Abstract
Background:
Sarcocystosis is a zoonotic disease worldwide caused by Sarcocystis spp., some of these species can show clinical and subclinical manifestations, resulting in financial losses. Our study was performed for identifying Sarcocystis sp., in slaughtered buffalo by PCR–RFLP based strategy with sequencing in Guilan, North of Iran.
Methods:
Overall, 400 fresh muscle samples were prepared via naked-eye observation from 100 buffaloes (esophagus, diaphragm, shoulder, and thigh), followed by the digestion of samples. The PCR was done to amplify partial parts of the 18S rRNA and mitochondrial cytochrome c oxidase subunit I (Cox1) genes. Then, the PCR products were digested by endonuclease SspI, DraI, and FokI. Sequencing of all species was done to confirm the RFLP results.
Results:
Five macroscopic cysts (1.25%) were visible in the sample by naked-eye examination. Furthermore, 293 samples (73.25%) were found to be Sarcocystis sp. positive through tissue digestion and microscopic observation, whereas 376 samples (94%) were positive by PCR. In addition, the findings of PCR-RFLP and nucleotide sequence samples exhibited the infection of buffaloes with S. cruzi.
Conclusion:
Based on the data presented herein, Bovine sarcocystosis caused by S. cruzi is very common in buffalo in the Guilan region. Regarding the high prevalence of sarcocystosis, developing disease control and prevention policies for buffaloes is necessary, and a change of attitude in traditional farming is recommended.
Keywords: Buffalo, Sarcocystis cruzi, Iran
Introduction
More than 200 known species of Sarcocystis worldwide are capable of infecting animals (1). Sarcocystis is one of the most common cyst-forming species of protozoan parasites with universal distribution in different types of hosts as definitive (carnivores) and intermediate hosts (herbivores) (2–5). Some species of Sarcocystis may contribute to clinical symptoms, leading to weight loss, anorexia, fever, diarrhea, anemia, weakness, and death (6). Consumption of undercooked or raw beef is an important risk factor for humans (7).
Human intestinal sarcocystosis is a zoonotic disease, caused by two species of S. fusiformis (by consumption of infected beef) and S. meischeriana (by consumption of raw infected pork), resulting in digestive disorders (8). In addition to economic losses in animal husbandry, Sarcocystis infection can be a threat to public health (9). This parasite has a global spread and can infect various species of domestic animals. There have been many validated species in buffaloes such as S. fusiformis, and S. buffalonesis (cats as definitive hosts), as well as S. levinei (canids are the definitive host), and S. dubeyii with an unknown definitive host (10).
Adult Sarcocystis of different species is distinguishable via distinct phenotypic characteristics, e.g., the shape, size, cyst wall thickness, etc. Nevertheless, these characteristics may be influenced by the location and stages of cyst, growth, parasite stage and host tissue type. Therefore, molecular methods are considered reliable for affirming species identification and differentiation (11). The variable regions of the 18S rRNA gene are a valuable target for identifying different species (12). Molecular markers including 18S rRNA, 28S rRNA, ITS, and Cox1 sequence have been effectively used to identify Sarcocystis (13). However, the Cox1 is capable of differentiating Sarcocystis spp. thus described to be a favorable marker when compared with 18S rRNA gene (14).
Protozoan parasites are common in Iranian domestic livestock (15). However, few studies are available in Iran for Sarcocystis spp. in water buffaloes (2).
We aimed at evaluating the phylogenetic and molecular data of 18S rRNA and Cox1 genes of Sarcocystis sp. in Guilan province, North of Iran.
Materials and Methods
Ethical considerations
This study was done on slaughtered buffaloes. Research Ethics committees of Islamic Azad University-Science and Research Branch approved the study with ID IR.IAU.SRB.REC.1401.126.
