Abstract
Mutation of the divIVB locus in Bacillus subtilis causes misplacement of the septum during cell division and allows the formation of anucleate minicells. The divIVB locus contains five open reading frames (ORFs). The last two ORFs (minCD) are homologous to minC and minD of Escherichia coli but a minE homolog is lacking in B. subtilis. There is some similarity between minicell formation and the asymmetric septation that normally occurs during sporulation in terms of polar septum localization. However, it has been proposed that MinCD has no essential role in sporulation septum formation. We have used electron microscopic studies to show septation events during sporulation in some minD strains. We have observed an unusually thin septum at the midcell position in minD and also in minD spoIIE71 mutant cells. Fluorescence microscopy also localized a SpoIIE-green fluorescent protein fusion protein at the midcell site in minD cells. We propose that the MinCD complex plays an important role in asymmetric septum formation during sporulation of B. subtilis cells.
Bacillus subtilis is a gram-positive endospore-forming bacterium that when grown vegetatively divides by precise placement of a septum at midcell. However, during sporulation the generation of two cells with different development fates is preceded by an asymmetric division. Vegetative division involves FtsZ, FtsA, DivIB, DivIC, FtsL, and PBP2B, in addition to other proteins. It is believed that the asymmetric division in sporulation uses the same division proteins. FtsZ appears to play a pivotal role in selection of the division site prior to cell division in both Escherichia coli and B. subtilis (7, 8); at the onset of sporulation, assembly of an FtsZ ring shifts from midcell to the two potential polar sites, and this shift is dependent on the transcription factor Spo0A (25).
Three min genes (minC, minD, and minE) are known to exist in E. coli. Homologous MinC and MinD proteins have been identified in B. subtilis, but a MinE equivalent has not yet been found (23, 24, 32). The MinE protein appears to play a role in topological specificity in E. coli (29). Because of the similarity between minicell formation and asymmetric septation during sporulation, it was proposed that there were two MinE-like B. subtilis proteins, with different topological properties (29). The vegetative specific MinE protein would activate septation at midcell, whereas sporulation-specific MinE would allow division at the polar position and simultaneously allow the division inhibitor to block septation at midcell. This model also predicts that these different MinE proteins will have different topological properties. The vegetative MinE would counteract the action of the MinCD division inhibitor at midcell but not at the cell poles. In contrast, sporulation-specific MinE would counteract the division inhibitor at the polar sites and would block the midcell septation site. The search for MinE-like proteins in B. subtilis led to the discovery of the DivIVA protein, which has some characteristics expected of a MinE-like protein (9, 14). Although genetic evidence supports the hypothesis that in B. subtilis DivIVA has a function analogous to that of MinE in E. coli, there are important differences between these two proteins. Cha and Stewart (9) also proposed that DivIVA is involved in activation of polar septation sites during the sporulation process.
MinC and MinD proteins are inhibitors of septation at medial and polar sites in both E. coli and B. subtilis, and their absence leads to minicell formation. However, the role of MinCD in sporulation is unclear, as minCD mutants sporulate very efficiently, in contrast with mutations at another locus associated with the minicell phenotype, divIVA. Here we report that the lack of active MinCD during sporulation causes the creation of thin sporulation-like septa at the midcell position. This is a surprising discovery that has likely not been reported previously because this phenotype has only a minimal influence on the level of sporulation efficiency. We also report that SpoIIE, a sporulation-specific protein which allows activation of ςF in the forespore, localizes in these cells not only in a bipolar manner but also at the midcell position.
MATERIALS AND METHODS
Bacterial strains, culture media, and genetic techniques.
