Abstract
Accumulating studies suggest that Huaier exerts anti-tumor effects through intricate mechanisms. Despite extensive research on its efficacy in lung cancer, further investigation is required to elucidate the molecular mechanism of Huaier. The involvement of long noncoding RNAs (lncRNAs) in the anti-lung cancer effects of Huaier remains unknown. In this study, we found Huaier suppressed cell viability, migration and invasion in non-small cell lung cancer (NSCLC) cells. LncRNA sequencing analysis revealed Deleted in lymphocytic leukemia 2 (DLEU2) to be significantly downregulated in Huaier-treated NSCLC cells. Furthermore, DLEU2 silencing was observed to suppress NSCLC progression, while DLEU2 overexpression attenuated the anti-tumor effects of Huaier in NSCLC, thereby promoting cell viability, migration and invasion of NSCLC. The ceRNA role of DLEU2 had been demonstrated in NSCLC, which directly interacted with miR-212-5p to rescue the repression of E74 Like ETS Transcription Factor 3 (ELF3) by this microRNA. Additionally, Huaier was found to regulate the expression of miR-212-5p and ELF3. Functionally, miR-212-5p inhibitor or ELF3 overexpression reversed the effects of DLEU2 silencing or Huaier treatment, resulting in increased colony formation, migration and invasion in NSCLC. Taken together, these results illuminate the mechanism underlying Huaier's anti-tumor effects via the DLEU2/miR-212-5p/ELF3 signaling pathway, which offers novel insights into the anti-tumor effects of Huaier and constitutes a promising therapeutic target for the treatment in NSCLC.
Keywords: Huaier, NSCLC, ncRNA, migration, invasion
Introduction
As indicated by the 2023 cancer statistics, lung cancer is a prevalent malignancy with the highest mortality rate1. Lung cancer can be categorized into two main types: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC accounts for approximately 85% of all lung cancer cases, encompassing subtypes such as lung adenocarcinoma, squamous cell carcinoma, and large cell lung cancer. Chemotherapy remains the primary treatment modality for NSCLC patients2. However, many effective chemotherapeutic agents are associated with systemic toxicity and multidrug resistance, which significantly limits their efficacy3. Several studies have demonstrated that combining conventional therapy with adjuvant Traditional Chinese Medicine (TCM) can mitigate treatment-related adverse effects, enhance the quality of life and reduce mortality in cancer patients4, 5.
Trametes robiniophila Murr is an official fungus that originates from the trunks of trees and has been utilized as TCM for approximately 1600 years6. The hot aqueous extraction of Trametes robiniophila Murr (Huaier) or granule is a commonly used pharmaceutical preparation. In recent years, an increasing number of studies have been conducted to explore the intricate mechanisms underlying the anti-tumor effects of Huaier6, 7. The available evidence indicated that Huaier could induce immunogenic cell death8, inhibit cell growth, migration and energy metabolism9, induce apoptosis10, activate cell autophagy induced ferroptosis11, enhance the sensitivity to chemotherapeutic agents12, and so on. Although some research have been performed on the anti-lung cancer mechanisms of Huaier13, 14, further research is imperative.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs that exceed 200 nucleotides in length. LncRNAs have been reported to act as oncogene or tumor suppressor in diverse biological processes15-17. With the development of RNA sequencing, more and more lncRNAs have been found to be dysregulated in lung cancer18. Several lncRNAs, such as LINC0114019, HNF1A-AS120 and SNHG621 have been demonstrated to be upregulated in lung cancer. Other lncRNAs, such as ZNRD1-AS122 and HITT23 are downregulated in lung cancer. These lncRNAs play a crucial role in the pathogenesis and therapeutic strategies of lung cancer. There are reports shown Huaier suppresses breast cancer progression via linc00339/miR-4656/CSNK2B signaling Pathway24. Since lncRNAs have been implicated in the anti-breast cancer effects of Huaier, it is worth investigating whether they also play a role in the anti-NSCLC effects of Huaier. So far, there have been no reports indicating the involvement of lncRNAs in Huaier's anti-NSCLC mechanisms.
In this study, we examined the expression patterns of lncRNAs in non-small cell lung cancer cells treated with Huaier and untreated controls using RNA sequencing, and subsequently investigated the potential role of specific lncRNA in mediating Huaier's anti-tumor effects. Our data provides innovative insights into the anti-tumor effects of Huaier and presents a promising therapeutic target for NSCLC treatment.
Materials and methods
Cells and cell culture
The human non-small cell lung cancer cell lines (A549, H1299), and the normal lung epithelial cell line BEAS-2B, and the human embryonic kidney (HEK) 293T cell were all obtained from American Type Culture Collection (ATCC). A549, H1299, BEAS-2B and HEK-293T cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, PAN, Eidenbach, Bagoria, Germany). Cells were grown in an incubator with 5% CO2 at 37 °C.
Preparation of Huaier
Huaier was provided by Qidong Gaitianli Pharmaceutical Co., Ltd (Jiangsu, China). The stock solution of Huaier (100 mg/mL) was prepared for use as previously described 25.
Cell counting kit-8 (CCK8) assay
Cell viability was detected by CCK8 assay. 8000 A549 cells/well and 5000 H1299 cells/well were respectively seeded into 96-well plate and cultured overnight. Then, medium was replaced with the fresh medium containing specific concentrations of Huaier (0, 1, 2, 4, 8, 12, 16 or 20 mg/mL). After incubation for 48h and 72h, the supernatant was removed and the 100 μL medium containing 10% CCK8 reagent (Beyotime, Nantong, China) was added to each well. Absorbance values were determined by an EnSpire multimode reader (PerkinElmer, Waltham, MA) at 450nm.
Colony formation assay
For colony formation assay, A549 and H1299 cells (500 cells/well) were seeded in 12-well plates and cultured for ten days. The formed colonies were washed with phosphate-buffered saline (PBS) twice and stained with 0.1% crystal violet (Beyotime). A colony containing at least 50 cells was counted as one colony to represent the malignant viability of a single cell.
Wound healing assay
For cell migration assay, A549 and H1299 cells were seeded into 6-well plates (3×105 cells/well) and cultured for 24 h. A sterile 200 μl pipette tip was used to scratch the confluent cell monolayer vertically to make an artificial wound. Then, cells were washed with PBS twice and maintained in DMEM containing 2% FBS or various concentrations of Huaier. The wound closure images (magnification, 100×) were captured at 0 h and 48 h by IX81 microscope (Olympus, Tokyo, Japan). The wound healing rate was analyzed by Image J software.
Transwell assay
For cell invasion assay, transwell chamber (Corning, NY, USA) with 8 µm pore filters was used. The upper chamber was precoated with 100 μl 10% Matrigel (BD Biosciences, CA, USA). After appropriate treatment, 4×104 cells were resuspended in 100 μl serum-free medium and seeded into the upper chamber. DMEM containing 15% FBS was added into the lower chamber. After incubation for 24 h, the non-invaded cells on the surface of the upper chambers were removed by cotton swab. The cells that invaded to the basal side of the membrane were fixed with 4% formaldehyde (Beyotime) and stained with 0.1% crystal violet (Beyotime). Five fields of view at 100× magnification of the stained cells were selected randomly and calculated the mean using IX81 microscope (Olympus).
lncRNA sequencing
A549 and H1299 were treated with 0 mg/mL and 4 mg/mL Huaier for 48 h. Then, the cells were collected and frozen in TRIzol (Invitrogen, Carlsbad, CA, USA). The experiment was repeated three times. Next, we pooled the cell lysate generated from three respective times of the same group for further RNA extraction. Total RNA was quantified and qualified using Agilent 2100 bioanalyzer (Thermo Fisher Scientific, MA, USA). Subsequently, the cDNA library was created and assessed before sequencing. Illumina (HiSeq X-Ten) sequencing was then performed to profile the expression of lncRNA according to the manufacturer's protocol. The raw data was analyzed using HISAT comparison software. Fold change was calculated to identify the deregulated lncRNAs. Heml software was used to draw the heatmap according to the lncRNA level in NSCLC cells.
