procedure includes a refolding step while the protein is immobilized on a red agarose matrix, a nucleotide binding fold might not have been formed (data not shown). Thus, whether any of the N-terminal fragments would be sufficient to exhibit ATPase activity is currently unknown.
Coproduced N1/C1 fragments and C1 alone display regulatory activity.
To test for the regulatory activity of MalK, the plasmids encoding the truncated MalK variants were introduced into E. coli SK1280. This strain carries a chromosomal malK-lacZ fusion that results in a maltose-negative phenotype but places the lacZ gene under the control of the pmalK promoter (15). Thus, the repressing activities of MalK variants expressed from introduced plasmids are conveniently monitored by assaying the β-galactosidase activity of the cells (Table 2). In the presence of the vector plasmid (pSE380), full enzymatic activity was obtained, while in the presence of a plasmid carrying the malK wild-type gene (pSW7), the expression of the fusion gene was repressed (6% of vector control). Data from four independent sets of experiments revealed that cells transformed with plasmids pGS94-8 (malKC1) and pGS95-19 (malKN1 malKC1) exhibited substantial repressing activity, as only 46 and 49%, respectively, of β-galactosidase activity could be recovered. In contrast, expression of other malK deletions, whether separately or as pairs, allowed recovery of between 82 (malKN2) and 104% (malKN3 malKC3), respectively, of β-galactosidase activity, indicating no interference with gene transcription. (The consistently observed minor inhibitory effect by malKN2 cannot be explained to our satisfaction at the present stage.) Thus, the data suggest that the C-terminal half molecule of MalK not only folds in the absence of the corresponding N-terminal half but is sufficient to cause half-maximal repression of maltose-regulated genes. The finding that the activity was not improved in the presence of N1 might be explained by a need to maintain a certain conformation that can only be brought about by a covalent linkage. This might also be reflected by the failure of the fragment pair to display full transport activity.
Construction of lacK′-′malK fusions.
The regulatory activity of MalK has been proposed to reside in the C-terminal 100 amino acid residues by which MalK and related proteins are extended relative to the prototype ABC subunits of other binding protein-dependent transport systems (2, 20, 23, 35). Consistent with this notion are the locations of missense mutations in E. coli MalK that abolish this function (W267G, G302D) (15) and the observation that an S. typhimurium MalK variant truncated by the C-terminal 51 residues also has lost the repressing activity (29). Evidence, however, that a fragment encompassing the C-terminal 100 amino acids would be sufficient is still lacking. As demonstrated above, the respective MalKC3 fragment did not exhibit such an activity even when coproduced with the corresponding N-terminal portion, most likely due to misfolding. To overcome this problem, we constructed by genetic manipulations LacK′-′MalK chimeras. The LacK protein represents the ABC subunit of the binding protein-dependent transport system for lactose in Agrobacterium radiobacter (35). MalK and LacK are 40% identical and comparable in length (Fig. 2), and LacK can partially substitute for MalK in maltose transport (34). Moreover, a MalK′-′LacK fusion protein, encompassing the N-terminal 140 amino acids of MalK fused to residues 141 to 363 of LacK proved to be superior to LacK in allowing maltose transport in a malK strain (34). However, LacK displayed absolutely no repression of maltose-regulated genes in E. coli (34). Thus, we reasoned that hooking C-terminal fragments of MalK to the corresponding N-terminal portions of LacK would be a useful approach in order to avoid folding problems.
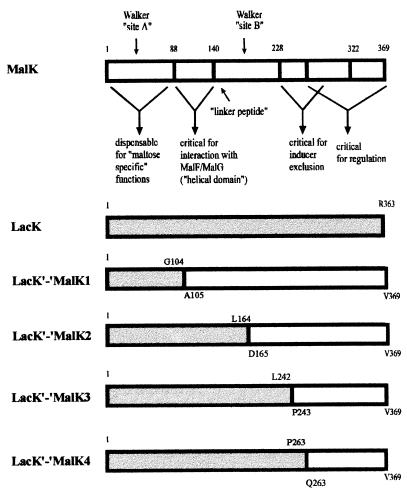
Consequently, we have constructed by a PCR-based method (11) two lacK′-′malK hybrids with joining points at codons 242 (lacK′-′malK3) and 263 (lacK′-′malK4) whose products encompass MalK fragments comparable in length to MalKC3 (Fig. 2 and 5). As controls to test for the functionality of the system, we additionally constructed by the same protocol the fusions lacK′-′malK1 and lacK′-′malK2, which contain substantially longer malK portions and should thus be active in both transport and regulation. The lacK′-′malK2 hybrid has both entities joined at codon 164, which is close to the split site that created the functional malKN1/malKC1 pair (Fig. 2). In lacK′-′malK1, a lacK fragment up to codon 104 replaces the corresponding malK portion, which has previously been shown to lack “maltose-specific” functions (29).
