Abstract
Tissue-resident macrophages (TRMs) are long-lived cells that maintain locally and can be phenotypically distinct from monocyte-derived macrophages (MDMs). Whether TRMs and MDMs have district roles under differing pathologies is not understood. Here, we showed that a significant portion of the macrophages that accumulated during pancreatitis and pancreatic cancer in mice had expanded from TRMs. Pancreas TRMs had an extracellular matrix remodeling phenotype that was important for maintaining tissue homeostasis during inflammation. Loss of TRMs led to exacerbation of severe pancreatitis and death, due to impaired acinar cell survival and recovery. During pancreatitis, TRMs elicited protective effects by triggering the accumulation and activation of fibroblasts, which was necessary for initiating fibrosis as a wound healing response. The same TRM-driven fibrosis, however, drove pancreas cancer pathogenesis and progression. Together, these findings indicate that TRMs play divergent roles in the pathogenesis of pancreatitis and cancer through regulation of stromagenesis.
Tissue-resident macrophages (TRMs) are established through embryonic and adult hematopoiesis. At birth, TRMs are derived from embryonic progenitors, but some TRMs are gradually replaced with monocyte-derived cells over time, to varying extents determined at a tissue-specific level1, 2, 3, 4, 5, 6, 7, 8, 9. “Closed” TRM populations, such as brain microglia, undergo little monocyte replacement, whereas “open” TRM populations, such as lung alveolar macrophages, undergo gradual replacement, the extent of which is determined at a tissue-specific level6, 9, 10. On the other hand, short-lived, monocyte-derived macrophages (MDMs) rely on continual replenishment from circulating monocytes11, 12. It is understood that both developmental origin and tissue microenvironment offer cues that contribute to macrophage phenotypes13, 14, 15. TRMs have a variety of roles in maintaining tissue homeostasis, extracellular matrix (ECM) remodeling and inflammation16. Across the fat, dermis, heart, lung and mesenteric membranes, it is thought that interstitial TRMs can be subdivided into at least two major subpopulations, LYVE1hiCX3CR1loMHCIIlo/Int or LYVE1loCX3CR1hiMHCIIhi cells, with LYVE1hiCX3CR1loMHCIIlo/Int TRMs reported to be involved in ECM remodeling17, 18, 19, 20. However, it is not fully understood how TRM subsets and MDMs might be uniquely poised to respond to tissue damage or pathologic conditions.
Studies have investigated the roles of TRMs and MDMs under tumor conditions13, 15, 21. TRMs have tumor-promoting roles in driving the responses of regulatory T cells (Treg cells) responses, contributing to fibrosis and supporting tumor cell growth13, 15, 21. In pancreatic ductal adenocarcinoma (PDAC), macrophages have divergent roles based on developmental origin, with embryonic-derived TRMs being particularly adept at driving fibrosis15. Inflammation and tissue damage during pancreatitis is not only a risk factor for PDAC, but also involves significant accumulation of desmoplastic stroma rich in myeloid cells22, 23. Macrophages contribute to inflammation during pancreatitis, but the specific contribution of TRMs or MDMs has not been well characterized 24, 25. While the unique ability of the pancreas to respond to injury with significant desmoplastic stroma is likely critical for preserving function, it is a feature that drives PDAC progression26, 27.
Here, we showed that pancreas TRMs and MDMs increased markedly during pancreatitis and displayed distinct transcriptional phenotypes. We identified a subset of LYVE1hi TRMs that expressed genes related to extracellular matrix remodeling, and depletion of TRMs negatively impacted mouse survival during severe acute pancreatitis by attenuating the fibrotic response. TRMs triggered the accumulation of fibroblasts, affected their activation and stimulated the production of collagen and ECM proteins. While TRM-driven fibrosis was protective in pancreatitis, it was co-opted by tumors to support their growth.
Results
Pancreatitis and PDAC display immune rich fibrotic stroma
Pancreatitis and PDAC are characterized by desmoplastic fibrotic stromal responses and significant immune infiltration23, 28, 29. Histological analysis of human pancreatitis and PDAC tissues both showed marked loss of acinar cell area and increased fibrotic stroma compared to normal tissues (Fig. 1a). Furthermore, we observed increased numbers of CD163+ macrophages, podoplanin+ fibroblasts and trichome+ collagen area in pancreatitis and PDAC tissues compared to normal pancreas (Fig. 1a–b). This indicated shared stromal alterations across the diseased states. To study this in detail we used serial injection of cerulein in mice, which causes overproduction of digestive enzymes leading to pancreatitis30. Compared to vehicle-treated mice, cerulein-treated mice showed widespread acinar-to-ductal metaplasia (ADM) by both histology and immunohistochemical (IHC) staining for CK19+, Sox9+ and Ki67+ epithelial cells (Fig. 1c–d, Extended Data Fig. 1a). Consistent with widespread inflammation during pancreatitis, we observed increased F4/80+MHCII+ macrophages, F4/80loMHCIIlo eosinophils, Ly6G+ granulocytes and Ly6C+ monocytes in cerulein-treated mice (Fig. 1c–d, Extended Data Fig. 1b–c). Together these observations highlight the accumulation of macrophages and fibrotic stroma during pancreatitis and PDAC.
Figure 1: Pancreatitis and PDAC display immune rich fibrotic stroma.
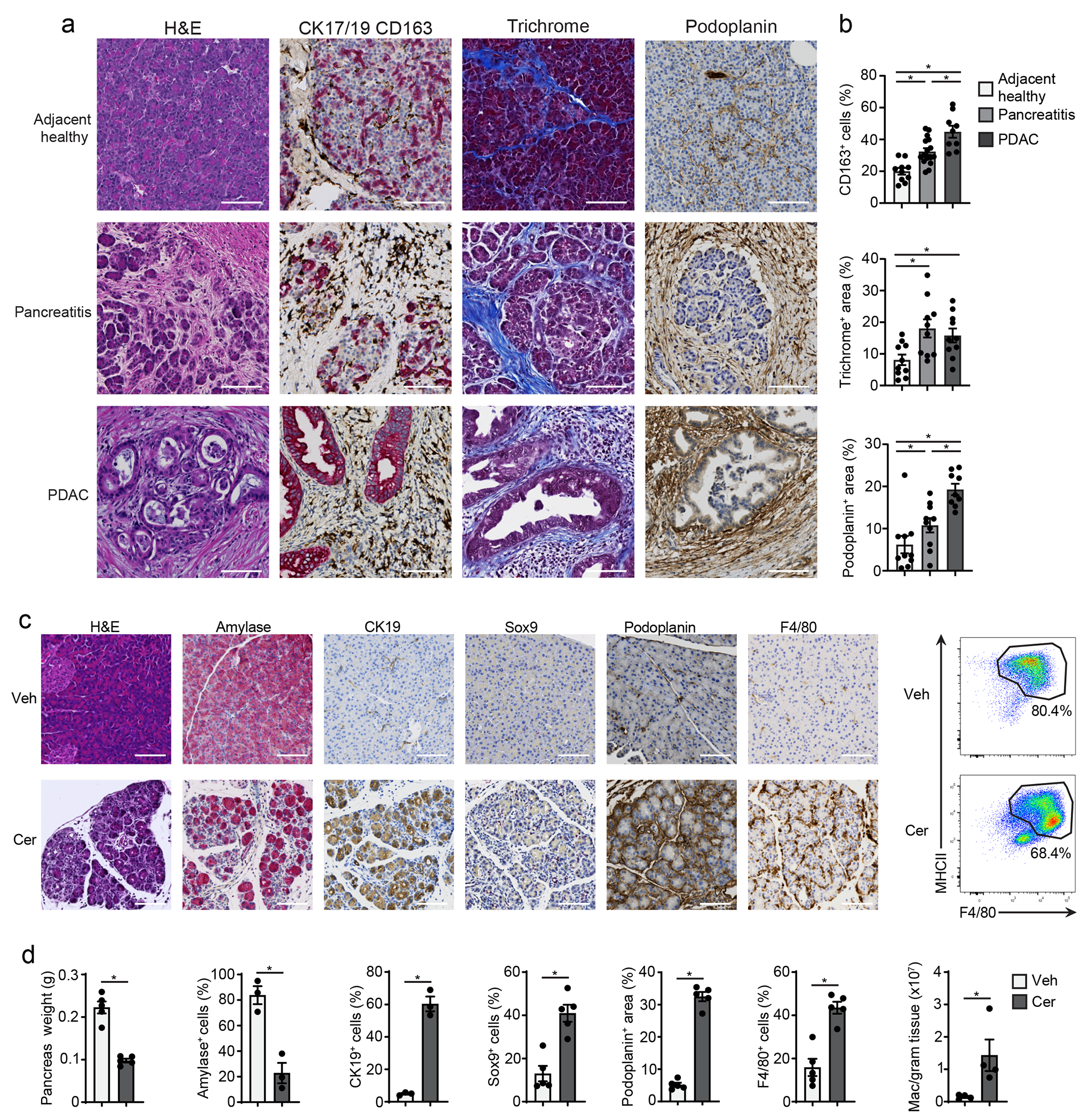
a. Representative immunohistochemistry (IHC) images of H&E, CK17/19, CD163, trichrome and podoplanin stains on human adjacent healthy pancreas, pancreatitis and PDAC tissue. Scale bars are 100μM. b. Quantification of CD163, trichrome collagen or podoplanin staining in human adjacent healthy pancreas, pancreatitis and PDAC represented as percentage of cells or percentage of tissue area. Adjacent healthy, n=10; pancreatitis, n=10; PDAC, n=9. CD163 quantification top to bottom and left to right *P<0.0001, *P=0.0039, and *P=0.0051, trichrome quantification top to bottom and left to right *P=0.0232, *P=0.0115, and podoplanin quantification top to bottom and left to right *P=0.0010, *P=0.0433, and *P=0.0015) c. Representative IHC images of H&E, amylase, CK19, Sox9, podoplanin and F4/80 and flow cytometry of F4/80+MHCIIhi/lo macrophages in the pancreatic tissue from mice treated with vehicle (Veh) or cerulein (Cer) by 6-hourly i.p. injections every other day for one week. Scale bars are 100μM. d. Quantification of pancreas weight, IHC staining with amylase, CK19, Sox9, podoplanin and F4/80 and flow cytometry analysis of F4/80+MHCIIhi/lo macrophages in the mouse pancreas as in c; (pancreas weight, n = 5 mice/group and *P=0.0079; amylase, n = 3 mice/group and *P=0.0046; CK19, n = 3 mice/group and *P=0.0003; Sox9, n = 5 mice/group and *P=0.0079; podoplanin, n = 5 mice/group and *P=0.0079; F4/80, n = 5 mice/group and *P=0.0079; F4/80+MHCIIhi/lo macrophages, n = 4 mice/group and *P=0.0286. Data are presented as mean ± SEM. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Cerulein treatment increases tissue-resident macrophages
To investigate whether the increase in the number of macrophages in pancreatitis was due to monocyte recruitment or the local expansion of tissue-resident macrophages, we crossed Flt3-Cre mice with R26-LSL-eYFP mice to generate Flt3-YFP mice. In this model, adult bone marrow-derived cells are YFP+, while embryonic-derived macrophages lack YFP expression31. While both the number of Flt3-YFP+ and Flt3-YFP− macrophages were increased in cerulein-treated mice compared to vehicle, the number of Flt3-YFP− macrophages was increased 13-fold, while Flt3-YFP+ macrophages increased 5-fold (Fig. 2a–c, Extended Data Fig. 2a). The frequency of Ki67+ Flt3-YFP− macrophages was also higher compared to Flt3-YFP+ in cerulein-treated mice (Fig. 2d), indicating TRMs proliferated in situ. To confirm these findings, we next treated CSF1R-mer-iCre-mer/LSL-tdTomato mice (hereafter CSF1R-tdTom) with a single dose of tamoxifen on embryonic day 9.5 (E9.5) to specifically label embryonic erythro-myeloid progenitor cells. As expected, brain microglia, which are yolk sac derived1, showed nearly 80% recombination in CSF1R-tdTom offspring (Fig. 2e–g), while Ly6Chi monocytes showed minimal tdTomato labeling (Fig. 2f). Nearly 10% of pancreas F4/80+MHCIIhi/lo macrophages were tdTomato+ (Fig. 2f), indicating a subpopulation of pancreas TRMs were derived from embryonic progenitors. Quantitation of tdTomato+ macrophages by flow cytometry and IHC staining indicated that this population expanded during pancreatitis (Fig. 2h–i).
Figure 2: Cerulein treatment increases tissue-resident macrophages.
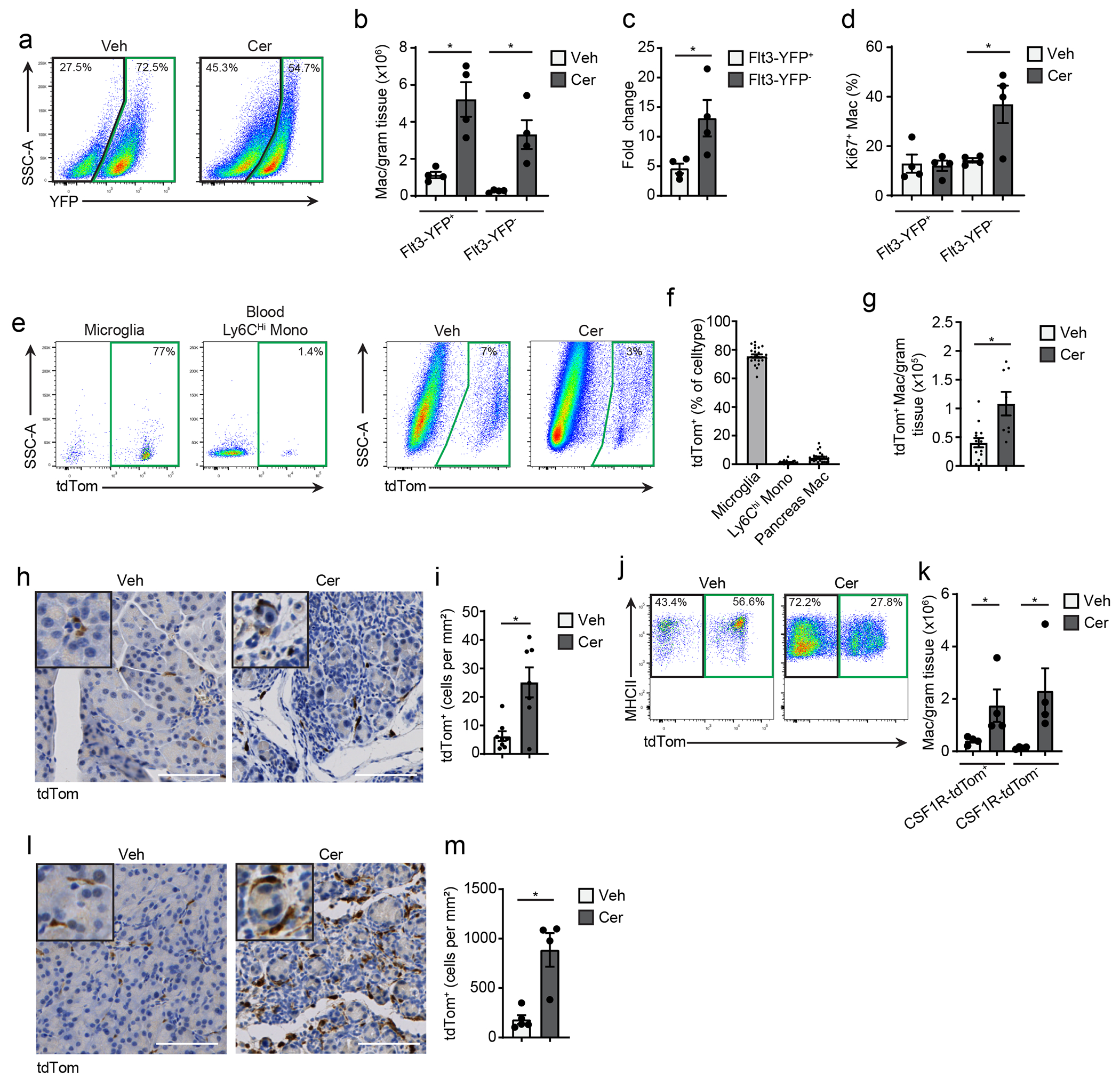
a. Flow cytometry staining of Flt3-YFP+ and Flt3-YFP− cells pre-gated on pancreas F4/80+MHCIIhi/lo macrophages in Flt3-Cre LSL-YFP (Flt3-YFP) mice. b. Density of Flt3-YFP+ and Flt3-YFP− macrophages in the pancreas of Flt3-YFP mice after 6 hourly i.p. injections with Veh or Cer every other day for one week; n = 4 mice/group, left to right *P=0.0052 and *P=0.0077. c. Fold-change in the density of Flt3-YFP+ and Flt3-YFP− macrophages in Veh or Cer-treated Flt3-YFP mice as in b; n = 4 mice/group, *P=0.0286. d. Percentage of Flt3-YFP+ and Flt3-YFP− macrophages that expressed Ki-67 in Veh or Cer-treated Flt3-YFP mice as in b; n = 4 mice/group, *P=0.0251. e. Flow cytometry of brain CD45+F4/80lo microglia, blood Ly6Chi monocytes and pancreas F4/80+MHCIIhi/lo macrophages in CSF1R-mer-iCre-mer LSL-tdTomato (CSF1R-tdTom) mice treated in utero with tamoxifen on embryonic day 9.5 (9 days post vaginal plug appearance) followed by treatment with Veh or Cer starting at 8-weeks age as in b. f. Percentage of brain CD45+F4/80lo microglia, blood Ly6Chi monocytes and pancreas F4/80+MHCIIhi/lo macrophages labeled by tdTomato in CSF1R-tdTom mice treated with tamoxifen followed by Veh or Cer as in e; n=24 mice/cell type analyzed. g. Density of pancreas F4/80+MHCIIhi/lo macrophages labeled by tdTomato in CSF1R-tdTom mice treated with tamoxifen followed by Veh or Cer as in e; Veh, n=14 mice; Cer, n=8 mice, *P=0.0022 . h. Representative images of IHC stain for tdTomato in pancreas tissue from CSF1R-tdTom mice treated with tamoxifen followed by Veh or Cer as in e i. Quantification of tdTomato+ cells in pancreas tissue of CSF1R-tdTom mice treated with tamoxifen followed by Veh or Cer as in e; Veh, n=8 mice; Cer, n=7 mice, *P=0.0289. j. Flow cytometry of tdTomato expression in pancreas F4/80+MHCIIhi/lo macrophages in CSF1R-tdTom mice administered tamoxifen by oral gavage for five consecutive days, followed by 10 weeks rest and then treated with Veh or Cer as in b; n = 4 mice/group. k. Density of pancreas tdTomato+ and tdTomato−macrophages in CSF1R-tdTom mice as in j; n = 4 mice/group, left to right *P=0.0286 and *P=0.0286. l. Representative images of IHC stain for tdTomato on pancreas from CSF1R-tdTom as in j; n = 4 mice/group. m. Quantification of pancreas tdTomato+ macrophages from CSF1R-tdTom mice as in j; n = 4 mice/group, *P=0.0159. Data are presented as mean ± SEM. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Some embryonically derived macrophages, such as heart or alveolar macrophages, are replaced by MDMs to varying degrees in a tissue-specific manner6, 8, 9. To measure the total TRMs irrespective of developmental origin, we administered tamoxifen by oral gavage in adult CSF1R-tdTom mice for 5 consecutive days, then stopped tamoxifen for 10 weeks to allow monocytes and short-lived MDMs to be washed out, while long-lived TRMs remain tdTomato+ (hereafter we refer to this regimen as pulse-chase, Extended Data Fig. 2b–c). The number of tdTomato+F4/80+MHCII+ TRMs in the pancreas was increased 4-fold in cerulein treated mice compared to vehicle (Fig. 2j–m). While CX3CR1-CreERT2 LSL-tdTomato mice (hereafter CX3CR1-tdTom) showed a similar increase in pancreas tdTomato+ TRMs in tamoxifen pulse-chase experiments, a maximum of 50% of pancreas macrophages were tdTomato+ (Extended Data Fig. 2d–k), as reported10. IHC staining of CX3CR1-tdTom mice indicated that most F4/80+ macrophages in the islets of Langerhans were tdTomato+ TRMs (Extended Data Fig. 2l). Islet macrophages were also tdTomato+ in CSF1R-tdTom tamoxifen pulse-chase mice (Extended Data Fig. 2m). These data suggested that pancreas TRMs consisted of both embryonic-derived and adult HSC-derived cells that expanded numerically during pancreas tissue damage.
Pancreas TRMs have distinct transcriptional phenotypes
Both ontogeny and tissue of residence can influence TRM phenotype13, 15, 18, 21, 32. To investigate the ontogeny-dependent roles of macrophages during pancreas injury and cancer, we performed bulk-RNA-seq analysis of pancreas F4/80+MHCII+ macrophages sorted from Flt3-YFP mice treated with vehicle or acute cerulein to induce pancreatitis or implanted orthotopically with PDAC tumors. Differential gene expression analysis identified several thousand differentially expressed genes (DEGs) between Flt3-YFP+ and Flt3-YFP− macrophages in pancreatitis and PDAC, but only several hundred in the vehicle-treated pancreas (Extended Data Fig. 3a–c). Flt3-YFP− macrophages highly expressed TRM and alternative-activation genes (e.g. Folr2, Cd163, Cd209, Mrc1), as well as genes related to ECM remodeling (Mmp9, Col4a3bp, Colec12) (Extended Data Fig. 3a). While Flt3-YFP− macrophages expressed genes linked to collagen production (Col3a1 and Col1a2) (Extended Data Fig. 3a), this could be derived from contaminating fibroblasts or mesenchymal cells. Conversely, Flt3-YFP+ macrophages had high expression of antigen-presentation genes, such as H2-Ab1, H2-Eb1 and Cd72 (Extended Data Fig. 3a). Gene set enrichment analysis (GSEA) indicated that genes upregulated in Flt3-YFP− macrophages were primarily enriched for pathways related to ECM remodeling and growth factor signaling, while Flt3-YFP+ macrophages were enriched for antigen presentation and T cell activation pathways (Extended Data Fig. 3b). Some of these differences were specific to tissue damage, rather than homeostasis (Extended Data Fig. 3c).
