Summary
Toll-like receptor (TLR) ligand-binding domains comprise 18–25 tandem copies of a 24-residue motif known as the leucine-rich repeat (LRR). Unlike other LRR proteins, TLRs contain significant numbers of non-consensus LRR sequences, which makes their identification by computer domain search programs problematic. Here, we provide methods for identifying non-consensus LRRs. Using the location of these LRRs, hypothetical models are constructed based on the known molecular structures of homologous LRR proteins. However, when a hypothetical model for TLR3 is compared with the molecular structure solved by x-ray crystallography, the solenoid curvature, planarity, and conformations of the LRR insertions are incorrectly predicted. These differences illustrate how non-consensus LRR motifs influence TLR structure. Since the determination of molecular structures by crystallography requires substantial amounts of protein, we describe methods for producing milligram amounts of TLR3 extracellular domain (ECD) protein. The recombinant TLR3-ECD previously used to solve the molecular structure of TLR3-ECD has also been used to study the binding of TLR3-ECD to its ligand, double-stranded RNA (dsRNA). In the last section, we describe the preparation of defined TLR3 ligands and present methods for characterizing their interaction with TLR3-ECD.
Keywords: Toll-like receptor, TLR, Homology-based modelling, Receptor specificity, Double-stranded RNA, dsRNA
1. Introduction
Pathogen recognition by Toll-like receptors (TLRs) is essential for initiating immune responses, but the molecular mechanism by which receptor:ligand interactions trigger this response remains poorly understood (1). TLR extracellular domains (ECDs) recognize and bind ligands through tandem copies of a motif known as the leucine-rich repeat (LRR) (2). The molecular structures of several LRR-containing proteins are known, making it possible to propose hypothetical structural models of TLR-ECDs from their amino acid sequences (3). These models may fail to accurately predict features such as solenoid curvature, planarity, and the conformations of non-consensus LRRs. Such limitations indicate that true TLR molecular structures can be determined only by high resolution x-ray crystallography, which requires several milligrams of recombinant protein. In addition, by using recombinant protein for solution-phase ligand-binding studies, the stochiometry, affinity, and binding kinetics of the receptor:ligand complex can be determined. These parameters are essential for the production of receptor:ligand co-crystals, thus completing the atomic-scale picture of ligand recognition. Recently, we and another group solved the molecular structure of the TLR3-ECD (4, 5), and here we describe some of the procedures we have used to propose hypothetical structural models, generate TLR3-ECD protein, and measure its interaction with its ligand, double-stranded RNA (dsRNA). Many of these procedures can likely be adapted to characterize other TLRs.
2. Materials
2.1. Modelling TLR-ECD Structures
2.1.1. Locating Leucine-Rich Repeats
TLR amino acid sequence.
SMART (Simple Modular Architecture Research Tool) web site (http://smart.embl-heidelberg.de/).
2.1.2. Using LRRs to Predict Structures of TLRs
TLR amino acid sequence with LRRs identified.
Amino acid sequence with individual LRRs identified and structural coordinates for homologous proteins to be used as model scaffold such as the Nogo receptor (PDB 1OZN [6]).
Molecular software program O from Uppsala Software Factory (http://xray.bmc.uu.se/usf).
2.2. TLR3-ECD Production and Purification
High Five™ cells (Invitrogen, Carlsbad, CA).
Express Five® SFM medium (Invitrogen) supplemented with 20 mM l-glutamine, 10 μg/mL gentamicin, or Hy-SFX medium (HyClone, Logan, UT) supplemented with 10 μg/ mL gentamicin media.
Complete EDTA-free protease inhibitor cocktail tablets (Roche Applied Science, Indianapolis, IN).
Immobilized metal affinity chromatography (IMAC) resin (NiNTA, Qiagen, Valencia, CA).
Binding buffer (for IMAC): phosphate-buffered saline (PBS), pH 7.4 supplemented with 350 mM NaCl, 5% glycerol, and 1 mM β-mercaptoethanol (BME).
Elution buffer (for IMAC): binding buffer + 500 mM imidazole.
Anti-FLAG® M2 agarose and FLAG® peptide (Sigma, St. Louis, MO).
Binding/wash buffer (for Anti-FLAG®): tris-buffered saline (TBS), pH 7.4.