Bacterial strains used in this work are listed in Table 1. Isolation of genomic and plasmid DNA, methods for transformation of B. subtilis and E. coli strains, selection of antibiotic resistance markers, and other standard genetic techniques were carried out as described previously (10). Standard enzymatic reagents, including restriction enzymes, DNA ligase, and Taq DNA polymerase, were purchased from Boehringer Mannheim, New England Biolabs, or Perkin-Elmer, Inc. Amplification of DNA by PCR was carried out using standard methods (19). To construct plasmids pBGCDM3 and pBGCDdel, PCR products were amplified from PY184 and PY79 chromosomal DNA, respectively, using oligonucleotides MINCECO (5′-GTGGAATTCGTGAAGACCAAAAAGCAGC-3′) and MINDBAM (5′-CGGTGGATCCAAGAACAAAGCAGGC-3′). These PCR products contained the nucleotide sequence coding for the entire minC and minD genes. The PCR fragment derived from PY184 was digested with EcoRI and BamHI and inserted into pGEM-3Zf(+)cat (35) digested with the same restriction enzymes, creating plasmid pBGCDM3. The PCR fragment derived from PY79 was inserted into plasmid pGEM-3Zf(+)cat as described above, creating plasmid pBGCD. This plasmid was cut with KpnI, and then a 5-kb backbone was purified by agarose gel electrophoresis and ligated, creating plasmid pBGCDdel. The presence of the divIVB1 mutation on plasmid pBGCDM3 was confirmed by DNA sequencing using the fmol DNA cycle sequencing system (Promega Corp.). Plasmids pBGCDM3 and pBGCDdel were used for transformation of PY79 cells by selection for chloramphenicol resistance, creating strains IB498 and IB515, respectively. The correct recombination of both plasmids in the minCD region of the chromosome was verified by Southern blot hybridization. Strain IB508 was prepared and its chromosomal structure was verified as described above for strain IB498. Plasmid pBGCDM3-kan, a derivative of plasmid pBGCDM3, was prepared by replacing the cat gene with the Kmr gene from plasmid pUK19 (a gift from W. Haldenwang). This plasmid was used for transformation of strain PY79, creating strain IB508. The chromosomal structure was also verified by Southern blot hybridization.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype, relevant characteristics, and/or construction | Reference or source |
|---|---|---|
| E. coli MM294 | endE thiA hsdR17 supE44 | 3 |
| B. subtilis | ||
| PY79 | Prototrophic | 36 |
| PY1165 | spoIIE71 trpC2 | 4 |
| PY180 | spoIIE::Tn917ΩHU7 | 30 |
| PY184 | thyA thyB metB divIVB1 | P. Youngman |
| PMF16 | spoIIE-gfp; transformation of PY79 to Cmr with pBsIIE-mutGFP plasmid DNA | P. Fawcett |
| IB498 | divIVB1; transformation of PY79 to Cmr with pBGCDM3 plasmid DNA | This work |
| IB508 | divIVB1; transformation of PY79 to Kmr with pBGCDM3-kan plasmid DNA | This work |
| IB506 | spoIIE::Tn917ΩHU7 divIVB1; transformation of PY180 to Cmr with IB498 chromosomal DNA | This work |
| IB507 | spoIIE71 divIVB1; transformation of PY1165 to Cmr with IB498 chromosomal DNA | This work |
| IB509 | divIVB1 spoIIE-gfp; transformation of PMF16 to Kmr with IB508 chromosomal DNA | This work |
| IB515 | PY79 divIVB::cat (at KpnI); transformation of PY79 to Cmr with pBGCDdel plasmid DNA | This work |
Electron microscopy.
Sporulation was induced by nutrient exhaustion in DSM (Difco sporulation medium) broth as described previously (10). At various times after cessation of growth, samples were harvested by centrifugation, washed in a buffer containing 0.1 M sodium cacodylate (pH 7.2), and fixed for 1 h at 4°C in the same buffer containing 2% glutaraldehyde and 1% OsO4 (17). Following fixation, cells were pelleted by centrifugation, washed in 0.1 M cacodylate buffer, and dehydrated by a series of washes and incubations (10 min each at ambient temperature) in graded concentrations of ethanol (30, 50, 70, 85, 95, and 100% [twice in 100% ethanol]). This was followed by washing (twice for 30 min each time) in acetone. Dehydrated cells were embedded in Spurr medium (31) and polymerized for 8 h at 70°C. Samples were sectioned (60- to 120-nm thickness) on a NOVA3 ultramicrotome (LKB) and floated onto copper grids. Sections were stained on uranyl acetate drops for 20 min and on lead citrate drops for 5 min. Stained thin sections were examined and photographed on a JEOL-100 CX or Tesla Brno (T541) electron microscope.
Scoring criteria.
Thickness of septa was scored from digitalized electron micrographs scanned with a Hewlett-Packard 4C scanner and using PowerPoint 7.0 software (Microsoft) or by direct measurement of thickness from photographs taken at higher magnification. For quantification of morphological classes by electron microscopy, at least 100 complete longitudinal sections were scored from random fields for each sample.
Fluorescence microscopy and light microscopy.
Cells were prepared for fluorescence microscopy as described previously (4). B. subtilis IB509, containing a copy of plasmid pBsIIE-mutGFP integrated at the chromosomal spoIIE locus, was cultured in 50 ml of DSM broth at 37°C. At selected times during growth and sporulation, 400-μl culture samples were transferred to 1.5-ml Eppendorf tubes and collected by centrifugation, and the pellet was resuspended in preservation buffer (40 mM NaN3, 50% [wt/vol] sucrose, 0.5 M Tris [pH 7.5], 0.77 M NaCl). Samples were examined or photographed after a few hours. Epifluorescence micrographs were recorded on Kodak transparency film with an Olympus B201 microscope using a filter set (420- to 480-nm excitation, DM500 dichroic reflector, and 515-nm emission filters). Film transparencies were digitalized, and the epifluorescence micrographs were prepared for printing by using CorelDraw 6.0 software.