Sampling
From January to May 2022, esophagus, diaphragm, shoulder, and thigh muscle samples of 100 buffalo carcasses from different slaughterhouses were collected in Guilan Province, northern Iran. Fifty grams of each muscle tissue was taken by veterinarians during postmortem inspections, transported to the laboratory through a cold chain, and then stored at −20°C until further evaluation.
Trypsin digestion
Briefly, 20 g of buffalo muscles were digested in 50 ml trypsin based on the optimized protocols for 16 h at 37 °C and then centrifuged for 5 min at 7000 rpm, followed by suspending the pellet in 5 ml PBS and centrifugation for 3 min at 5000 rpm (16, 17). The pellet was suspended in 5 ml of PBS and the resulting suspension was finally applied for DNA extraction.
DNA extraction and molecular detection of Sarcocystis spp
DNA extraction was performed using a commercial DNA extraction Kit (MBST, IRAN), according to the Kit instruction (18). Amplification of 18S rRNA and Cox1 genes were done by primers of the 18S rRNA gene (f: 5-TCAGGGAGGTAGTGACAAGA-3; R: 5-ATGTCTGG ACCTGGTGAGTT-3) and Cox1 gene (f: 5-CTTTAGCGTTGTTGGTAC-3; R: 5- CCCGTAGGAATGGCAAT-3). The primers applied for 18S rRNA (JQ713824), and Cox1 genes (KU247899) were designed based on published sequences, and synthesized by Sinaclon Biotech Co, Tehran, Iran. PCR was performed by the standard method, except for denaturation temperature (94°C). Each 25 μl PCR reaction consisted of 0.5 μM of each primer, 0.2 mM of each dNTP, 0.2 u/μl Dream Taq buffer, 1 μg DNA template, and distilled water. The cycling conditions consisted of initial denaturation at 95 °C for 5 min, followed by 40 cycles of 94 °C for 45 s, 56 °C for 45 s and 72 °C for 45 s, and a final extension cycle at 72 °C for 10 min. Negative control of the H2o was used as a sample. PCR products were finally electrophoresed on a 1.5% (w/v) agarose gel for 90 minutes (19).
Sequencing and phylogenetic analysis
The amplified 18S rRNA and Cox1 genes of Sarcocystis were sequenced based on a specific primer set to achieve species identification. The obtained sequences were blasted, and ClastalW V.2 software was applied to align the 18S rRNA and Cox1 genes sequences (20). MEGA 6.06 software was applied to generate a phylogenetic tree using Neighbor-Jinning (NJ) phylograms (with 1000 replicates) of both gene sequences (21, 22).
Results
RFLP-PCR
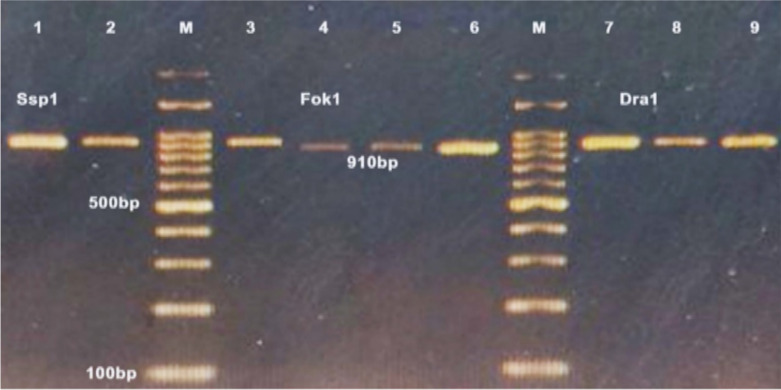
PCR amplification of 18S rRNA and Cox1 genes from of Sarcocystis species produced DNA fragments with the expected size of about 860 and 950 bps, respectively (Fig.1 and 2). Interestingly, RFLP analysis of PCR products with endonucleases exhibited the pattern of S. cruzi. As shown in Fig. 1, digestion with Dra1 and Fok1 produced 790 and 600 bps fragments of S. cruzi, respectively. The endonuclease Ssp1 did not result in any digestion band pattern of the 18S rRNA gene.