To determine whether embryonically- and HSC-derived TRMs defined phenotypically unique or mixed populations, we performed single-cell RNA-sequencing (scRNA-seq). We sorted Flt3-YFP+ and Flt3-YFP− F4/80+MHCII+ macrophages from vehicle-treated (healthy pancreas), cerulein-treated (pancreatitis) and PDAC tumors. We also analyzed tumor-bearing liver and lung, as a positive control for an embryonic TRM population (liver Kupffer cells) and mixed TRM-MDM (alveolar macrophages)4, 6, 7, 9. As expected, liver and lung macrophages showed origin-specific clustering in scRNA-seq analysis (Fig. 3a–b, Extended Data Fig. 3d–f). By contrast, in the pancreas, most macrophage scRNA-seq clusters were of mixed origin, with contribution from Flt3-YFP+ and YFP− macrophages (Fig. 3c–d). The only exception was Lyve1hi clusters (vehicle cluster 0, cerulein cluster 2 and PDAC cluster 5, Fig. 3e, Extended Data Fig. 4a–d), which showed up to 90% enrichment for Flt3-YFP− macrophages (Fig. 3c–e, Extended Data Fig. 4a–h). Some Lyve1lo clusters (vehicle clusters 2 and 4, and cerulein cluster 4) showed a bias towards Flt3-YFP+ cells and were marked by high expression of Ccr2 and MHCII family genes (Fig. 3c–e). Other mixed-origin Lyve1lo clusters expressed Trem2 and Cx3cr1 (Extended Data Fig. 4e–h). This suggested that aside from Lyve1hi TRMs, most TRMs were of mixed origin, and their transcriptional phenotypes might be defined by tissue cues.
Figure 3: Pancreas TRMs have distinct transcriptional phenotypes.
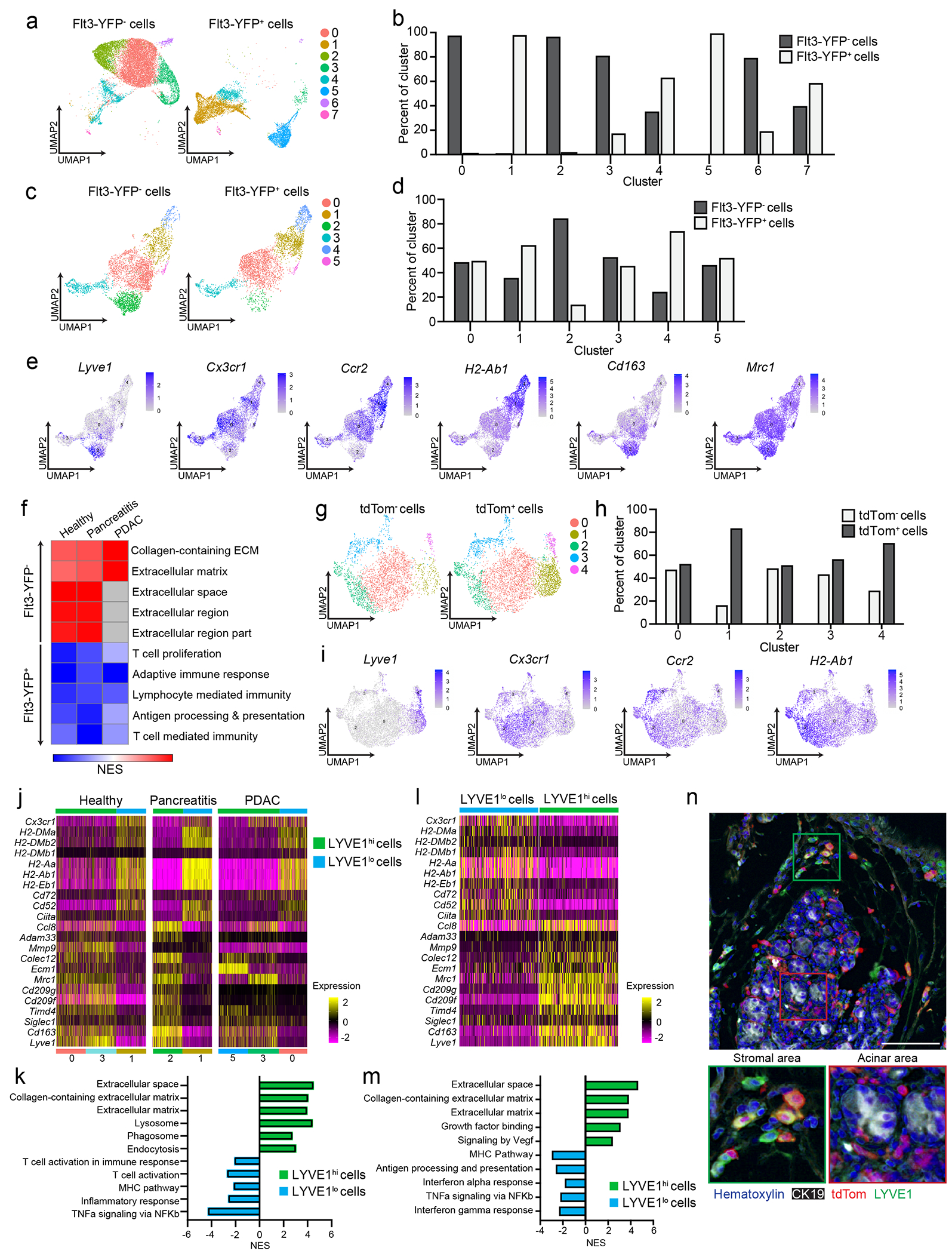
a. UMAP plot from single-cell RNA-sequencing (scRNA-seq) analysis of F4/80+MHCIIhi/lo macrophages sorted from livers of Flt3-YFP mice on day 14 post-implantation with the KP2 pancreatic cancer cell line. b. Quantification of Flt3-YFP+ and Flt3-YFP− macrophages by cluster from UMAP in a, displayed as percentage of each cluster. c. UMAP plot of scRNA-seq analysis of F4/80+MHCIIhi/lo macrophages sorted from the pancreas of Flt3-YFP mice treated with 6 hourly i.p. injections of Cer every other day for one week. d. Quantification of Flt3-YFP+ and Flt3-YFP− macrophages by cluster from UMAP in c, displayed as percentage of each cluster. e. UMAP plots displaying Lyve1, Cx3cr1, Ccr2, H2-Ab1, Cd163 and Mrc1 gene expression in Csf1r and C1qa expressing pancreas macrophages from Flt3-YFP mice as in c f. Heatmap displaying normalized enrichment score (NES) of significantly enriched gene sets comparing all Flt3-YFP− macrophages versus Flt3-YFP+ macrophages in indicated single-cell data sets; pathways selected by FDR < 0.05. g. UMAP plot of scRNA-seq analysis of F4/80+MHCIIhi/lo macrophages sorted from the pancreas of CSF1R-tdTom mice administered tamoxifen by oral gavage for 5 consecutive days, followed by a rest period of 10 weeks and a regimen of 6 hourly Cer i.p. injections every other day for one week. h. Quantification of tdTom− and tdTom+ macrophages by cluster from UMAP in g, displayed as percentage of each cluster. i. UMAP plots of Lyve1, Cx3cr1, Ccr2 and H2-Ab1 expression in Csf1r and C1qa expressing pancreas macrophages from CSF1R-tdTom mice as in g. j. Heatmap of DEGs upregulated in either LYVE1hi or LYVE1lo macrophages across healthy pancreas, pancreatitis and PDAC samples from Flt3-YFP mice treated with vehicle (Healthy) or cerulein (Pancreatitis) as in c, or orthotopically implanted with the KP1 pancreatic cancer cell line (PDAC). k. Bar graph of NES values of gene sets in LYVE1hi and LYVE1lo macrophages as in j. l. Heatmap of DEGs upregulated in either LYVE1hi or LYVE1lo macrophages from the pancreas of CSF1R-tdTom mice treated as in g. m. Bar graph of NES values of gene sets in LYVE1hi to LYVE1lo macrophages as in l. n. Representative images from multiplex immunohistochemistry (mIHC) staining for hematoxylin, F4/80, tdTomato, LYVE1 and CK19 in the pancreas of cerulein-treated CSF1R-tdTom mice treated as in g, staining repeated for 4 Veh- and 4 Cer-treated mice. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used, except for j and l, where Bonferroni correction was used, and f, k, and m, where FDR was used.
Because most pancreas clusters were of mixed origin, we investigated the impact of origin on macrophage phenotype. Pseudobulk analysis of the scRNA-seq data, irrespective of clusters, showed enrichment in ECM/ECM remodeling signatures and antigen processing and presentation in Flt3-YFP− and Flt3-YFP+ macrophages, respectively (Fig. 3f). Of note, the Flt3-YFP− macrophages in the scRNA-seq clusters did not show expression of collagen genes, indicating that expression of collagen in the bulk RNA-seq might have been due to contamination with other cell types. Next, we assessed how origin might impact phenotype within a single cluster. Comparison of Flt3-YFP− and Flt3-YFP+ cells in pancreatitis cluster 0 indicated that Flt3-YFP− cells were enriched for TNF signaling by NF-κB, inflammatory responses and TGF-β signaling, while Flt3-YFP+ cells showed higher expression of oxidative phosphorylation and Myc targets (Extended Data Fig. 4g). These data suggest that even in mixed origin macrophage subsets, origin may yield subtle phenotypic differences.
We next investigated whether Lyve1hi macrophages consisted predominantly of long-lived TRMs. scRNA-seq on F4/80+MHCII+ macrophages sorted from pancreas of CSF1R-tdTom mice pulse-chased with tamoxifen showed that clusters of Lyve1hi macrophages were predominantly tdTomato+, while Lyve1lo clusters showed mixed labeling (Fig. 3g–i), suggesting pancreas Lyve1hi macrophages were predominantly embryonically-derived, long-lived TRMs. As before, Lyve1hi macrophage clusters expressed scavenger receptors and alternative-activation markers (Cd163, Siglec1, Timd4, Cd209, and Mrc1) and showed enrichment in ECM and ECM remodeling genes and gene sets (Fig. 3j–m, Extended Data Fig. 5a–b). Lyve1lo macrophage clusters highly expressed H2-Aa, H2-Ab1, H2-Eb1, Cd52 and Cd72 and showed enrichment in T cell activation, MHC and NF-κB signaling pathways (Fig. 3j–m). Pseudobulk analysis comparing all tdTomato+ and tdTomato− cells, irrespective of cluster, identified similar gene sets (Extended Data Fig. 5a–b). This analysis suggested that Lyve1hi macrophages were embryonic TRMs with unique transcriptional phenotype.
To determine possible localization differences in LYVE1+ TRMs we performed multiplex IHC (mIHC). Pancreas from CSF1R-tdTom tamoxifen pulse-chase mice treated with cerulein for one week were stained for F4/80, LYVE1, tdTomato, and CK19, and showed more than 80% of F4/80+LYVE1hi cells were tdTomato+ and were located between the lobules of the pancreas and rarely in close contact with acinar cells or within islets (Fig. 3n, Extended Data Fig. 5c). This distribution was not observed for other tdTomato+ TRMs, some of which showed infiltration between clusters of acinar cells (Fig 3n), suggesting that LYVE1hi TRMs had distinct regional distributions within the pancreas. Finally, to test the tissue residence of the LYVE1+ macrophages, healthy CD45.1 and CD45.2 mice were surgically joined for 6 weeks, then parabiotic pairs were treated with vehicle or cerulein for one week while still joined. followed by treatment with vehicle or cerulein. After one week of cerulein or vehicle treatment, Ly6C+ blood monocytes were 20-30% of parabiotic donor origin, while LYVE1+CD163+ pancreas macrophages were 4% of donor origin (Extended Data Fig. 5d–f). 15-20% of LYVE1−CD163− and MHCII+ pancreas macrophages were of donor origin (Extended Data Fig. 5d–f), suggesting they were MDMs. While most recruited monocytes had an LYVE1−CD163− or MHCIIhi MDM profile, 11% were CD163+, indicating a TRM phenotype (Extended Data Fig. 5f). In total, these data suggested that LYVE1+ macrophage phenotypes and localization might contribute to pathologic fibrosis during pancreas injury.
Human LYVE1+ TRMs display similar phenotype and localization
To compare macrophage subsets in human pancreas tissues33, 34, we identified clusters of macrophages and monocytes in publicly available scRNA-seq data from three healthy and nine chronic pancreatitis tissues34. We first mapped the gene expression signature of mouse Lyve1hi macrophages using an averaged z-score. This signature was most highly expressed in human cluster 2, which was the only human cluster with significant expression of the LYVE1 gene (Fig 4a–b). Human cluster 2 expressed similar marker genes as mouse Lyve1hi macrophages, including FOLR2, MRC1 and PDGFC (Fig. 4c). By GSEA analysis, human LYVE1hi macrophages (cluster 2) had enrichment in ECM remodeling, TGFβ signaling and VEGF pathways (Fig. 4d). In publicly available human PDAC scRNA-seq datasets33, cluster 0 was enriched for the mouse Lyve1hi signature, had LYVE1 gene expression, and had similar phenotypes to both mouse and human pancreatitis Lyve1hi clusters by GSEA (Fig. 4e–h). The mouse Lyve1lo macrophage signature was found in most other human macrophage clusters (Extended Data Fig. 6a–b). mIHC for CD163, LYVE1 and CK19 on human healthy pancreas, pancreatitis and PDAC tissues showed that CD163 expression was more ubiquitous across macrophage subsets in humans than in mice (Fig. 4i). However, expression of LYVE1 was limited to a subset of CSF1R+CD68+CD163+ cells (Extended Data Fig. 6c–f). In human pancreatitis and PDAC tissues, LYVE1+CD163+ macrophages were only detected within the stroma and outside of acinar areas, while LYVE1−CD163+ macrophages were localized in both compartments (Fig. 4i). These data suggested conservation in phenotype and localization between mouse and human LYVE1+ pancreas macrophages.
Figure 4: Human LYVE1+ TRMs display similar phenotype and localization.
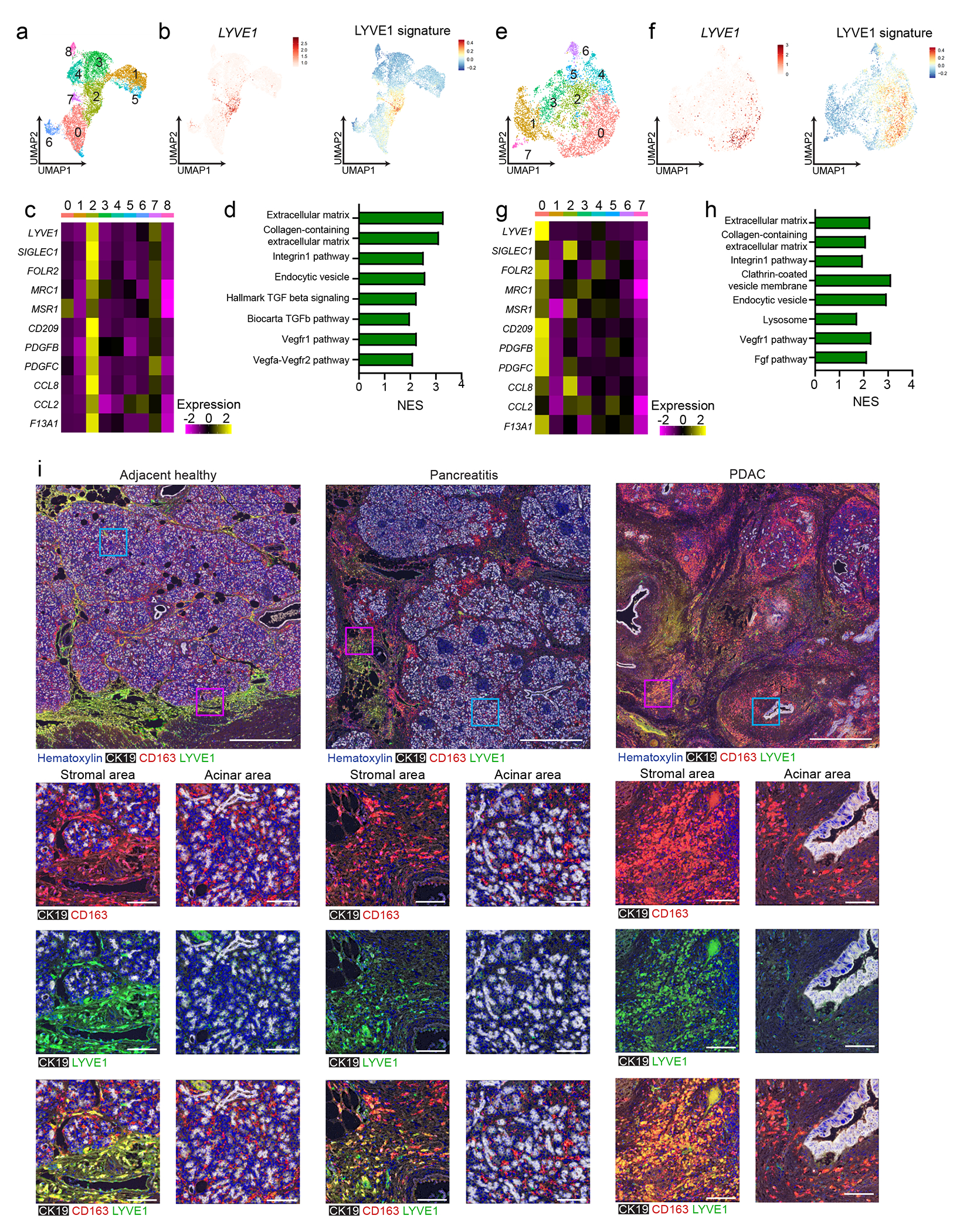
a. UMAP of scRNA-seq analysis in monocytes/macrophages from published human chronic pancreatitis dataset34, including 3 healthy pancreas, 4 idiopathic and 5 hereditary chronic pancreatitis samples. b. UMAP plots showing LYVE1 gene expression and mouse LYVE1hi macrophage scRNA-seq signature using top 100 DEGs from mouse LYVE1hi macrophages mapped into healthy and chronic pancreatitis samples from a. c. Heatmap of select marker genes differentially expressed in human chronic pancreatitis cluster 2. Data are p < 0.05 significant using Bonferroni correction. d. Bar graph of NES values of gene sets enriched in LYVE1hi macrophages from human chronic pancreatitis (cluster 2) compared to all other clusters. Data are FDR < 0.05 significant. e. UMAP of scRNA-seq analysis in monocytes/macrophages from 16 human PDAC samples from published PDAC dataset33. f. UMAP plots of LYVE1 gene expression and mouse LYVE1hi macrophage scRNA-seq signature using top 100 DEGs from mouse LYVE1hi macrophages mapped into PDAC samples from f. g. Heatmap of select marker genes differentially expressed in the human PDAC cluster 0. Data are p < 0.05 significant using Bonferroni correction. h. Bar graph of NES values of gene sets enriched in human PDAC LYVE1hi macrophages (cluster 0) compared to all other clusters. Data are FDR < 0.05 significant. i. Representative multiplex IHC images from adjacent healthy pancreas or pancreatitis and PDAC tumor samples obtained from patients at Barnes-Jewish Hospital stained for hematoxylin, CK19, CD163 and LYVE1. Highlighted areas of stromal or acinar area showing CK19 and CD163, CK19 and Lyve1, or CK19, CD163 and LYVE1 merged. Scale bars are 1mm (top row) or 100μM (bottom 3 rows).
TRMs maintain tissue integrity during pancreatitis
We next asked whether TRMs and MDMs have a distinct functional impact during pancreatitis. To deplete TRMs, mice were treated with either the combination of CSF1 neutralizing antibodies (CSF1 Abs) and clodronate-loaded liposomes (CLD)15, 35 or control antibodies and PBS-loaded liposomes (IgG-PBS). Mice were then rested for 10 days to allow the recovery of blood Ly6Chi monocytes and MDMs, and then treated with cerulein or vehicle by i.p injection for 7 days. At day 10 post-CSF1 Ab-CLD, before the cerulein treatment, blood Ly6Clo monocytes were reduced compared to IgG-PBS control mice (Fig. 5a–b), while the total number of pancreas F4/80+MHCII+ macrophages was significantly reduced (Fig. 5b, Extended Data Fig. 7a–c). At day 7 of cerulein treatment, CSF1Ab-CLD mice still had reduced numbers of pancreas F4/80+MHCII+ macrophages compared to IgG-PBS controls, despite having normal numbers of circulating Ly6Chi monocytes (Fig. 5b). mIHC indicated that F4/80+LYVE1+CD163+ TRMs were reduced more than 90% in CSF1Ab-CLD+cerulein mice (Fig. 5c). To investigate the effect of TRM loss on pathogenesis, CSF1Ab-CLD or IgG-PBS mice were implanted with osmotic pumps that administered cerulein or vehicle for up to 14 days. In IgG-PBS control mice, we observed acute body weight loss at days 2-5 post-cerulein, followed by recovery at days 6-9 post-cerulein (Fig. 5d). By contrast, CSF1Ab-CLD-treated mice continually lost body weight after cerulein pump implantation and reached humane survival endpoints by week 1 (Fig. 5d–e). Similar observations were made with both 8μg/day and 10μg/day doses of cerulein and FvB and C57BL/6 mice (Fig. 5d–e, Extended Data Fig. 7d–e). Along with impaired survival, CSF1Ab-CLD-treated mice had a significant decrease in blood glucose and an increase in serum amylase during pancreatitis compared to IgG-PBS mice (Extended Data Fig. 7f–i). Histologic analysis of the pancreas tissues indicated loss of pancreas tissue integrity, significant tissue necrosis and 90% loss of amylase+ acinar cells and CK19+ ductal cells and an increase in cleaved caspase-3+ cells in CSF1Ab-CLD compared to IgG-PBS mice (Fig. 5f–h). In contrast, we observed no histologic difference in the livers of CSF1Ab-CLD and IgG-PBS mice (Extended Data Fig. 7i), suggesting a pancreas-specific defect.