RESULTS AND DISCUSSION
Sporulation of minD mutants.
We prepared divIVB1 mutant (IB498) and minD deletion (IB515) strains as described in Materials and Methods. The divIVB1 strain contains two mutations in the last gene of the divIVB operon, which is minD. The first mutation results in substitution of lysine for methionine at amino acid residue 87, and the second results in an isoleucine-to-threonine change at amino acid 147 of MinD (32). The minD deletion is at a KpnI site which leaves only the first 23 amino acid residues at the N terminus of MinD.
A modest reduction in sporulation of the divIVB1 strain was observed (Table 2). While Levin et al. (24) reported that a divIVB1 mutant strain sporulates at a level approximating that of the wild type, other authors reported a slight reduction after 24 h (9, 23). This reduction may have been caused by the change in number of sites potentially available for septation during sporulation in such strains and/or by a delay of sporulation due to improper localization of some sporulation-specific proteins at the same midcell septa.
TABLE 2.
Sporulation efficiencies of various strains
| Strain | Spore titer (relative efficiencya)
|
|
|---|---|---|
| 24 h | 48 h | |
| PY79 (wild type) | 3.0 × 108 (1) | 4 × 108 (1) |
| IB498 divIVB1 | 2.6 × 107 (0.9) | 6 × 107 (1) |
| IB515 divIVB::cat | 4.0 × 107 (0.7) | 5 × 107 (0.9) |
Defined as (CFU of spores/total viable CFU)/(viable PY79 spores/PY79 total viable CFU).
Ultrastructure of minD mutant cells.
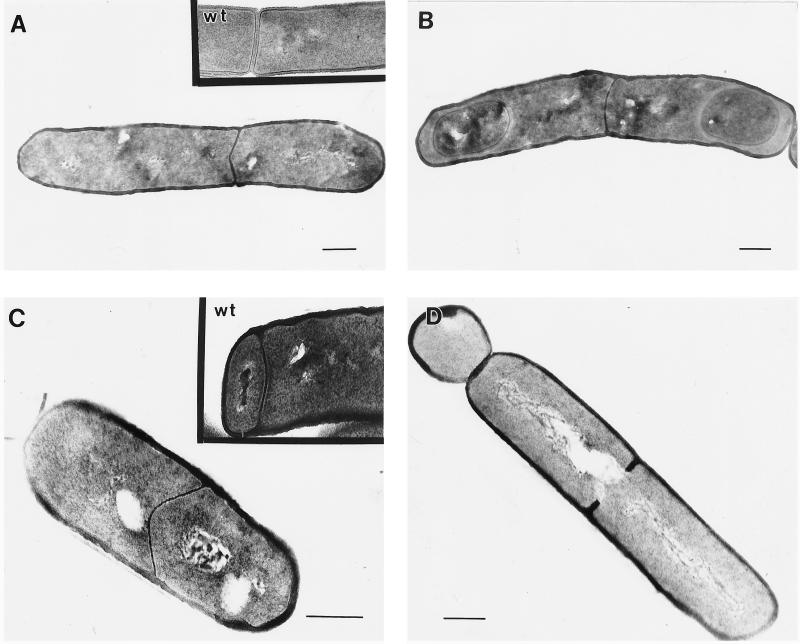
Sporulating cultures of the divIVB1 mutant (IB498), the minD deletion strain (IB515), and the wild-type strain (PY79) were harvested at various times during sporulation and examined by electron microscopy to test whether the small decrease in sporulation efficiency in divIVB1 cells was caused by release of MinCD repression at all possible sites of septation (middle and both polar sites). We used samples harvested at t4 (4 h after the beginning of sporulation) for quantitative determinations of different types and localization of septa. As expected, in the wild-type strain we observed two kinds of septa: asymmetrically positioned thin sporulation septa and thick vegetative septa at the midcell site (Fig. 1A and C). In strains IB498 and IB515, we observed cells with unusually thin septa at the midcell position, which we call sporulation-like septa (Fig. 1A to C). We also observed these thin septa at the midcell position in different stages of forespore development. Figures 1A and C show cells with a thin midcell septum at a very early time in sporulation. Some midcell thin septa also were observed in later stages of forespore development (Fig. 1B). In a few cross sections, cells with multiple septa were observed. Some of the thin septa are not positioned at the precise midpoint of the cell (Fig. 1A), possibly a consequence of the nature of the specific minD mutation carried by the cells. Heterogeneity of cell length was observed previously in E. coli minD mutants, and it is believed that this is caused by the role of MinD protein in nucleoid partition, which is coupled with septation (12). Dassain and Bouché (11), in contrast, proposed that polar division in minD mutants causes an abrupt and temporary decrease in the amount of FtsZ available for nonpolar division, which in turn has effects such as the cell length heterogeneity noted above.