Fig. 1:
PCR-RFLP analysis of 18S rRNA gene of Sarcocystis isolated from Iranian water buffalo demonstrating species-specific fragments of S. cruzi: Lane 2, 3 and 4 digested whit Ssp1(a), Lane 2 and 4 digested whit Dra1, Lane 3 and 5 digested whit Fok1(b), M marker of 100 bp, lane1(a and b) uncut (control)
Fig. 2:
PCR-RFLP analysis of Cox1 gene of Sarcocystis, demonstrating species-specific fragments of S. cruzi: Lane1and 2 digested whit Ssp1, Lane 4, 5 and 6 digested whit Fok1, Lane 8 and 9 digested whit Dra1, M marker of 100 bp, lane 3 and 7 uncut (control)
In the mitochondrial gene Cox1, Digestion with Fok1 produced a DNA fragment of 910 bp, while the endonuclease Ssp1 and Dra1 did not result in any digestion band pattern of Cox1, representing S. cruzi (Fig. 2).
PCR-RFLP
For PCR-RFLP analysis, the 18S rRNA and Cox1 products of different Sarcocystis species were digested (19). Digestion of amplified PCR products was separately perfumed by three restriction enzymes: Dra1, Ssp1, and Fok1. Briefly, reaction mixtures were applied in a total volume of 50 μl, consisting of 10 units of restriction enzyme, 10–20 μl of PCR product, and 5 ml of buffer. All restriction mixtures were incubated for 16 h at 37 °C (Dra1, Ssp1) and 5 min at 37 °C (Fok1), followed by inactivation at 65 °C for 20 min (Dra1 and Ssp1) and at 65°C for 5 min (Fok1), (Thermo Scientific, USA). A 100-bp DNA ladder was utilized to estimate the sizes of the restriction fragments (Armanbiotech, cat no. YT8503).
DNA sequencing and analysis
The amplified PCR products of both genes were successfully sequenced for sampled S. cruzi. Assembly of DNA sequence on both strands was obtained from fragments consisting 773 and 858 consensus nucleotides of rRNA and Cox1, respectively. All Iranian 18S rRNA and Cox1 sequences were completely similar to each other as registered in GenBank under accession no. OP278729, and OP609867, respectively. The Iranian sequences were compared to eight corresponding sequences obtained from 18S rRNA gene (AF176935, KT306827, KM434885, and KJ917944), and Cox1 gene of S. cruzi (MT796928, MT796935, MW490605, KC209598). Based on the multiple alignments of Iranian 18S rRNA (Fig. 3) and Cox1 genes sequences with other S. cruzi strains (Fig. 4), different isolates were characterized by single nucleotide polymorphisms (SNPs) and indels in the present study.
Fig. 3:
A sequence alignment of the 18S rRNA (680 bp) derived from S. cruzi of the Iranian water buffalo and other S. cruzi strains. Nucleotide polymorphism in the sequenced 18S rRNA genes, at positions 444, 459, 511, 518, 526, 527, 539, and 664.
Fig. 4:
A part of multiple sequence alignment of the Cox1 (855 bp) derived from S. cruzi of the Iranian water buffalo and other S. cruzi strains derived. Nucleotide polymorphism at positions 585, 599, 708, and 845
According to accession numbers OP278729 and OP609867 (Figs 3, and 4), these sequencing results also showed nucleotide polymorphism in the sequenced 18S rRNA and Cox1 genes at positions 444, 459, 511, 518, 526, 527 and 539, as well as at 664 of the 18S rRNA and 585, 599, 708, and 845 of the Cox1, respectively. Sequencing of 18S rRNA gene of the Iranian isolates demonstrated Sequencing of the Iranian isolates of 18S rRNA demonstrated four nucleotide positions (444, 459, 539, and 664) are responsible for double peaks on the chromatogram. Two possible nucleotide characters were present for each of the four locations (Fig. 3); also, sequencing of Cox1 revealed two nucleotide positions (585, and 845) responsible for double peaks, where two possible nucleotide characters were observed in each of these locations (Fig. 4).