Figure 5: TRMs maintain tissue integrity during pancreatitis.
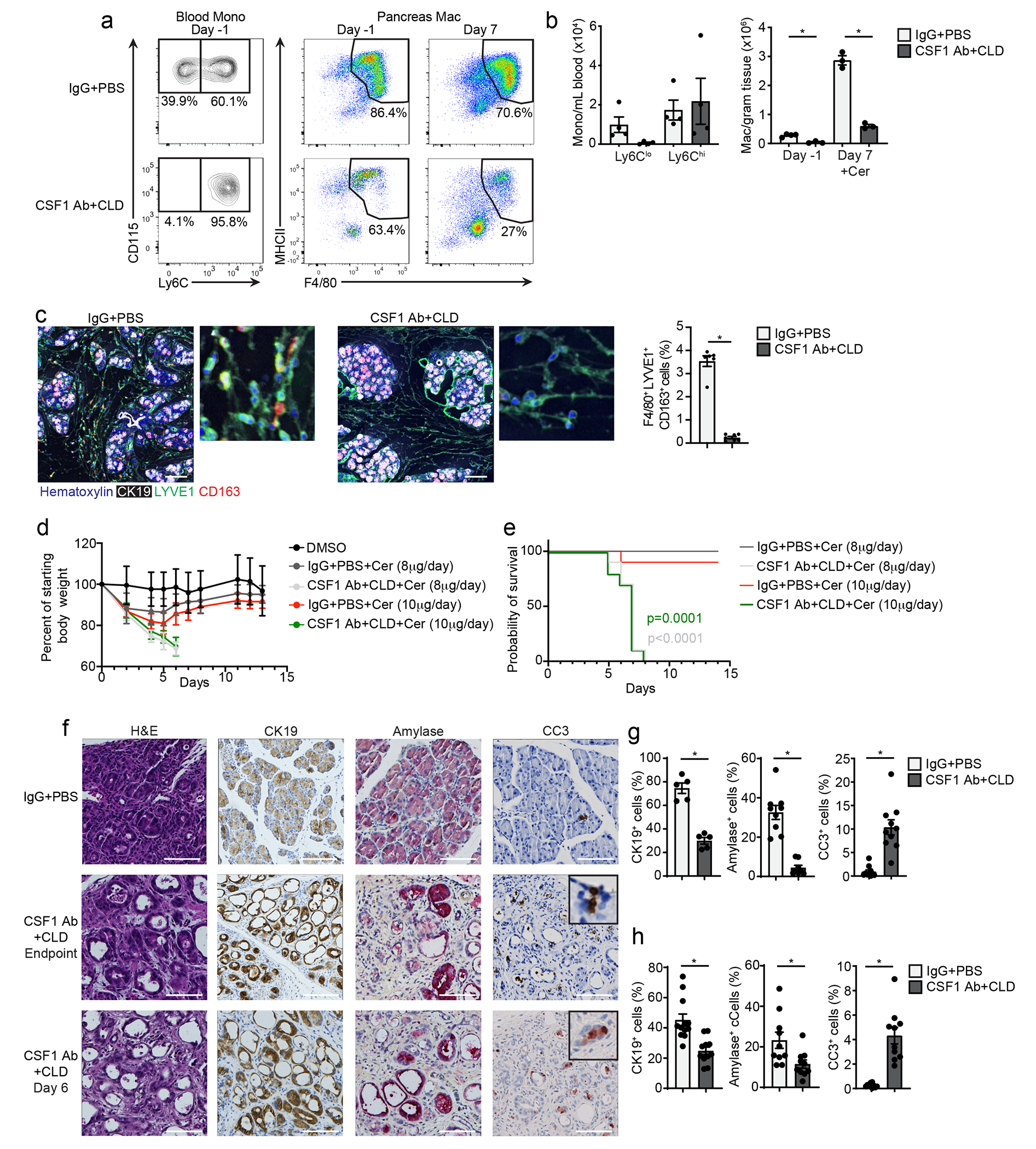
a. Flow cytometry plots of blood Ly6Chi monocytes and pancreas F4/80+MHCIIhi/lo macrophages from wild-type mice that received CSF1 Ab and clodronate-loaded liposomes (CSF1 Ab+CLD) or IgG and PBS-loaded liposomes (IgG+PBS) by i.p. injection, then allowed to recover for 10 days (day −1), followed by 6 hourly i.p. injections of Cer every other day for one week (day 7). b. Quantification of blood Ly6Clo and Ly6Chi monocytes after the recovery period (day −1) in mice as in a, and pancreas F4/80+MHCIIhi/lo macrophages after the recovery period (day −1) and after Cer treatment (day 7) in mice treated as in a. blood Ly6Clo and Ly6Chi, n=4 mice/group; pancreas F4/80+MHCIIhi/lo macrophages IgG+PBS day −1, n=4 mice/group, all other, n=3mice/group, left to right *P=0.0012 and *P=0.0002. c. Representative mIHC of staining for hematoxylin, F4/80, LYVE1, CD163, and CK19 in the pancreas and quantification of pancreas F4/80+LYVE1+CD163+ macrophages, displayed as the percentage of cells, in mice as in a; IgG+PBS, n=6mice/group, CSF1 Ab+CLD, n=7mice/group, *P=0.0006. Scale bars, 100μM, d. Body weight measurement in mice treated with IgG+PBS or CSF1 Ab+CLD and implanted 10 days later with peritoneal cavity osmotic pumps for the delivery of DMSO (control), 8μg/day cerulein or 10μg/day cerulein. Measurements starting on day of osmotic pump implantation; DMSO, n=5 mice, all other, n=10 mice. e. Kaplan-Meier survival curve showing mice treated with IgG+PBS and CSF1 Ab+CLD followed by osmotic pump implantation as in e; n=10mice/group. IgG+PBS versus CSF1 Ab+CLD with 10μg/day cerulein, p=0.0001; IgG+PBS versus CSF1 Ab+CLD with 8μg/day cerulein, p<0.0001. f. Representative images of pancreas tissue stained for H&E, CK19, amylase or cleaved-caspase-3 (CC3) from mice treated with IgG+PBS or CSF1 Ab+CLD followed by osmotic pump delivery of 10μg/day Cer, as in e, taken at humane survival endpoint (top two rows) or at day 6 post-osmotic pump implantation (bottom row). Scale bars are 100μM. g. Quantification of CK19, amylase, and CC3 IHC stains in pancreas tissue from IgG+PBS or CSF1 Ab+CLD-treated mice administered 10μg/day Cer by osmotic pump, as in e, at humane survival endpoint, displayed as the percentage of cells; CK19, n=5mice/group and *P=0.0079; amylase, IgG+PBS, n=9 mice, amylase CSF1 Ab+CLD, n=8 mice, *P<0.0001; CC3, n=10mice/group and *P<0.0001. h. Quantification of CK19, amylase and CC3 IHC stains in pancreas tissue from IgG+PBS or CSF1 Ab+CLD-treated mice administered 10μg/day Cer by osmotic pump, as in e, at day 6 post-osmotic pump implantation, displayed as the percentage of cells; IgG+PBS, n=12 mice; CSF1 Ab+CLD, n=11 mice; CK19, *P<0.0001; amylase, n=10mice/group, *P=0.0089; CC3, n=10mice/group and *P<0.0001. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
To test the role of MDMs in pancreatitis progression, we used CCR2-deficient mice (CCR2-KO), which have impaired egress of Ly6Chi monocytes from the bone marrow36 resulting in reduced numbers of blood Ly6Chi monocytes (Extended Data Fig. 7j). We observed normal numbers of total F4/80+ macrophages in steady-state pancreas tissues compared to wild-type mice (Extended Data Fig. 7j). Cerulein-treated CCR2-KO mice had no change in total pancreas F4/80+ macrophages (Extended Data Fig. 7j), but a decrease in the number of pancreas MHCIIhi macrophages compared to wild-type mice (Extended Data Fig. 7j). CCR2-KO and wild-type mice had similar survival and pancreas weight following cerulein-loaded osmotic pump implantation (Extended Data Fig. 7k). Together, this suggested that TRMs were critical in restoring pancreas tissue homeostasis after acute damage, whereas MDMs were largely dispensable.
Depletion of TRMs attenuates fibrotic responses
To address whether the protective effect of TRMs was due to changes in the ADM process, we injected mice with CSF1Ab-CLD or IgG-PBS, waited 10 days, then injected cerulein or vehicle by 6-hourly i.p. injections every other day for one week. By IHC, CSF1Ab-CLD mice had increased percentage of CK19+ cells in the pancreas compared to IgG-PBS controls (Fig. 6a). Close examination indicated CSF1Ab-CLD mice had significantly fewer stromal cells and stromal area between the acinar cell clusters (Fig 6a). mIHC using the ADM markers CK19, Sox9 and Ki-67 showed no difference in the percentage of amylase+ acinar cells expressing CK19, Sox9 or Ki-67 between CSF1Ab-CLD and IgG-PBS mice (Fig. 6b–c), indicating that TRM-depletion did not affect the ability of acinar cells to undergo ADM. In contrast, CSF1Ab-CLD mice had a >50% reduction in the number of podoplanin+ fibroblasts compared to IgG-PBS mice (Fig. 6b–d). Together, these data suggested that TRMs may impact recovery from pancreatitis by promoting stromagenesis.
Figure 6: Depletion of TRMs attenuates fibrotic responses.
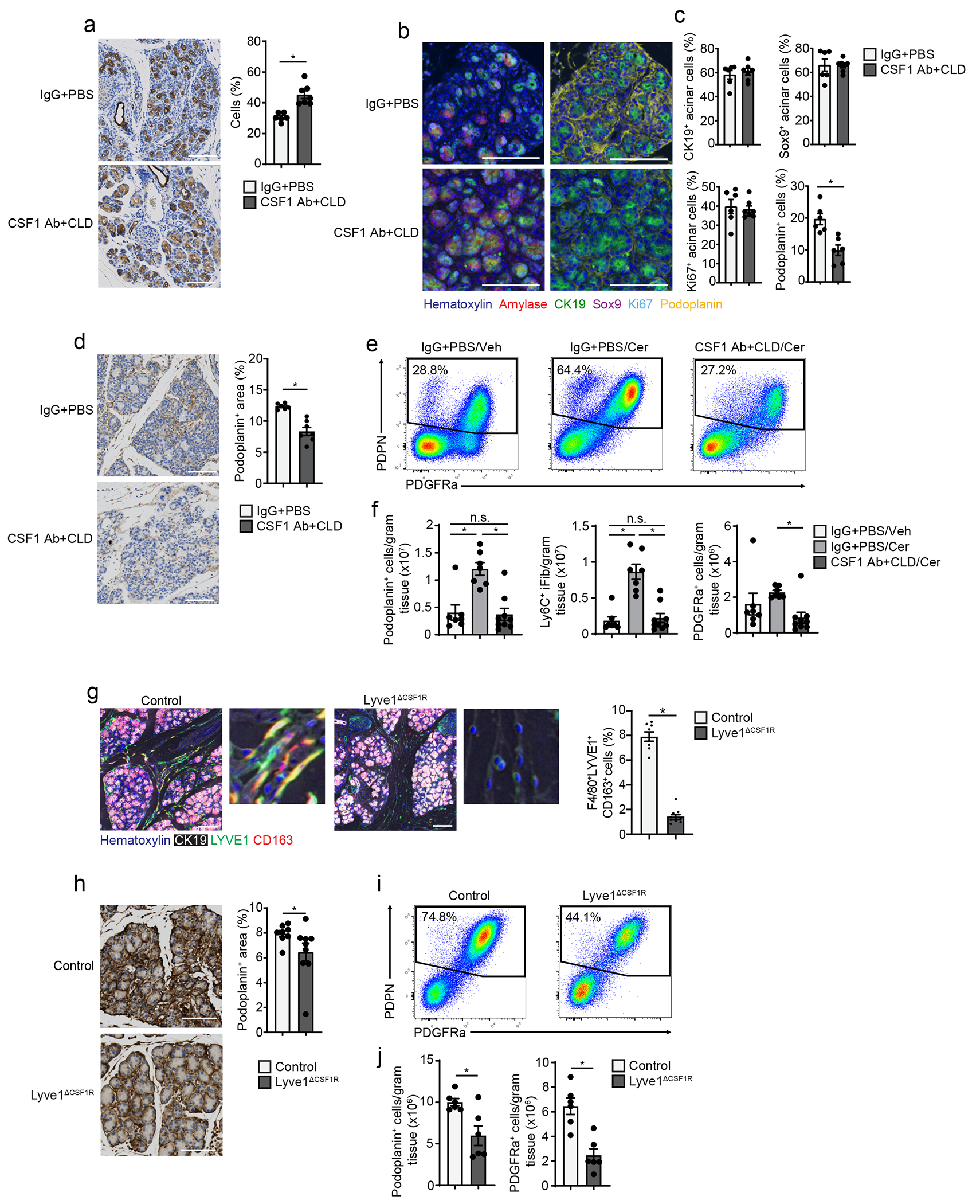
a. Representative images and quantification of CK19 IHC stain on pancreas tissue of mice treated with IgG+PBS or CSF1 Ab+CLD, followed by a 10-day recovery period, then 6 hourly i.p. injections with Cer every other day for one week; IgG+PBS, n=6 mice; CSF1 Ab+CLD, n=7 mice and *P=0.0012. b. Representative mIHC images of pancreas tissue from mice treated as in a, stained for hematoxylin, amylase, CK19, Sox9, Ki-67, and podoplanin, scale bars are 100μM. c. Quantification of amylase+ acinar cells expressing CK19, Sox9 or Ki-67, displayed as percentage of acinar cells, and podoplanin+ cells, displayed as percentage of total cells; IgG+PBS, n=6 mice; CSF1 Ab+CLD, n=7 mice; and *P=0.0022 for podoplanin analysis. d. Representative images of pancreas tissue stained for podoplanin and quantification of podoplanin+ area in pancreas from mice treated as in a; IgG+PBS , n=6 mice; CSF1 Ab+CLD, n=7 mice and *P=0.0012. e. Flow cytometry plots of IgG+PBS + pancreas fibroblasts from IgG+PBS-treated mice injected i.p with Veh (IgG+PBS/Veh), or IgG+PBS- or CSF1 Ab+CLD-treated mice injected i.p. with Cer (IgG+PBS/Cer or CSF1 Ab+CLD/Cer), as in a. f. Density of pancreas IgG+PBS + fibroblasts, Ly6C+ inflammatory fibroblasts (iFibs) and PDGFRa+ fibroblasts from IgG+PBS/Veh, IgG+PBS/Cer or CSF1 Ab+CLD/Cer mice as in e; IgG+PBS/Veh, n=7 mice; IgG+PBS/Cer, n=7 mice; CSF1 Ab+CLD/Cer, n=9 mice; top to bottom and left to right, *P=0.0005 and *P=0.0003 in podoplanin analysis, *P<0.0001 and *P<0.0001 in Ly6C+ iFib analysis, and *P=0.0409 in PDGFRa analysis. g. Representative mIHC images of pancreas tissue from Lyve1-Cre− mice (Control) or Lyve1-CreCSF1Rflox/flox mice (Lyve1ΔCSF1R) treated with 6 hourly i.p. injections of Cer every other day for one week, stained for hematoxylin, CK19, F4/80, LYVE1 and CD163 and quantification of F4/80+LYVE1+CD163+ pancreas macrophages from Control or Lyve1ΔCSF1R mice treated with Cer, displayed as the percentage of cells; Control, n=8 mice; Lyve1ΔCSF1R, n=9; *P<0.0001. Scale bar, 100μM h. Representative images of podoplanin-stained pancreas tissue and quantification of podoplanin+ area from Control or Lyve1ΔCSF1R mice as in g; Control, n=8 mice; Lyve1ΔCSF1R, n=9, *P<0.0360. i. Flow cytometry plots of podoplanin+ pancreas fibroblasts from Control and Lyve1ΔCSF1R mice as in g. j. Density of pancreas podoplanin+ fibroblasts and PDGFRa+ fibroblasts from Control and Lyve1ΔCSF1R mice as in g; n=6 mice/group, *P=0.0411 in podoplanin analysis and *P=0.0043 in PDGFRa analysis. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
To profile how TRMs were involved in driving the fibrotic response during pancreatitis, we examined the changes in fibroblast and ECM deposition during pancreatitis progression. CSF1Ab-CLD or IgG-PBS treated mice were treated with cerulein for 3, 7, and 17 days. CSF1Ab-CLD mice had reduced numbers of podoplanin+ fibroblasts compared to IgG-PBS mice at all timepoints (Fig. 6d, Extended Data Fig. 8a). The ECM molecule fibronectin was reduced at early time points (day 3 and 7 post-cerulein injections), but markedly reduced at day 17 compared to IgG-PBS control (Extended Data Fig. 8a). In the absence of pancreatitis, we observed no difference in podoplanin+ fibroblasts between CSF1Ab-CLD and IgG-PBS mice (Extended Data Fig. 8b–c). Flow cytometry indicated total podoplanin+ fibroblasts were markedly increased in IgG-PBS mice, but not in CSF1Ab-CLD mice during pancreatitis (Fig. 6e–f). Four distinct subsets of fibroblasts, including Ly6C+ inflammatory fibroblasts (iFibs), MHCII+ antigen-presenting fibroblasts (apFibs), αSMA+ myofibroblasts (myFibs) and PDGFRα+ fibroblasts (Extended Data Fig. 8d) have been described in homeostatic pancreas and PDAC 37, 38. Of these, we detected an increase in the number of Ly6C+ and PDGFRα+ fibroblasts in IgG-PBS mice, but not in cerulein-treated CSF1Ab-CLD mice (Fig. 6e–f, Extended Data Fig. 8e).
To test whether LYVE1+ macrophages had a role in the initiation of the fibrotic response, we crossed Lyve1-Cre mice with CSF1Rfl/fl mice (hereafter Lyve1ΔCSF1R), to specifically delete the CSF1R gene in LYVE1+ cells18. Lyve1ΔCSF1R mice treated with cerulein for 7 days had >80% reduction in F4/80+LYVE1+CD163+ TRMs compared to Cre− littermates (Fig. 6g, Extended Data Fig. 8f), but no change in the number of total F4/80+MHCII+ or LYVE1− macrophages (Extended Data Fig. 8f). Cerulein-treated Lyve1ΔCSF1R mice had reduced expansion of podoplanin+ fibroblasts and PDGFRα+ fibroblasts compared to cerulein-treated Cre− littermates, as measured by IHC and flow cytometry (Fig. 6h–j). Notably, we saw no increase in iFibs or a survival difference in cerulein-treated Lyve1ΔCSF1R mice compared to Cre− counterparts (Extended Data Fig. 8f–g), suggesting other TRMs could partially compensate. To check the role of MDMs, we treated CCR2-KO and wild-type mice with cerulein for 7 days and found no difference in the number of fibroblasts in the pancreas or in ECM deposition (Extended Data Fig. 8h–i). These data suggested TRMs, including LYVE1+ TRMs, coordinated fibroblast activation and expansion during pancreas injury.
TRMs shape the tissue-protective fibrotic response
To investigate how TRMs impacted fibroblast phenotype, we performed scRNA-seq on sorted podoplanin+ fibroblasts from CSF1Ab-CLD and IgG-PBS mice treated with cerulein by 6-hourly i.p. injections every other day for one week. UMAP clustering resulted in 10 distinct clusters that broadly fit into the iFib (based on the expression of Ly6c1 and Clec3b), myFib (Acta2, Col8a1,Cxcl14,) and apFib (H2-Ab1, Cd74, and Slpi) subsets (Fig. 7a–c, Supplementary Table 3)37, 38. DGE and GSEA analyses indicated Ly6c1+ iFibs from IgG-PBS mice were enriched in gene sets related to ECM production and organization and collagen production, while CSF1Ab-CLD Ly6c1+ iFibs were enriched in interferon responses and NF-κB signaling, which are known drivers of pancreatitis severity39, 40(Fig. 7d–e, Supplementary Table 4). Enrichment in these gene sets was also detected in Acta2+ myFibs and H2-Ab1+ apFibs (Fig. 7e–f). Fibroblasts sorted from pancreas of IgG-PBS+vehicle (steady-state) mice clustered separately from cerulein-treated IgG-PBS or CSF1Ab-CLD fibroblasts, while cerulein-treated IgG-PBS fibroblasts upregulated collagen and ECM genes more than cerulein-treated CSF1Ab-CLD fibroblasts (Extended Data Fig. 9a–d). This suggested that fibroblasts upregulated genes related to collagen and ECM in the presence of TRMs, but were reduced in number and expressed genes linked to an inflammatory phenotype rather than protective fibrosis genes, in the absence of TRMs.
Figure 7: TRMs shape the tissue-protective fibrotic response.