FIG. 1.
Electron micrographs of representative examples of septum morphologies in strains IB498 (divIVB1) (A to C), IB507 (spoIIE::Tn917ΩHU7 divIVB1) (D), and IB506 (spoIIE71 divIVB1) (E). The insets to panels A and C show examples of wild-type (wt) vegetative and sporulation septa, respectively. Samples were harvested at 2 (A and C), 4 (D and E), and 7 h after cessation of logarithmic growth (B). Each scale bar represents 0.3 μm.
We suggest that the midcell-positioned thin septa in B. subtilis minD cells are not used as proper sporulation septa, even though they are similar in thickness to sporulation septa (Table 3). Septa were considered thin if they measured 14 to 26 nm and thick if they were 40 to 46 nm (Table 3). All scored septa fell into these two classes except those sporulation-like septa which proceeded further in forespore development, beyond stage IIi according to the classification of Illing and Errington (18). Both kinds of septa, vegetative and sporulation, are of uniform thickness when they start to form at the site of septation in wild-type cells. The midcell division septum is a participant in cytokinesis, with the peptodoglycan component splitting bilaterally, cleaved by autolysins as daughter cells separate from one another (22). In contrast, the asymmetrically positioned sporulation septum is a participant in the engulfment process, during which most if not all of the peptidoglycan is removed.
TABLE 3.
Thickness of septa in IB498 cells
| Classification (no. scored) | Thicknessa (nm)
|
|
|---|---|---|
| Range | Avg | |
| Thick midcell (20) | 42–46 | 44 |
| Thick polar (27) | 40–45 | 44 |
| Thin midcell (40) | 14–26 | 20 |
| Thin polar (33) | 18–26 | 22 |
Measured at the middle of the septa. Sporulation-like septa from cells which proceeded in development beyond stage IIi were not scored.
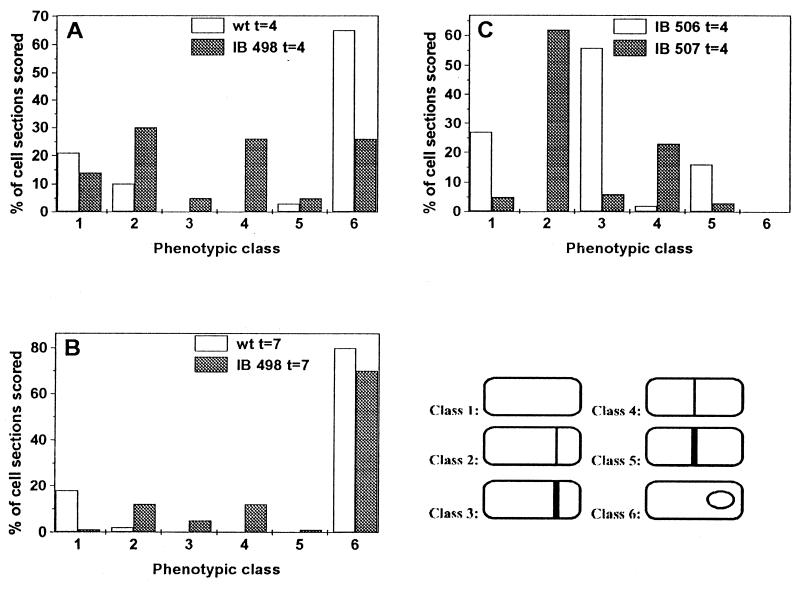
Statistical studies (Fig. 2A and B) revealed that forespore development in strain IB498 was significantly delayed compared with that in the wild-type strain. As discussed previously, vegetative cells lacking MinCD function should produce 67% minicells, with only 33% of cells undergoing midcell division (9). These cells actually produced about 30% or less minicells, and it was proposed that other systems could be involved in septation site selection during vegetative growth (9). During sporulation, the total number of minC minD cells with asymmetrically positioned septa at t4 was approximately two times higher (61%) than the number of cells with septa at the midcell position (31%) (Fig. 2A). The total number of polar septa also includes the thick vegetative-like septa of minicells, as well as a number of forespores at stage III and later of sporulation; these forespores had to originate from asymmetrically positioned septa. These results suggest that all three possible sites of septation (two polar and one midcell) are equally used by the division machinery at the beginning of the sporulation process. It also suggests that the MinCD complex is probably the main septation repressor of the midcell position during stage I of sporulation, when polar septation is initiated. Mutations in minD might thus reduce the sporulation efficiency, by about 30%; a decrease is difficult to measure in commonly used sporulation efficiency assays. A delay in forespore development in strains IB498 and IB515 might also be caused by localization of some sporulation-specific proteins at the thin midcell septa, thus allowing some of these proteins to function in the wrong compartment of the sporulating cell.