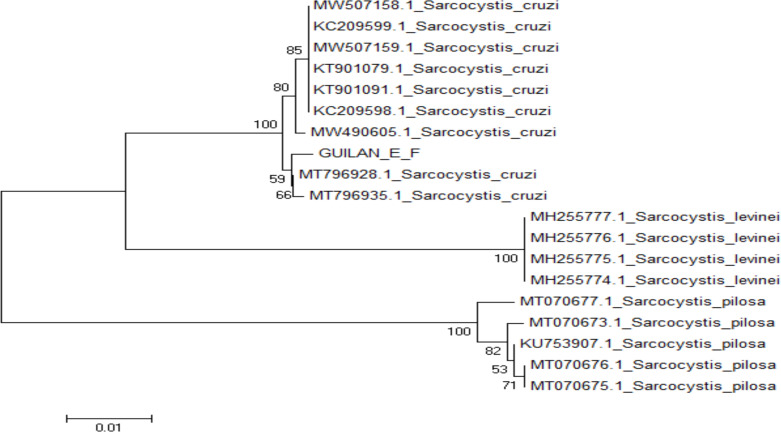
The phylogenetic analysis of 18S rRNA and Cox1 sequences by the maximum likelihood (ML) method (Fig. 5 and 6) exhibited that the Iranian isolates of Sarcocystis 18S rRNA sequences were in a group with three strains of S. cruzi (AF176935, KT306827 and KM434885), while Cox1 sequences were grouped with two previously reported S. cruzi strains (MT796928 and MT796935). Furthermore, the cladogram of 18S rRNA and Cox1 genes demonstrated that these groups were more linked to S. Levinei (KU247921, and KU247920) and S. cruzi (MW490605), respectively.
Fig. 5:
Phylogenetic tree of partial 18S rRNA sequence of Iranian water buffalo strain and other Sarcocystis species based on maximum likelihood (ML) method. The scale bar indicates distance
Fig. 6:
Phylogenetic tree based on partial Cox1 sequence of Iranian water buffalo strain and other Sarcocystis species using maximum likelihood (ML) method. The scale bar indicates the distance
Discussion
Sarcocystis infection has been reported among vertebrates, including humans (23). The morphological evaluation of Sarcocystis, especially the structure of the cyst wall and sporocysts, has been used to identify Sarcocystis species in livestock. Sarcocystis may show quite a few variabilities, depending on the location and stage of development and other parameters of the parasite cell. Molecular investigations have been carried out for morphologically identified species (24). Today, available molecular methods are capable of identifying morphologically similar sarcocysts or belonging to other species (25). Cox1 is the preferred target for taxonomic differentiation of Sarcocystis species affected ruminant intermediate hosts (e.g., cows, sheep, goats, deer, etc.), (26). Although 18S rRNA sequences are mostly registered in public databases due to their frequent use for identification, the mitochondrial Cox1 is the most useful tool for differentiating closely related Sarcocystis spp. in venomous mammals (27).
According to clinical, morphological, and pathological characteristics, S. cruzi has been identified in infected calves after experimentally infecting the calves with excreted oocysts of puppies (27). This protozoan may result in clinical manifestations such as weight loss, and abortion in calves. On the other hand, it probably causes nausea, stomachache, and diarrhea in humans after consuming half-cooked meat (28). Identification of Sarcocystis species has been performed through host characteristics, cyst morphology, and cyst wall ultrastructure, as well as molecular and biochemical features (29). To detect different species of Sarcocystis in meat, common techniques such as methylene blue staining, dob smear, and digestion have been applied in studies (30).