RNA isolation and qRT‑PCR
The total RNA was extracted from NSCLC cells with TRIzol reagent (Invitrogen). The concentration was measured by Nanodrop One (Thermo Fisher Scientific). The reverse transcription of RNA was conducted using HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China). The qPCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme). U6 and GAPDH were used as the internal control respectively for miR-212-5p and lncRNA detection. The relative expression values were calculated using the 2-ΔΔCt method for target genes. The primers for miR-212-5p and U6 were purchased from RiboBio Co., Ltd (Guangzhou, China). The primers for lncRNAs were as follows: DLEU2: sense, GTC TTT TGA AAG GTG TAC TGC AAG, and antisense, TGG TAT CAA AAT TTA CAC TAG AGC C. DNLZ: sense, TCG TCT ACA CCT GCA AGC CAG, and antisense, CTC TTC CCA TTC AGG TCC GAG. SNAPIN: sense, ACG TAC ACG CCG TCA GAA C, and antisense, GGT TTA GCC GTC TCA GTC GT. CDK2AP1: sense, ACA CCA CTC GCC ACC ATT TT, and antisense, CTC CGC GTA TTT GCT TTG GG. LMNTD2-AS1: sense, CCT TTG CCT GTG ATG TGG TG, antisense, AGT GAG GCT TCA GTT TGG GG. SNHG5: sense, GAA GAT GCA AAG ATA CAC G, antisense, CAA CAG TCA AGT AAA CCT CG. FGD5-AS1: sense, CTC GCT TAG CCA AGG CTT CT, antisense, ACA AAA AGC ATA TTC TAC AGA CGC T. CMAS: sense, GGA GTT CGT GAA GTG ACC GA, antisense, GAG ATT AGC CTC ACC TTA ATA CTC T. GAPDH: sense, GGG AGC CAA AAG GGT CAT, and antisense, GAG TCC TTC CAC GAT ACC AA.
Plasmid construction and cell transfection
LncRNA DLEU2 short interfering RNA (siRNAs), miR-212-5p mimic, miR-212-5p inhibitor and their corresponding controls were synthesized by Ribo Co., Ltd (Guangdong, China). SiRNAs and miR-212-5p inhibitor were transfected at a final concentration of 100nM using ExFect® Transfection reagent (Vazyme). The miR-212-5p mimic was transfected at a final concentration of 50nM. The DLEU2 targeting sequences were as follows: siDLEU2-1: GGT GAG AAC TGA CTA AAC T, siDLEU2-2: GGT AGA AGC TTG AAG GAA A, and siDLEU2-3: TGT CAG CAA AGA ACT GTA A. The sequence of DLEU2 and the full-length of ELF3 CDS region were respectively cloned into pcDNA-3.1 vector (Invitrogen). The efficiency of overexpression of DLEU2 or ELF3 was detected by qRT-PCR or western blot.
Western blot assay
Western blot analysis was performed as previously described25. The primary antibodies were as follows: mouse monoclonal anti-E-cadherin (1:2000, Proteintech, Rosemont, USA), mouse monoclonal anti-Vimentin (1:10000, Proteintech), rabbit polyclonal anti-MMP9 (1:500, Proteintech), rabbit polyclonal anti-VEGF (1:1000, Proteintech), rabbit polyclonal anti-ELF3 (1:500, Abcam, Cambridge, MA), rabbit polyclonal anti-Notch3 (1:500, Proteintech). Rabbit polyclonal anti-GAPDH (1:5000, Proteintech) was used as an endogenous control.
Dual-luciferase reporter assay
The sequence of DLEU2 and ELF3 3'UTR region, which containing the putative or mutative miR-212-5p binding sites was respectively inserted into the downstream of the reporter gene of the pRL-TK vector (Promega, Madison, WI). The above constructed plasmids were confirmed using DNA sequencing. Luciferase reporter assays were conducted in HEK-293T as described previously25. In brief, For DLEU2 luciferase reporter assay, pRL-TK-DLEU2 or pRL-TK-DLEU2 mutant vectors and pGL3 control (Promega) were transfected into HEK-293T along with miR-212-5p mimic or miR-212-5p inhibitor by ExFect® Transfection reagent (Vazyme) according to the protocol. For miR-212-5p target gene ELF3 luciferase reporter assay, pRL-TK-ELF3-3΄UTR or pRL-TK-ELF3-3΄UTR mutant vectors and pGL3 control (Promega) were transfected into HEK-293T along with miR-212-5p mimic or miR-212-5p inhibitor. The luciferase activity was detected 48 h post transfection using the Dual-Lumi luciferase reporter gene assay kit (Beyotime).
Statistical analysis
All statistical analyses were conducted by SPSS 22.0 software. Statistical analysis was performed with Student's t-test (for two-group comparisons). All experiments were performed in triplicate and data are presented as the mean ± S.D. A p-value < 0.05 represented statistically significant. *P < 0.05, ** P < 0.01, ***P < 0.001.
Results
Huaier suppresses cell viability, migration and invasion in A549 and H1299 cells
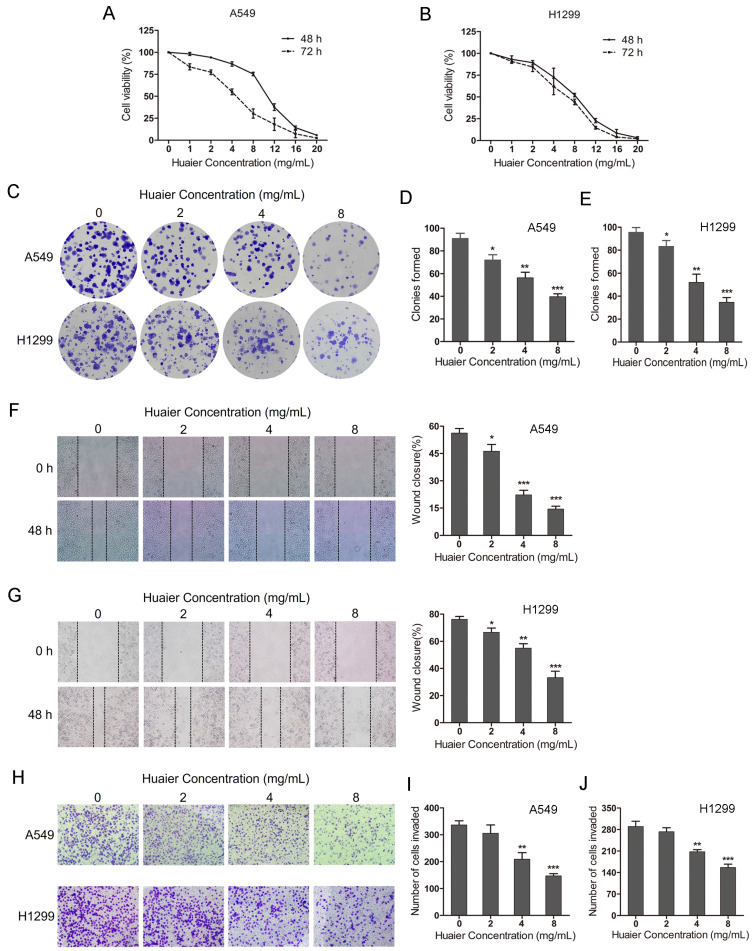
To explore the effects of Huaier on NSCLC cells malignant viability, CCK8 assay and colony formation assay were used. The CCK8 assay showed that Huaier treatment (0-20 mg/mL for 48-72 h) inhibited A549 and H1299 cells growth and the inhibitory effects exhibited a concentration- and time-dependent manner (Figure 1A and B). The colony formation assay showed that the colonies were significantly reduced in A549 and H1299 cells with the increased concentration of Huaier treatment (2, 4, 8 mg/mL) compared with the control group (Figure 1C, D and E). Moreover, the wound healing assay suggested that Huaier treatment obviously decreased the migration of NSCLC cells (Figure 1F and G). The transwell assay demonstrated that the invaded cells were markedly decreased after Huaier treatment (4, 8 mg/mL) compared with the control group, while 2 mg/mL Huaier treatment had no significant effects on cell invasion (Figure 1H, I and J). These results reveal that Huaier suppresses cell viability, migration and invasion in NSCLC cells. According to these results, we chose the treatment (4 mg/mL Huaier for 48 h) as the optimal concentration and time for the subsequent experiments.
