Abstract
The thalamus plays a central coordinating role in the brain. Thalamic neurons are organized into spatially distinct nuclei, but the molecular architecture of thalamic development is poorly understood, especially in humans. To begin to delineate the molecular trajectories of cell fate specification and organization in the developing human thalamus, we used single cell and multiplexed spatial transcriptomics. Here we show that molecularly defined thalamic neurons differentiate in the second trimester of human development, and that these neurons organize into spatially and molecularly distinct nuclei. We identified major subtypes of glutamatergic neuron subtypes that are differentially enriched in anatomically distinct nuclei and six subtypes of GABAergic neurons that are shared and distinct across thalamic nuclei.
One-Sentence Summary:
Single cell and spatial profiling of the developing thalamus in the first and second trimester yields molecular mechanisms of thalamic nuclei development.
Thalamus can be anatomically subdivided into multiple nuclei with distinct patterns of neuronal projections (1). Many thalamic subregions are hypothesized to have undergone extensive remodeling and reorganization in recent evolution, including the pulvinar nucleus that is extensively expanded in humans and primates compared to mice (2, 3). Thalamic neurons emerge from progenitor cells located in the diencephalon of the prenatally developing nervous system (4). Recent studies have begun to apply single cell genomics approaches to uncover the molecular dynamics of their differentiation in mice (5), but our understanding of thalamic neuron differentiation in humans is less well-characterized. To better understand their molecular and spatial organization, we investigated developing human thalamus during the first and second trimester of development (gestational weeks 6–25) using single cell and spatial transcriptomics.
Neurogenesis in the first trimester thalamus