Serological methods have their own limitations in domestic animals such as significant antigenic similarity and subsequent cross-reactivity with other Sarcocystis species. In addition, serological tools are not useful for species identification (31). Molecular methods have been used to classify Sarcocystis spp. isolated from diverse samples (32). Highly conserved 18S ribosomal subunit variable regions are suitable for distinguishing Sarcocystis species in hosts. According to recent studies, Cox1 is preferred for identifying Sarcocystis species in domestic animals (5, 33, 34). Based on the data presented herein, S. cruzi was found among buffaloes in Guilan Province, but no infection with other Sarcocystis species was found. Sarcocystis was reported mainly in the muscles of the esophagus, larynx, and tongue (35, 36). Our findings showed the highest rate of infection in the diaphragm. We utilized 18S rRNA and Cox1 genes to identify Sarcocystis sp. According to the gene sequence data collected by analysis of morphological criteria, it can be concluded that the tested isolates collected from water buffalo belong to S. cruzi.
The phylogenetic analyzes performed based on the 18S rRNA gene showed a close evolutionary relationship between the current S. cruzi isolate and the China S. cruzi isolate (accession no. AF176935) (32), the Indian isolate (accession no. KT306827) (36) shown to have a little more than 99% sequence similarity with only five nucleotide substitution and 100% query coverage. Also, according to Cox1 gene showed a close evolutionary relationship between the current S. cruzi isolate and the Lithuania S. cruzi isolate (accession no. MT796928, MT796935) (37), which was shown to have more than 99.48% sequence similarity with only two nucleotide substitution and 100% query coverage.
By comparing the recorded sequences, the polymorphisms of the 18S rRNA and Cox1genes of the Iranian S. cruzi strain in our study may indicate geographical stability isolation. Intraspecific polymorphisms in these target genes can be usefully applied for phylogenetic assessment and the genetic structure of S. cruzi. Studying sarcocystosis from wider geographical areas and sequencing more loci, such as the rRNA large subunit gene, ITS1 or ITS2, can contribute to our deeper understanding of the differences between each species.
Sarcocystis isolates of sheep by 18S rDNA - RFLP and macroscopic cysts were reported to be S. gigantea and S. arieticanis, respectively (39). S. cameli was reported in camels for the first time in Iran based on an electron microscope and PCR-RFLP method (30). Available epidemiological data confirm the increase of sarcocystosis cases and the geographical spread of Sarcocystis in buffaloes of the Khuzestan province of Iran (2).
Buffalo breeding in Iran is mainly limited to the south and northwest regions. Sarcocystosis may be associated with decreased production of water buffaloes (10). Another study (2) revealed macroscopic forms of Sarcocystis in 3% of water buffaloes, while 83% exhibited microscopic forms. Our study is the first molecular description of S. cruzi in water buffalo in Iran. The findings of another study (19) are in accordance with the hypothesis that S. cruzi is capable of infecting water buffalo and is not limited to cattle. On the other hand, water buffalo were infected with S. fusiformis, S. cruzi, S. hominis, and S. hirsute based on RFLP of the 18S rRNA gene. They reported that Sarcocystis spp. in infected cattle is capable of infecting water buffaloes (19).
Despite the presence of polymorphisms in the 18S rRNA and Cox1 variable regions of S. cruzi, complete identity for the FokI restriction site was determined in all strains from various regions, (i.e., China, India, Iran for 18S rRNA; Iran and Lithuania for Cox1), suggesting the suitability of these conserved regions for FokI digestion as a suitable method for easy differentiation of S. cruzi from other species infecting water buffalo worldwide (Fig. 2 & 3).
Furthermore, all the double peaks in the chromatograms may be associated with at least two distinct variants of 18S rRNA and Cox1 genes in DNA isolated from cyst merozoites. The distinct variants might result from the 18S rRNA and Cox1 genes being a multi-copy gene. They might be expressed in the Sarcocystis due to differences between gene copies from different merozoites. Unrelated double peaks for SNPs may be linked to the variants of the gene in various isolates of any global species.