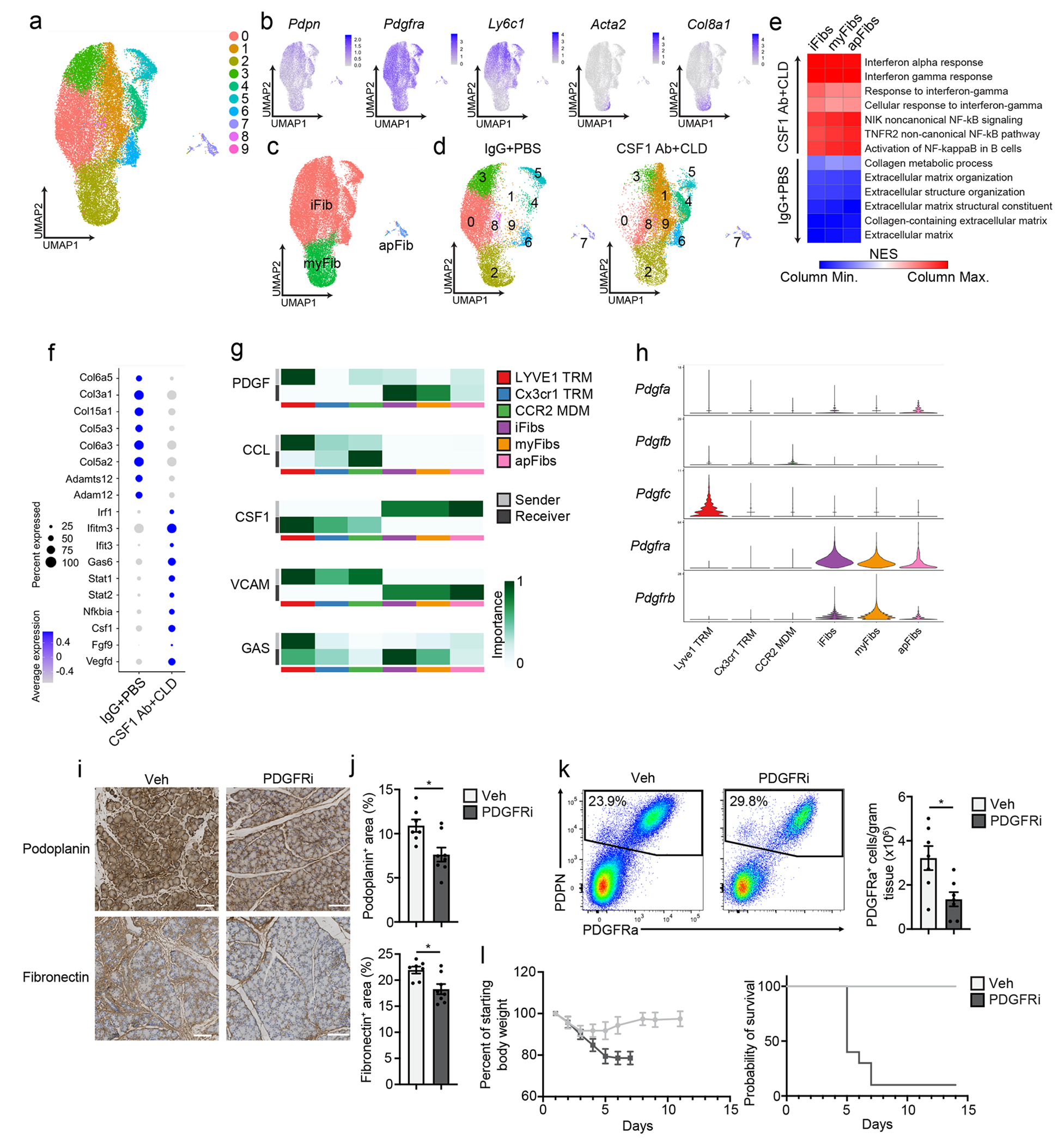
a. UMAP plot of pancreas podoplanin+ fibroblasts sorted from IgG+PBS or CSF1 Ab+CLD treated mice rested for 10 days then administered 6 hourly i.p. injections of Cer every other day for one week. b. UMAP plots displaying Podoplanin, Pdgfra, Ly6c1, Acta2 and Col8a1 expression in podoplanin+ pancreas fibroblasts of IgG+PBS and CSF1 Ab+CLD-treated mice as in a. c. UMAP plot showing the classification of fibroblast clusters into Ly6C+ iFib, αSMA+ myFib and MHCII+ apFib subtypes in pancreas podoplanin+ fibroblasts of IgG+PBS and CSF1 Ab+CLD treated mice as in a. d. UMAP plot of pancreas podoplanin+ fibroblasts from IgG+PBS or CSF1 Ab+CLD mice as in a. e. Heatmap displaying NES of gene sets significantly enriched across podoplanin+ fibroblast subtypes (as in c) comparing IgG+PBS to CSF1 Ab+CLD samples. f. Dot plot of DEGs upregulated in podoplanin+ fibroblasts from IgG+PBS or CSF1 Ab+CLD-treated mice as in a. g. Heatmaps displaying relative importance of network centrality measures for sender (ligand signals) and receiver (receptors) across macrophage clusters from Flt3-YFP mice treated with Cer by 6 hourly i.p. injections every other day for one week (from Fig. 3c) and fibroblast clusters as in a for PDGF, CCL, CSF1, VCAM and GAS signaling pathway networks from CellChat analysis49. h. Violin plots showing expression of PDGF signaling pathway genes Pdgfa, Pdgfb, Pdgfc, Pdgfra and Pdgfrb across macrophage and fibroblast clusters as in g. i. Representative images of podoplanin or fibronectin-stained pancreas tissue from mice injected i.p with Veh or PDGFRi once per day along with 6 hourly Cer i.p. injections every other day for one week. Scale bars, 100μM. j. Quantification of podoplanin+ and fibronectin+ area in pancreas tissue of mice treated with Veh or PDGFRi and cerulein as in i; Veh, n=7 mice; PDGFRi, n=8 mice; *P=0.0205 in podoplanin analysis and *P=0.0140 in fibronectin analysis. k. Flow cytometry of podoplanin+ fibroblasts and density of PDGFRa+ fibroblasts from mice treated with Veh or PDGFRi along with Cer as in i,; n=7 mice/group and *P=0.0175. l. Body weight measurement and Kaplan-Meier survival curve in mice treated with Veh or PDGFRi along with 10μg/day Cer delivered by peritoneal osmotic pumps. Measurements start on day of osmotic pump implantation; n=10 mice/group, *P<0.0001 comparing vehicle and PDGFRi in survival curve. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used, except for f, where Bonferroni correction was used, and e, where FDR was used.
Next, to identify TRM-fibroblast signaling molecules, we merged macrophage and fibroblast single cell datasets and queried LYVE1hiCX3CR1loMHCIIlo TRMs, LYVE1loCX3CR1hiMHCIIhi TRMs, CCR2hi MDMs and Ly6C+ iFibs, αSMA+ myFibs and MHCII+ apFibs for enriched receptor and ligand genes (Extended Data Fig. 9e–g). We identified 60 pathways with statistical significance across clusters (Extended Data Fig. 9h). Across all pathways between macrophages and fibroblasts, LYVE1hi TRMs had the highest aggregate score (Extended Data Fig. 9g–i), suggesting they were likely producing the most signals received by fibroblasts. Among these, PDGF signaling was enriched between LYVE1hi TRMs and both iFibs and myFibs (Fig. 7g). The LYVE1hi cluster had the highest expression of Pdgfc, while fibroblasts had high expression Pdgfra and Pdgfrb (Fig. 7h). Notably, all fibroblast populations produced Csf1, which signals through Csf1r, expressed by all macrophage clusters (Fig. 7g). To determine if PDGF-PDGFR signaling was required for fibroblast accumulation during pancreatitis, we treated mice with the PDGFR inhibitor imatinib mesylate (hereafter PDGFRi) or vehicle at the time of cerulein administration. PDGFRi-treated mice had reduced podoplanin+ fibroblasts and fibronectin (Fig. 7i–j) and decreased numbers of PDGFRα+ fibroblasts (Fig. 7k), but similar number of LYVE1hi and other TRMs (Extended Data Fig. 9j) compared to vehicle-treated mice. Mice treated with PDGFRi concurrent with cerulein-loaded osmotic pump implantation did not recover body weight and showed 90% lethality (Fig. 7l). These data suggested that fibroblast accumulation during pancreas injury might rely on signals from TRMs, including PDGF-PDGFR signaling.
TRMs drive fibrosis and pancreatitis-accelerated PDAC progression
To assess whether TRM regulated fibrosis in PDAC, we used three genetic models. These models are driven by pancreas-specific p48-Cre activation of KRASG12D expression either constitutively or under control of doxycycline (iKRAS* mice). Two of the models have Cre-mediated deletion of one (KPC mice) or both p53 alleles (KPPC mice)41. 2.5-months old KPC mice were treated with CSF1Ab-CLD and IgG-PBS, rested for 10 days and treated with cerulein by 6-hourly i.p. injections every other day for 5 days (Extended Data Fig. 10a). Three weeks after cerulein-treatment, a timepoint when microscopic carcinomas can be observed in IgG-PBS control mice, CSF1Ab-CLD-treated mice had an 80% reduction in the number of F4/80+LYVE1+CD163+ TRMs compared to IgG-PBS controls (Fig. 8a). By histology, CSF1 Ab-CLD-treated KPC mice displayed significantly less high-grade PDAC tissue (Fig. 8b–c), compared to IgG-PBS-treated mice, and instead exhibited well-differentiated cyst-like desmoplastic areas characterized by low stromal desmoplasia and fewer podoplanin+ and αSMA+ fibroblasts (Fig. 8c–d). These data suggest that TRMs might drive PDAC progression in part through facilitating stromal desmoplasia. In the more aggressive KPPC model, we also observed a reduction in podoplanin and fibronectin staining, and a reduced tumor burden in CSF1Ab-CLD compared to IgG-PBS mice at 2 months of age (Fig. 8e–f)15. Finally, to induce depletion of TRMs prior to KRAS oncogene induction, we treated iKRAS* mice with CSF1Ab-CLD or IgG-PBS, rested for 10 days, then treated with cerulein by i.p. injection for two days and administered doxycycline in their drinking water for three weeks (Extended Data Fig. 10b)41, 42. At week 3 post KRASG12D induction, CSF1Ab-CLD-treated iKRAS* mice had a significant reduction in F4/80+LYVE1+CD163+ TRMs, as well as reduced pre-malignant pancreas weight and stromal area compared to IgG-PBS-treated mice (Extended Data Fig. 10c–e). Implantation of tumors in CCR2-KO and wild-type mice resulted in no change in tumor weight, size or podoplanin+ fibroblasts (Extended Data 10f–g). These data indicated that total TRMs facilitated the expansion of the fibrotic stroma during tumor development.
Figure 8: TRMs drive fibrosis and pancreatitis-accelerated PDAC progression.
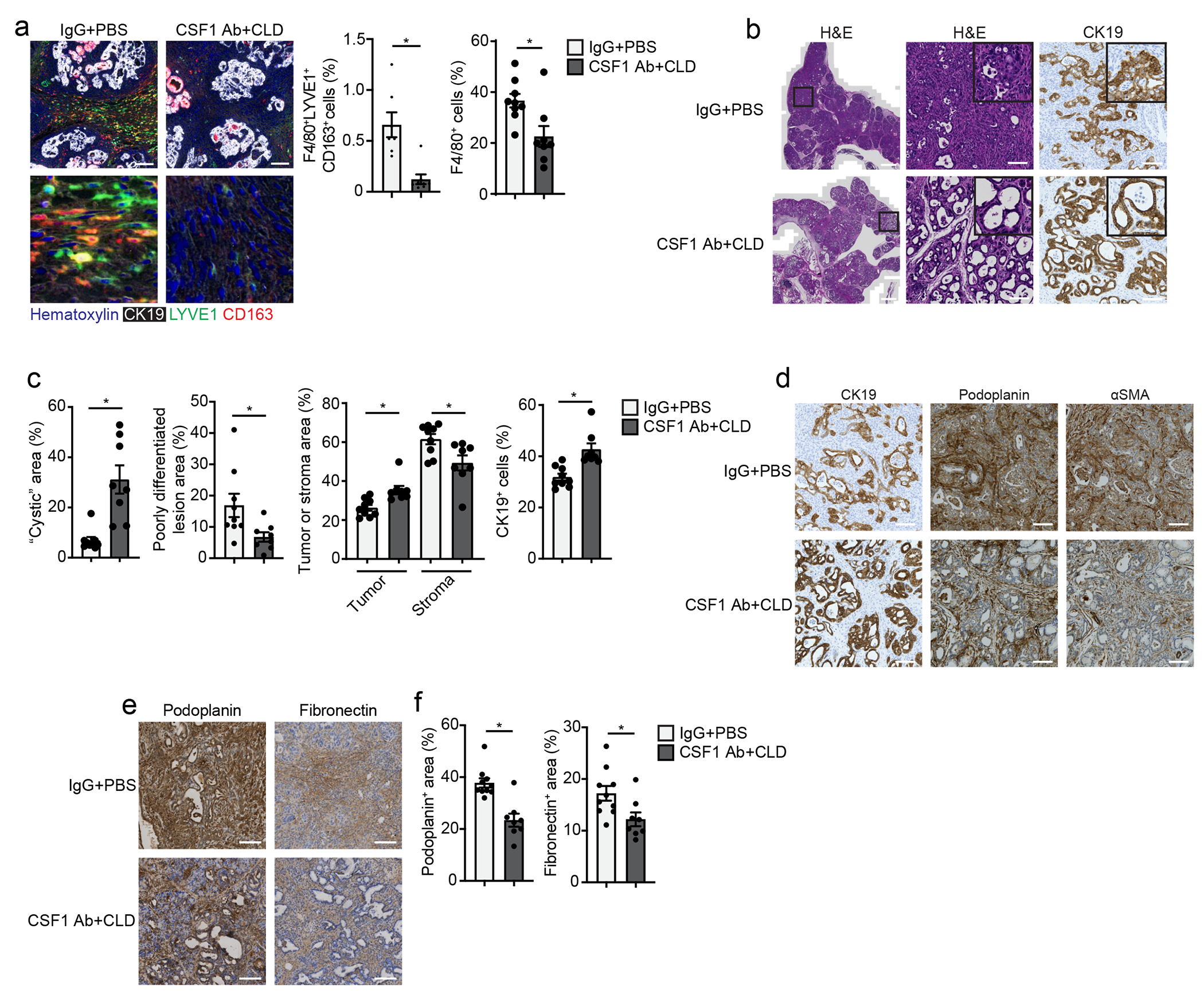
a. Representative mIHC images of pancreas tissue stained for hematoxylin, CK19, F4/80, LYVE1 and CD163 and quantification of F4/80+LYVE1+CD163+ macrophages and F4/80+ macrophages, displayed as the percentage of cells, from 2.5 month old p48-Cre LSL-KRASG12D p53fl/+ (hereafter KPC) mice treated with IgG+PBS or CSF1 Ab+CLD, recovered for 10 days then treated with 6 hourly i.p. injections of Cer every other day for 5 days; F4/80+LYVE1+CD163+ macrophage analysis: IgG+PBS, n=7; CSF1 Ab+CLD, n=8 mice; *P=0.0012; F4/80+ macrophage analysis: IgG+PBS, n=9; CSF1 Ab+CLD, n=8 mice; *P=0.0079. Scale bar, 100μM. b. Representative images of pancreas tissue stained for H&E or CK19 from IgG+PBS or CSF1 Ab+CLD-treated KPC mice as in a. Scale bars are 1mm (left column) or 100μM (right two columns). c. Quantification of tumor area presenting cyst-like appearance, poorly differentiated/high grade invasive pancreatic tumor lesions, tumor lesions and stromal areas as percent of total pancreas area, and CK19+ cells as percentage of total cells in IgG+PBS or CSF1 Ab+CLD-treated KPC mice as in a; IgG+PBS, n=9 mice; CSF1 Ab+CLD, n=8, left to right, *P=0.0003 for cystic area analysis, *P=0.0079 for poorly differentiated area analysis, *P=0.0010 and *P=0.0152 for tumor and stromal area analysis, and *P=0.0003 for CK19 analysis. d. Representative images of pancreas tissue stained for CK19, podoplanin or αSMA from IgG+PBS or CSF1 Ab+CLD-treated KPC mice as in a, scale bars are 100μM. e. Representative images of pancreas tissue from 2 month old p48-Cre LSL-KRASG12D p53fl/fl (KPPC) mice treated with IgG+PBS or CSF1 Ab+CLD at 1 month of age and rested for 4 weeks. Scale bars are 100μM. f. Quantification of podoplanin+ or fibronectin+ area in KPPC mice treated with IgG+PBS or CSF1 Ab+CLD as in e. IgG+PBS, n=10 mice; CSF1 Ab+CLD, n=8, *P=0.0014 for podoplanin analysis and *P=0.0205 for fibronectin analysis. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Discussion
Here we showed that a significant portion of pancreas macrophages were embryonic-derived. TRMs in the pancreas did not rely on continual replenishment from monocytes and displayed distinct transcriptional phenotype and location compared to MDMs. The LYVE1hiCX3CR1loMHCIIlo TRM population expressed gene sets indicative of ECM remodeling and alternative-activation. Depletion of the LYVE1hi subset impacted the accumulation and phenotype of fibroblasts during pancreatitis and diminished the effects of protective fibrosis.
It is not fully understood to what extent macrophage origin versus tissue residence or microenvironment impact their phenotype. Lineage-tracing studies have shown monocyte-derived cells can adopt similar phenotype as TRMs over time6. Transcriptional and epigenetic profiling has shown macrophages retain tremendous plasticity, and adoptive transfer of TRMs into a different tissue can cause the reprogramming of up to 70% of all genes14. This could align with the preferential localization of LYVE1+ TRMs within areas of higher fibrosis, suggesting their role in coordinating fibrosis may be driven by their specific microenvironment within the stroma. Further, pancreas TRMs can be distinguished as LYVE1hiCX3CR1loMHCIIlo or LYVE1loCX3CR1hiMHCIIhi, in agreement with expression of phenotypic markers in TRMs in fat, lung, mesenteric membranes and other tissues17, 18, 19, 20, 43. Studies on LYVE1hiMHCIIlo TRMs in the lung, arteries and mesenteric membranes report their involvement in ECM remodeling, and display a strikingly similar transcriptional profile as LYVE1hiMHCIIlo TRMs in the pancreas.
TRM-driven tissue remodeling also led to distinct outcomes depending on the type of tissue injury. TRM-driven fibrosis was critically important for survival and recovery during pancreatitis through protective effects that limited tissue damage. It is thought that pancreatic stellate cell (PSC) activation is critical for recovery following inflammatory injury, both by providing growth factors and possibly driving ductal cell proliferation and regeneration26, 27. PSC activation for wound healing must, however, be transient. In a chronic setting, prolonged PSC activation is detrimental and can induce extensive fibrosis, which can interfere with pancreas function23, 27. Our study was limited to acute pancreatitis, where inflammation resolves upon cerulein withdrawal, but it would be important to understand how TRM-fibroblast interactions change in a chronic inflammation setting.
Similar to chronic pancreatitis, tumors involve prolonged inflammation and wound healing responses. We showed that TRM-driven fibrosis was coopted by tumors to promote tumor growth. While still debated, studies have reported that fibrosis, and even macrophage-induced fibrosis, can play a tumor-supportive role18, 29, 44, 45. Fibrotic pathologies have been linked to higher risk of cancer, with chronic pancreatitis being a significant risk factor for PDAC46, 47. TRMs have also been reported to support tumor growth through other mechanisms. In lung adenocarcinoma, TRMs coordinate Treg cell responses, and when depleted, tumors grow more slowly21. In metastatic ovarian cancer models, TRMs promote tumor cell growth and metastatic spreading18, 48. While the pro-tumor mechanisms of TRMs vary by tumor model, most TRMs display an alternative-activation phenotype and secrete growth factors and cytokines to either directly or indirectly promote tumor progression.
MDMs were reported to promote inflammation and tissue damage during pancreatitis25. Our transcriptional profiling indicated the upregulation of inflammatory pathways in MDMs, but we were not able to detect a functional outcome by targeting them. CCR2-KO mice showed no changes in ADM, tissue damage or mouse survival, perhaps due to CCR2-independent mechanisms by which inflammatory macrophages could accumulate. In summary, our study demonstrated that LYVE1hi TRMs expand in number to help coordinate a fibrotic response critical in protecting the exocrine pancreas from inflammatory damage, but this program is coopted by tumors to support their growth.
Methods
Mouse models
The following mouse models were purchased from Jackson Laboratories: CCR2-KO, Rosa26-LSL-eYFP, Csf1r-mer-iCre-mer. Lyve1-Cre;CSF1Rfl/fl (Lyve1ΔCSF1R) were a gift from G. Randolph at Washington University in St. Louis. LSL-tdTomato mice in the FVB/NJ background were a gift from G. Longmore at Washington University in St. Louis, and were crossed to Csf1r-mer-iCre-mer for lineage tracing studies. Flt3-Cre mice were a gift from T. Boehm at Max Planck Institute, and were crossed to Rosa26-LSL-eYFP. For Flt3-Cre;Rose26-LSL-eYFP experiments, male mice were used between ages of 8-16 weeks. All other experiments were performed on both male and female mice between the ages of 8-16 weeks. Mice were maintained in the Laboratory for Animal Care barrier facility at the Washington University School of Medicine under 12-hour light/dark cycle. Washington University School of Medicine Institutional Animal Studies Committee approved all animal studies. Survival end points were scored when mice lost >15% body weight or tumors reached >1.8cm in diameter by palpation.