FIG. 2.
Quantitation of morphological classes in sporulating cells of strains PY79, IB498, IB506, and IB507. Cells were cultured in DSM broth at 37°C and harvested for examination by electron microscopy 4 or 7 h after cessation of logarithmic growth. Morphological classes were used as indicated at the right: 1, no septa; 2, thin septum at polar position; 3, thick septum at polar position; 4, thin septum at midcell position; 5, thick septum at midcell position; 6, cell with forespore at stage III or later.
Localization of SpoIIE-GFP fusion in the minD strain.
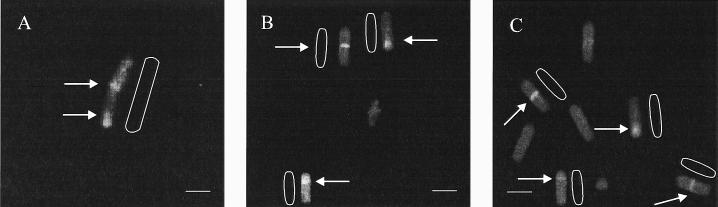
A few sporulation-specific proteins are known to be associated specifically with the sporulation septum: SpoIIE (2, 4), SpoIIGB (20), SpoIIGA (15), and SpoIIIE (34). The SpoIIE protein has an important role as a serine phosphatase that releases ςF activity specifically in the forespore compartment (13). We investigated the localization of SpoIIE protein in minD cells by using a SpoIIE-green fluorescent protein (GFP) fusion in strain IB509. The main pattern of GFP localization at t2 was bipolar (Table 4), as observed previously in a MinCD+ strain (2, 4). In addition to polar localization of SpoIIE, we expected to find cells in which the signal localized at the midcell position. Indeed, in some cells we observed a signal that represents the localization of SpoIIE at the unusual midcell position (Fig. 3), although the number of such cells was lower (9% of total cells) (Table 4) than we expected (30% of total cells). The unexpectedly low frequency of cells with SpoIIE localized to the midcell may well have been caused by the fast disappearance of the protein from this position, as earlier work has shown that SpoIIE rapidly disappears from the sporangium, first from the mother cell pole and then from the forespore pole (28). Another possibility for the low level of cells with SpoIIE at the midcell position could be lower affinity of the protein for this site than for polar sites at onset of sporulation. However, localization of SpoIIE phosphatase at the midcell position might inhibit sporulation and thus cause a delay or arrest of further forespore development.
TABLE 4.
Comparison of SpoIIE-GFP localization patterns in IB509 and PMF16 cellsa
| Fluorescence pattern | No. of cells showing pattern
|
|
|---|---|---|
| Wild type (PMF16) | minCD (IB509) | |
| Midcellb | 0 | 14 |
| Polarc | 122 | 114 |
| No localizationd | 14 | 20 |
Cells were grown in DSM and harvested 2 h after the cessation of logarithmic growth. All scoring was performed with cells from several independent experiments.
Includes cells showing all midcell SpoIIE-GFP localization, including cells with both midcell and polar signals.
Includes cells showing bipolar and unipolar SpoIIE-GFP localization with or without an additional midcell signal.
Includes cells with unlocalized or no signal.
FIG. 3.
Fluorescence microscopy of cells with SpoIIE-GFP-associated fluorescence at sites of septation in strain IB509 from cultures harvested 2 h after cessation of logarithmic growth, photographed as described in Materials and Methods. Cells with SpoIIE-GFP localization are marked. Arrows indicate the position of SpoIIE-GFP signals. Each scale bar represents 2 μm.
Ultrastructure of minD and spoIIE double-mutant cells.