However, changes in this gene can be studied directly by DNA sequencing, as has been done previously (6, 33), but this is expensive for identification and epidemiological research.
Jehle et al evaluated the partial sequence of the 18S rDNA by modified RFLP analysis to identify Sarcocystis spp. in cattle and water buffaloes in Vietnam. This method was called a cost-effective technique for identifying Sarcocystis species. In addition, the PCR amplicon sequence analysis of the collected samples was performed in comparison with the results of registered sequences. Different species such as S. hirsute, S. cruzi, and S. hominis were detected in cattle, while infection of water buffaloes with S. fusiformis, S. cruzi, S. hominis, and S. hirsute has been reported. Most Sarcocystis species of cattle were capable of infecting water buffalo (39).
Based on the molecular results of the present study, it seems that the microscopic forms are probably early stages of developing cysts of S. cruzi and are not different species of Sarcocystis.
Conclusion
This study is the first report of S. cruzi in water buffalo in Guilan Province North of Iran. Water buffalo could be considered as a replacement intermediate host for S. cruzi. In addition to the complete sequence of the 18S rRNA gene and the mitochondrial Cox1 gene, sequence analysis of other genetic loci will be useful to better identify the genotypes of different Sarcocystis species isolated worldwide.
Acknowledgements
Sincerely thanks to people in the investigated regions who assisted in collecting buffalo muscle samples, and others participating in this work. No funding was received.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
Data availability
The nucleotide sequence generated in the present study has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/) under accession numbers OP278729 and OP609867.
References
- 1.Ayazian Mavi S, Teimouri A, Mohebali M, et al. Sarcocystis infection in beef and industrial raw beef burgers from butcheries and retail stores: A molecular microscopic study. Heliyon. 2020; 6(6): e04171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oryan A, Ahmadi N, Mousavi SM. Prevalence, biology, and distribution pattern of Sarcocystis infection in water buffalo (Bubalus bubalis) in Iran. Trop Anim Health Prod. 2010; 42(10):1513–8. [DOI] [PubMed] [Google Scholar]
- 3.Kirillova V, Prakas P, Calero-Bernal R, et al. Identification and genetic characterization of Sarcocystis arctica and Sarcocystis lutrae in red foxes (Vulpes vulpes) from Baltic States and Spain. Parasites Vectors. 2018; 11(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-kady AM, Hussein NM, Hassan AA. First molecular characterization of Sarcocystis spp. in cattle in qena governorate, upper Egypt. J Parasit Dis. 2018; 42(1):114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gjerde B, De la Fuente C, Alunda JM, et al. Molecular characterization of five Sarcocystis species in domestic sheep (Ovis aries) from Spain. Parasitol Res. 2020; 119(1):215–31. [DOI] [PubMed] [Google Scholar]
- 6.Yang ZQ, Li QQ, Zuo YX, et al. Characterization of Sarcocystis species in domestic animals using PCR-RFLP analysis of variation in the 18S rRNA gene: A cost-effective and simple technique for routine species identification. Exp Parasitol. 2002; 102(3–4):212–7. [DOI] [PubMed] [Google Scholar]