Figure 1.
Effects of Huaier on lung cancer cell viability, migration and invasion. (A and B) Cell viability of A549 and H1299 cells treated with 0, 1, 2, 4, 8, 12, 16, 20 mg/mL Huaier for 48 h and 72 h. (C, D and E) Colony formation assay for A549 and H1299 cells treated with 0, 2, 4, 8 mg/mL Huaier. (F and G) Wound healing assay reflecting cell migration with 0, 2, 4, 8 mg/mL Huaier treatment for 48 h. (H, I and J) Transwell assay reflecting cell invasion with 0, 2, 4, 8 mg/mL Huaier treatment for 48 h.
Huaier downregulates the highly expressed lncRNA DLEU2 in NSCLC cell lines
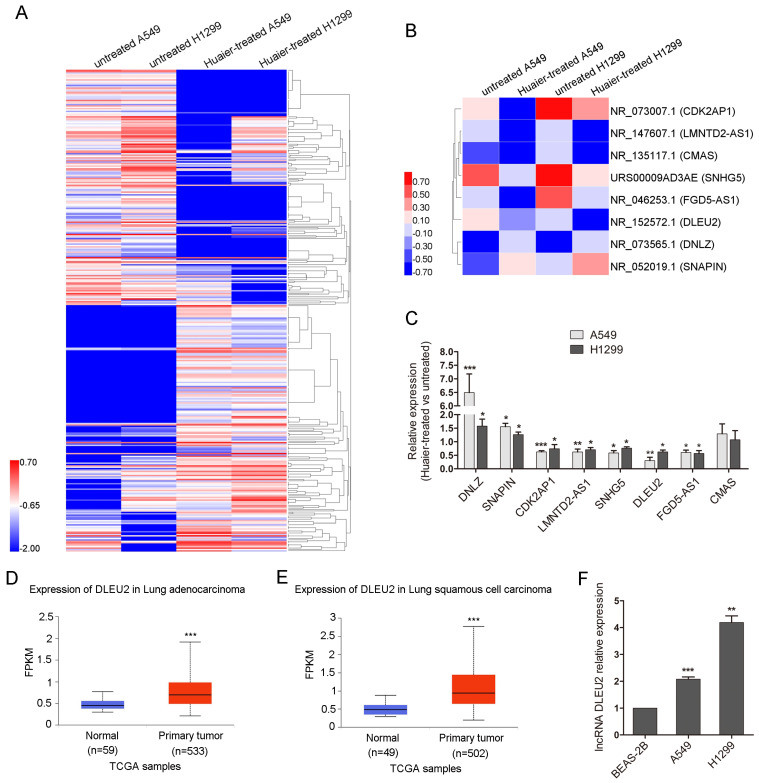
To investigate the mechanisms by which Huaier suppresses cell viability, migration and invasion in NSCLC cells, lncRNA sequencing was conducted. Heatmap showed 180 known significantly co-upregulated and 172 co-downregulated lncRNAs of 4 mg/mL Huaier-treated A549 and H1299 cells compared with the untreated group (Figure 2A). Heatmap also partially displayed six co-downregulated lncRNAs (CDK2AP1, LMNTD2-AS1, CMAS, SNHG5, FGD5-AS1, DLEU2) and two co-upregulated lncRNAs (DNLZ, SNAPIN) (Figure 2B). The qRT-PCR was used to verify the significantly differently expressed lncRNAs in Figure 2B. The expression of the selected lncRNAs by qRT-PCR assay was consistent with the results of lncRNA sequencing, while lncRNA CMAS exhibited no difference (Figure 2C). The occasional inconsistency may be related to the different detection principles of qRT-PCR and lncRNA sequencing. Among the differently expressed lncRNAs, Deleted in lymphocytic leukemia 2 (DLEU2) has been reported to be related to the tumorigenesis and progression of various malignant tumors and the upregulation of DLEU2 expression was associated with poor survival in patients with NSCLC26, 27. So, we chose DLEU2 for the further study. We analyzed the expression of DLEU2 in lung cancer tissues in UALCAN cancer database28. In UALCAN cancer database, DLEU2 expression was higher in 533 lung adenocarcinoma tissues than in 59 noncancerous lung tissues (Figure 2D). So did that in 502 lung squamous carcinoma tissues compared to 49 noncancerous lung tissues (Figure 2E). DLEU2 was also highly expressed in A549 and H1299 cells than in BEAS-2B (Figure 2F). These results suggest that Huaier can inhibit the overexpressed DLEU2 in NSCLC cell lines.
Figure 2.
The overexpressed lncRNA DLEU2 in lung cancer cell lines was downregulated after Huaier treatment. (A) Heatmap showing the significantly co-upregulated and co-downregulated lncRNAs of A549 and H1299 cells treated with 4 mg/mL Huaier for 48 h. The lncRNA expression profile was based on Huaier-treated versus untreated A549 and H1299 cells at fold change > 2 and false discovery rate (FDR) < 0.001. Red represented high expression and blue represented low expression. (B) Heatmap displaying the indicated differentially expressed lncRNAs. Six co-downregulated lncRNAs (CDK2AP1, LMNTD2-AS1, CMAS, SNHG5, FGD5-AS1, DLEU2) and two co-upregulated lncRNAs (DNLZ, SNAPIN). (C) The qRT-PCR to verify the co-upregulated and co-downregulated lncRNAs. (D and E) DLEU2 expression in lung adenocarcinoma and lung squamous carcinoma compared to the noncancerous lung tissues from UALCAN cancer database. (F) The qRT-PCR for DLEU2 expression in NSCLC cells and the noncancerous lung epithelial cell line BEAS-2B.