It was shown previously that SpoIIE is essential for normal formation of the asymmetric sporulation septum and that a spoIIE null mutant produced cells containing thick asymmetric septa similar to those formed at the midcell site during normal vegetative growth (5, 16, 18, 27); in addition, the formation of these thick asymmetric septa is usually delayed (5, 16). Two strains with missense mutations, spoIIE64 and spoIIE71, that block sporulation at stage II have also been identified, but they have a strikingly different phenotype, characterized by the presence of only thin asymmetric septa, frequently at both polar positions (5, 16, 27). These two mutations are in the region of the spoIIE domain important for encoding the protein’s phosphatase activity, but they have no effect on the morphology of sporulation septa (1, 5, 16). We have investigated septation defects in double-mutant strains, IB506 and IB507, each with the divIVB1 mutation and with one of the two different kinds of spoIIE mutations, i.e., the null mutation spoIIE::Tn917ΩHU7 and the missense mutation spoIIE71, respectively. Sporulating cells of strain IB506 displayed characteristic phenotypic features of both minD and spoIIE null mutations. We observed only thick asymmetric septa, many cells having thick septa at the midcell position (about 16% of total cells examined) (Fig. 2C). Figure 1D shows a cross section of an IB506 cell, harvested 4 h after the beginning of sporulation, with minicell and vegetative septum beginning to be formed. Approximately 2% of the cells examined in cross sections contained midcell septa that by their thickness (14 to 26 nm) were classified as thin septa. An interesting feature of this double mutant was that we observed no significant delay of asymmetric septation (Fig. 2C) as was described previously for the spoIIE null mutant (5, 16). This observation suggests that the SpoIIE protein can be a part of the control mechanism that activates septation at the polar sites, probably through action of the MinCD complex. When SpoIIE was expressed earlier in development from an isopropyl-β-d-thiogalactopyranoside-inducible Pspac promoter, cells produced many septation abnormalities and also an increase in the number of septa at different positions (6). However, it is not likely that SpoIIE is able to directly recognize the MinCD complex at the poles because, as has been shown recently, localization of SpoIIE is FtsZ dependent (26) and thus FtsZ and probably other division proteins are directed to possible sites of septation earlier than SpoIIE itself. In contrast with this observation, Khvorova et al. (21) demonstrated that SpoIIE is involved in the Spo0A-dependent switch in the location of FtsZ rings.
We observed in approximately 60% of strain IB507 cells thin sporulation-like asymmetric septa (Fig. 2C), a normal feature of spoIIE71 mutant cells. Thick asymmetric septa likely represent minicell structures caused by the divIVB1 mutation. In contrast with cells of strain IB506, almost all septa at the midcell position in IB507 cells were thin septa (Fig. 1E and 2C). This finding supports the notion that SpoIIE is crucial for formation of thin sporulation septa (5, 16) and also suggests that SpoIIE allows formation of an unusually thin septum at midcell position when this site is not blocked by the MinCD complex.
Model of MinCD function during asymmetric septation.
The main contribution of this work is the discovery that the MinCD complex blocks the midcell site of septation during sporulation. There are two main lines of evidence that support this finding. First, in the absence of functional minD gene products, about 30% of cells create thin sporulation-like septa at the midcell position, although the polar septation event and subsequent stages of spore morphogenesis proceed normally. Our findings confirm the model proposed recently by Rothfield and Zhao (29), which predicts that the midcell position is released from inhibition in the absence of MinC and MinD products during sporulation. Second, the SpoIIE protein, normally localized in a bipolar manner in wild-type sporulating cells, can also be recruited to the midcell septal position in minD cells. Previous work has shown that SpoIIE localization is FtsZ dependent and that SpoIIE may be recruited to the potential division sites directly by FtsZ or may interact with other FtsZ-associated division proteins (26). Localization of SpoIIE protein to the midcell position in minD cells clearly indicates that this position can be recognized not only by division proteins involved in binary fission but also by SpoIIE and probably other sporulation-specific proteins as well. It was shown previously that SpoIIE expressed during vegetative growth can be localized to the midcell division site (26) where unusually thin septa are formed (6). Levin and Losick (25) have shown that Spo0A protein switches the localization of FtsZ from medial to a bipolar pattern. Additionally, these authors showed that a mutation in minD does not restore bipolar localization of FtsZ in sporulating cells of a spo0A mutant. They observed bands of FtsZ positioned at poles as well as at medial locations (but only one band per cell length, although theoretically all three sites are available) and argued that Spo0A is not acting through the MinCD proteins. In contrast, our results indicate that bipolar localization of SpoIIE is influenced by MinCD, and FtsZ should be localized earlier than SpoIIE in a similar manner. The mechanism that serves to sequester FtsZ to a single division site, in the absence of active Spo0A and MinCD, is likely to be regulated by the level of FtsZ. This suggestion is supported by the finding that in E. coli, increased levels of FtsZ can override the inhibition imposed by the min system at the cell poles (33). One explanation for the increase in concentration of FtsZ at the onset of sporulation is that Spo0A, as a transcription activator, positively regulates its transcription. This regulation also can be indirectly mediated by AbrB and/or Spo0H proteins.