To identify the molecular trajectories of cell types in the developing human thalamus during neurogenesis, we analyzed scRNA-seq datasets generated from samples from the first trimester of human development (6, 7). Across 5 samples (3 males and 2 females) between gestational week (GW) 6 to 10, data re-processing retained 27,362 cell transcriptomes passing quality control metrics generated using droplet-based scRNA-seq (10X Chromium v2 assay) (fig. S1A). Initial preprocessing steps include ambient RNA removal using FastCAR (8), doublet filtering with scds (9), and further cell filtering using spliced/unspliced ratio with DropletQC (10). We then performed SCTransform normalization (11) with Harmony (12) for batch correction between different samples and Louvain clustering to identify cell types (Fig. 1, A and B). We recovered ten clusters representing all of the major classes in the thalamus, including radial glia (RG1-2), intermediate progenitor cells (IPC1-2), GABAergic inhibitory neurons (IN1-3), glutamatergic excitatory neurons (EN1-2), and neural crest-derived mesenchymal stem cells (MES) that develop to form the cerebral vasculature (Fig. 1B)(13).
Fig. 1. Neurogenesis in the first trimester thalamus.
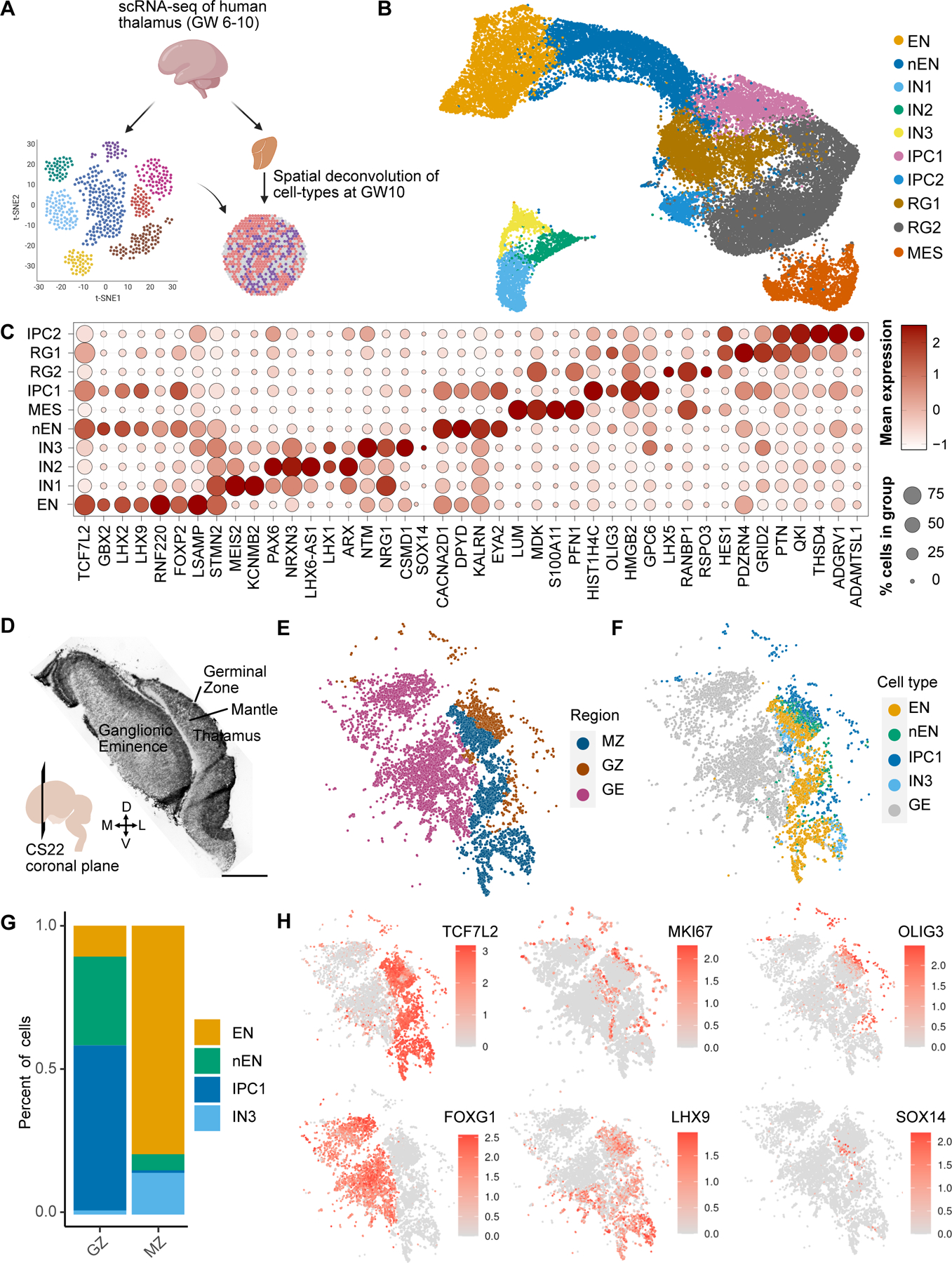
A. Schematic overview of figure 1. We analyzed scRNA-seq data collected from three biologically independent first trimester thalamus samples (GW6-10). Spatial transcriptomics using MERFISH was performed to spatially map cell types identified by scRNA-seq. B. UMAP embedding of cell type annotations on the Harmony batch corrected space: EN - excitatory neuron, nEN – newborn excitatory neuron, IN1/2/3 - inhibitory neuron 1/2/3, RG1/2 - radial glia 1/2, IPC1/2 - intermediate progenitor cells 1/2, and MES - mesenchyme. C. Dot plot of marker genes across cell types/states. D. H&E of a MERFISH adjacent coronal section of CS22 (GW10) E. MERFISH region annotations. MZ - Thalamic Mantle Zone, GZ - Thalamic Germinal Zone, GE - Ganglionic Eminences. F. Spatially projected cell type annotations. G. Relative proportion of cell types within regions from E. H. Spatially projected expression of select marker genes, including telencephalic (FOXG1) and diencephalic (TCF7L2, LHX9) identity, dividing cells (MKI67), radial glia and IPCs (OLIG3), and rostral thalamus-derived GABAergic neurons (SOX14).