- 7.Fayer R, Esposito DH, Dubey JP. Human infections with Sarcocystis species. Clin Microbiol Rev. 2015; 28(2): 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubey JP, Speer CA, Fayer R. Sarcocystosis in animals and man. 1st ed. CRC, Boca Raton, FL. 1989a; 1–145. [Google Scholar]
- 9.Blagojevic B, Antic D. Assessment of the potential contribution of official meat inspection and abattoir process hygiene to biological safety assurance of final beef and pork carcasses. Food Control. 2014; 36:174–82. [Google Scholar]
- 10.Dubey JP, Speer CA, Shah HL. Ultrastructure of sarcocysts from water buffalo in India. Vet Parasitol. 1989b; 34(1–2):149–52. [DOI] [PubMed] [Google Scholar]
- 11.Kalantari N, Bayani M, Ghaffari S. Sarcocystis cruzi: first molecular identification from cattle in Iran. Int J Mol Cell Med. 2013; 2(3): 125–30. [PMC free article] [PubMed] [Google Scholar]
- 12.Borji H, Azizzadeh M, Kamelli M. A retrospective study of abattoir condemnation due to parasitic infections: economic importance in Ahwaz, southwestern Iran. J Parasitol. 2012; 98(5): 954–7. [DOI] [PubMed] [Google Scholar]
- 13.Gjerde B, Josefsen TD. Molecular characterization of Sarcocystis lutrae n. sp. and Toxoplasma gondii from the musculature of two Eurasian otters (Lutra lutra) in Norway. Parasitol Res. 2015; 114(3):873–6. [DOI] [PubMed] [Google Scholar]
- 14.Gjerde B. Sarcocystis species in red deer revisited: with a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial Cox1 sequences. Parasitology. 2014; 141(3):441–52. [DOI] [PubMed] [Google Scholar]
- 15.Nourollahi Fard SR, Asghari M, Nouri F. Survey of Sarcocystis infection in slaughtered cattle in Kerman, Iran. Trop Anim Health Prod. 2009; 41(8):1633–6. [DOI] [PubMed] [Google Scholar]
- 16.Verma SK, Lindsay DS, Grigg ME, et al. Isolation, culture and cryopreservation of Sarcocystis species. Curr Protoc Microbiol. 2017; 45: 20D.1.1–20D.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiesa F, Muratore E, Dalmasso A, et al. A new molecular approach to assess the occurrence of Sarcocystis spp. in cattle and products thereof: preliminary data. Ital J Food Saf. 2013; 2:148–51. [Google Scholar]
- 18.Al Qassab T, Shayan P, Kamkar Ab, et al. Reverse line blot hybridization assay as a suitable method for the determination of food adulteration in an example of sausage samples. European Food Research and Technology. 2019; 245:1677–83. [Google Scholar]
- 19.Li QQ, Yang ZQ, Zuo YX, et al. A PCR-based RFLP analysis of Sarcocystis cruzi (Protozoa: Sarcocystidae) in Yunnan province, PR China, reveals the water buffalo (Bubalus bubalis) as a natural intermediate host. J Parasitol. 2002; 88(6):1259–61. [DOI] [PubMed] [Google Scholar]
- 20.Larkin MA, Blackshields G, Brown NP, et al. ClustalW and Clustal X version 2.0. Bioinformatics. 2007; 23(21):2947–8. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28(10):2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdel-Ghaffar F, Mehlhorn H, Bashtar AR, et al. Life cycle of Sarcocystis camelicanis infecting the camel (Camelus dromedarius) and the dog (Canis familiaris), light and electron microscopic study. Parasitol Res. 2009; 106(1):189–95. [DOI] [PubMed] [Google Scholar]
- 23.Kia E B, Mirhendi H, Rezaeian M, et al. First molecular identification of Sarcocystis meischeriana (Protozoa, Apicomplexa) from wild boar (Sus scrofa) in Iran. Exp Parasitol. 2011; 127(3): 724–6. [DOI] [PubMed] [Google Scholar]
- 24.Dahlgren S, Gjerde B. Genetic characterization of six Sarcocystis species from reindeer (Rangifer tarandus tarandus) in Norway based on the small subunit rRNA gene. Vet Parasitol. 2007; 146(3–4):204–13. [DOI] [PubMed] [Google Scholar]
