Abstract
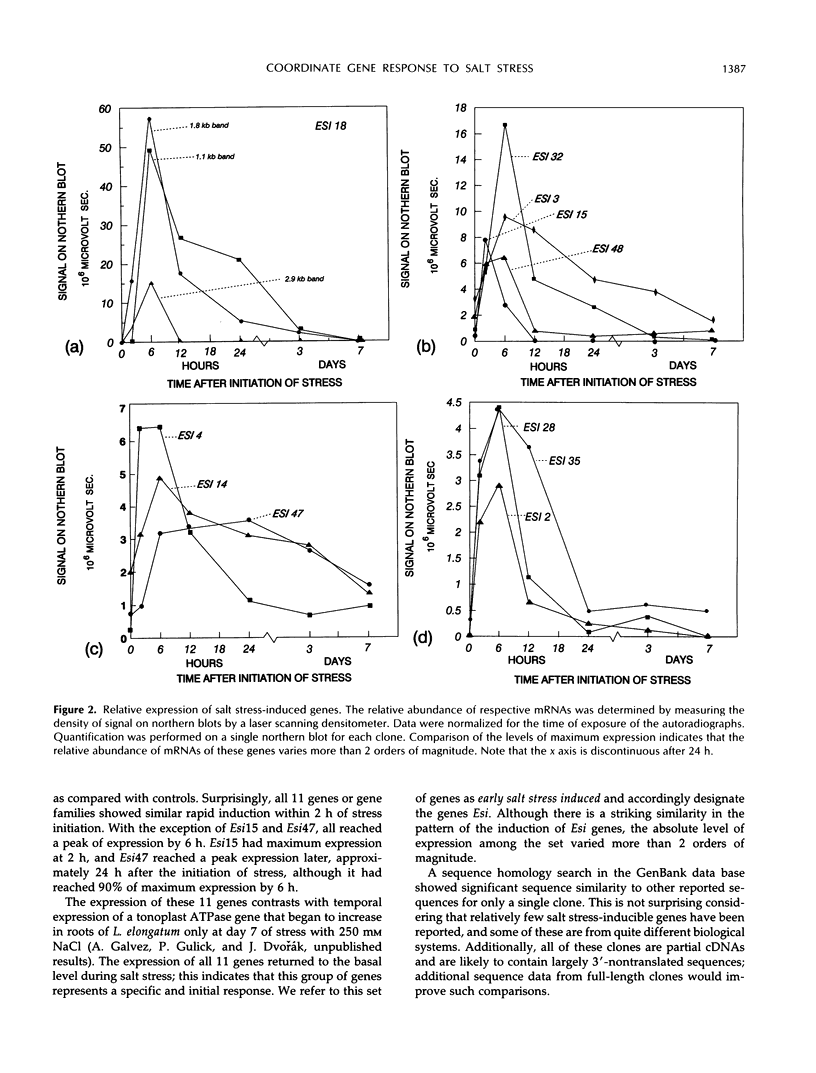
Lophopyrum elongatum is a highly salt-tolerant relative of wheat. A previous study showed that the abundance of a number of mRNA species is enhanced or reduced in the roots of the L. elongatum × Triticum aestivum amphiploid by salt stress. Eleven genes with enhanced expression in the roots of salt-stressed L. elongatum plants have been cloned as cDNAs. The clones were used as probes to characterize temporal expression of these genes in roots after initiation of salt (250 mm NaCl) stress. All 11 genes are induced within 2 h after exposure to 250 mm NaCl and reached peak expression after 6 h. The decline of gene expression distinguished two groups, one in which mRNA concentrations returned to basal levels by 24 h and the other in which this occurred between 3 and 7 d. One of the 11 clones was found to be homologous to a multigene family of abscisic acid-induced genes, rab and dhn, identified in other species. We suggest that the coordinate expression of this large number of genes reflects the existence of a highly specific early response to salt stress. We refer to this response as the “early salt stress response.”
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cathala G., Savouret J. F., Mendez B., West B. L., Karin M., Martial J. A., Baxter J. D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2(4):329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- Claes B., Dekeyser R., Villarroel R., Van den Bulcke M., Bauw G., Van Montagu M., Caplan A. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell. 1990 Jan;2(1):19–27. doi: 10.1105/tpc.2.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T. J., Kortt A. A., Chandler P. M. A cDNA-based comparison of dehydration-induced proteins (dehydrins) in barley and corn. Plant Mol Biol. 1989 Jul;13(1):95–108. doi: 10.1007/BF00027338. [DOI] [PubMed] [Google Scholar]
- Dvorák J., Edge M., Ross K. On the evolution of the adaptation of Lophopyrum elongatum to growth in saline environments. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3805–3809. doi: 10.1073/pnas.85.11.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick P. J., Dvorák J. Selective enrichment of cDNAs from salt-stress-induced genes in the wheatgrass, Lophopyrum elongatum, by the formamide-phenol emulsion reassociation technique. Gene. 1990 Nov 15;95(2):173–177. doi: 10.1016/0378-1119(90)90359-y. [DOI] [PubMed] [Google Scholar]
- Gulick P., Dvorák J. Gene induction and repression by salt treatment in roots of the salinity-sensitive Chinese Spring wheat and the salinity-tolerant Chinese Spring x Elytrigia elongata amphiploid. Proc Natl Acad Sci U S A. 1987 Jan;84(1):99–103. doi: 10.1073/pnas.84.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M., Walbot V. Effects of cold-treatment on protein synthesis and mRNA levels in rice leaves. Plant Physiol. 1989 Nov;91(3):930–938. doi: 10.1104/pp.91.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman W. J., Fornari C. S., Tanaka C. K. A Comparison of the Effect of Salt on Polypeptides and Translatable mRNAs in Roots of a Salt-Tolerant and a Salt-Sensitive Cultivar of Barley. Plant Physiol. 1989 Aug;90(4):1444–1456. doi: 10.1104/pp.90.4.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman W. J., Tanaka C. K., Dupont F. M. The effects of salt stress on polypeptides in membrane fractions from barley roots. Plant Physiol. 1988 Dec;88(4):1263–1273. doi: 10.1104/pp.88.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman W. J., Tanaka C. K. The effects of salt on the pattern of protein synthesis in barley roots. Plant Physiol. 1987 Mar;83(3):517–524. doi: 10.1104/pp.83.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy J., Chua N. H. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988 Aug;7(8):2279–2286. doi: 10.1002/j.1460-2075.1988.tb03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkowski D., Schneider K., Salamini F., Bartels D. Characterization of Five Abscisic Acid-Responsive cDNA Clones Isolated from the Desiccation-Tolerant Plant Craterostigma plantagineum and Their Relationship to Other Water-Stress Genes. Plant Physiol. 1990 Dec;94(4):1682–1688. doi: 10.1104/pp.94.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopal S. Differential mRNA transcription during salinity stress in barley. Proc Natl Acad Sci U S A. 1987 Jan;84(1):94–98. doi: 10.1073/pnas.84.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. K., Bracker C. A., Hasegawa P. M., Handa A. K., Buckel S., Hermodson M. A., Pfankoch E., Regnier F. E., Bressan R. A. Characterization of osmotin : a thaumatin-like protein associated with osmotic adaptation in plant cells. Plant Physiol. 1987 Oct;85(2):529–536. doi: 10.1104/pp.85.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Mundy J., Chua N. H. Four tightly linked rab genes are differentially expressed in rice. Plant Mol Biol. 1990 Jan;14(1):29–39. doi: 10.1007/BF00015652. [DOI] [PubMed] [Google Scholar]