Huaier suppresses NSCLC cells viability, migration and invasion partially by downregulating DLEU2
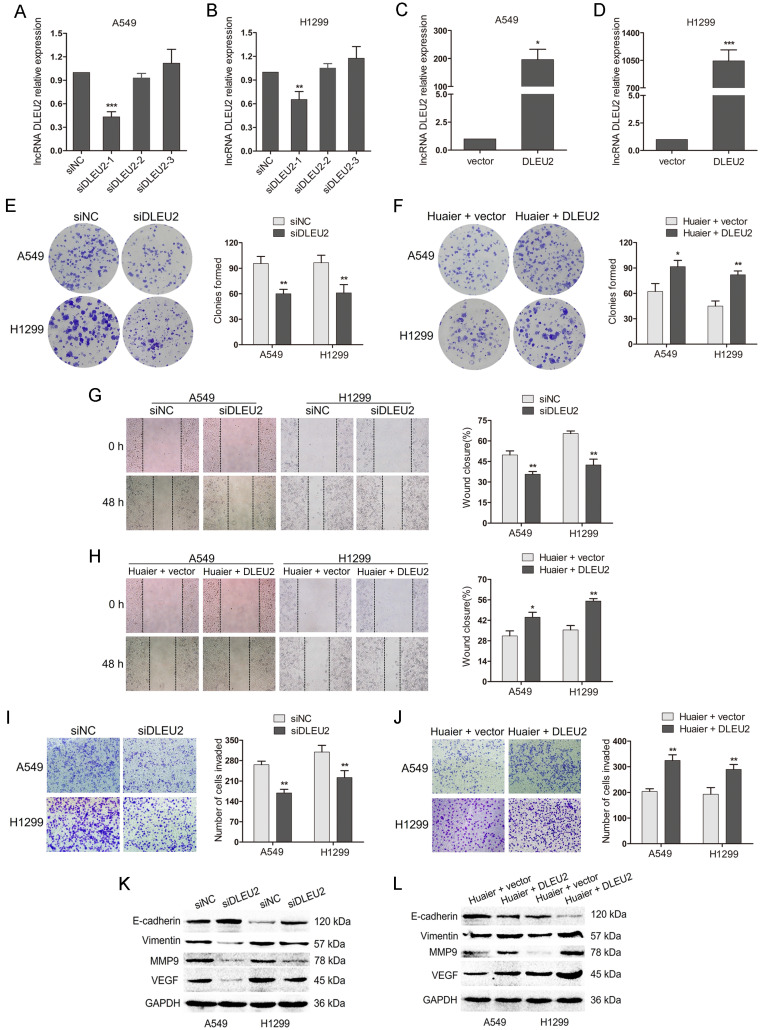
To further investigate the functions of DLEU2 in NSCLC cells, DLEU2 siRNAs (siDLEU2-1, siDLEU2-2, siDLEU2-3) were transfected into A549 and H1299 cells. The qRT-PCR showed that DLEU2 siRNA (siDLEU2-1) markedly inhibited DLEU2 expression, while siDLEU2-2 and siDLEU2-3 showed no effects (Figure 3A and B). Therefore, siDLEU2-1 was employed in the subsequent experiments to interfere with DLEU2 expression. In addition, DLEU2 was obviously increased in A549 and H1299 cells transfected with pcDNA3.1-DLEU2 compared with control (Figure 3C and D). We then transfected A549 and H1299 cells with siDLEU2 or siNC. DLEU2 silencing resulted in decreased colony formation, slower wound healing and less invaded cells in NSCLC cells (Figure 3E, G and I). DLEU2 silencing also led to the upregulation of the epithelial marker E-cadherin and the downregulation of mesenchymal marker Vimentin, the cell matrix metalloproteinase MMP9 and the vascular endothelial growth factor VEGF (Figure 3K). We further transfected pcDNA3.1-DLEU2 or control into Huaier-treated A549 and H1299 cells. The reduced colony formation medicated by Huaier treatment was reversed by DLEU2 overexpression (Figure 3F). Wound healing and transwell assays demonstrated that Huaier treatment-mediated inhibition in migration and invasion of both A549 and H1299 cells were overturned by DLEU2 overexpression simultaneously (Figure 3F, H and J). The western blot assay showed that the upregulation of E-cadherin and the downregulation of Vimentin, MMP9, VEGF by Huaier treatment were inverted in A549 and H1299 cells by highly expressed DLEU2 (Figure 3K). Collectively, these results suggest that inhibition of DLEU2 restrain NSCLC progression and Huaier suppresses NSCLC cells viability, migration and invasion partially by downregulating DLEU2.
Figure 3.
The effects of inhibition or overexpression of DLEU2 on cell viability, migration and invasion in both A549 and H1299 or Huaier-treated A549 and H1299 cells. (A and B) The qRT-PCR assay for DLEU2 expression in A549 and H1299 cells transfected with DLEU2 siRNA (siDLEU2-1, siDLEU2-2, siDLEU2-3) or control (siNC). (C and D) The qRT-PCR to detect the DLEU2 level in NSCLC cells transfected with pcDNA3.1-DLEU2 (DLEU2) or vector control (vector). (E, G, I and K) A549 and H1299 cells transfected with DLEU2 siRNA (siDLEU2) or control (siNC) for 48 h. Colony formation assay reflected cell viability (E). Wound healing assay reflected cell migration (G). Transwell assay reflected cell invasion (I). Western blotting detected the protein expression of E-cadherin, Vimentin, MMP9 and VEGF (K). (F, H, J and L) A549 and H1299 cells treated with 4 mg/mL Huaier and pcDNA3.1-DLEU2 (Huaier + DLEU2) or Huaier and vector control (Huaier + vector) for 48 h. Colony formation assay (F), wound healing assay (H), transwell assay (J) and western blot assay(L) were performed.
MiR-212-5p is the downstream of DLEU2 and miR-212-5p inhibition weakens the anti-tumor effects of DLEU2 downregulation and Huaier treatment
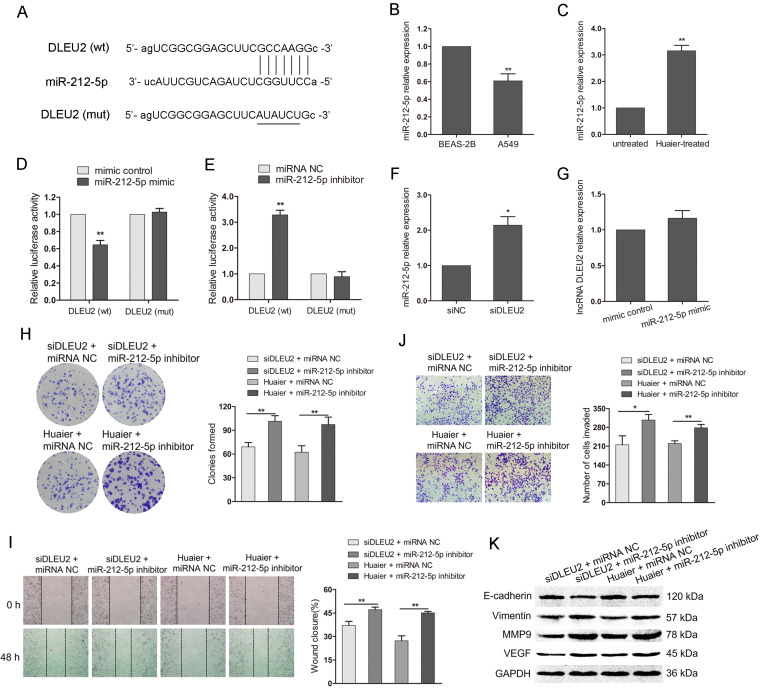
DLEU2 has been reported to involve in the tumorigenesis by playing as competing endogenous RNAs (ceRNAs) through competitively binding miRNAs27, 29. We then predicted miRNA that possibly bound to DLEU2 using starBase v3.0 (http://starbase.sysu.edu.cn/). The research showed that miR-212-5p could bind to DLEU2 with high conservation (Figure 4A). The qRT-PCR indicated that miR-212-5p was low expressed in A549 cells compared with BEAS-2B (Figure 4B).
Figure 4.