Elution buffer (for Anti-FLAG®): TBS + 100 μg/mL FLAG® peptide.
Centriprep YM-30 concentrator (Millipore, Billerica, MA).
Akta™ system with Superdex 200 (2.6 × 100 cm) size exclusion chromatography (SEC) column (GE Healthcare, Piscataway, NJ).
SEC buffer: 20 mM buffer (dependent upon application), 150 mM NaCl, 5% glycerol, 1 mM BME.
2.3. Ligand Binding to TLR3-ECD
2.3.1. dsRNA Synthesis
dsRNA template vector: pGEM-T Easy (Promega, Madison, WI). Store at −20°C.
NdeI restriction enzyme + 10X buffer (New England Biolabs (NEB), Ipswich, MA). Store at −20°C.
QIAquick PCR purification kit (Qiagen).
dsRNA synthesis kit: T7 Ribomax (Promega), includes Ribomax Express buffer (2X), T7 Express enzyme mix, RQ1 DNAse, RNAse A, nuclease-free water, 3.2 M NaOAc. Store enzyme and buffer components at −20°C (store RNAse A at room temperature), and minimize freeze-thawing the buffer by aliquotting after the first thaw.
PIPES-buffered saline (PiBS): 20 mM PIPES (Sigma), 150 mM NaCl, pH 6.0 (unless otherwise indicated). Store at room temperature.
Akta™ liquid chromatography system with Superdex 200 10/300 GL column (GE Healthcare).
Absolute ethanol.
2.3.2. Ligand Biotinylation
Antarctic phosphatase + 10X buffer (NEB). Store at −20°C.
Adenosine 5′-(gamma-thio) triphosphate (ATP-γS, Sigma). Dissolve stock in water to 10 mM, and store aliquots at −20°C.
Polynucleotide kinase (PNK) + 10X buffer (NEB). Store at −20°C.
No-weigh maleimide-PEO2-biotin (Pierce, Rockford, IL). Store at 4°C, and dissolve in water immediately prior to use.
Saturated phenol.
2.3.3. TLR3 Binding ELISAs
Reacti-Bind Streptavidin HBC strip-wells (Pierce). Store at 4°C.
PiBST: PiBS + 0.1% Tween-20 (Sigma). Store at room temperature.
PolyI:C: to prepare stocks, dissolve a mixture of polyinosinic and polycytidylic acids (GE Healthcare) in PBS to 2 mg/mL, heat to 70°C for 10 min, then cool slowly to room temperature to anneal strands. Store aliquots at −20°C.
Anti-FLAG® M2-HRP monoclonal antibody (mAb) (Sigma).
Bovine serum albumin (BSA) (Sigma).
HRP substrate reagent (R&D systems, Minneapolis, MN).
H2SO4 (1 M).
3. Methods
3.1. Modelling TLR-ECD Structures
3.1.1. Locating Leucine-Rich Repeat
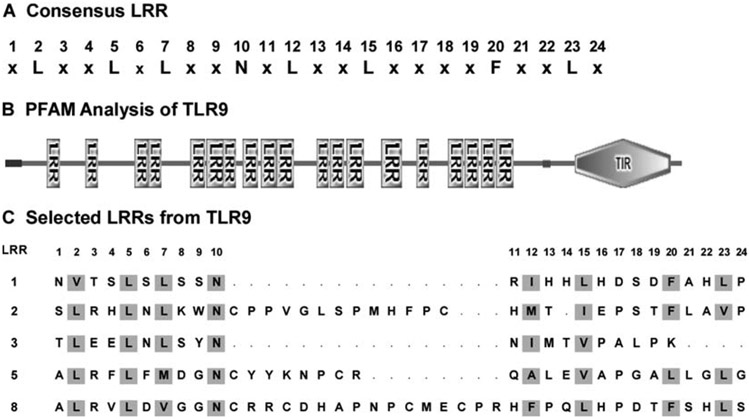
TLR-ECDs consist of 18–25 consecutive LRR motifs. The consensus LRR found in these receptors contains 24 amino acid residues (Fig. 1A). However, TLRs typically contain several non-consensus LRRs, which are not recognized by domain search programs such as protein families (Pfam) (7). Therefore, LRRs that deviate from the consensus sequence must be located manually. Such an approach was used to locate the LRRs in the ten human TLRs (3), which, in the case of TLR3, was shown to be valid at the structural level (4, 5).