Murine tumor models
KPC mice (p48-Cre;LSL-KRASG12D;p53fl/+) used in these studies have been previously described. They were backcrossed to C57BL/6 mice and tested for congenic markers. iKRAS* mice (Pft1a-Cre;LSL-rtTa;TetO-KRASG12D) were a kind gift from Dr. Marina Pasca di Magliano, and have been previously characterized. Briefly, KrasG12D expression was induced by administration of doxycycline in drinking water at a concentration of 0.5mg/mL, supplemented with 3% sucrose w/v. Upon start of doxycycline, mice were treated with acute cerulein on two consecutive days, each day injecting 100ug/kg of cerulein every hour for 6 hours. Doxycycline administration was continued for 3 weeks, replacing drinking water every 3 days. KPC-1 cell line was derived from a 2.2 month old KPPC mouse (p48-Cre;LSL-KRASG12D;p53fl/fl), and grown on collagen-coated tissue culture flasks for < 12 generations. To establish orthotopic pancreas KPC tumor models, Flt3-YFP or C57BL/6 mice were implanted with 100,000 KPC-1 cells in 5uL serum-free DME/F-12 and 45uL Cultrex (Trevigen) by injecting cells directly into the pancreas.
For lung tumors, the KPL86 cell line was derived from lung tissues of the 9-month-old KrasLSL-G12D/p53flox/+ mouse treated with adenovirus-delivered Cre-recombinase. Cells were grown on collagen-coated tissue culture flasks for < 10 passages. To establish orthotopic NSCLC models, 1,000,000 KPL86 cells in 100 μL of phosphate-buffered saline (PBS, Trevigen, Gaithersburg, MD, USA) were retro-orbitally injected into 8-12-week-old Flt3-YFP mice.
For liver metastasis of PDAC cells, hemispleen injection of 500,000 KPC-2 (aka “KP2”) tumor cells was performed using an adapted procedure from published studies1, 2. The KP2 cell line was derived from pancreatic tumors of a 6-month-old p48-CRE+/LSL-KRASG12D/p53flox/+ mouse, and tumor cells additionally were modified to express mCherry and click beetle red luciferase. To implant cells in the liver, the mice were anesthetized under continuously-monitored isoflurane, and the spleen was exteriorized from the peritoneal cavity under sterile conditions through a left flank laparotomy. The spleen was ligated using medium titanium Horizon ligation clips (Teleflex, Morrisville, NC) and divided in half. Tumor single-cell suspension in sterile PBS (100 uL), followed by 150uL of sterile PBS flush, was injected into the inferior pole of the spleen with a 30-gauge needle (BD, Franklin Lakes, NJ), monitoring for blanching of the spleen parenchyma and splenic vein to ensure injection efficacy. Once the cells and PBS flush were injected, the splenic vein was ligated, and the inferior pole of the spleen was excised. Once hemostasis was ensured, the peritoneum was closed with 5-0 ligature (Ethicon, Raritan, NJ), and the skin was closed with the AutoClip system (Braintree Scientific, Braintree, MA). Mice were post-operatively monitored and managed per IACUC guidelines for survival surgery. Livers were harvested at day 14 for cell sorting and single cell RNA sequencing analysis.
Human samples
Human PDAC (with adjacent non-tumor “normal” pancreatic tissue) and pancreatitis (with adjacent “normal” pancreatic tissue) samples were obtained from consenting patients diagnosed at Barnes-Jewish Hospital. The studies described were approved by the Washington University Institutional Review Board under protocol numbers 201704078 and 201908148. Adjacent healthy pancreas was identified by pathologist by H&E-stained slides.
Experimental pancreatitis
Acute pancreatitis was induced in mice by administering 6 hourly intraperitoneal injections (i.e. once per hour for six hours) of cerulein at a dose of 100ug/kg given every other day for one week.
For severe acute pancreatitis via osmotic pump delivery of cerulein, osmotic pumps (Alzet Osmotic Pumps, Cupertino, CA) were loaded with 100uL of concentrated cerulein (8ug cerulein per day was loaded with 1.33mg/mL and 10ug cerulein per day was loaded with 1.67mg/mL). Pumps were then implanted into the peritoneal cavity of mice, and body weight was monitored every day for up to 14 days.
Macrophage depletion
Tissue-resident macrophages were depleted with a combination of CSF1 neutralizing antibodies and clodronate-loaded liposomes. 8-12 week old FVB/NJ mice were treated with two doses of CSF1 antibody (BioXcell, 5A1, 1mg and 0.5mg on days −14 and −11 respectively), followed by two doses of clodronate-loaded liposomes (Liposoma, 200uL each dose) on days −13 and −10. Mice were then allowed to recover for 10 days to allow monocytes and monocyte-derived macrophages to reconstitute. On day 0, mice were either treated with cerulein by IP injection (described above) or implanted with osmotic pump to deliver cerulein.
Similarly, KPC mice were treated with two doses of CSF1 Ab (1mg and 0.5mg) separated by two days and starting at 10 weeks of age. Subsequent days after CSF1 Ab treatment, mice were injected with 100uL of clodronate-loaded liposomes. Mice were then allowed to recover for ~2 weeks, then given brief cerulein treatment starting at 12 weeks of age by giving 6 hourly injections of 100ug/kg cerulein every other day for 5 days. Following cerulein, tumors were allowed to grow for ~3 weeks and mice were sacrificed at 15 weeks of age. KPPC mice were similarly treated with two doses of CSF1 Ab (both 0.5mg) and two doses of clodronate-loaded liposomes (100uL each) starting at 1-month age, as previously described3. Tissue was then taken and analyzed at 2-months of age.
iKRAS* mice were treated with a similar regimen of two doses of CSF1 Ab (1mg and 0.5mg) 14 and 11 days before starting doxycycline administration. Following CSF1 Ab treatment, mice were injected with clodronate-loaded liposomes (100uL) on subsequent days. After CSF1 Ab and clodronate treatment, mice were allowed to recover for 10 days, after which doxycycline was given in drinking water (0.5mg/mL with 3% sucrose replaced every 3 days, as above) for ~3 weeks. Additionally, on days 0 and 1, iKRAS* mice were treated with acute cerulein (100ug/kg) by 6 hourly injections.
Lineage tracing
For lineage tracing of embryonically derived macrophages, timed breeding pairs were set up by crossing Csf1r-mer-iCre-mer mice with Rosa26-LSL-tdTomato mice (both on FVB/NJ background). Embryonic timeline was estimated by observation of vaginal plug, with 12 pm on the day of plug formation being 0.5 days post coitum (dpc). Pregnant mice were then treated with tamoxifen (75ug/g) supplemented with progesterone (37.5ug/g) dissolved in corn oil at ~9.5dpc. Offspring of tamoxifen-pulsed mice were then treated with vehicle or acute cerulein (as described above) at 6 weeks of age and sacrificed for fate mapping analyses.
For lineage tracing of resident macrophages, adult Cx3cr1-CreERT2;LSL-tdTomato and Csf1r-mer-iCre-mer;LSL-tdTomato mice were pulsed with tamoxifen (75mg/kg) dissolved in corn oil on five consecutive days. Mice were then allowed a chase period or 3-10 weeks, or until blood monocytes lost tdTomato labeling. Following chase period, mice were treated with vehicle or acute cerulein as described above, then sacrificed for flow cytometry or immunohistochemistry analyses.
Parabiosis
Parabiotic pairs were established based on previously described protocols3, 4. Briefly, age and weight matched female CD45.2+ and CD45.1+ mice were surgically joined. Mice were injected with Buprenex subcutaneously after surgery. Sulfatrim was continuously added in drinking water for 10 days post-surgery to minimize infections at surgical wounds. Mice were separated and perfused with phosphate buffered saline (PBS) containing 0.2% heparin. Single cell suspensions from tissues were stained with antibodies for flow cytometry analyses 6 weeks after the establishment of parabiosis.
PDGFR inhibition with imatinib mesylate
Mice were treated once daily with IP injection of 100mg/kg imatinib mesylate (MCE, HY-50946). Imatinib was dissolved in 10% DMSO in PBS, then diluted 1:10 for final concentration of 100mg/kg in 1% DMSO solution. Concurrent with imatinib treatment, mice were either treated with standard IP injection of cerulein for flow cytometry and histology experiments, or were implanted with osmotic pumps loaded with 1.67mg/mL cerulein to deliver 10ug/day (as described above).
Mouse tissue isolation and flow cytometry
Mice were sacrificed by cardiac perfusion with ~10-15mL PBS-heparin under isofluorane anesthesia. Pancreas or tumor tissue was then isolated, minced with scissors, and digested in ~15-25mL Dulbecco’s Modified Eagle Medium (DMEM) with 2mg/mL Collagenase A and 1x DNase I for either 15 minutes (steady-state pancreas and cerulein treated pancreas) or 25 minutes (tumor tissue) at 37°C with constant stirring. Digestion buffer was quenched with 3mL fetal bovine serum (FBS) and filtered through 100uM Nylon mesh, pelleted by centrifugation (2000rmp for 5 min) and resuspended in FACS buffer (1% BSA in PBS). Brain tissue was minced and triturated through 40uM Nylon mesh and resuspended in FACS buffer to obtain single-cell suspension.
Cell suspensions were then blocked with rat anti-mouse CD16/CD32 antibodies for 10 minutes on ice, pelleted by centrifugation, and stained with fluorophore-conjugated extracellular antibodies for 25 minutes on ice. For samples with endogenous fluorescent protein labeling, cells were then washed twice with FACS Buffer, then run on BD Fortessa X20 immediately. For unlabeled cells, samples were washed twice following antibody staining, fixed with BD Fixation Buffer for 30 minutes on ice, washed, and resuspended in FACS Buffer. Fixed samples were stored at 4°C and run on BD Fortessa X20 within two weeks. For samples requiring intracellular staining, cells were washed following extracellular staining, and stained using eBioscience FoxP3 Transcription Factor Staining Kit according to instructions by manufacturer.
To quantify proliferating cells in Flt3-YFP mice, tissue was digested and extracellular staining was conducted as described above. Cells were then fixed in 4% paraformaldehyde for 10 minutes on ice, then permeabilized in ice-cold 70% ethanol for 3 hours, and stained with Ki-67 antibody diluted in FACS Buffer for 20 minutes on ice. Cells were then washed twice and immediately run on BD Fortessa X20.
To quantitate blood monocytes, 100uL blood was obtained by cardiac puncture prior to perfusions and deposited into PBS-heparin. Blood was then pelleted by centrifugation, resuspended in 5mL red blood cell lysis buffer for 10 minutes at room temperature, cells pelleted by centrifugation, then stained with fluorophore-conjugated antibodies for 25 minutes on ice. Cells were then fixed in BD Fixation Buffer for 30 minutes on ice, washed twice with FACS Buffer, and stored at 4°C and run on BD Fortessa X20 within two weeks. For experiments requiring blood quantitation prior to mice being sacrificed, blood was obtained by tail vein bleeding, resuspended in RBS Lysis Buffer, stained with extracellular antibodies as described above.
Fluorescence-activated cell sorting (FACS)
Single-cell suspensions were obtained as described above. Cells were then resuspended in FACS Buffer containing CD16/CD32 antibodies to block for 10 minutes on ice, pelleted by centrifugation, then resuspended in fluorophore-conjugated antibodies, stained for 25 minutes on ice, then washed with FACS Buffer. Cells were then immediately sorted on Aria-II (BD Biosciences).
For homeostatic pancreas, pancreatitis, and pancreatic tumors, live macrophages (CD45+CD11b+CD3-CD19-B220-SiglecF-Ly6G-Ly6C-7AAD-F4/80+MHCIIHI/Lo) were sorted. For Bulk RNAseq analyses, Flt3-YFP positive and negative macrophages were sorted directly into RNA Lysis Buffer (Omega Biotek) and RNA isolated using EZNA Kit (Omega Biotek). For single-cell RNA-sequencing (scRNAseq), cells were sorted into FACS buffer and library preparation was conducted as described below.
Liver single-cell suspensions were obtained as described above. Cells were then resuspended in anti-CD45 magnetic beads (BD Biosciences), incubated for 15 minutes on ice, washed in FACS buffer, and applied to LS magnetic column to enrich for CD45+ cells. Following magnetic isolation, cells were spun down, resuspended in fluorophore-conjugated antibodies, stained for 25 minutes on ice, washed with FACS buffer, then immediately sorted. Flt3-YFP positive and negative liver macrophages were sorted (CD45+CD11b+CD3-CD19-B220-SiglecF-Ly6G-7AAD-) into FACS buffer, then scRNAseq library preparation was conducted as described below.
Lung single-cell suspensions were obtained as described above. Cells were then resuspended in FACS Buffer containing CD16/CD32 antibodies to block for 10 minutes on ice, pelleted by centrifugation, then resuspended in fluorophore-conjugated antibodies, stained for 25 minutes on ice, then washed with FACS Buffer. Cells were then immediately sorted on Aria-II (BD Biosciences). Flt3-YFP positive and negative macrophages were sorted (CD45+CD3-CD19-B220-7AAD-CD11b+CD11cHi/LoF4/80+) into FACS buffer, then scRNAseq library preparation was conducted as described below.
For fibroblasts, homeostatic or cerulein treated pancreas tissue was taken, and single-cell suspensions were isolated as described above. Cells were then resuspended in FACS Buffer containing CD16/CD32 antibodies to block for 10 minutes on ice, pelleted by centrifugation, then resuspended in fluorophore-conjugated antibodies, stained for 25 minutes on ice, then washed with FACS Buffer. Cells were then immediately sorted on Aria-II (BD Biosciences). Total fibroblasts were sorted (CD45-EpCAM-CD31-PODOPLANIN+) into FACS Buffer, then scRNAseq library preparation was conducted as described below.
After cells were sorted into FACS Buffer, they were pelleted by centrifugation, resuspended in 0.04% BSA in PBS, and cell counts were obtained. Single-cell RNA-sequencing library preparation was then conducted by the Genome Technology Access Center at Washington University. Briefly, cells from each sample were encapsulated into droplets, and libraries were prepared using Chromium Single Cell 3’v3 Reagent kits according to the manufacturer’s protocol (10x Genomics, Pleasanton, CA, USA). The generated libraries were sequenced by a NovaSeq 6000 sequencing system (Illumina, San Diego, CA, USA) to an average of 50,000 mean reads per cell. Cellranger mkfastq pipeline (10X Genomics) was used to demultiplex illumine base call files to FASTQ files. Afterward, fastq files from each sample were processed with Cellranger counts and aligned to the mouse mm10 reference genome (Cellranger v.4.0.0, 10X Genomics, mouse reference mm10-2020-A from https://cf.10xgenomics.com/supp/cell-exp/refdata-gex-mm10-2020-A.tar.gz) to generate feature barcode matrix.
Mouse bulk RNAseq analysis
Pancreas macrophages were sorted as described above, directly into TRK Lysis buffer (Omega Bio-Tek). RNA was isolated by E.Z.N.A. Total RNA isolation kit per manufacturer’s recommendations (Omega Bio-Tek). Following quality control by bioanalyzer, libraries were prepared using the Clontech SMARTer kit (Takara Bio) at the Washington University Genome Technology Access Center (GTAC). Libraries were then indexed, pooled, and sequenced on Illumina HiSeq 3000 (Illumina). Illumina’s bcl2fastq software was then used for basecalls and demultiplexing. Reads were then aligned to the Ensembl release 76 top-level assembly with STAR version 2.0.4b, and gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version1.4.5. Gene counts were then imported into Edge,R and TMM normalization size factors were calculated to adjust for differences in library size. Limma and voomWithQualityWeights was then used to calculate weighted likelihoods based on the observed mean-variance relationship of every gene and sample. Differential expression analysis was then performed to analyze for differences between Flt3-YFP lineages and results filtered by Benjamini-Hochberg false discovery rate adjusted p-values less than or equal to 0.05. Global perturbations in known Gene Ontology (GO) terms and KEGG pathways were detected using GAGE to test for changes in expression of reported log 2 fold-changes reported by Limma in each term versus background log 2 fold-changes of all genes outside the respective term. GSEA was also performed using GO terms, KEGG pathways, Reactome, and MSigDB gene sets with Benjamini-Hochberg FDR < 0.05. Differentially expressed genes and gene sets were displayed using pHeatmap or the Phantasus online tool (Artyomov, https://artyomovlab.wustl.edu/phantasus/).
Mouse single-cell RNAseq analysis
The filtered feature barcode matrix from each sample was then loaded into Seurat as Seurat objects (Seurat v.3). For each Seurat object, genes that were expressed in less than three cells and cells that expressed less than 1,000 or more than 8,000 genes, were excluded. Cells with greater than 10% mitochondrial RNA content were also excluded, resulting in between 2297 and 17880 cells, as indicated in Supplementary Table 1. SCTransform with default parameters was used on each individual sample to normalize and scale the expression matrix against the sequence depths and percentages of mitochondrial genes 5. Principle component analysis (PCA) was performed (function RunPCA). A UMAP dimensional reduction was performed on the scaled matrix using the first 25 PCA components to obtain a two-dimensional representation of cell states. Then, these defined 25 dimensionalities were used to refine the edge weights between any two cells based on Jaccard similarity (FindNeighbors), and were used to cluster cells through FindClusters functions.
To characterize clusters, the FindAllMarkers function with logfold threshold = 0.25 and minimum 0.25-fold difference and MAST test were used to identify signatures alone with each cluster. For macrophage/monocyte samples, the macrophage/monocytes clusters were selected, and the top 3,000 variable features were recalculated to recluster to a higher resolution. Macrophages were selected based on clusters with high expressions of known macrophage marker genes, including Csf1r, C1qa, and C1qb, and confirmed by the absence of Cd3e, Ms4a1, Krt19, Zbtb46, and Flt3, and further confirmed by identifying DEGs associated with potential macrophage clusters, when compared to known macrophage specific marker genes. For fibroblast samples, the fibroblast clusters were selected and similarly reclustered. Fibroblasts were selected based on clusters with high expression of known fibroblast marker genes, including Podoplanin, Pdgfra, Col1a1, and confirmed by the absence of Krt19, Prss2, Amy1a. For GSEA comparisons, the log2 (fold-change) of all genes detected with min.pct > 0.1 and past MAST test was used as a ranking metric. GSEA was performed using GO terms, KEGG pathways, Reactome, and MSigDB gene sets with Benjamini-Hochberg FDR < 0.05 in ClusterProfiler (Wu et al., 2021). For DEGs between the two groups in each library, we filtered genes with a Bonferroni-corrected p-value < 0.05 and fold-change >1.2 or <0.8.
Human single-cell RNAseq analysis
For the human datasets 6, cells with greater than 15% mitochondrial genes were excluded and cells that expressed less than 500 genes were excluded. SCTransform with default parameters was used on each individual sample to normalize and scale the expression matrix against sequence depth and percentage of mitochondrial genes 5. Cell cycle scores and corresponding cell cycle phases were then calculated and assigned after SCTransform based on S and G2/M genes (CellCycleScoring). Differences between the S phase and G2/M phase were then regressed out by SCTransform on individual samples. Variable features were calculated for every sample in the dataset independently and randed based on the number of samples they were independently identified (SelectIntegrationFeatures). The top 3,000 variable features were then used for PCA (RunPCA). The calculated PCA embedding of each cell was then used as an input for the soft k-means clustering algorithm. Briefly, through iteration, the algorithm designated the cluster-specific centroids and cell-specific correction factors corresponding to batch effects. The correction factors were used to assign cells into clusters until the assignment was stable (RunHarmony). The first 20 PCA components were then used to refine the edge weights between any two cells based on Jaccard similarity (FindNeighbors), and were used to cluster cells through FindClusters function at a resolution of 0.3, resulting in 24 clusters. To characterize clusters, the FindAllMarkers function with logfold threshold = 0.25 and minimum 0.25-fold difference and MAST test were used to identify signatures alone with each cluster. Macrophage/monocyte clusters were then selected, and the top 3,000 variable features were recalculated to recluster to a higher resolution, resulting in 5970 total cells and 8 clusters. Macrophages were selected based on clusters with high expressions of known macrophage marker genes, including CSF1R, C1QA, and C1QB, CD68, and confirmed by the absence of CD3E, MS4A1, KRT19, CD1C, and further confirmed by identifying DEGs associated with potential macrophage clusters, when compared to known macrophage specific marker genes.
Immunohistochemistry and immunofluorescence
Mouse tissues were fixed in 10% formalin overnight at 4°C, dehydrated through graded ethanol washes of 30% ethanol, 50% ethanol, and 70% ethanol for 20 minutes each. Tissue was then run through Leica ASP 6025 tissue processor and embedded in paraffin, then sectioned at 6-μm thick tissue sections. Immunohistochemistry staining was then performed on Bond Rxm autostainer (Leica Biosystems) according to manufacturer’s recommendations. Briefly, paraffin slides were loaded onto machine, baked at 60°C for 15 minutes, dewaxed, rehydrated, antigens were retrieved with either citrate or EDTA antigen retrieval solution (Leica Biosystems). Endogenous peroxidase activity was blocked with a peroxide block, primary antibodies were then applied for 60 minutes. Either species-specific or biotinylated secondary antibodies were then applied to the slides. Next, either DAB or Fast Red chromogens were applied, finally followed by hematoxylin counterstain. Slides stained with DAB were then dehydrated through graded ethanols, xylene, then mounted with Cytoseal XYL (Thermo Fisher), and slides stained with Fast Red were air dried for 15 minutes then mounted with Vectamount Permanent Mounting Medium (Vector Laboratories).