The interaction between DLEU2 and miR-212-5p in the anti-tumor effects of Huaier. (A) DLEU2 wild-type (wt) sequence containing the putative miR-212-5p binding sites and DLEU2 sequence with the mutant (mut) miR-212-5p binding sites. (B) The miR-212-5p level in A549 cells and the noncancerous lung epithelial cell line BEAS-2B. (C) The miR-212-5p level in Huaier-treated and untreated A549 cells. (D and E) Luciferase reporter assay in HEK-293T. HEK-293T was cotransfected with pRL-TK carrying wild-type or mutant DLEU2 sequence and the miR-212-5p mimic or miR-212-5p inhibitor, and the luciferase activity was detected 48 h after transfection. (F) The miR-212-5p level in NSCLC cells transfected with DLEU2 siRNA (siDLEU2) or control (siNC). (G) The expression of DLEU2 in NSCLC cells transfected with miR-212-5p mimic or mimic control. (H) Colony formation assay in DLEU2-silenced A549 cells transfected with miR-212-5p inhibitor (siDLEU2 + miR-212-5p inhibitor) or inhibitor control (siDLEU2 + miRNA NC). Colony formation assay in Huaier-treated A549 cells transfected with miR-212-5p inhibitor (Huaier + miR-212-5p inhibitor) or inhibitor control (Huaier + miRNA NC). (I, J and K) A549 cells were treated as in (H). wound healing assay (I), transwell assay (J) and western blot assay (K) were performed.
Additionally, miR-212-5p was upregulated in Huaier-treated A549 cells (Figure 4C). The level of miR-212-5p and DLEU2 exhibited the opposite expression in NSCLC cells or Huaier-treated NSCLC cells. To further clarify whether miR-212-5p could directly bind to DLEU2, we constructed luciferase reporters containing DLEU2 sequence with either wild-type or mutant miR-212-5p binding site. The luciferase reporter assay in HEK-293T showed that the overexpression of miR-212-5p negatively regulated the luciferase activity in a DLEU2 wild-type construct but not in a mutant (Figure 4D). Inhibition of miR-212-5p obviously increased the luciferase activity with wild-type DLEU2 sequence but not the mutant (Figure 4E). Furthermore, the expression of miR-212-5p was significantly increased with DLEU2 inhibition in A549 cells (Figure 4F). But, the expression of DLEU2 was not obviously changed by miR-212-5p overexpression in A549 cells (Figure 4G). These results suggest that there is a direct binding between miR-212-5p and DLEU2 and miR-212-5p is the downstream of DLEU2.
To explore the involvement of miR-212-5p in the anti-tumor effects of Huaier mediated by DLEU2, we cotransfected DLEU2 siRNA and miR-212-5p inhibitor or DLEU2 siRNA and inhibitor control into A549 cells. At the same time, we also transfected miR-212-5p inhibitor or control into Huaier-treated A549 cells. As shown in Figure 4H, the decreased colony formation caused by DLEU2 inhibition or Huaier treatment was reversed after cotransfection with miR-212-5p inhibitor. Wound healing and transwell assay showed that, in the presence of miR-212-5p inhibitor, the wound closure was increased and the invaded cells were much more despite DLEU2 inhibition or Huaier treatment (Figure 4I and J). The downregulation of E-cadherin and the upregulation of Vimentin, MMP9 and VEGF were induced in group (siDLEU2 + miR-212-5p inhibitor, Huaier + miR-212-5p inhibitor) compared to the respective control group (Figure 4K). These findings suggest that the inhibition of miR-212-5p attenuates the anti-tumor effects of DLEU2 downregulation and Huaier treatment, highlighting the pivotal role of DLEU2/miR-212-5p in mediating the anti-tumor effects of Huaier.
DLEU2, miR-212-5p and ELF3 constitutes a ceRNA network and DLEU2/miR-212-5p/ELF3 axis mediates the anti-tumor effects of Huaier
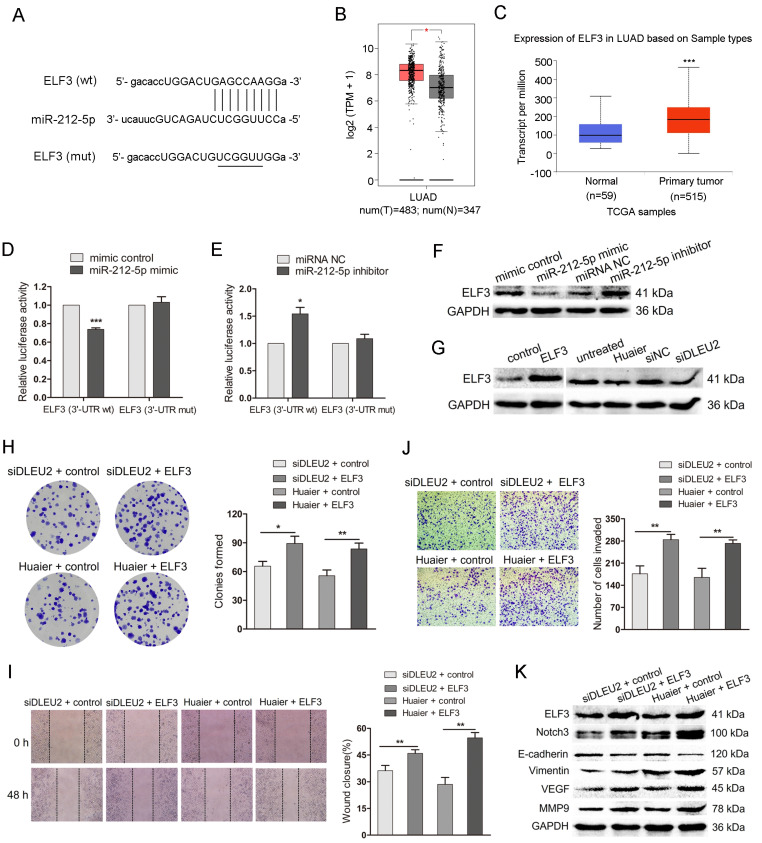
To further elucidate the underlying biological mechanisms mediated by DLEU2/miR-212-5p in Huaier-treated NSCLC cells, we searched for the potential targets of miR-212-5p by using TargetScan 8.0 (https://www.targetscan.org) and miRDB (http://www.mirdb.org/) database. MiR-212-5p was predicted to have putative binding to E74 Like ETS Transcription Factor 3 (ELF3) 3′UTR in both TargetScan8.0 and miRDB (Figure 5A). ELF3 acts as an oncogene and a promising target for therapeutic intervention in lung adenocarcinoma, with genetic, functional, and clinical evidence that supports the specificity of its subtypes30. ELF3 also promotes cell growth and metastasis in non-small cell lung cancer via the PI3K/Akt and ERK signaling pathways31, 32. In UALCAN cancer database, the expression of ELF3 was significantly elevated in lung adenocarcinoma tissues (n = 483), compared to the noncancerous lung tissues (n = 347; Figure 5B). Besides, GEPIA database also indicated that ELF3 level was higher in lung adenocarcinoma tissues (n = 515) than in normal control tissues (n = 59) (Figure 5C). The luciferase reporter assay in HEK-293T exhibited that miR-212-5p mimic repressed the luciferase activity from ELF3 wild-type 3′ UTR, but not from mutant 3′UTR (Figure 5D). Conversely, when miR-212-5p was downregulated, there was a significant elevation in luciferase activity in HEK-293T with the wild-type 3′UTR, but not the mutant (Figure 5E). These results indicated that ELF3 was a direct target of miR-212-5p. In A549 cells, miR-212-5p mimic significantly inhibited ELF3 protein level, while miR-212-5p inhibitor led to the increased protein level (Figure 5F). Moreover, the transfection with pcDNA3.1-ELF3 obviously upregulated the ELF3 protein level, while Huaier treatment or siDLEU2 transfection resulted in the decreased the protein level in A549(Figure 5G). These results suggested that ELF3 expression was regulated by the miR-212-5p, DLEU2 and Huaier and DLEU2, MiR-212-5p and ELF3 constitute a ceRNA network.