Fig. 1.
Location of leucine-rich repeats (LRR) in Toll-like receptors (TLRs). (A) The consensus 24-residue sequence found in TLRs and some other LRR proteins such as the Nogo receptor and CD42b. (B) Domain organization of TLR9 as determined by protein families (Pfam) computer analysis. TIR refers to the cytoplasmic (Toll/IL-1R/Resistance) signalling domain, and the LRRs are the consensus LRRs located by the search algorithm. A signal sequence is located at the N-terminus, and a transmembrane segment separates the extracellular domain (ECD) from the cytoplasmic domain. (C) Representative LRRs from TLR9. LRR1 is a consensus LRR, while LRRs 2, 5, and 8 contain insertions of 13, 8, and 16 residues, respectively, following the consensus asparagine residue at position 10. By contrast, LRR3 is four residues shorter than the consensus.
The TLR amino acid sequence is copied into the SMART web site (http://smart.embl-heidelberg.de/) and searched for Pfam domains and signal sequences. This will locate the ECD and the consensus LRRs, as shown for TLR9 in Fig. 1B. Note also the intervening, undefined sequences between the consensus LRRs.
Deviations from the consensus LRR sequence cause search programs to miss LRRs. To find these missed LRRs, the regions between the consensus LRRs should be examined for L-x-x-L-x-L-x-x-N- sequences. Note that leucine residues can be replaced by other hydrophobic amino acids such as I, V, M, and F, and that N is frequently replaced by C, S, and T. The L-x-x-L-x-L-x-x-N- motif is present in the N-terminal portions of most LRRs. By contrast, the C-terminal portions are more variable in both length and sequence.
To locate the C-terminal ends of LRRs, look for the next L-x-x-L-x-L-x-x-N- motif. As a guide, the C-terminal portions of LRRs typically contain F-x-x-L or L-x-x-L sequences one or two residues before the beginning of the next LRR. LRRs are usually 24 residues long, but this can vary considerably. For example, Fig. 1C shows several non-consensus LRRs from TLR9, which were missed by domain search algorithms but were identified by their N-terminal L-x-x-L-x-L-x-x-N- sequence and the next LRR.
3.1.2. Using LRRs to Predict Structures of TLRs
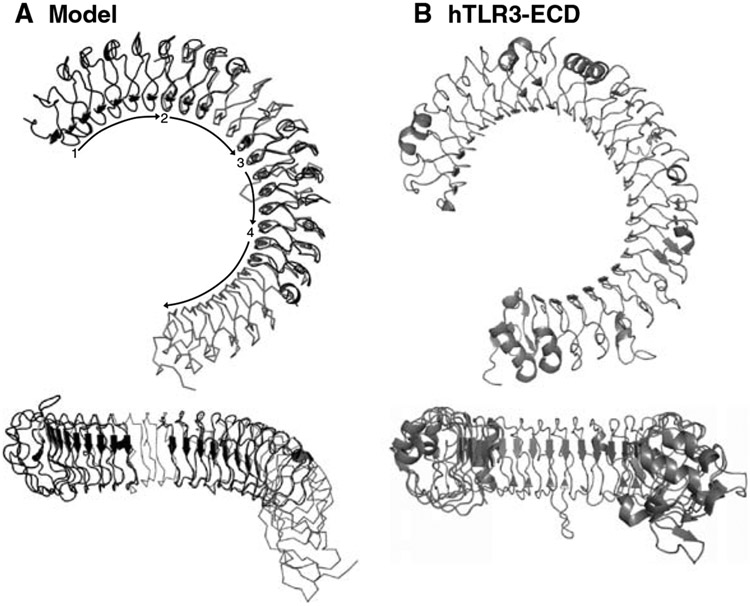
It is instructive to create a hypothetical model for the TLR-ECDs based upon the known molecular structures of other LRR proteins and the number of LRRs within TLR-ECDs (3, 8). The Nogo receptor contains LRR sequences that conform to the TLR consensus motif shown in Fig. 1A. In addition, the nine contiguous LRRs in the Nogo crystal structure provide a large building block for constructing models of TLRs, which contain 18–25 LRRs. Below is an example of how a model structure for the TLR3-ECD was constructed:
The N-terminal portion of the TLR3-ECD model is composed of Nogo receptor residues 26–248 (LRR-NT-LRR9) (Fig. 2A, segment 1). Coordinates of LRRs 2–9 of the Nogo receptor are extracted from its Protein Data Bank (PDB, http://www.rcsb.org/pdb/home/home.do) file to be used as a “building unit.” The next section of the TLR3-ECD model is composed of one building unit (Nogo LRRs 2–9). To align the first two sections of the model, LRRs 2–4 of the building unit are overlayed with LRRs 7–9 of the N-terminal unit (Fig. 2A, segment 2) using a molecular software package such as the program O.