The delay in asymmetric septum formation observed in spoIIE null mutants (5, 16) was not observed in the minD spoIIE null double-mutant strain. Suppression of this delay by the absence of MinD would suggest a role for SpoIIE in activation of this septation process, probably indirectly through the MinCD complex. The SpoIIE protein may have not only a structural role in formation of a proper sporulation septum but also an important role in progression of septum formation (21).
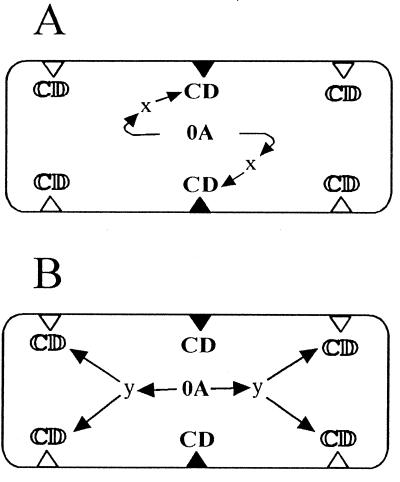
We propose three general models to explain how the MinCD complex could control the switch from midcell division to bipolar initiation of septation during the onset of sporulation, keeping in mind that this switch is under the control of Spo0A protein. Two of these models are illustrated in Fig. 4 The midcell-stabilizing model (Fig. 4A) predicts instability of the MinCD complex at the onset of sporulation and the existence of factor x, which specifically recognizes the midcell position, stabilizes the MinCD complex, and thus prevents initiation of septation at this position. The MinCD complex at the polar sites then can be eliminated without any stabilizing effect of factor x at these positions. In minCD mutant cells then, all three possible sites are available for septation. In cells without active Spo0A, initiation of septation is similar to that seen in vegetatively growing cells and is carried out at the midcell position. Although a model has been proposed for DivIVA function in the control of MinCD division inhibition during vegetative growth (14), it is still not clear how the midcell site of septation is activated in binary fission. The second, bipolar activation model, outlined in Fig. 4B, predicts the opposite scenario: the MinCD complex is very stable at the onset of sporulation, and its inhibition activity is released by factor y, which specifically recognizes only polar division sites and is under the control of the Spo0A protein. This model is very similar to the model proposed earlier by Cha and Stewart (9), in which DivIVA protein initiates sporulation septum formation by contact with a sporulation-specific protein. Our third model, a combination of the two models described above, predicts the existence of both factors x and y. Although it is premature to propose exactly how the asymmetric septum forms, intensive genetic and biochemical research in the near future should give us the answer to this crucial question.
FIG. 4.
Possible models demonstrating how the MinCD complex controls the switch from midcell division to bipolar initiation of septation during the onset of sporulation. Triangles represent possible sites of septation; open triangles represent sites not blocked by MinCD, and filled triangles represent sites blocked by active MinCD complex (CD). 0A, Spo0A protein. (A) The midcell-stabilizing model, whereby Spo0A controls factor x, which specifically stabilizes the MinCD complex at midcell position. (B) The bipolar activation model. Here Spo0A controls factor y, which specifically releases MinCD inhibition at both polar positions. For details, see Results and Discussion.
ACKNOWLEDGMENTS
We thank Phil Youngman for many years of support and for comments on the manuscript. We thank Patrick Piggot for communicating results before publication. In addition, we thank Paul Fawcett for supplying strain PMF16, Jozef Krištín for use of his electron microscope, Jozef Nosek for use of his Olympus microscope, and L’uboš Kl’učár for processing of digitized images. We thank DeEtte Walker, Danka Valková, Dušan Perečko, and Katarína Muchová for additional comments on the manuscript.
This work was supported by grant 2027 from the Slovak Academy of Sciences.