Figure 5.
The DLEU2/miR-212-5p/ELF3 axis in the anti-tumor effects of Huaier. (A) ELF3 3'UTR wild-type (wt) sequence containing the putative miR-212-5p binding sites and ELF3 3'UTR sequence with the mutant (mut) miR-212-5p binding sites. (B) ELF3 expression in lung adenocarcinoma compared to the noncancerous lung tissues from GEPIA database. (C) ELF3 expression in lung adenocarcinoma from UALCAN cancer database. (D and E) Luciferase reporter assay in HEK-293T. HEK-293T was cotransfected with pRL-TK carrying wild-type or mutant ELF3 3'UTR sequence and the miR-212-5p mimic or miR-212-5p inhibitor, and the luciferase activity was detected 48 h after transfection. (F) The western blot assay for ELF3 in A549 transfected with miR-212-5p mimic or miR-212-5p inhibitor. (G) The western blot assay for ELF3 in A549 cells transfected with pcDNA3.1-ELF3 (ELF3), DLEU2 siRNA (siDLEU2) or Huaier and the respective control. (H) Colony formation assay in DLEU2-silenced A549 cells transfected with pcDNA3.1-ELF3 (siDLEU2 + ELF3) or vector control (siDLEU2 + control). Colony formation assay in Huaier-treated A549 transfected with pcDNA3.1-ELF3 (Huaier + ELF3) or vector control (Huaier + control). (I and J) A549 cells were treated as in (H). wound healing assay (I), transwell assay (J) were performed. (K) The western blot assay for ELF3, Notch3, E-cadherin, Vimentin, MMP9 and VEGF in A549 cells treated as in (H).
To clarify the ELF3 in the anti-tumor effects of Huaier mediated by DLEU2/MiR-212-5p, we cotransfected DLEU2 siRNA and pcDNA3.1-ELF3 or DLEU2 siRNA and vector control into A549 cells. Meanwhile, we also transfected pcDNA3.1-ELF3 and vector control into Huaier-treated A549 cells. The colony formation assay showed that the colonies were increased in group (siDLEU2 + ELF3, Huaier + ELF3) compared with that in the control group (Figure 5H). As shown in Figure 5I and J, the impaired wound closure and reduced cell invasion induced by DLEU2 siRNA or Huaier treatment were restored upon co-transfection with pcDNA3.1-ELF3. DLEU2-silenced and Huaier-treated A549 cells transfected with pcDNA3.1-ELF3 exhibited elevated protein levels of ELF3, Notch3, Vimentin, MMP9 and VEGF, as well as reduced E-cadherin level (Figure 5K). The findings suggest that the DLEU2/miR-212-5p/ELF3 axis plays a crucial role in mediating the anti-tumor effects of Huaier.
Discussion
Lung cancer is the leading cause of cancer-related mortality, with metastasis being a significant contributor to treatment failure in clinical patients33. The exploration of more optimized drugs and the development of more suitable strategies are still necessary to enhance the survival rate and expand treatment options for lung cancer. TCM, characterized by unique pharmacological mechanisms, minimal side effects and low toxicity, has demonstrated efficacy against lung cancer34. We have previously reported that Huaier inhibits proliferation and induces apoptosis in human lung cancer cells via a miR-26b-5p-EZH2-mediated approach25. LncRNAs play biological role in NSCLC through different pathways, such as lncRNA-protein interaction, lncRNA-ceRNA network, and binding to promoter regions of encoding genes35. Since lncRNAs are implicated in the pathogenesis of NSCLC, it remains to be determined whether they contribute to the anti-NSCLC effects of Huaier. This is the inaugural article to document that lncRNA serves as a mediator of Huaier's anti-tumor effects in NSCLC.
In the present study, we discovered Huaier inhibited the viability, migratory capacity, and invasive potential of NSCLC cells in a concentration- and time-dependent manner. LncRNA DLEU2 was significantly downregulated in NSCLC cells after Huaier treatment. DLEU2 was initially identified through a comprehensive analysis of the chromosome 13q14 region, a genomic locus that is frequently subjected to deletions in B-cell chronic lymphocytic leukemia (BCLL)36. Subsequent investigations have demonstrated the upregulation and oncogenic effects of DLEU2 across various cancer types37. DLEU2 captured our attention, prompting us to focus on this particular lncRNA. Moreover, DLEU2 exhibited high expression levels in primary lung tumors and cancer cells. Inhibition of DLEU2 in A549 and H1299 cells resulted in a significant decrease in cell viability, migration, and invasion. Conversely, overexpression of DLEU2 in Huaier-treated NSCLC cells counteracted the anti-tumor effects of Huaier, leading to increased colony formation capacity, migratory potential, and invasive ability.
Emerging evidence suggests that DLEU2 may disrupt miRNA function and their interplay is involved in the regulation of numerous types of cancer. We then identified miRNAs that bound to DLEU2. We discovered that StarBase V3.0 predicted an interaction between miR-212-5p and DLEU2. MiR-212-5p functions as a tumor-suppressor in various cancers, such as prostate cancer38, breast cancer39, and so on. However, miR-212-5p is also reported to be a tumor-promoter in certain cancers, such as colorectal cancer40 and lung adenocarcinoma cells41. The expression and function of miR-212-5p in cancers exhibit inconsistency across different genetic contexts and specific cell type. In our study, miR-212-5p was low expressed in A549 cells and was then upregulated after huaier treatment. Luciferase reporter assay showed that miR-212-5p directly bound to DLEU2. MiR-212-5p functions as a downstream effector of DLEU2, and inhibition of miR-212-5p attenuates the anti-tumor effects elicited by downregulation of DLEU2 and treatment with Huaier. The DLEU2/miR-212-5p pathway mediates the anti-tumor effects of Huaier.
To investigate the mechanisms mediated by DLEU2/miR-212-5p in Huaier-treated NSCLC cells, we identified a miR-212-5p target gene, ELF3. ELF3 was found to have a direct interaction with miR-212-5p and its expression was regulated by both miR-212-5p and Huaier, as well as DLEU2. These findings elucidated a complex ceRNA network involving DLEU2, miR-212-5p, and ELF3. Moreover, the upregulation of ELF3 counteracted the inhibitory effects induced by DLEU2 knockdown or Huaier treatment, leading to enhanced colony formation, migration and invasion in NSCLC. The protein kinase C iota activates the level of Notch3 via phosphorylating the transcription factor ELF3 and subsequent occupancy of ELF3 on the Notch3 promoter in lung adenocarcinoma42. The Notch3 upregulation facilitates the autologous invasion and migration of lung adenocarcinoma cells43. The modulation of Notch3 signaling is a pivotal factor in the progression and pathogenesis of lung cancer44, 45. In the current study, overexpression of ELF3 reversed the effects of DLEU2 silencing and Huaier treatment by promoting Notch3 expression, which subsequently upregulated the downstream proteins Vimentin, MMP9 and VEGF while downregulating E-cadherin. Collectively, the DLEU2/miR-212-5p/ELF3 pathway mediates the anti-tumor effects of Huaier by ultimately regulating proteins involved in migration and invasion.