This procedure is repeated by aligning LRRs 2–4 of a second building unit with LRRs 7–9 of the first building unit (Fig. 2A, segment 3). To complete the C-terminal portion of the TLR3-ECD model, Nogo receptor residues 56–309 (LRR2-LRR-CT) are overlayed with the second building unit. In this case, the first three LRRs of the Nogo receptor (residues 56–309) are aligned with LRRs 6–9 of building unit two (Fig. 2A, segment 4).
LRRs that were duplicated in the alignment process (N-terminal unit LRRs 7–9, building unit 1, LRRs 7–9, building unit 2, LRRs 6–9) are removed.
Fig. 2.
Cartoon representations of the TLR3-extracellular domain (ECD) structure. Upper figures show lateral views of the Toll-like receptor (TLR) “horseshoe,” and lower figures look into the open end of the horseshoe. (A) A model based upon the Nogo receptor structure. Segment 1, the N-terminal region, consisting of residues 26–248 of the Nogo receptor; segment 2, building unit 1; segment 3, building unit 2; segment 4, C-terminal unit (residues 56–309 of the Nogo receptor). (B) The TLR3-ECD crystallographic structure. Note that the model correctly predicts the overall shape of the TLR3-ECD but fails to predict the planarity of the TLR structure (bottom views). Moreover, the actual structure exhibits greater curvature than expected from the model.
A comparison of this hypothetical TLR3-ECD model with the known crystal structure is shown in Fig. 2A, B. Although the model grossly reflects the correct “horseshoe” shape of the TLR3-ECD molecule, the curvature and planarity of the horseshoe are not predicted correctly. Since the model was constructed using consensus LRRs, this comparison suggests that the curvature and planarity of the actual TLR3 structure derive from non-consensus LRRs.
3.2. TLR3-ECD Production and Purification
To determine the molecular structures of TLR-ECDs and to determine how they interact with pathogens at the molecular level, milligram amounts of pure protein are required. In our experience, a baculovirus secretion system has proven to be the best method for producing large amounts of correctly folded protein. However, there is great variation in yield between the different TLRs, and the conditions for expression need to be optimized for each TLR. Here, we describe the method used for expression and purification of TLR3-ECD protein, which is engineered to contain both FLAG® and His6 affinity tags fused to its C-terminus.
High Five™ cells (2 × 106 cells/mL) are infected with TLR3-ECD baculovirus at a multiplicity of infection of 3 at 27°C. Four hours after infection, the temperature is lowered to 21°C (see Note 1).
Forty-eight hours postinfection, cells are removed by centrifugation (12,000 × g, 20 min).
The supernatant is concentrated by ultrafiltration across a 10 kDa molecular weight cut-off membrane and diafiltered into IMAC binding buffer using a SartoJet diaphragm pump (Sartorius, Goettingen, Germany). A complete EDTA-free protease inhibitor tablet is added at this step.
Supernatant is applied to a Ni2+-charged IMAC column that has been pre-equilibrated with IMAC binding buffer, and the column is washed with the same buffer until the A280 nm approaches the baseline. A gradient of 0–500 mM imidazole in IMAC binding buffer is applied, and fractions containing TLR3-ECD, as identified by SDS-PAGE and Western blotting with Anti-FLAG® HRP, are pooled.
Pooled TLR3-ECD IMAC fractions are applied to a 10 mL Anti-FLAG®M2 agarose column pre-equilibrated with TBS (see Note 2). The column is washed with 20 column volumes of TBS, and the TLR3-ECD is eluted with 5 column volumes of TBS containing 100 μg/mL FLAG®peptide. Protein containing fractions are pooled and concentrated using a Centriprep YM-30 concentrator.