FIG. 5.
Structures of LacK′-′MalK chimeras. MalK- and LacK-derived sequences of the chimeras are represented by open and shaded bars, respectively. At the joining points, the last amino acid residue originating from LacK and the first amino acid residue derived from MalK are shown above and below the bars, respectively. MalK is shown with the assignment of putative functional domains (see also the legend to Fig. 1).
Complementation studies.
The plasmid-borne hybrids were introduced into strain ES25 and analyzed for growth on minimal-maltose plates. As anticipated, cells harboring fusions 1 and 2 basically grew as rapidly as bacteria expressing the wild-type gene from plasmid pSW7. In contrast, fusions 3 and 4 failed to allow growth. These results were confirmed by transport assays (Table 3). Compared to the wild-type control, chimera 1 allowed a slightly higher rate of uptake of [14C]maltose, while cells expressing chimera 2 exhibited 26% of the control rate. In agreement with our recent findings (34), this was slightly higher than the transport rate measured with cells harboring the plasmid-borne lacK gene. In contrast, chimeras 3 and 4 lacked the capability to support the transport of maltose above the background level.
TABLE 3.
Effects of lacK′-′malK hybrids on maltose transport and mal gene expressiona
| Plasmid | Protein synthesized | Maltose uptake (% of control) | β-Galactosidase activity (% recovered) |
|---|---|---|---|
| pSE380 | None | 3 ± 0.3 | 100 |
| pSW7 | MalK | 100 | 6 ± 0.5 |
| pGS96-22 | LacK | 17 ± 2.8 | 101 ± 1.6 |
| pGS97-30 | LacK′-′MalK1 | 125 ± 6.9 | 15 ± 2.4 |
| pGS97-14 | LacK′-′MalK2 | 26 ± 3.8 | 22 ± 0.6 |
| pGS97-24 | LacK′-′MalK3 | 3 ± 0.3 | 36 ± 1.4 |
| pGS97-31 | LacK′-′MalK4 | 3 ± 0.3 | 50 ± 0.3 |
The effect of plasmid-encoded LacK′-′MalK chimeras on transport activity was measured for S. typhimurium ES25 (malK) (100% = 1.17 ± 0.14 mol/min/109 cells). The effect on the expression of malK-lacZ was measured for E. coli SK1280 (100% = 229 ± 5.5 Miller units). Values are means ± standard deviations of three and four independent experiments, respectively.
Immunochemical detection of chimeras.
Since the failure of chimeras 3 and 4 to substitute for MalK in transport could be due to a lack of gene expression, cells harboring the respective plasmids were subjected to SDS-PAGE followed by transfer of proteins to nitrocellulose. Subsequently, the protein content was probed with a polyclonal antiserum raised against the C1 fragment of MalK, in order to account for the observed poor detection of C-terminal epitopes by the serum raised against mature MalK. As shown in Fig. 6, all chimeras could be detected, although the amount of protein apparently decreased concomitant with the length of the MalK-derived peptide fragment. Nevertheless, the failure of chimeras 3 and 4 to support maltose uptake cannot be attributed to a lack of gene expression.
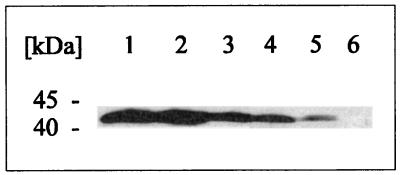
FIG. 6.
Immunoblot analysis of LacK′-′MalK chimeras. Cells of strain ES25, harboring the respective plasmids, were grown and further treated as described in the legend to Fig. 3 but were probed with a polyclonal antiserum raised against the purified MalKC1 fragment. Lanes: 1, MalK (pSW7); 2, LacK′-′MalK1 (pGS97-30); 3, LacK′-′MalK2 (pGS97-14); 4, LacK′-′MalK3 (pGS97-24); 5, LacK′-′MalK4 (pGS97-31); 6, control (pSE380).
Regulatory activity of the lacK′-′malK hybrids.