REFERENCES
- 1.Adler E, Donella-Deana A, Arigoni F, Pinna L A, Stragier P. Structural relationship between a bacterial developmental protein and eucaryotic PP2C protein phosphatases. Mol Microbiol. 1997;23:57–62. doi: 10.1046/j.1365-2958.1997.1801552.x. [DOI] [PubMed] [Google Scholar]
- 2.Arigoni F, Pogliano K, Webb C, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- 3.Backman K, Ptashne M, Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci USA. 1976;73:4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barák I, Behari J, Olmedo G, Guzmán P, Brown D P, Castro E, Walker D, Westpheling J, Youngman P. Structure and function of the Bacillus SpoIIE protein and its localization to sites of sporulation septum assembly. Mol Microbiol. 1996;19:1047–1060. doi: 10.1046/j.1365-2958.1996.433963.x. [DOI] [PubMed] [Google Scholar]
- 5.Barák I, Youngman P. SpoIIE mutants of Bacillus subtilis comprise two distinct phenotypic classes consistent with a dual functional role for the SpoIIE protein. J Bacteriol. 1996;178:4984–4989. doi: 10.1128/jb.178.16.4984-4989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barák I, Olmedo G, Youngman P. Abstracts of the 9th International Conference on Bacilli, Lausanne, Switzerland. 1997. Role of SpoIIE in the septum assembly and in the initiation of septation during development in Bacillus subtilis; p. 41. [Google Scholar]
- 7.Bi E, Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA) J Bacteriol. 1990;172:5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi E, Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990;172:5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha J H, Stewart G C. The divIVA locus of Bacillus subtilis. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutting S M, Youngman P. Gene transfer in gram-positive bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 348–364. [Google Scholar]
- 11.Dassain M, Bouché J P. The min locus, which confers topological specificity to cell division, is not involved in its coupling with nucleoid separation. J Bacteriol. 1994;176:6143–6145. doi: 10.1128/jb.176.19.6143-6145.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Boer P A J, Crossley R E, Hand A R, Rothfield L I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science. 1995;270:641–644. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- 14.Edwards D H, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 15.Fawcett P, Barák I, Olmedo G, Youngman P. Presented at the 12th International Spores Conference, Madison, Wis. 1996. A dual role for the SpoIIE protein in regulating morphogenesis and gene expression during sporulation in Bacillus subtilis. [Google Scholar]
- 16.Feucht A, Magnin T, Yudkin M D, Errington J. Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis. Genes Dev. 1996;10:794–803. doi: 10.1101/gad.10.7.794. [DOI] [PubMed] [Google Scholar]
- 17.Hayat M A. Fixation for electron microscopy. San Diego, Calif: Academic Press; 1981. [Google Scholar]
- 18.Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of ςE and ςF in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR protocols. San Diego, Calif: Academic Press; 1990. [Google Scholar]
- 20.Ju J, Luo T, Haldenwang W G. Bacillus subtilis Pro-ςE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J Bacteriol. 1997;179:4888–4893. doi: 10.1128/jb.179.15.4888-4893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khvorova A, Zhang L, Higgins M L, Piggot P. The spoIIE locus is involved in the Spo0A-dependent switch in the location of the FtsZ rings in Bacillus subtilis. J Bacteriol. 1998;180:1256–1260. doi: 10.1128/jb.180.5.1256-1260.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchner G, Kemper M A, Koch A L, Doyle R J. Zonal turnover of cell poles of Bacillus subtilis. Ann Inst Pasteur Microbiol. 1988;139:645–654. doi: 10.1016/0769-2609(88)90069-5. [DOI] [PubMed] [Google Scholar]
- 23.Lee S, Price C W. The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division. Mol Microbiol. 1993;7:601–610. doi: 10.1111/j.1365-2958.1993.tb01151.x. [DOI] [PubMed] [Google Scholar]
- 24.Levin P A, Margolis P S, Setlow P, Losick R, Sun D. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin P A, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 26.Levin P A, Losick R, Stragier P, Arigoni F. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol Microbiol. 1997;25:839–846. doi: 10.1111/j.1365-2958.1997.mmi505.x. [DOI] [PubMed] [Google Scholar]
- 27.Piggot P J. Mapping of asporogenous mutations of Bacillus subtilis: a minimum estimate of the number of sporulation operons. J Bacteriol. 1973;114:1241–1253. doi: 10.1128/jb.114.3.1241-1253.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogliano K, Hofmeister A E M, Losick R. Disappearance of the ςE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothfield L I, Zhao C R. How do bacteria decide where to divide? Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 30.Sandman K, Losick R, Youngman P. Genetic analysis of Bacillus subtilis spo mutations generated by Tn917-mediated insertional mutagenesis. Genetics. 1987;117:603–617. doi: 10.1093/genetics/117.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spurr A R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 32.Varley A W, Stewart G C. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J Bacteriol. 1992;174:6729–6742. doi: 10.1128/jb.174.21.6729-6742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward J E, Lutkenhaus J F. Overproduction of FtsZ indices minicells in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 34.Wu L J, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youngman P. Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus species. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 221–266. [Google Scholar]
- 36.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]