The concentrated TLR3-ECD is eluted on a 2.6 × 100 cm Superdex 200 column pre-equilibrated with SEC buffer. Fractions in the major peak, as detected by absorbance at 280 nM, are pooled, concentrated, and stored at 4°C (see Note 3).
3.3. Ligand Binding to TLR3-ECD
To test whether TLR3 binds its dsRNA ligand saturably and specifically, we developed an ELISA in which the protein binds to an immobilized ligand and is then detected using antibodies to the C-terminal tags. A similar approach should be applicable for characterizing other TLRs.
3.3.1. dsRNA Synthesis
To synthesize dsRNA enzymatically, a dsDNA template is first created by PCR. Since TLR3 recognizes dsRNA molecules of any sequence, any PCR template may be used, but the length of the PCR product determines the length of the final dsRNA molecules. The PCR product is ligated into the pGEM-T Easy vector, which contains a T7 RNA polymerase promoter upstream and a number of restriction sites downstream of the insertion site (see Note 4). Since the PCR product can insert in either direction, DNA sequencing is used to isolate one clone each having forward (sense) and reverse (antisense) orientations.
Plasmid DNA is isolated and linearized by digesting 50 μg DNA with 10 μL NdeI and 20 μL 10X buffer in 200 μL total overnight at 37°C (see Note 5). Linearized templates are purified using a QIAquick PCR purification kit.
Sense and antisense ssRNAs are synthesized simultaneously by mixing 40 μL Ribomax Express T7 2X buffer, 6 μg each of linearized sense and antisense templates, 8 μL T7 Express enzyme mix, and nuclease-free water to bring the total volume to 80 μL. The mix is incubated 2 h at 37°C (see Note 6).
RNA strands are annealed by heating to 70°C for 10 min in a heat block, turning off the heat, and allowing the block and RNA to cool slowly to room temperature (~1 h).
Digestion of DNA template and ssRNA (both un-annealed ssRNA and unpaired overhangs due to non-complementary pGEM-derived sequences that are present in ssRNAs) is accomplished by adding 4 μL each of RNAse A (freshly diluted in water to 20 ng/mL) and RQ1 DNAse, then incubating at 37°C for 30 min. Note that when using pGEM-T-based templates, four bases of self-complementary vector-derived sequence on each end of the PCR template (GATT-template-AATC) will survive RNAse A digestion.
To remove unwanted by-products of synthesis, the dsRNA is purified by gel filtration using a Superdex 200 10/300 GL column. The column is equilibrated in PiBS, loaded with up to 100 μL of the synthesis reaction, and dsRNA is eluted in PiBS following the manufacturer’s instructions (see Note 7). If multiple peaks are observed, each peak is pooled separately and compared with a dsRNA ladder using gel electrophoresis to determine which peak corresponds to the desired size of dsRNA.
Isolated dsRNA is precipitated by adding 2.5 volumes of ethanol and 0.1 volume of 3.2 M NaOAc. After incubation at −80°C for at least 2 h, the sample is pelleted in a microfuge at 4°C at maximum speed for 20 min. Liquid is removed, and pellets are washed with 500 μL of 70% ethanol in water and dried for 15 min. dsRNA is resuspended in PiBS or water and quantified by absorbance at 260 nm (1 OD = 45 μg/mL dsRNA, when pathlength is 1 cm).
3.3.2. Ligand Biotinylation
In the first step, phosphate groups are removed from the ends of dsRNA. 125 μL dsRNA in water (~100 μg) is mixed with 75 μL water, 25 μL 10X Antarctic phosphatase buffer, and 25 μL Antarctic phosphatase, and the mix is incubated at 37°C for 40 min. Enzyme is inactivated by heating to 65°C for 10 min.
Thiophosphate groups are transferred to the 5′ ends by adding 125 μL water, 50 μL PNK buffer, 25 μL PNK, and 50 μL of ATP-γS (10 mM) to the phosphatase-treated dsRNA, followed by incubation at 37°C for 30 min.