Multiplex immunohistochemistry staining
Formalin-fixed paraffin-embedded (FFPE) tissue was sectioned at 6-μm, loaded onto Bond Rxm autostainer, baked for 60 minutes at 60°C, dewaxed and rehydrated. Series of staining was performed with multiple markers as indicated, based on previously published study 7. Briefly, slides were first blocked with peroxide block, goat serum block, and species-specific F’ab block (Jackson Laboratories), then primary antibodies were incubated for 60 minutes, followed by species-specific secondaries conjugated to horseradish peroxidase (HRP). For primary antibodies raised in rabbit, Polymer (anti-rabbit poly-HRP, Leica Biosystems) was used as a secondary antibody, for primary antibodies raised in mouse, Post-Primary (rabbit anti-mouse, Leica Biosystems) secondary was used, followed by Polymer, for other primary antibodies, biotinylated species-specific secondaries were used, followed by streptavidin-HRP (SA-HRP, Leica Biosystems)(anti-rabbit poly-HRP and SA-HRP are provided in Intense R and Polymer Refine Detection Kits respectively, Leica Biosystems). Slides were then chromogenically visualized with AEC (Invitrogen) and counterstained with hematoxylin (Dako). Stained slides were then mounted with Vectamount Aqueous Mounting Medium (Vector Laboratories) and imaged on Zeiss Axio Scan.Z1 (Zeiss) at 20X magnification. Before the next round of staining, coverslips and mounting medium was removed by soaking slides in TBST (1X TBS with 0.05% Tween-20) for several hours, then destained by graded ethanols (50% ethanol, 70% ethanol with 1% HCl, 100% ethanol, 70% ethanol, 50% ethanol, and DI water for 5-10 minutes each) then loaded onto Bond Rxm for staining of next marker. Images were cropped and exported using Zeiss ZEN software (Zeiss, v3.2.0) then analyzed using HALO software (Indica labs).
Quantification and statistical analysis
Number of mice and statistical tests used are reported in each figure legend. Statistical analyses were performed by Graphpad Prism v9, using unpaired Student’s t-test, ANOVA analysis (Bonferroni multiple comparisons), or unpaired non-parametric Mann-Whitney U test as appropriate for normality of data. Log-Rank (Mantel-Cox) test was used for survival experiments. Data displayed are mean ± SEM, unless otherwise noted. p<0.05 is considered statistically significant for all studies; *p<0.05, n.s. denotes not significant. Mice genotyped as positive for strain-specific alleles were allocated randomly into treatment groups, littermate controls that were genotyped negative were subsequently allocated randomly into control groups when appropriate. All mice were assigned numbers to allow researchers to be blinded to mouse genotype or treatment group when possible. During data collection by either flow cytometry, IHC analysis, or pathological grading, mice were only referred to by their number to avoid bias towards genotype or treatment group. Data were then organized according to group or treatment once data collection was complete.
Data availability
Sequencing data (bulk and single-cell RNAseq) has been made publicly available under Gene Expression Omnibus (GEO) repository accession numbers GSE196778 (scRNAseq), and GSE203005 (bulk RNAseq). All other data are available upon request to the lead contact, David G. DeNardo (ddenardo@wustl.edu).
Code availability
No custom code was generated for this study. Code used to generate figures are available upon request to the lead contact, David G. DeNardo (ddenardo@wustl.edu).
Extended Data
Extended Data Figure 1:
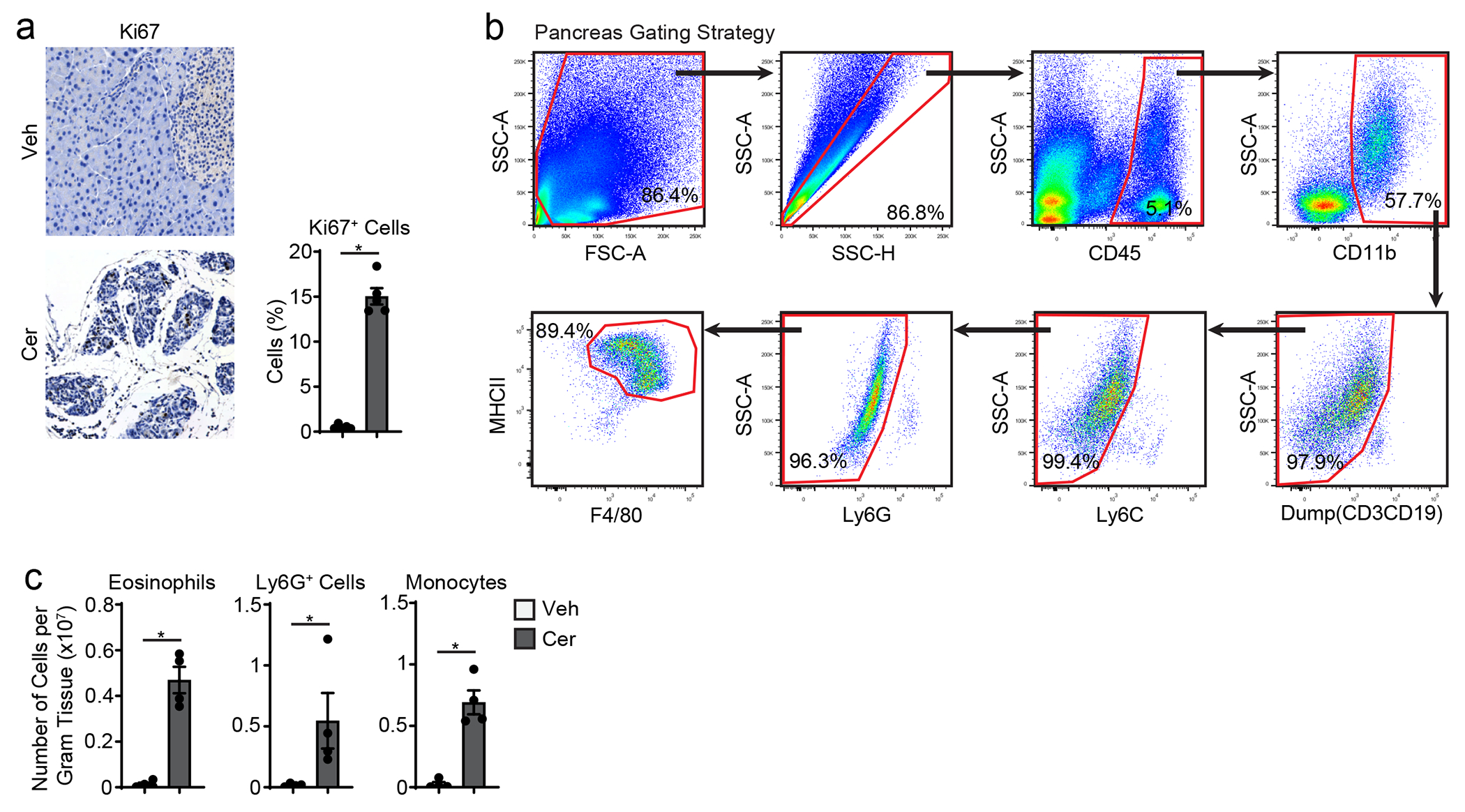
a. Representative immunohistochemistry images and quantification of Ki67 staining in pancreas tissue of vehicle and cerulein treated mice as in a; n=4 mice/group, *P=0.0079. b. Flow cytometry plots of gating strategy used for pancreas F4/80+MHCIIhi/lo macrophages. c. Density of F4/80loMHCII− eosinophils, Ly6G+ granulocytes, and Ly6C+ monocytes in pancreas of mice treated with vehicle or cerulein by 6 hourly i.p. injections every other day for one week; n=4 mice/group, *P=0.0286 for eosinophil analysis, *P=0.0286 for granulocyte analysis, and *P=0.0286 for monocyte analysis. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Extended Data Figure 2:
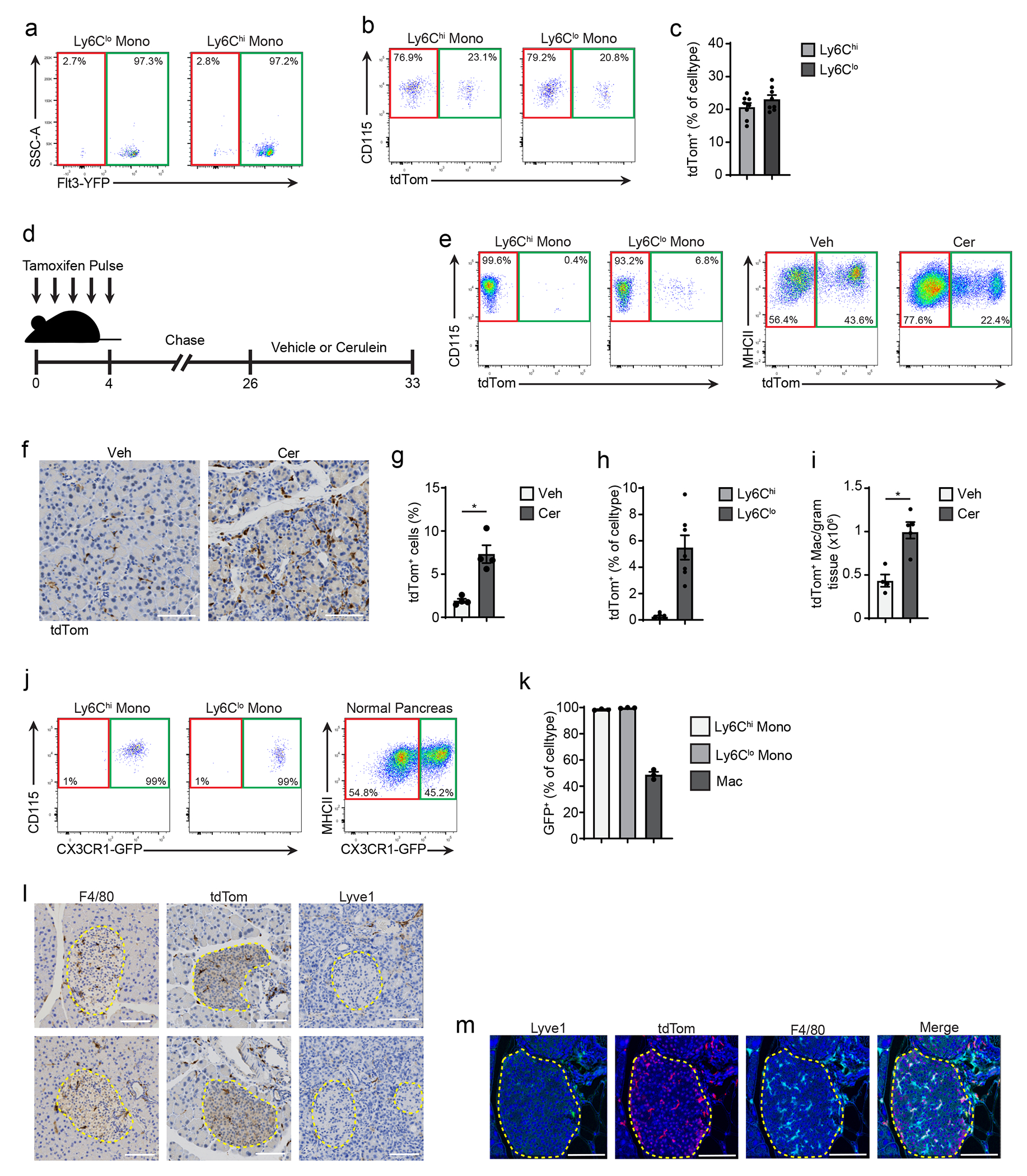
a. Flow cytometry of Flt3-YFP+/− cells pre-gated on blood Ly6Clo/hi monocytes. b. Flow cytometry of CSF1R-tdTom mice given tamoxifen by oral-gavage for five consecutive days, then tamoxifen was stopped for 10 weeks, pre-gated on Ly6Chi/lo blood monocytes. c. Quantification of tdTomato+ Ly6Chi/lo blood monocytes after treatment as in b, displayed as percentage of each cell type; n=8mice/group. d. Schematic of tamoxifen treatment in CX3CR1-CreERT2;LSL-tdTomato mice (hereafter CX3CR1-tdTom mice). e. Flow cytometry of tdTomato−/+ cells in CX3CR1-tdTom mice treated as in d followed by vehicle or cerulein by 6 hourly i.p. injections every other day for one week, pre-gated on Ly6Chi/lo blood monocytes, and F4/80+MHCIIhi/lo pancreas macrophages. f. Representative images of IHC staining of tdTomato in pancreas tissue of CX3CR1-tdTom mice treated as in e, scale bars, 100μM. g. Quantification of tdTomato+ cells from pancreas tissue of CX3CR1-tdTom mice treated as in e, displayed as percentage of cells; n=4mice/group, *P=0.0286. h. Quantification of tdTomato+ blood Ly6Chi/lo monocytes from CX3CR1-tdTom mice treated as in e, displayed as percentage of each cell type; n=7mice/group. i. Density of tdTomato+ pancreas F4/80+MHCIIhi/lo macrophages from CX3CR1-tdTom mice treated as in e; vehicle, n=4 mice; cerulein, n=5 mice, *P=0.0159. j. Flow cytometry of GFP in CX3CR1-GFP mice, pre-gated on Ly6Chi/lo blood monocytes, and pancreas F4/80+MHCIIhi/lo macrophages. k. Quantification of GFP+ blood Ly6Chi/lo monocytes and pancreas F4/80+MHCIIhi/lo pancreas macrophages from CX3CR1-GFP mice, displayed as percentage of each cell type; n=3mice/group. l. Representative images of IHC staining of pancreas tissue from CX3CR1-tdTom mice treated as in e and stained for F4/80, tdTomato, and LYVE1, islets of Langerhans outlined in yellow, scale bars, 100μM, staining was repeated for 8 mice. m. Representative images of multiplex-IHC (mIHC) staining of pancreas tissue from CSF1R-tdTom mice treated with tamoxifen as in b, followed by cerulein treatment as in e, and stained for LYVE1, tdTomato, and F4/80, islet of Langerhans outlined in yellow dash, scale bars, 100μM, staining was repeated for 4 vehicle- and 4 cerulein-treated mice. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Extended Data Figure 3:
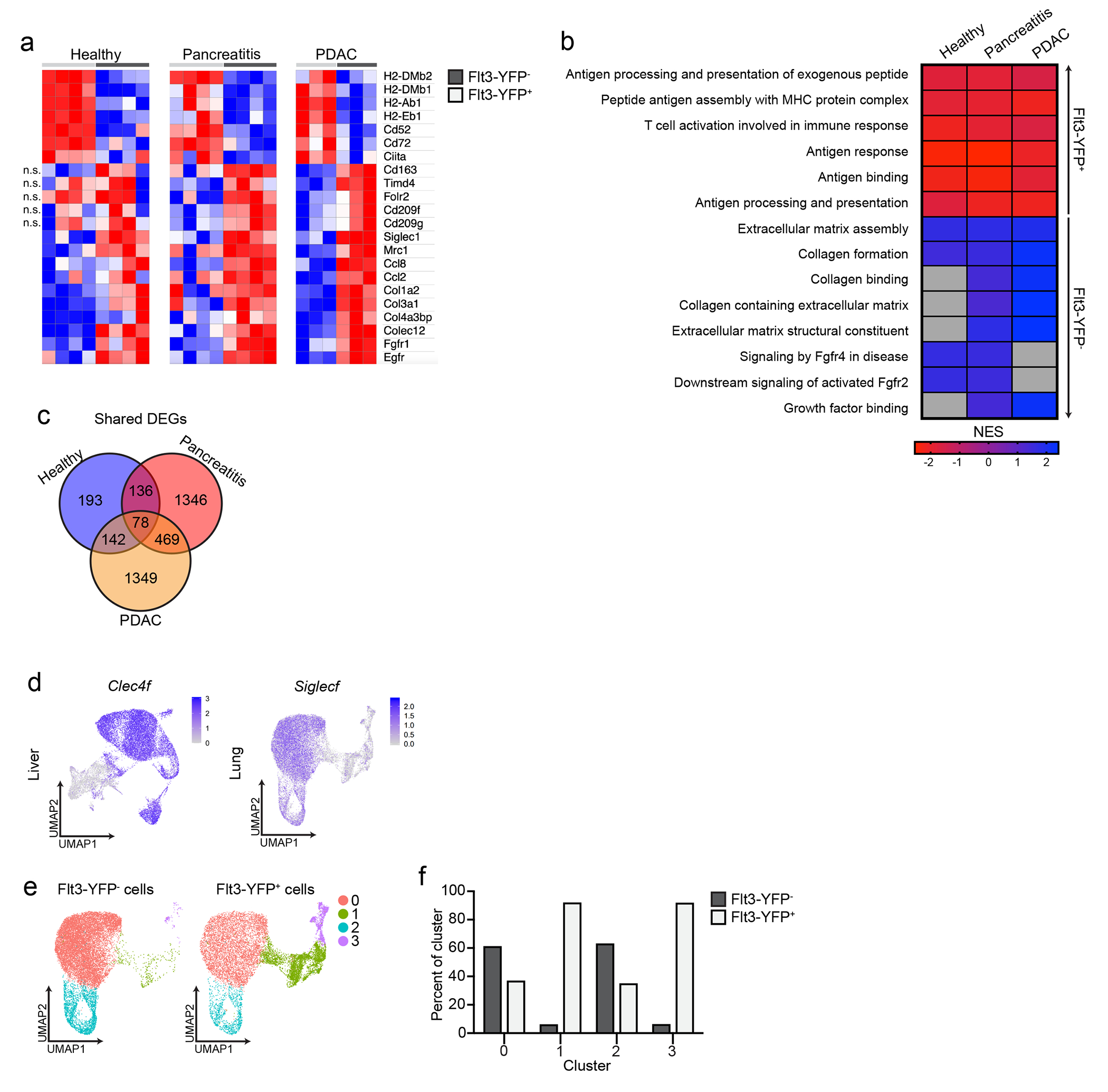
a. Heatmap displaying DEGs in bulk RNAseq data upregulated in Flt3-YFP+ or Flt3-YFP− pancreas macrophages from mice treated with 6 hourly i.p. injections every other day for one week of vehicle (healthy), cerulein (pancreatitis), or orthotopically implanted with the KP1 pancreatic cancer cell line (PDAC). b. Heatmap displaying normalized enrichment score (NES) of significantly enriched gene sets in bulk RNAseq data upregulated in Flt3-YFP+ or Flt3-YFP− pancreas macrophages of mice treated as in a, pathways selected by FDR < 0.05. c. Venn diagram of overlap in number of DEGs upregulated in Flt3-YFP+ or Flt3-YFP− pancreas macrophages from mice treated as in a. d. UMAP plot displaying Clec4f expression in F4/80+MHCIIhi/lo macrophages sorted from livers of Flt3-YFP mice 14 days after implantation with the KP2 pancreatic cancer cell line, and UMAP plot displaying Siglecf expression in CD11bint/hiCD11cint/hiF4/80+ macrophages from lungs of Flt3-YFP mice 15 days after i.v. injection of the KPL86 lung cancer cell line. e. UMAP plot from scRNAseq analysis of CD11bint/hiCD11cint/hiF4/80+ macrophages sorted from Flt3-YFP lungs implanted with KPL86 lung cancer cell line as in a. f. Quantification of Flt3-YFP+ and Flt3-YFP− lung macrophages by cluster from UMAP in e, displayed as percentage of each cluster.
Extended Data Figure 4:
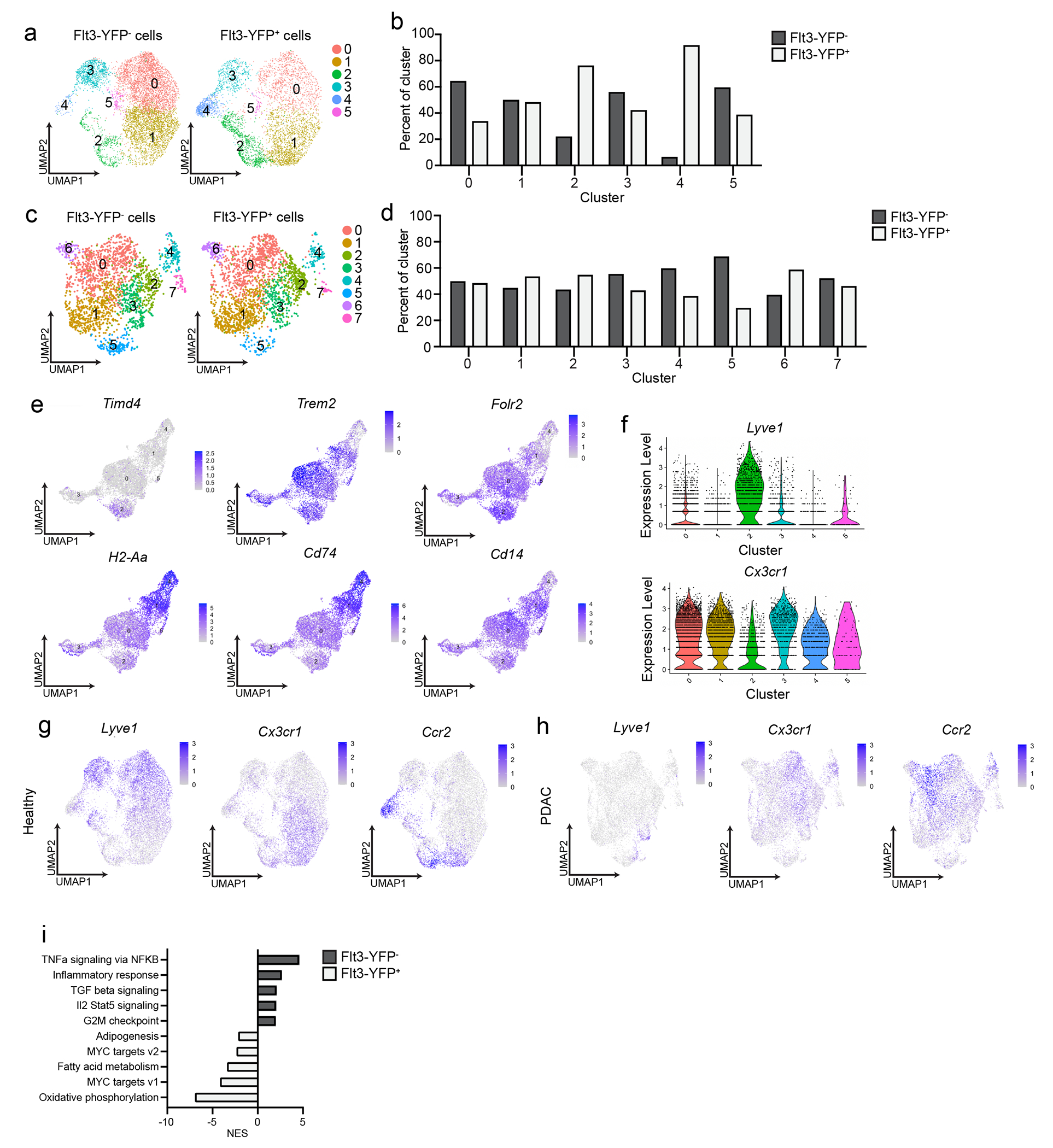
a. UMAP plot of scRNAseq analysis of F4/80+MHCIIhi/lo macrophages sorted from pancreas of Flt3-YFP mice treated with vehicle by 6 hourly i.p. injections every other day for one week. b. Quantification of Flt3-YFP+ and Flt3-YFP− pancreas macrophages by cluster from UMAP in a, displayed as percentage of each cluster. c. UMAP plot of scRNAseq analysis of F4/80+MHCIIhi/lo macrophages sorted from pancreas of Flt3-YFP mice orthotopically implanted with the KP1 pancreatic cancer cell line. d. Quantification of Flt3-YFP+ and Flt3-YFP− pancreas macrophages by cluster from UMAP in c, displayed as percentage of each cluster. e. UMAP plots displaying Timd4, Trem2, Folr2, H2-Aa, Cd74, and Cd14 expression in Csf1r and C1qa expressing pancreas macrophages from Flt3-YFP mice treated with cerulein by 6 hourly i.p. injections every other day for one week. f. Violin plots displaying expression of Lyve1 and Cx3cr1 for each cluster of Csf1r and C1qa expressing pancreas macrophages from Flt3-YFP mice treated with cerulein as in a. g. UMAP plots displaying Lyve1, Cx3cr1, and Ccr2 expression in F4/80+MHCIIhi/lo macrophages sorted from pancreas of Flt3-YFP mice treated as in a. h. UMAP plots displaying Lyve1, Cx3cr1, and Ccr2 expression in F4/80+MHCIIhi/lo macrophages sorted from pancreas of Flt3-YFP mice implanted with tumors as in c. i. Bar graph of normalized enrichment score (NES) values of gene sets upregulated in Flt3-YFP+ or Flt3-YFP− pancreas macrophages within pancreatitis cluster 0 (from Fig. 3c). Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used, except i where FDR was used.