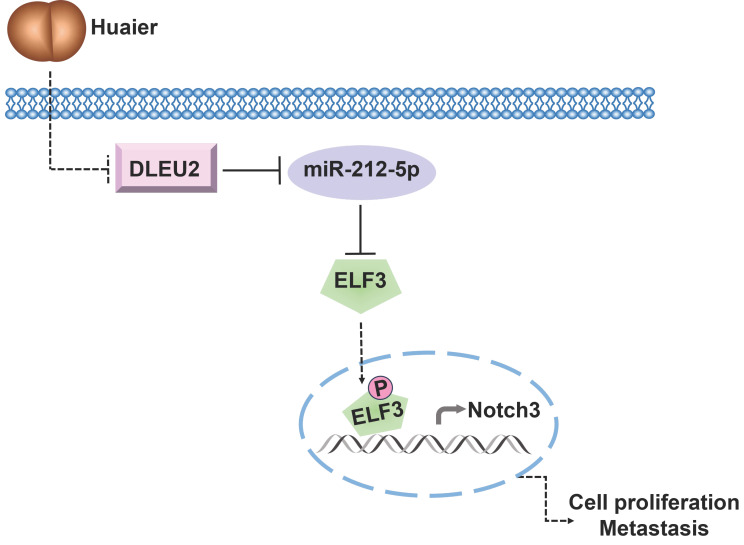
In conclusion, our study provides novel insights into the underlying mechanism of Huaier's anti-tumor effects in NSCLC. The ceRNA network involving DLEU2/miR-212-5p/ELF3 axis emerges as a crucial signaling pathway through which Huaier effectively suppresses NSCLC viability, migration, and invasion (Figure 6). These findings lay the foundation for considering the DLEU2/miR-212-5p/ELF3 axis as a promising therapeutic target for NSCLC treatment.
Figure 6.
Schematic representation of the underlying mechanisms governing Huaier's anti-tumor effects via modulation of the DLEU2/miR-212-5p/ELF3 signaling pathway.
Acknowledgments
We would like to thank all laboratory members for their critical discussion of this manuscript.
Funding
This project was supported by grants from the Yellow Crane Talent Plan Foundation, Research Fund of Wuhan Public Health Bureau (WZ22Q44).
Data Availability Statement
All data generated or analyzed during this study are included in this published article. Further inquiries can be directed to the corresponding authors.
Author Contributions
WTW: study design, project development and manuscript writing. SYL: project development and data collection. WQC and DZ: data analysis and coordination. ZXL: study design, supervision and manuscript editing. All authors have read and approved the final version of this manuscript to be published.
Abbreviations
- lncRNAs
long noncoding RNAs
- NSCLC
non-small cell lung cancer
- DLEU2
Deleted in lymphocytic leukemia 2
- ceRNA
competing endogenous RNAs
- ELF3
E74 Like ETS Transcription Factor 3
- SCLC
small cell lung cancer
- TCM
Traditional Chinese Medicine
- LINC01140
Long intergenic non-protein coding RNA 1140
- HNF1A-AS1
Hepatocyte nuclear factor 1 homeobox A antisense RNA 1
- SNHG6
small nucleolar RNA host gene 6
- ZNRD1-AS1
zinc ribbon domain-containing 1-antisense 1
- HITT
HIF-1α inhibitor at translation level
- CSNK2B
Casein kinase 2 beta
- ATCC
American Type Culture Collection
- HEK-293T
Human embryonic kidney 293T
- FBS
fetal bovine serum
- CCK8
Cell counting kit-8
- PBS
phosphate-buffered saline
- qRT‑PCR
quantitative reverse transcription polymerase chain reaction
- DNLZ
DNL-type zinc finger
- SNAPIN
synaptosomal-associated protein associated protein
- CDK2AP1
cyclin dependent kinase 2 associated protein 1
- LMNTD2-AS1
lamin tail domain containing 2 antisense RNA 1
- SNHG5
small nucleolar RNA host gene 5
- FGD5-AS1
PH domain containing 5 antisense RNA 1
- CMAS
cytidine monophosphate N-acetylneuraminic acid synthetase
- MMP9
matrix metallopeptidase 9
- VEGF
vascular endothelial cell growth factor
- Notch3
neurogenic locus notch homolog protein 3
- PI3K
phosphatidylinositol 3-kinase
- Akt
serine/threonine kinase
- ERK
extracellular-regulated kinase
- EZH2
enhancer of zeste 2 polycomb repressive complex 2 subunit
- BCLL
B-cell chronic lymphocytic leukemia
References
- 1.Siegel RL, Miller KD, Wagle NS. et al. Cancer statistics, 2023. CA: a cancer journal for clinicians. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Pirker R. Chemotherapy remains a cornerstone in the treatment of nonsmall cell lung cancer. Curr Opin Oncol. 2020;32(1):63–67. doi: 10.1097/CCO.0000000000000592. [DOI] [PubMed] [Google Scholar]
- 3.Tsiouda T, Sardeli C, Porpodis K. et al. Sex Differences and Adverse Effects between Chemotherapy and Immunotherapy for Non-Small Cell Lung Cancer. J Cancer. 2020;11(11):3407–3415. doi: 10.7150/jca.40196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao Z, Chen Z, Han R. et al. Comprehensive TCM treatments combined with chemotherapy for advanced non-small cell lung cancer: A randomized, controlled trial. Medicine (Baltimore) 2021;100(18):e25690. doi: 10.1097/MD.0000000000025690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang XQ, Zhang Y, Hou W. et al. Association between Chinese Medicine Therapy and Survival Outcomes in Postoperative Patients with NSCLC: A Multicenter, Prospective, Cohort Study. Chin J Integr Med. 2019;25(11):812–819. doi: 10.1007/s11655-019-3168-6. [DOI] [PubMed] [Google Scholar]
- 6.Pan J, Yang C, Jiang Z. et al. Trametes robiniophila Murr: a traditional Chinese medicine with potent anti-tumor effects. Cancer Manag Res. 2019;11:1541–1549. doi: 10.2147/CMAR.S193174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi T, Dong Y, Gao Z. et al. Research Progress on the Anti-Cancer Molecular Mechanisms of Huaier. Onco Targets Ther. 2020;13:12587–12599. doi: 10.2147/OTT.S281328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, Wang X, Chen T. et al. Huaier Induces Immunogenic Cell Death Via CircCLASP1/PKR/eIF2alpha Signaling Pathway in Triple Negative Breast Cancer. Front Cell Dev Biol. 2022;10:913824. doi: 10.3389/fcell.2022.913824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Liu L, Chen K. et al. Huaier shows anti-cancer activities by inhibition of cell growth, migration and energy metabolism in lung cancer through PI3K/AKT/HIF-1alpha pathway. J Cell Mol Med. 2021;25(4):2228–2237. doi: 10.1111/jcmm.16215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu DQ, Yuan XJ, Hirayama M. et al. Huaier Extract Induces Apoptosis in Hepatoblastoma Cells Via the MEK/ERK Signaling Pathway. In Vivo. 2020;34(5):2381–2388. doi: 10.21873/invivo.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Z, Wang X, Zhang W. et al. Huaier suppresses pancreatic cancer progression via activating cell autophagy induced ferroptosis. Front Oncol. 2022;12:960858. doi: 10.3389/fonc.2022.960858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gou Y, Zheng X, Li W. et al. Polysaccharides Produced by the Mushroom Trametes robiniophila Murr Boosts the Sensitivity of Hepatoma Cells to Oxaliplatin via the miR-224-5p/ABCB1/P-gp Axis. Integr Cancer Ther. 2022;21:15347354221090221. doi: 10.1177/15347354221090221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv F, Li X, Wang Y. An extraction from Trametes robiniophila Murr. (Huaier) inhibits non-small cell lung cancer proliferation via targeting to epidermal growth factor receptor. Bioengineered. 2022;13(4):10931–10943. doi: 10.1080/21655979.2022.2066757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan H, Xu X, Bai Y. Trametes robiniophila represses angiogenesis and tumor growth of lung cancer via strengthening let-7d-5p and targeting NAP1L1. Bioengineered. 2022;13(3):6698–6710. doi: 10.1080/21655979.2021.2012619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. 2022;82(12):2252–2266. doi: 10.1016/j.molcel.2022.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Q, Li Y, Zhang Y, LncRNA NRON promotes tumorigenesis by enhancing MDM2 activity toward tumor suppressor substrates. EMBO J. 2023:e112414. [DOI] [PMC free article] [PubMed]