Fifty microlitres of water are added to each of two maleimide-PEO2-biotin microtubes (2 mg/tube) immediately prior to use. After the maleimide-PEO2-biotin is dissolved, it is added to the thiophosphate derivatized dsRNA, and the reaction mixture is incubated at 65 °C for 30 min, which covalently conjugates PEO2-biotin to the ends of the dsRNA.
Proteins are removed by extraction with 600 μL saturated phenol, and the aqueous phase is precipitated as in Subheading 3.3.1, step 7.
Biotinylated ligands are re-purified by gel filtration and concentrated as in Subheading 3.3.1, steps 6 and 7.
3.3.3. TLR3 Binding ELISAs
The following ELISAs are used to characterize the binding of TLR3-ECD to dsRNA and related ligands (see Note 8):
Direct Binding ELISA
Reacti-Bind Streptavidin HBC strip-wells are washed three times with 300 μL PiBST and are incubated with biotinylated dsRNA (1 μg/mL in PiBS, 100 μL/well) overnight at 4°C in a humidified box (see Note 9).
The biotinylated dsRNA solution is discarded, wells are washed as above, and a dilution series of TLR3-ECD in PiBS (100 μL/well) is added and incubated at room temperature for 2 h. Each condition should be performed in duplicate or triplicate, and one set containing zero TLR3-ECD should be included as a blank.
Solution is discarded, wells are washed as above, and Anti-FLAG® HRP mAb (diluted 1:8,000 in PiBS + 0.1% BSA) is added (100 μL/well) and incubated at room temperature for 1 h.
HRP substrate reagents A and B are mixed 1:1 at room temperature immediately prior to use. mAb solution is discarded, wells are washed as above, and mixed HRP substrate is added (100 μL/well). The reaction is allowed to progress for about 10 min or until any well begins to turn dark blue, at which point all wells are halted by adding 1 M H2SO4 (50 μL/well).
Absorbance at 450 nm is read with a 96-well plate reader, and the absorbances of the blank wells are averaged and subtracted as background. Figure 3A illustrates that the binding of TLR3-ECD to immobilized dsRNA is saturable.
Fig. 3.
Binding of Toll-like receptor 3 (TLR3)-extracellular domain (ECD) to immobilized double-stranded RNA (dsRNA). (A) Direct binding of TLR3-ECD to immobilized dsRNA as measured by ELISA. Binding reaches a plateau at high TLR3-ECD concentrations, and no binding is observed in the absence of immobilized dsRNA, indicating that binding is saturable and specific. (B) Inhibition of TLR3-ECD (0.2 μg/mL) binding to immobilized dsRNA. Soluble dsRNA and polyI:C compete with immobilized dsRNA for TLR3-ECD. The direct binding and inhibition ELISAs were run at pH 6.0 and 5.0, respectively. Both the soluble and immobilized dsRNA used in both ELISAs were 139 bp long.
Inhibition ELISA
Conversely, an inhibition ELISA is used to probe for binding specificity; this is accomplished by slightly modifying the direct binding ELISA protocol. A dilution series of non-biotinylated dsRNA or polyI:C (50 μL/well) is added to wells containing immobilized dsRNA (after the washes as given in the subheading Direct Binding ELISA, step 2), followed by 50 μL/well of a subsaturating concentration of TLR3-ECD (e.g., 0.2 μg/mL). Plates are incubated at room temperature for 2 h.
Samples are washed, labeled with Anti-FLAG® HRP, and assayed as in the subheading Direct Binding ELISA, steps 3–5. As seen in Fig. 3B, both ligands compete for TLR3-ECD, demonstrating that binding is specific.
4. Notes
Baculovirus was prepared using Gateway entry and bacmid vectors from Invitrogen. The TLR3-ECD construct contained an N-terminal baculovirus gp67 secretion signal, and at the C-terminal end, a tobacco etch virus (tev) protease cleavage site, followed by FLAG® and His6 epitope tags. Expression was driven by a polyhedron promoter. For production, 10 L cultures of High Five cells were used. This can be done in-house or commercially (e.g., Kemp Biotechnologies, Frederick, MD).
If significant amounts of the protein fail to adhere to the resin, the flow-through fraction can be reapplied to the column. We have also observed that more efficient binding occurs at lower flow rates.
TLR3-ECD should be stored at 2–3.5 mg/mL, pH 5.5, and at 4°C for maximum stability. Under these conditions, human TLR3-ECD is stable for at least 6 months. The protein tends to aggregate at higher concentrations.