Radial glia are neural stem cells in the developing brain. In the embryonic diencephalon, radial glia line the third ventricle and are partitioned into three zones: prosomere 1, which gives rise to the pretectum, prosomere 2, which gives rise to the thalamus and epithalamus, and prosomere 3, which gives rise to the prethalamus (14). They express PAX6 but lack the expression of IPC markers NEUROD1, NEUROG1, NEUROG2, and ASCL1, or neuronal markers such as SLC17A6 and GAD1 (15–17). We identified two clusters of radial glia (RG1-2) (Fig. 1B). RG1 are prosomere 2-derived thalamic radial glia based on their expression of OLIG3, a marker for radial glia and IPCs of thalamic glutamatergic lineage (Fig. 1C) (18). RG1 was enriched for genes associated with the Wnt pathway, including the Wnt ligands WNT2B and WNT3, the Wnt co-receptor LRP6, and a negative feedback regulator ZNRF3, consistent with the major role of Wnt signaling in thalamus development (Fig. 1C and fig. S1D) (19, 20). Other enriched genes included GLI2, a mediator of sonic hedgehog signaling, and the Rho GTPase CPNE8 (fig. S1D). Despite the lack of canonical IPC marker expression, these cells also express LHX9, a marker associated with thalamic IPCs and glutamatergic neurons in mouse and zebrafish (Fig. 1C) (17, 21). RG2 are likely radial glia of GABAergic lineage with representation from the prosomere 3-derived prethalamus based on the expression of FOXD1 and LHX5 (22), and prosomere 2-derived rostral thalamus based on expression of NKX2.2 and OLIG2 (Fig. 1C; fig. S1, C and D) (23–28). Other distinguishing marker genes include ID3, SFRP2, GPC3, and the antisense RNA gene LHX5-AS1 (Fig. 1C). ID3 was previously described as a pan-radial glial marker in mouse diencephalon including the thalamus (29), but ID3 appears to be specific to RG2 and MES in humans (Fig. 1C). A sparse population of PAX3/LMO1-expressing prosomere 1/pretectal progenitors (5, 24, 30) and SHH/SIM2-expressing zona limitans intrathalamica (ZLI) progenitors that reside between the thalamus and prethalamus (5, 31, 32) also cluster with RG2, suggesting that there was not enough representation from these populations to cluster separately (fig. S1, C and D). We also identified two IPC populations. IPC1 represents thalamic glutamatergic IPCs that express canonical markers that include OLIG3, NEUROD1, NEUROG1, NEUROG2, and NEUROD4 (Fig. 1C and fig. S1C) (17, 33). We found additional markers for IPC1, including KIF18B, C21ORF58, APOLD1, EYA2, and ARHGAP11B, a human-specific gene that has been implicated in the evolutionary expansion of the human neocortex (Fig. 1C) (34–38). Preferential expression of ARHGAP11B in thalamic IPCs suggests that it may be involved in the elaboration of the human thalamus, similar to its known role in the cortex. IPCs of GABAergic lineage (IPC2) from prosomeres 2 and 3 express the GABAergic proneural transcription factor ASCL1 (Fig. 1C) (17). This cluster was additionally enriched for the Wnt ligand WNT7B, the NMDA receptor subunit GRIN2A, the transcription factor GLIS3, the EGFR regulator LRIG1, and the tumor suppressor gene FAM107A (Fig. 1C).
Glutamatergic and GABAergic neurons were identified based on the expression of SLC17A6 and GAD1, respectively. We identified two glutamatergic neuron populations in the scRNAseq dataset, EN and nEN, which pertain to thalamic neurons based on their expression of canonical markers LHX9, LHX2, and GBX2 (Fig. 1C and fig. S1D) (39, 40). EN appear to be mature neurons based on their enrichment for GAP43 (fig. S1D). These cells are additionally enriched for markers including SGCZ, PLPPR1, and SNCA (Fig. 1C). nEN are newborn neurons based on co-expression of GBX2, a marker expressed in post-mitotic glutamatergic neurons in the thalamus (39–41) and IPC markers NEUROD1 and NEUROG2 (Fig. 1C and fig. S1D). These cells are enriched for CACNA2D1, DPYD, KALRN, and EYA2 (Fig. 1C and fig. S1D). Three clusters represented GABAergic inhibitory neurons (IN1-3). IN1 and IN2 represent prosomere 3-derived prethalamic neurons, given their expression of canonical markers PAX6, DLX5, SIX3, and ARX (16, 39, 42–44) and absence of FOXG1 expression, which is a pan-telencephalic marker (45) (Fig. 1C and fig. S1D). IN1 is additionally enriched for ZNF804B, KCNMB2, ISL1, and SST, a gene encoding for the neurotransmitter somatostatin (Fig. 1C and fig. S1D). IN2 is a LHX1-positive cluster enriched for the antisense RNA gene DLX6-AS1 and RELN (Fig. 1C). IN3 are likely prosomere 2/rostral thalamus (rTh)-derived GABAergic neurons, based on the expression of LHX1, NXK2.2, OTX2, SOX14, and genes encoding for the neurotransmitters neuropeptide Y (NPY) and enkephalin (PENK) (fig. S1D) (18, 46–49). Whereas OTX2 and SOX14 are also expressed in GABAergic neurons derived from midbrain and from prosomere 1-derived pretectum, NKX2.2 expression is absent in pretectal and midbrain-derived GABAergic neurons (18, 32, 50). Furthermore, these cells lack expression of LMO1 and IRX3, markers of caudal diencephalon also expressed in midbrain-derived GABAergic neurons, suggesting that the migratory stream of interneurons from the midbrain has not emerged during the first trimester (fig. S1C and fig. S3D) (5, 51, 52).