The respective plasmids were then introduced into strain SK1280 to test for the ability of the hybrid genes to down-regulate the β-galactosidase activity. The results, as summarized in Table 3, revealed that the percentage of enzymatic activity recovered increased with a decrease in the portion of MalK present in the chimera. The LacK′-′MalK4 protein, encompassing only the C-terminal 103 residues of MalK, still exhibited 50% of the regulatory activity of native MalK. Most notably, and in agreement with the results obtained with the truncated MalK proteins (Table 2), transport activity is not a prerequisite for the regulatory function of MalK.
DISCUSSION
We have carried out a functional analysis of truncated MalK proteins and of LacK′-′MalK chimeras to address the question of whether MalK is organized in structurally distinct domains. Such a view was implied by the observation that MalK functions in transport and regulation could be separated by mutations (15). By studying N- and C-terminal half molecules of MalK, we have demonstrated that a functional transport complex was assembled from fragments encompassing residues 1 to 179 (N1) and 179 to 369 (C1), respectively. Thus, this result indicates that both fragments can functionally interact with each other in the absence of a covalent linkage, suggesting a two-domain structure of the protein. This view is consistent with the occurrence of a transiently stable peptide fragment obtained by limited trypsinolysis that encompasses residues 185 to 369 (30).
In contrast, separate expression of the N1 fragment was not sufficient to constitute an active transport complex together with MalF and MalG. This was anticipated due to the lack of a conserved histidine residue at position 192, which has been shown for several ABC proteins, including MalK, to be crucial for activity (6, 28, 33). Strikingly, the C1 fragment displayed partial repressing activity, when expressed separately, that was not enhanced in the presence of the N1 fragment. Thus, C1 seems to acquire some secondary or tertiary structural elements independent of the corresponding N-terminal half of MalK that are sufficient for it to recognize its target. Although the mechanism of MalK-mediated regulation is still poorly understood, there is evidence for a direct interaction with positive regulator MalT of the maltose regulon (3). Interestingly, analyses of suppressor mutations have led to the identification of other proteins in E. coli that are unrelated to MalK but that display a similar repressing phenotype when overproduced (21, 22). This finding prompted the authors to speculate that a secondary structural motif common to these proteins rather than a recognizable sequence might interact with MalT. Our results obtained with N1/C1 could be interpreted along these lines.
The result that none of the other pairs displayed any activity at all might be due to misfolded protein fragments. Consistent with this notion is the lack of a dominant-negative phenotype of the malK deletions when expressed in a wild-type cell. This indicates that the encoded truncated MalK proteins failed to productively interact with MalF and MalG. Consequently, the C1 fragment, when coexpressed, probably assists in the folding process of MalKN1.
Thus, in order to define the boundaries of the putative regulatory domain of MalK more precisely, possible folding problems had to be minimized. To this end, we constructed and functionally analyzed chimeras that have C-terminal fragments of MalK varying in length fused to the corresponding N-terminal fragments of close homolog LacK of A. radiobacter. As shown previously, LacK can partially substitute for MalK in transport but does not exhibit repressing activity despite the presence of a similar C-terminal extension (34) (Table 3). Our results clearly demonstrate that the C-terminal extension of MalK, when fused to the corresponding N-terminal portion of LacK (LacK′-′MalK4), has retained substantial repressing activity (50% relative to the wild type). Moreover, the data also show that this activity increases with the length of the MalK fragment, thereby strongly indicating that the C-terminal extension is essential, albeit not sufficient, for the function.
Strikingly, a much stronger repressing activity was observed with cells of the tester strain expressing lack′-′malK2 (22%) than with cells expressing malKC1 (46%). Since the C-terminal MalK portions present in both constructs have about the same length, this result suggests that a covalent linkage between N- and C-terminal portions is required to obtain full regulatory activity. Possibly, only a covalent bond allows the N-terminal peptide to impose a specific conformational constraint on the C-terminal extension, as a prerequisite for optimal binding to its target.