Other dsDNA templates may also be used for RNA synthesis. For example, a T7 promoter sequence can be incorporated at the 5′ end of a PCR primer, as described in the T7 Ribomax manual (Promega). In this case, an ssRNA is generated directly from the PCR product, and the complementary ssRNA is generated from a separate PCR product in which the T7 promoter is instead incorporated into the reverse primer. For short dsR-NAs, a single DNA oligonucleotide encoding both the T7 promoter and the entire template sequence can be synthesized chemically (be sure to incorporate a G transcription start site immediately following the promoter). Since T7 polymerase works more efficiently from a dsDNA template, the coding strand is annealed to its complementary oligonucleotide. Separate dsDNA templates are required to make sense and antisense ssRNA oligonucleotides, which are then annealed as in Subheading 3.3.1, step 4. RNAse A cleaves ssRNA at the 3′ side of C and U residues, and this activity is compatible with the described use of the pGEM-T Easy vector. If the template used results in an RNA molecule with an unpaired G residue adjacent to the 5′ side of the double-stranded region, RNAse T1 may be used to cleave ssRNA at the 3′ side of the G residue.
The pGEM-T Easy vector contains restriction sites other than NdeI that may be used to linearize plasmid templates, but digests that produce 3′ overhangs should be avoided, since they can lead to undesired synthesis by-products.
For templates shorter than ~200 bp, incubation for up to 6 h has been found to increase yields significantly.
Gel filtration may be run in buffers other than PiBS, but phosphate-containing buffers should be avoided, since they promote bacterial contamination. Other methods for removing unincorporated nucleotides, such as MicroSpin G-25 columns (GE Healthcare), do not efficiently separate by-products and unincorporated nucleotides from the desired dsRNA; this problem becomes more apparent when synthesizing dsRNA smaller than ~200 bp. Note that unincorporated nucleotides cannot be detected using ethidium bromide in agarose gels, but they absorb at 260 nm, so failure to remove these will lead to errors in calculating dsRNA concentration.
If a Biacore surface plasmon resonance instrument (GE Healthcare) is available, direct binding of TLR3-ECD to immobilized ligand can be measured quantitatively and in real time using a similar strategy.
A mock series lacking immobilized ligand should also be included, especially during initial characterizations. This will test whether the receptor binds non-specifically to the plate (see Fig. 3A). In cases where non-specific binding is observed, it should be subtracted from the test samples. Non-specific binding might also be reduced by including 5 mg/mL BSA while coating overnight with biotinylated ligand (see Direct Binding ELISA, step 1).
Acknowledgements
This work was supported by the Intramural Research Program of the NIH (NCI and NIDDK) and by a Trans-NIH/FDA Intramural Biodefense Award from NIAID.
References
- 1.Gay NJ and Gangloff M (2007) Structure and function of toll receptors and their ligands. Annu. Rev. Biochem 76, 141–165. [DOI] [PubMed] [Google Scholar]
- 2.Kobe B and Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol 11, 725–732. [DOI] [PubMed] [Google Scholar]
- 3.Bell JK, Mullen GED, Leifer CA, Mazzoni A, Davies DR, and Segal DM (2003) Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 24, 528–533. [DOI] [PubMed] [Google Scholar]
- 4.Bell JK, Botos I, Hall PR, Askins J, Shiloach J, Segal DM, and Davies DR (2005) The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc. Natl. Acad. Sci. U.S.A 102, 10976–10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe J, Kelker MS, and Wilson IA (2005) Crystal structure of human Toll-like receptor 3 (TLR3) ectodomain. Science 309, 581–585. [DOI] [PubMed] [Google Scholar]
- 6.He XL, Bazan JF, McDermott G, Park JB, Wang K, Tessier-Lavigne M, He Z, and Garcia KC (2003) Structure of the Nogo receptor ectodomain: a recognition module implicated in myelin inhibition. Neuron 38, 177–185. [DOI] [PubMed] [Google Scholar]
- 7.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, and Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34, D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, Arditi M Hemming VG, Blanco JC, Segal DM, and Vogel SN (2006) Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J. Immunol 177, 322–332. [DOI] [PubMed] [Google Scholar]