Extended Data Figure 5:
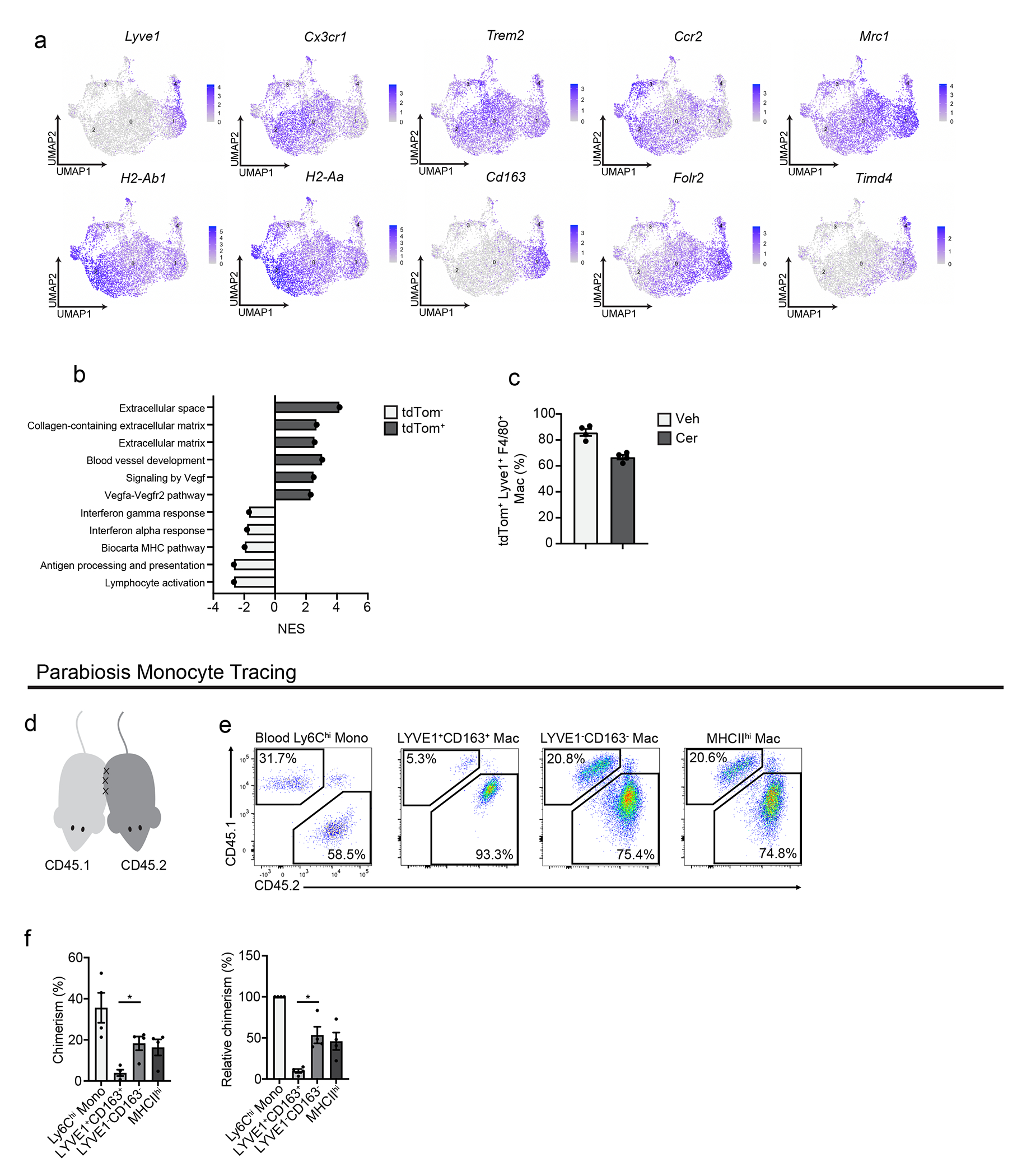
a. UMAP plots displaying Lyve1, Cx3cr1, Trem2, Ccr2, Mrc1, H2-Ab1, H2-Aa, Cd163, Folr2, and Timd4 expression in Csf1r and C1qa expressing pancreas macrophages from CSF1R- tdTom mice given tamoxifen by oral gavage for five consecutive days, then tamoxifen was stopped for 10 weeks, then cerulein (cer) was given by 6 hourly i.p. injections every other day for one week. b. Bar graph of NES values of gene sets upregulated in tdTomato+ or tdTomato− cells from CSF1R-tdTom mice treated as in a. c. Quantification of LYVE1+F4/80+tdTomato+ pancreas macrophages from CSF1R-tdTom mice treated with tamoxifen as in a, followed by vehicle (veh) or cerulein (cer) treatment as in a, displayed as percentage of LYVE1+F4/80+ macrophages; n=4 mice/group. d. Schematic of surgical joining of parabiotic pairs of CD45.1+ and CD45.2+ C57BL/6 mice. e. Flow cytometry staining of CD45.1 and CD45.2 in parabiotic pairs of mice, following 6 weeks of surgical joining, pre-gated on blood Ly6C+ monocytes or F4/80+MHCIIhi/lo pancreas macrophages. f. Quantification of percent chimerism (left) and chimerism normalized to blood Ly6Chi monocyte chimerism (relative chimerism, right) for blood Ly6Chi monocytes, LYVE1+CD163+ pancreas macrophages, LYVE1+CD163+ pancreas macrophages, and MHCIIhi pancreas macrophages; n=4 mice/group, *P=0.0286 for chimerism analysis and *P=0.0286 for relative chimerism analysis. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used, except b where FDR was used.
Extended Data Figure 6:
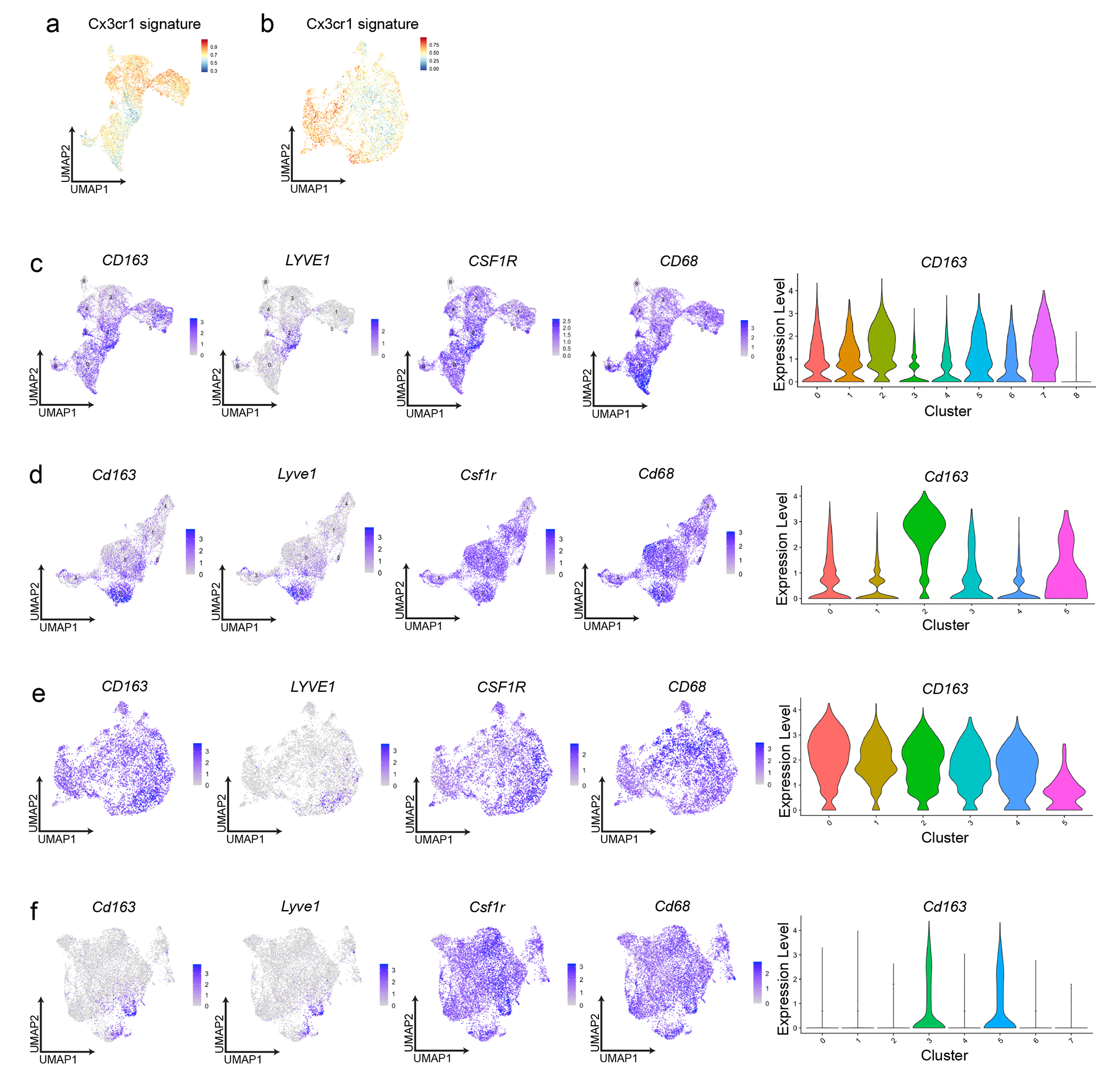
a. UMAP plot displaying mouse LYVE1lo pancreas macrophage scRNA-seq signature using top 100 DEGs from mouse LYVE1lo macrophages mapped into human chronic pancreatitis data set8. b. UMAP plot displaying mouse LYVE1lo pancreas macrophage scRNA-seq signature using top 100 DEGs from mouse LYVE1lo macrophages mapped into published human PDAC data set6. c. UMAP plots displaying CD163, LYVE1, CSF1R, and CD68 expression in human healthy and chronic pancreatitis samples from a, and violin plot showing CD163 gene expression across human pancreatitis macrophage clusters. d. UMAP plots displaying Cd163, Lyve1, Csf1r, and Cd68 expression in scRNA-seq of F4/80+MHCIIhi/lo macrophages sorted from the pancreas of Flt3-YFP mice treated with cerulein by 6 hourly i.p. injections every other day for one week, and violin plot showing Cd163 gene expression across same dataset of mouse pancreas macrophage clusters. e. UMAP plots displaying CD163, LYVE1, CSF1R, and CD68 expression in macrophages from human PDAC data set as in b, and violin plot showing CD163 gene expression across human PDAC macrophage clusters. f. UMAP plots displaying Cd163, Lyve1, Csf1r, and Cd68 expression in mouse F4/80+MHCIIhi/lo macrophages sorted from pancreas of Flt3-YFP mice orthotopically implanted with the KP1 pancreatic cancer cell line, and violin plot showing Cd163 gene expression across same dataset of mouse PDAC macrophage clusters.
Extended Data Figure 7:
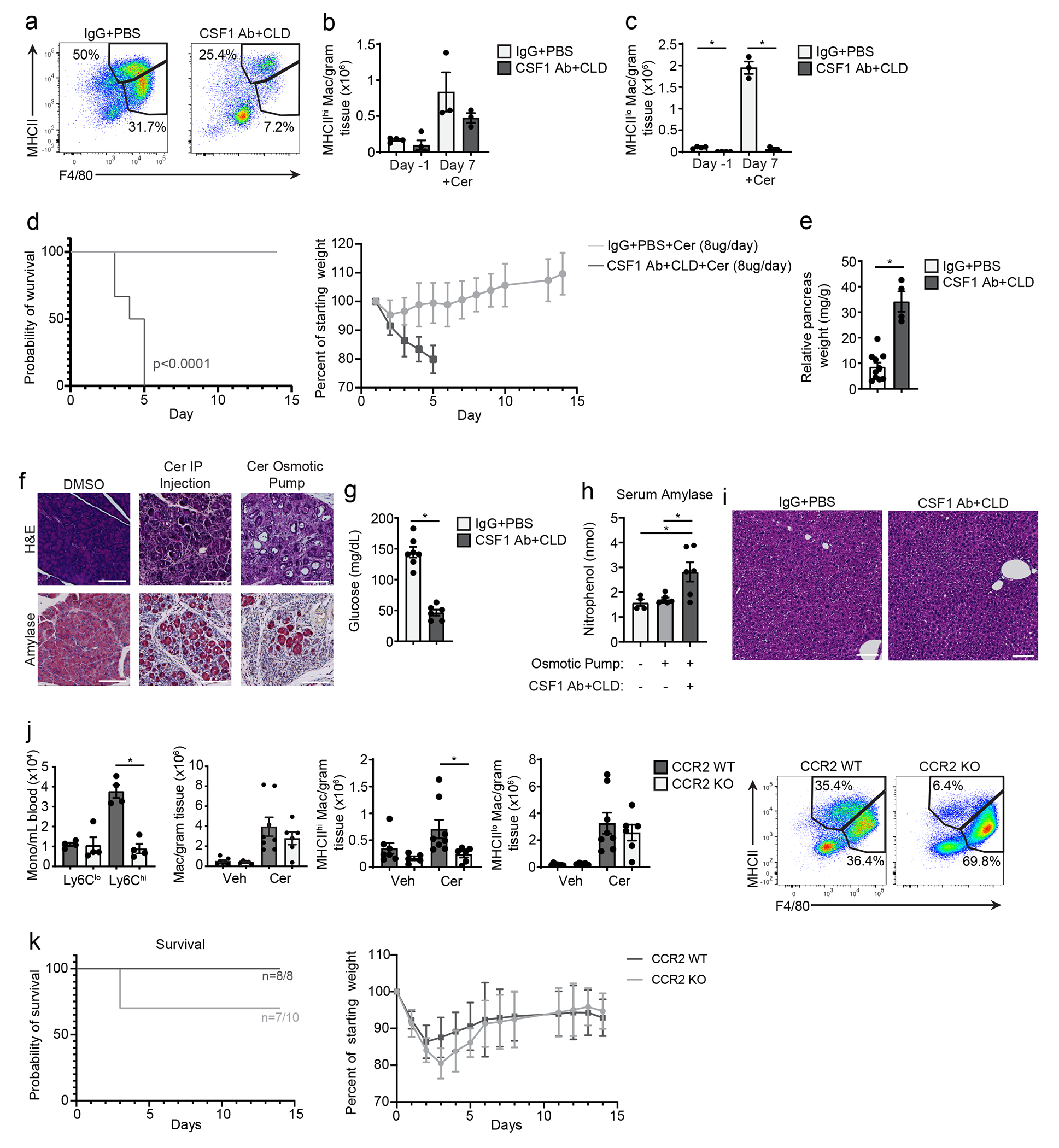
a. Flow cytometry of pancreas F4/80+MHCIIhi/lo macrophages after treatment with CSF1Ab+CLD or IgG+PBS, then a 10-day recovery period (day −1), then cerulein (cer) treatment by 6 hourly i.p. injections every other day for one week (day 7). b. Density of MHCIIhi pancreas macrophages from mice treated as in a; day −1, n=4mice/group; day 7, n=3mice/group. c. Density of MHCIIlo pancreas macrophages from mice treated as in a; day −1, n=4mice/group; day 7, n=3mice/group, left to right *P=0.0002 and *P=0.0002. d. Kaplan-Meier survival curve and body weight measurement of C57BL/6 mice treated with IgG+PBS or CSF1Ab+CLD as in a, followed by cerulein-loaded osmotic pump (10μg/day cerulein) implantation; IgG+PBS, n=10mice; CSF1 Ab+CLD, n=6mice, *P<0.0001 in survival analysis. e. Relative pancreas weight in C57BL/6 mice treated as in d; IgG+PBS, n=10mice, CSF1Ab+CLD, n=4mice, *P<0.0020. f. IHC images of H&E and amylase stain in pancreas from mice implanted with DMSO-loaded osmotic pump, cerulein treatment as in a, or cerulein-loaded osmotic pump, scale bars, 100μM. g. Blood glucose concentration at humane survival endpoint in mice treated as in d; IgG+PBS, n=7mice; CSF1Ab+CLD, n=6mice, *P=0.0012. h. Serum amylase level in mice treated as in f; left to right n=4mice, n=6mice, and n=6mice, and *P=0.0381 and *P=0.0390. i. H&E-stained liver from mice treated as in f, scale bars, 100μM. j. Blood Ly6Clo and Ly6Chi monocytes, and pancreas F4/80+, F4/80+MHCIIhi, and F4/80+MHCIIlo macrophages, and flow cytometry of F4/80+MHCIIhi/lo pancreas macrophages in CCR2-WT and CCR2-KO mice treated with cerulein by i.p. injection as in a; n=4 mice/group and *P=0.0286 in blood Ly6Clo and Ly6Chi analysis, n=7 mice in veh CCR2-WT group, n=5 mice in veh CCR2-KO group, n=8 mice in cer CCR2-WT group, and n=6 in cer CCR2-KO group in F4/80+, F4/80+MHCIIhi, and F4/80+MHCIIlo analyses, and *P=0.0047 in F4/80+MHCIIhi macrophage analysis. k. Kaplan-Meier survival curve and body weight measurement of CCR2-WT and CCR2-KO mice implanted with cerulein-loaded osmotic pumps, as in d; n=8 mice in CCR2-WT group, n=10 mice in CCR2-KO group. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Extended Data Figure 8:
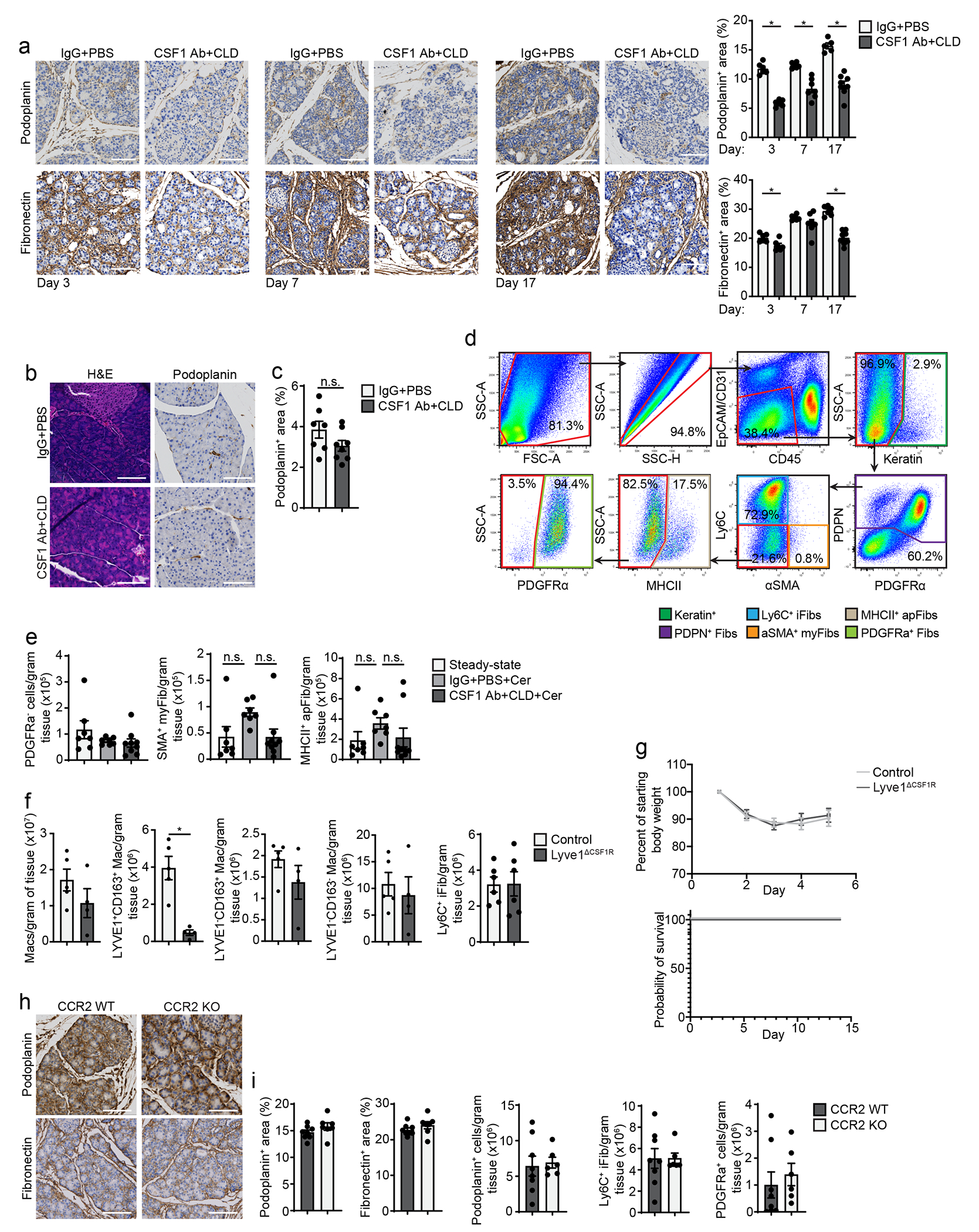
a. IHC staining and quantification for podoplanin and fibronectin on pancreas tissue from mice treated with CSF1Ab+CLD or IgG+PBS, then a 10-day recovery period, then cerulein (cer) treatment by 6 hourly i.p. injections every other day for 3, 7, or 17 days, scale bars, 100μM, IgG+PBS, CSF1Ab+CLD day 3, and IgG+PBS day 7, n=6mice; CSF1Ab+CLD day 7, n=7mice; IgG+PBS day 17, n=5mice; CSF1Ab+CLD day 17, n=8mice; podoplanin left to right, *P=0.0022, *P=0.0012, and *P=0.0016; fibronectin left to right, *P=0.0130 and *P=0.0007. b. H&E and podoplanin stained pancreas tissue from mice treated as in a, with no cerulein treatment, scale bars, 100μM. c. Podoplanin IHC quantification on pancreas tissue of mice treated as in b; IgG+PBS, n=7mice; CSF1Ab+CLD, n=8mice. d. Flow cytometry gating for pancreas fibroblast subsets. e. Density of pancreas PDGFRα−, α-SMA+, and MHCII+ fibroblasts from mice treated with IgG+PBS followed by vehicle (steady-state), IgG+PBS followed by cerulein (IgG+PBS+Cer), and CSF1Ab+CLD followed by cerulein (CSF1 AB+CLD+Cer) as in a; steady-state and IgG+PBS+Cer, n=7mice, and CSF1Ab+CLD+Cer, n=9mice. f. Density of pancreas F4/80+MHCIIhi/lo, LYVE1+CD163+, LYVE1−CD163+, LYVE1−CD163− macrophages, and Ly6C+ iFibs in Lyve1-Cre− littermate controls (control), or Lyve1ΔCSF1R mice treated with cerulein by i.p. injections every other day for one week; control, n=5mice; Lyve1ΔCSF1R, n=4mice; LYVE1+CD163+, *P=0.0159, and Ly6C+ iFib, n=6mice/group. g. Mouse body weight measurement and Kaplan-Meier survival curve following implantation of osmotic pump for delivery of 10μg/day cerulein in control or Lyve1ΔCSF1R mice; n=9 mice in control and n=8 mice in Lyve1ΔCSF1R group. h. Podoplanin and fibronectin IHC staining on pancreas tissue of CCR2-WT and CCR2-KO mice treated with cerulein as in f; scale bars, 100μM. i. Quantification of podoplanin and fibronectin IHC stains and density of podoplanin+ fibroblasts, Ly6C+ iFibs, and PDGFRα+ fibroblasts in pancreas tissue of CCR2-WT and CCR2-KO mice treated with cerulein as in f; CCR2-WT, n=8mice; CCR2-KO, n=7mice for podoplanin and fibronectin IHC analyses, and CCR2-WT, n=8mice; CCR2-KO, n=6mice for fibroblast flow cytometry analyses. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Extended Data Figure 9:
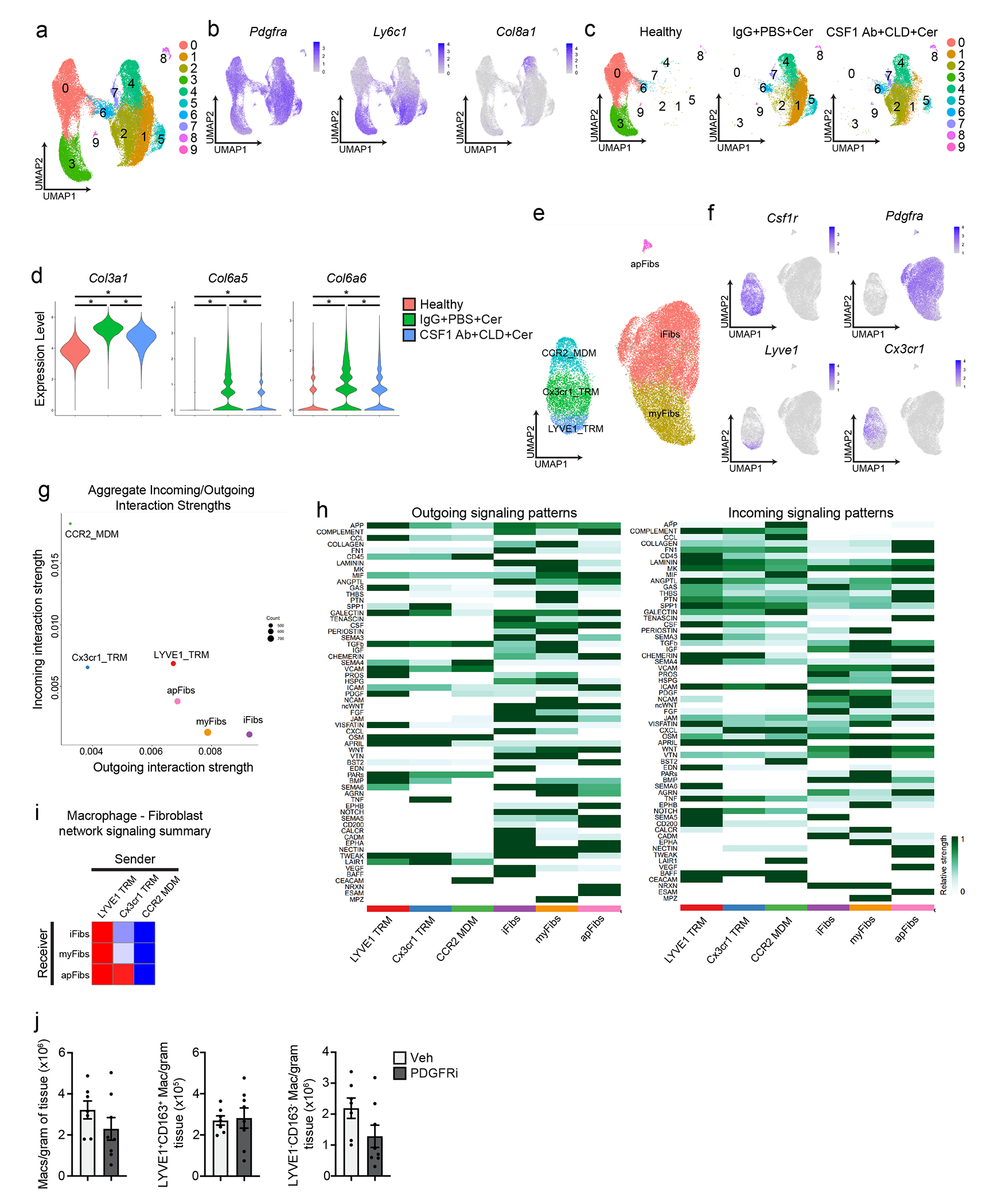
a. UMAP plot of scRNA-seq analysis of PDPN+ pancreas fibroblasts sorted from mice treated with IgG+PBS followed by vehicle (healthy), IgG+PBS followed by cerulein (IgG+PBS+Cer), and CSF1 Ab+CLD followed by cerulein (CSF1 Ab+CLD+Cer). b. UMAP plots displaying expression of Pdgfra, Ly6c1, and Col8a1 in PDPN+ fibroblasts from a. c. UMAP plot of pancreas PDPN+ fibroblasts separated by healthy, IgG+PBS+Cer, or CSF1 Ab+CLD+Cer samples as in a. d. Violin plot of Col3a1, Col6a5, and Col6a6 expression across samples (healthy, IgG+PBS+Cer, or CSF1 Ab+CLD+Cer); for all comparisons *P<0.0001. e. UMAP plot of scRNA-seq analysis combining macrophages from Flt3-YFP mice treated with cerulein and fibroblasts from IgG+PBS+Cer and CSF1 Ab+CLD+Cer treated mice. f. UMAP plots displaying Csf1r, Pdgfra, Lyve1, and Cx3cr1 expression in combined macrophage and fibroblast scRNA-seq analysis from e. g. Dot plot showing aggregate score of all incoming (receptor) and outgoing (ligand) interactions summarized for each macrophage and fibroblast cluster from e, as measured by CellChat9. h. Heatmap displaying relative strength of network centrality measures for outgoing (ligands, left heatmap) and incoming (receptors, right heatmap) signaling patterns of each intercellular signaling pathway. i. Heatmap displaying aggregate score across all differentially enriched receptor (receiver) or ligand (sender) pathways between macrophage and fibroblast clusters from e. j. Density of pancreas F4/80+MHCIIhi/lo, LYVE1+CD163+, and LYVE1−CD163− macrophages from mice treated with vehicle (veh) or PDGFRi once per day by i.p. injection along with cerulein treatment by 6 hourly i.p. injections every other day for one week; n=7 mice in vehicle and n=8 mice in PDGFRi groups. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used, except for d where Bonferroni correction was used.
Extended Data Figure 10:
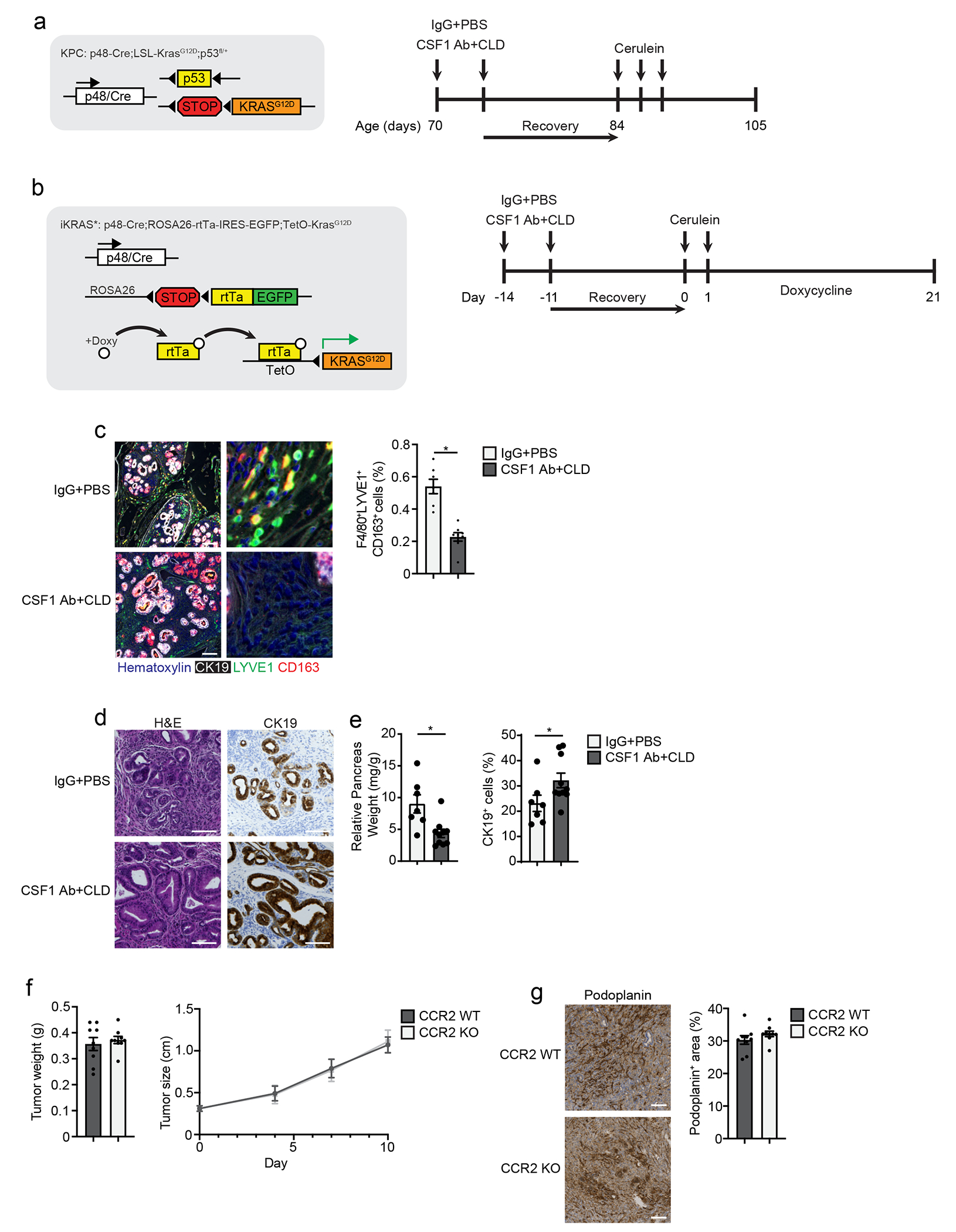