Most surprisingly, and in contrast to what was found for LacK, chimeras 3 and 4 failed to substitute for MalK in transport. Although misfolding cannot be completely excluded, our finding that, unlike the truncated fragments C2 and C3, both chimeras exhibited substantial regulatory activity argues against such a mechanism. Rather, it is tempting to speculate that the joining points (Fig. 2 and 5) are located within a region (approximately encompassing residues 243 to 263) that is highly sensitive to alterations in the tertiary structure of the successive C-terminal domain. The following observations might be taken as evidence in favor of such a view. (i) The primary structures of MalK and LacK display a significant drop in sequence identity after position 243 (Fig. 2). (ii) The region between residues 243 and 263 is the most variable between the MalK proteins from S. typhimurium and E. coli, which overall have 94% identical amino acid residues (Fig. 2). (iii) In SmoK, another close homolog of MalK and LacK (40% sequence identity), which is involved in polyol transport in Rhodobacter sphaeroides, the respective peptide fragment is largely deleted (31). (iv) The insertion of a peptide linker at codon 245 in the E. coli malK gene resulted in a highly unstable protein that could not be detected immunochemically (17). (v) Finally, substitution of leucine for proline at position 259 in the MalK protein of S. typhimurium resulted in a defective transport complex (13).
We have for the first time provided evidence that, taken together, supports the notion that the C-terminal extension of MalK is sufficient to display substantial regulatory activity. Moreover, our data led us to suggest that the MalK protein is composed of two structurally distinct N- and C-terminal domains, almost equal in length. Clearly, data on the tertiary structure of MalK will be required to confirm or disprove this hypothesis. Such information is within sight since crystals of MalK that diffract to a resolution of 3 Å have recently become available (27). Finally, our constructs might prove to be useful tools in attempts to further elucidate the mechanism of MalK-triggered repression of maltose-regulated genes by biochemical approaches.
ACKNOWLEDGMENTS
We thank Heidi Landmesser and Birgit Sattler for excellent technical assistance. The contribution of Anke Stein in detecting the chimeras on immunoblots is gratefully acknowledged. We also thank W. Boos (Konstanz) for providing strain SK1280.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB171, TP C12; SCHN274/6-1/6-2) and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Ames F-L G, Mimura C S, Shyamala V. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia coli to human: traffic ATPases. FEMS Microbiol Rev. 1990;75:429–446. doi: 10.1111/j.1574-6968.1990.tb04110.x. [DOI] [PubMed] [Google Scholar]
- 2.Boos W, Lucht J M. Periplasmic binding protein-dependent ABC transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1175–1209. [Google Scholar]
- 3.Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukau B, Ehrmann M, Boos W. Osmoregulation of the maltose regulon in Escherichia coli. J Bacteriol. 1986;166:884–891. doi: 10.1128/jb.166.3.884-891.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson A L, Nikaido H. Purification and characterization of the membrane-associated components of the maltose transport system from Escherichia coli. J Biol Chem. 1991;266:8946–8951. [PubMed] [Google Scholar]
- 6.Davidson A L, Sharma S. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J Bacteriol. 1997;179:5458–5464. doi: 10.1128/jb.179.17.5458-5464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean D A, Reizer J, Nikaido H, Saier M H. Regulation of the maltose transport system of Escherichia coli by the glucose-specific enzyme III of the phosphoenolpyruvate-sugar phosphotransferase system; characterization of inducer exclusion-resistant mutants and reconstitution of inducer exclusion in proteoliposomes. J Biol Chem. 1990;265:21005–21010. [PubMed] [Google Scholar]
- 8.Decker K, Peist R, Reidl J, Kossmann M, Brand B, Boos W. Maltose and maltotriose can be formed endogenously in Escherichia coli from glucose and glucose-1-phosphate independently of enzymes of the maltose system. J Bacteriol. 1993;175:5655–5665. doi: 10.1128/jb.175.17.5655-5665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giacomini A, Corich V, Olero F J, Squartini A, Nuti M P. Experimental conditions may affect reproducibility of the β-galactosidase assay. FEMS Microbiol Lett. 1992;100:87–90. doi: 10.1111/j.1574-6968.1992.tb14024.x. [DOI] [PubMed] [Google Scholar]
- 10.Higgins C F. ABC transporter: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 11.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Hofnung M, Hatfield D, Schwartz M. malB region in Escherichia coli K-12: characterization of new mutations. J Bacteriol. 1974;117:40–47. doi: 10.1128/jb.117.1.40-47.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunke, S., and E. Schneider. Unpublished data.