- 17.Dong S, Ma M, Li M. et al. LncRNA MEG3 regulates breast cancer proliferation and apoptosis through miR-141-3p/RBMS3 axis. Genomics. 2021;113(4):1689–1704. doi: 10.1016/j.ygeno.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Qian X, Yang J, Qiu Q. et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol. 2021;14(1):112. doi: 10.1186/s13045-021-01123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia R, Geng G, Yu X, LINC01140 promotes the progression and tumor immune escape in lung cancer by sponging multiple microRNAs. J Immunother Cancer. 2021. 9(8) [DOI] [PMC free article] [PubMed]
- 20.Wang Z, Liu L, Du Y. et al. The HNF1A-AS1/miR-92a-3p axis affects the radiosensitivity of non-small cell lung cancer by competitively regulating the JNK pathway. Cell Biol Toxicol. 2021;37(5):715–729. doi: 10.1007/s10565-021-09595-z. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Zhang W, Yin D. et al. Gene amplification-driven lncRNA SNHG6 promotes tumorigenesis via epigenetically suppressing p27 expression and regulating cell cycle in non-small cell lung cancer. Cell Death Discov. 2022;8(1):485. doi: 10.1038/s41420-022-01276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Tan L, Yu X. et al. lncRNA ZNRD1-AS1 promotes malignant lung cell proliferation, migration, and angiogenesis via the miR-942/TNS1 axis and is positively regulated by the m(6)A reader YTHDC2. Mol Cancer. 2022;21(1):229. doi: 10.1186/s12943-022-01705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Zheng S, Yang F. et al. lncRNA HITT inhibits metastasis by attenuating Rab5-mediated endocytosis in lung adenocarcinoma. Mol Ther. 2022;30(3):1071–1088. doi: 10.1016/j.ymthe.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Wang X, Li C. et al. Huaier Suppresses Breast Cancer Progression via linc00339/miR-4656/CSNK2B Signaling Pathway. Front Oncol. 2019;9:1195. doi: 10.3389/fonc.2019.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T, Chen W, Liu S. et al. Huaier suppresses proliferation and induces apoptosis in human pulmonary cancer cells via upregulation of miR-26b-5p. FEBS Lett. 2014;588(12):2107–2114. doi: 10.1016/j.febslet.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Bai M, Lin L. et al. LncRNA DLEU2 aggravates the progression of hepatocellular carcinoma through binding to EZH2. Biomed Pharmacother. 2019;118:109272. doi: 10.1016/j.biopha.2019.109272. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Shi H, Du Y. et al. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/SOX9 axis. Aging (Albany NY) 2019;11(18):7386–7401. doi: 10.18632/aging.102226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandrashekar DS, Karthikeyan SK, Korla PK. et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu B, Gong X, Zi L. et al. Silencing of DLEU2 suppresses pancreatic cancer cell proliferation and invasion by upregulating microRNA-455. Cancer Sci. 2019;110(5):1676–1685. doi: 10.1111/cas.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enfield KSS, Marshall EA, Anderson C. et al. Epithelial tumor suppressor ELF3 is a lineage-specific amplified oncogene in lung adenocarcinoma. Nat Commun. 2019;10(1):5438. doi: 10.1038/s41467-019-13295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Yu Z, Huo S. et al. Overexpression of ELF3 facilitates cell growth and metastasis through PI3K/Akt and ERK signaling pathways in non-small cell lung cancer. Int J Biochem Cell Biol. 2018;94:98–106. doi: 10.1016/j.biocel.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W, Sun Q, Yu Z. et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene. 2018;670:31–37. doi: 10.1016/j.gene.2018.05.100. [DOI] [PubMed] [Google Scholar]
- 33.Xie S, Wu Z, Qi Y. et al. The metastasizing mechanisms of lung cancer: Recent advances and therapeutic challenges. Biomed Pharmacother. 2021;138:111450. doi: 10.1016/j.biopha.2021.111450. [DOI] [PubMed] [Google Scholar]
- 34.Wei Z, Chen J, Zuo F. et al. Traditional Chinese Medicine has great potential as candidate drugs for lung cancer: A review. J Ethnopharmacol. 2023;300:115748. doi: 10.1016/j.jep.2022.115748. [DOI] [PubMed] [Google Scholar]
- 35.Hu Q, Ma H, Chen H. et al. LncRNA in tumorigenesis of non-small-cell lung cancer: From bench to bedside. Cell Death Discov. 2022;8(1):359. doi: 10.1038/s41420-022-01157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Corcoran M, Rasool O. et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15(20):2463–2473. doi: 10.1038/sj.onc.1201643. [DOI] [PubMed] [Google Scholar]
- 37.Ghafouri-Fard S, Dashti S, Farsi M. et al. Deleted in lymphocytic leukemia 2 (DLEU2): An lncRNA with dissimilar roles in different cancers. Biomed Pharmacother. 2021;133:111093. doi: 10.1016/j.biopha.2020.111093. [DOI] [PubMed] [Google Scholar]
- 38.Qi JC, Yang Z, Lin T. et al. CDK13 upregulation-induced formation of the positive feedback loop among circCDK13, miR-212-5p/miR-449a and E2F5 contributes to prostate carcinogenesis. J Exp Clin Cancer Res. 2021;40(1):2. doi: 10.1186/s13046-020-01814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv ZD, Yang DX, Liu XP. et al. MiR-212-5p Suppresses the Epithelial-Mesenchymal Transition in Triple-Negative Breast Cancer by Targeting Prrx2. Cell Physiol Biochem. 2017;44(5):1785–1795. doi: 10.1159/000485785. [DOI] [PubMed] [Google Scholar]
- 40.Du F, Li Z, Zhang G. et al. SIRT2, a direct target of miR-212-5p, suppresses the proliferation and metastasis of colorectal cancer cells. J Cell Mol Med. 2020;24(17):9985–9998. doi: 10.1111/jcmm.15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen FF, Sun N, Wang Y. et al. miR-212-5p exerts tumor promoter function by regulating the Id3/PI3K/Akt axis in lung adenocarcinoma cells. J Cell Physiol. 2020;235(10):7273–7282. doi: 10.1002/jcp.29627. [DOI] [PubMed] [Google Scholar]
- 42.Ali SA, Justilien V, Jamieson L. et al. Protein Kinase Ciota Drives a NOTCH3-dependent Stem-like Phenotype in Mutant KRAS Lung Adenocarcinoma. Cancer Cell. 2016;29(3):367–378. doi: 10.1016/j.ccell.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng W, Sheng Y, Xiao H. et al. Lung Adenocarcinoma Cells Promote Self-Migration and Self-Invasion by Activating Neutrophils to Upregulate Notch3 Expression of Cancer Cells. Front Mol Biosci. 2021;8:762729. doi: 10.3389/fmolb.2021.762729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bodas M, Subramaniyan B, Karmouty-Quintana H. et al. The emerging role of NOTCH3 receptor signalling in human lung diseases. Expert Rev Mol Med. 2022;24:e33. doi: 10.1017/erm.2022.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie K, Ye Y, Zeng Y. et al. Polymorphisms in genes related to epithelial-mesenchymal transition and risk of non-small cell lung cancer. Carcinogenesis. 2017;38(10):1029–1035. doi: 10.1093/carcin/bgx079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. Further inquiries can be directed to the corresponding authors.