- 25.Gjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol. 2013; 43(7): 579–91. [DOI] [PubMed] [Google Scholar]
- 26.Gjerde B. Molecular characterisation of Sarcocystis bovifelis, Sarcocystis bovini n. sp., Sarcocystis hirsuta and Sarcocystis cruzi from cattle (Bos taurus) and Sarcocystis sinensis from water buffaloes (Bubalus bubalis). Parasitol Res. 2016; 115(4):1473–92. [DOI] [PubMed] [Google Scholar]
- 27.Naghibi A, Razmi G, Ghasemifard M. Identification of Sarcocystis cruzi in cattle by using of experimental infection in final and intermediate hosts (In Persian). J Fac Vet Med Univ Tehran. 2002; 57:67–9. [Google Scholar]
- 28.Pescador CA, Corbellini LG, Oliveira ECD, et al. Aborto ovino associado com infecção por Sarcocystis sp. Pesquisa Veterin′aria Brasileira. 2007; 27(10):393–7. [Google Scholar]
- 29.Motamedi GR, Dalimi A, Nouri A, et al. Ultrastructural and molecular characterization of Sarcocystis isolated from camel (Camelus dromedarius) in Iran. Parasitol Res. 2011; 108(4): 949–54. [DOI] [PubMed] [Google Scholar]
- 30.Holmdahl OJ, Mattsson JG, Uggla A, et al. Oligonucleotide probes complementary to variable regions of 18S rRNA from Sarcocystis species. Mol Cell Probes. 1993; 7(6):481–6. [DOI] [PubMed] [Google Scholar]
- 31.Moré G, Bacigalupe D, Basso W, et al. Serologic profiles for Sarcocystis sp. and Neospora caninum and productive performance in naturally infected beef calves. Parasitol Res. 2010; 106(3):689–93. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, Zuo YX, Yao YG, et al. Analysis of the 18S rRNA genes of Sarcocystis species suggests that the morphologically similar organisms from cattle and water buffalo should be considered the same species. Mol Biochem Parasitol. 2001; 115(2):283–8. [DOI] [PubMed] [Google Scholar]
- 33.Ellis JT, Luton K, Baverstock PR, et al. Phylogenetic relationships between Toxoplasma and Sarcocystis deduced from a comparison of 18S rDNA sequences. Parasitology. 1995; 110(5): 521–8. [DOI] [PubMed] [Google Scholar]
- 34.Oryan A, Moghaddar N, Gaur SNS. The distribution pattern of Sarcocystis species, their transmission, and pathogenesis in sheep in the Fars Province of Iran. Vet Res Commun. 1996; 20(3): 243–3. [DOI] [PubMed] [Google Scholar]
- 35.Heckeroth AR, Tenter AM. Comparison of immunological and molecular methods for the diagnosis of infections with pathogenic Sarcocystis species in sheep. The Tokai J Exp Clin Med. 1998; 23(6):293–302. [PubMed] [Google Scholar]
- 36.Daptardarkar M, Singh BB, Gill JPS, et al. Prevalence and first molecular identification of Sarcocystis species in cattle and water buffaloes in Punjab, India. Acta Parasitol. 2016; 61(3): 523–8. [DOI] [PubMed] [Google Scholar]
- 37.Prakas P, Strazdaite-Zieliene Z, Januskevicius V, et al. Molecular identification of four macrocystis species in cattle from Lithuania, including S. hominis, and development of a rapid molecular detection method. Parasit Vectors. 2020;13(1):610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalimi A, Paykari A, Esmaeilzadeh M. Identification of Sarcocystis species of infected sheep in Ziaran abattoir, Qazvin, using PCR-RFLP (In Persian). Modares Med Sci. 2008; 11:65–72. [Google Scholar]
- 39.Jehle C, Dinkel A, Sander A. Diagnosis of Sarcocystis spp. in cattle (Bos Taurus) and water buffalo (Bubalus bubalis) in northern Vietnam. Vet Parasitol. 2009; 166(3–4):314–32. [DOI] [PubMed] [Google Scholar]