- 14.Hyde S C, Emsley P, Hartshorn M J, Mimmack M M, Gileadi U, Pearce S R, Gallagher M P, Gill D R, Hubbard R E, Higgins C F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 15.Kühnau S, Reyes M, Sievertsen A, Shuman H A, Boos W. The activities of the Escherichia coli MalK protein in maltose transport, regulation, and inducer exclusion can be separated by mutations. J Bacteriol. 1991;173:2180–2186. doi: 10.1128/jb.173.7.2180-2186.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippincott J, Traxler B. MalFGK complex assembly and transport and regulatory characteristics of MalK insertion mutants. J Bacteriol. 1997;179:1337–1343. doi: 10.1128/jb.179.4.1337-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 19.Morbach S, Tebbe S, Schneider E. The ATP-binding cassette (ABC) transporter for maltose/maltodextrins of Salmonella typhimurium. Characterization of the ATPase activity associated with the purified MalK subunit. J Biol Chem. 1993;268:18617–18621. [PubMed] [Google Scholar]
- 20.Overduin P, Boos W, Tommassen J. Nucleotide sequence of the ugp genes of Escherichia coli K-12: homology to the maltose system. Mol Microbiol. 1988;2:767–775. doi: 10.1111/j.1365-2958.1988.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 21.Peist R, Koch A, Bolek P, Sewitz S, Kolbus T, Boos W. Characterization of the aes gene of Escherichia coli encoding an enzyme with esterase activity. J Bacteriol. 1997;179:7679–7686. doi: 10.1128/jb.179.24.7679-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reidl J, Boos W. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognising glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J Bacteriol. 1991;173:4862–4876. doi: 10.1128/jb.173.15.4862-4876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes M, Shuman H A. Overproduction of MalK protein prevents expression of the Escherichia coli mal regulon. J Bacteriol. 1988;170:4598–4602. doi: 10.1128/jb.170.10.4598-4602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth J R. Genetic techniques in studies of bacterial metabolism. Methods Enzymol. 1970;17:3–35. [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning. a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Schauder B, Blöcker H, Frank R, McCarthy J E G. Inducible expression vectors incorporating the Escherichia coli atpE translational initiation region. Gene. 1987;52:279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- 27.Schmees, G., K. Hoener zu Bentrup, E. Schneider, C. Vinzenz, and U. Ermler. Crystallization and preliminary X-ray analysis of the bacterial ATP-binding-cassette (ABC)-protein MalK. Acta Crystallogr. Sect. D, in press. [DOI] [PubMed]
- 28.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 29.Schneider E, Walter C. A chimeric nucleotide-binding protein, encoded by a hisP-malK hybrid gene, is functional in maltose transport in Salmonella typhimurium. Mol Microbiol. 1991;5:1375–1383. doi: 10.1111/j.1365-2958.1991.tb00784.x. [DOI] [PubMed] [Google Scholar]
- 30.Schneider E, Wilken S, Schmid R. Nucleotide-induced conformational changes of MalK, a bacterial ATP binding cassette transporter protein. J Biol Chem. 1994;269:20456–20461. [PubMed] [Google Scholar]
- 31.Stein M A, Schäfer A, Giffhorn F. Cloning, nucleotide sequence, and overexpression of smoS, a component of a novel operon encoding an ABC transporter and polyol dehydrogenases of Rhodobacter sphaeroides Si4. J Bacteriol. 1997;179:6335–6340. doi: 10.1128/jb.179.20.6335-6340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter C, Höner zu Bentrup K, Schneider E. Large-scale purification, nucleotide binding properties and ATPase activity of the MalK subunit of S. typhimurium maltose transport complex. J Biol Chem. 1992;267:8863–8869. [PubMed] [Google Scholar]
- 33.Walter C, Wilken S, Schneider E. Characterization of site-directed mutations in conserved domains of MalK, a bacterial member of the ATP-binding cassette (ABC) family. FEBS Lett. 1992;303:41–44. doi: 10.1016/0014-5793(92)80473-t. [DOI] [PubMed] [Google Scholar]
- 34.Wilken S, Schmees G, Schneider E. A putative helical domain in the MalK subunit of the ATP-binding-cassette transport system for maltose of Salmonella typhimurium (MalFGK2) is crucial for interaction with MalF and MalG. A study using the LacK protein of Agrobacterium radiobacter as a tool. Mol Microbiol. 1996;22:655–666. doi: 10.1046/j.1365-2958.1996.d01-1724.x. [DOI] [PubMed] [Google Scholar]
- 35.Williams S G, Greenwood J A, Jones C W. Molecular analysis of the lac operon encoding the binding-protein-dependent lactose transport system and β-galactosidase in Agrobacterium radiobacter. Mol Microbiol. 1992;6:1755–1768. doi: 10.1111/j.1365-2958.1992.tb01348.x. [DOI] [PubMed] [Google Scholar]