a. Genetic loci for p48-Cre;LSL-KRASG12D;p53fl/+ (KPC) model and treatment scheme for IgG+PBS and CSF1 Ab+CLD followed by cerulein by 6 hourly i.p injections every other day for 5 days. b. Genetic loci for p48-Cre;ROSA26-rtTa-IRES-EGFP;TetO-KRASG12D (iKRAS*) model and treatment scheme for IgG+PBS and CSF1 Ab+CLD followed by cerulein by 6 hourly i.p injections on two consecutive days, followed by doxycycline administration in drinking water. c. Representative mIHC images of pancreas tissue from iKRAS* mice treated with IgG+PBS or CSF1 Ab+CLD as in b, stained for hematoxylin, CK19, F4/80, LYVE1, and CD163 (scale bars are 100μM), and quantification of F4/80+LYVE1+CD163+ macrophages, displayed as the percentage of cells; n=7 mice in IgG+PBS group, n=8 mice in CSF1 Ab+CLD group, and *P=0.0003. d. Representative images of pancreas tissue stained for H&E and CK19 from iKRAS* mice treated with IgG+PBS or CSF1 Ab+CLD as in b, scale bars are 100μM. e. Quantification of relative pancreas weight and CK19+ cells displayed as percentage of total cells from iKRAS* mice treated with IgG+PBS or CSF1 Ab+CLD as in b; n=7 mice in IgG+PBS group, n=10 mice in CSF1 Ab+CLD group, *P=0.0097 in relative pancreas weight analysis, and *P=0.0250 in CK19 analysis. f. Tumor weight and size measurements of CCR2-WT and CCR2-KO mice orthotopically implanted with the KP2 pancreatic cancer cell line; n=9 mice/group. g. Representative images and quantification of pancreas tissue from CCR2-WT and CCR2-KO mice orthotopically implanted with the KP2 pancreatic cancer cell line stained for podoplanin, scale bars are 100μM; n=9 mice/group. Data are presented as mean ± SEM unless otherwise indicated. n.s., not significant; *p <0.05. For comparisons between two groups, Student’s two-tailed t-test was used.
Supplementary Material
Acknowledgments
J.M.B. was funded by pre-doctoral fellowship F31 DK122633. D.G.D. and study costs were supported by NCI R01CA273190, R01CA177670, P50CA196510, P30CA09184215, and the BJC Cancer Frontier Fund. Collaborative studies with G.J.R. were supported by RO1AI049653. L.I.K. was funded by 5T32EB021955 2019-2021. K.W.K. was funded by NIDDK R01DK126753. J.C.M was funded by R01DK105129 and R01CA239645. We thank the Washington University Center for Cellular Imaging, the Flow Cytometry & Fluorescence Activated Cell Sorting Core and Genome Technology Access Center which are funded by D.G.D. fund P50CA196510.
Footnotes
Competing Interests
Authors declare no competing interests.
References:
- 1.Ginhoux F et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoeffel G et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epelman S et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz C et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto D et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z et al. Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell 178, 1509–1525.e1519 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Yona S et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bain CC et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat Commun 7, ncomms11852 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginhoux F & Guilliams M Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Calderon B et al. The pancreas anatomy conditions the origin and properties of resident macrophages. J Exp Med 212, 1497–1512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bain CC et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 15, 929–937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K-W et al. MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes. Journal of Experimental Medicine 213, 1951–1959 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loyher P-L et al. Macrophages of distinct origins contribute to tumor development in the lung. The Journal of Experimental Medicine 215, 2536–2553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavin Y et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 47, 323–338 e326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginhoux F & Jung S Monocytes and macrophages: developmental pathways and tissue homeostasis. Nature Reviews Immunology 14, 392–404 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Chakarov S et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Zhang N et al. LYVE1+ macrophages of murine peritoneal mesothelium promote omentum-independent ovarian tumor growth. Journal of Experimental Medicine 218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim HY et al. Hyaluronan Receptor LYVE-1-Expressing Macrophages Maintain Arterial Tone through Hyaluronan-Mediated Regulation of Smooth Muscle Cell Collagen. Immunity 49, 326–341.e327 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Dick SA et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Science Immunology 7 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Casanova-Acebes M et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature 595, 578–584 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apte M, Pirola R & Wilson J The Fibrosis of Chronic Pancreatitis: New Insights into the Role of Pancreatic Stellate Cells. Antioxidants & Redox Signaling 15, 2711–2722 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Klӧppel G, Detlefsen S & Feyerabend B Fibrosis of the pancreas: the initial tissue damage and the resulting pattern. Virchows Archiv − 1, 1–1 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Liou G-Y et al. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-κB and MMPs. Journal of Cell Biology 202, 563–577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeki K et al. CCL2-Induced Migration and SOCS3-Mediated Activation of Macrophages Are Involved in Cerulein-Induced Pancreatitis in Mice. Gastroenterology 142, 1010–1020.e1019 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann A Pancreatic stellate cells contribute to regeneration early after acute necrotising pancreatitis in humans. Gut 51, 574–578 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omary MB, Lugea A, Lowe AW & Pandol SJ The pancreatic stellate cell: a star on the rise in pancreatic diseases. Journal of Clinical Investigation 117, 50–59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosein AN, Brekken RA & Maitra A Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nature Reviews Gastroenterology & Hepatology 17, 487–505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandler C, Liu T, Buckanovich R & Coffman LG The double edge sword of fibrosis in cancer. Translational Research 209, 55–67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lerch MM & Gorelick FS Models of Acute and Chronic Pancreatitis. Gastroenterology 144, 1180–1193 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Boyer SW, Schroeder AV, Smith-Berdan S & Forsberg EC All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell 9, 64–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman RL et al. Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies. Cell Rep 17, 2445–2459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou DC et al. Spatial drivers and pre-cancer populations collaborate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Cold Spring Harbor Laboratory; 2021. [Google Scholar]
- 34.Lee B et al. Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis. Gut, gutjnl-2021–2324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nature Medicine 27, 152–164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serbina NV & Pamer EG Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nature Immunology 7, 311–317 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Elyada E et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discovery 9, 1102–1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Öhlund D et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. Journal of Experimental Medicine 214, 579–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manohar M, Verma AK, Venkateshaiah SU, Sanders NL & Mishra A Pathogenic mechanisms of pancreatitis. World J Gastrointest Pharmacol Ther 8, 10–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Habtezion A Inflammation in acute and chronic pancreatitis. Current Opinion in Gastroenterology 31, 395–399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerra C et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Collins MA et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 122, 639–653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbings SL et al. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. American Journal of Respiratory Cell and Molecular Biology 57, 66–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeNardo DG & Ruffell B Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang H et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med 22, 851–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vujasinovic M et al. Risk of Developing Pancreatic Cancer in Patients with Chronic Pancreatitis. Journal of Clinical Medicine 9, 3720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowenfels AB et al. Pancreatitis and the Risk of Pancreatic Cancer. New England Journal of Medicine 328, 1433–1437 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Etzerodt A et al. Tissue-resident macrophages in omentum promote metastatic spread of ovarian cancer. Journal of Experimental Medicine 217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin S et al. Inference and analysis of cell-cell communication using CellChat. Nature Communications 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods references:
- 1.Soares KC et al. A preclinical murine model of hepatic metastases. J Vis Exp, 51677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JW et al. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature 567, 249–252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 47, 323–338 e326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng H et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. Journal of Clinical Investigation 123, 1444–1456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafemeister C & Satija R Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou DC et al. Spatial drivers and pre-cancer populations collaborate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Cold Spring Harbor Laboratory; 2021. [Google Scholar]
- 7.Tsujikawa T et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Reports 19, 203–217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee B et al. Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis. Gut, gutjnl-2021–2324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin S et al. Inference and analysis of cell-cell communication using CellChat. Nature Communications 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data (bulk and single-cell RNAseq) has been made publicly available under Gene Expression Omnibus (GEO) repository accession numbers GSE196778 (scRNAseq), and GSE203005 (bulk RNAseq). All other data are available upon request to the lead contact, David G. DeNardo (ddenardo@wustl.edu).