We performed MERFISH spatial transcriptomics on a coronal section from the GW10 rostral human forebrain to spatially identify cells from the glutamatergic thalamic lineage identified from scRNA-seq (Fig. 1D). We identified three distinct regions based on an H&E stain of an adjacent slice: the medial and lateral ganglionic eminences (GE), the thalamic germinal zone (GZ), and the thalamic mantle zone (MZ) (Fig. 1D). Our MERFISH analysis confirmed regional identity based on mutually exclusive expression of TCF7L2 and FOXG1 in the thalamus and GE, respectively (Fig. 1H). The GZ is enriched for OLIG3, and the MZ is enriched for SLC17A6 (Fig. 1H and fig. S1F). MKI67 expression segregated the ventricular zone (VZ) from the subventricular zone (SVZ) (Fig. 1H). We performed clustering analysis of our MERFISH dataset in order to relate the scRNA-seq populations to the regional annotations (Fig. 1, E and F). We identified four clusters that corresponded to IPC1, EN1, EN2, and IN3 from our single cell sequencing dataset (Fig. 1F). IPC1, which co-express OLIG3 and EYA2, reside in the thalamic GZ (Fig. 1, G and H; fig. S1F). Newborn neurons (EN2) expressing GBX2 and EYA2 are localized to the GZ and MZ, with bias for the GZ (Fig. 1F–H; fig. S1F). Mature LHX9/SLC17A6-expressing glutamatergic neurons (EN1) are predominantly found in the MZ (Fig. 1F–H; fig. S1F). We also observed the presence of SOX14/NKX2.2-expressing GABAergic neurons (IN3) associated with rostral thalamus origin in the MZ (Fig. 1F–H; fig. S1F). PAX6/OLIG3-expressing radial glia are observed at the VZ (Fig. 1H and fig. S1F). However, due to damage at the VZ, there were likely too few of these cells in our MERFISH data to form a discrete cluster.
Collectively, our analysis uncovers the cellular diversity of the human thalamus and their spatial distribution during the first trimester. We identify OLIG3-positive thalamic radial glia and IPCs, and the glutamatergic neurons that they likely give rise to that classify broadly into two molecular subtypes. In addition, we define ID3/ASCL1-expressing neural progenitors, and their presumed GABAergic neurons progeny, which can be classified into three subtypes of diencephalon-derived GABAergic neurons. These cell types express similar patterns of marker genes compared to what has been previously observed in other species, in addition to genes not previously associated with thalamic cell type-enriched expression. These findings suggest that the overall molecular blueprint of first trimester diencephalic cell types is conserved in humans. The majority of GABAergic neurons in the thalamus has been reported to originate from extra-thalamic regions, including the midbrain in mice (50, 53) and possibly the ganglionic eminences in humans (54, 55). However, we do not observe evidence for their presence in the first trimester, suggesting that these migratory streams develop later. Spatially, the thalamus has not yet partitioned into nuclei, based on cytoarchitecture revealed by Nissl staining or by molecular profiles revealed by MERFISH (Fig. 1D–F) (56). As expected for this stage of development, thalamic cells segregate into layers that relate to the germinal zones containing radial glia and intermediate progenitor cells, and the mantle zone containing migrating and maturing neurons.
Differentiation of thalamic neurons in the second trimester
Next, to gain insight into the molecular mechanisms and cellular composition of early thalamic nuclei, we analyzed scRNA-seq and snRNA-seq datasets from the second trimester of development, which were derived from 10 specimens (4 female and 6 male) between GW16 and GW25. This analysis retained 137,761 high quality cells or nuclei. For a subset of the samples, metadata information for thalamic subdivisions had been retained from dissections (6) based on microdissections along the dorsal-ventral and rostral-caudal axes, and included the pulvinar nucleus (fig. S4A). We identified 23 cell clusters, including all major cell classes: glutamatergic neurons (EN1-2), GABAergic neurons (IN1-6), intermediate progenitor cells (IPC), astrocytes (AC1-2), glial progenitor cells (GL), dividing cells (DIV), microglia (MG), ependymal cells (EP), oligodendrocyte progenitor cells (OPC), and oligodendrocytes (OL) (Fig. 2A). We also identified several vascular cell classes including endothelial cells (EC), pericytes (PC), and fibroblasts (FB) that were annotated based on our recent transcriptomic atlas of the human cerebrovasculature (Fig. 2A) (23).
Fig. 2. Cellular diversity in the second trimester thalamus.
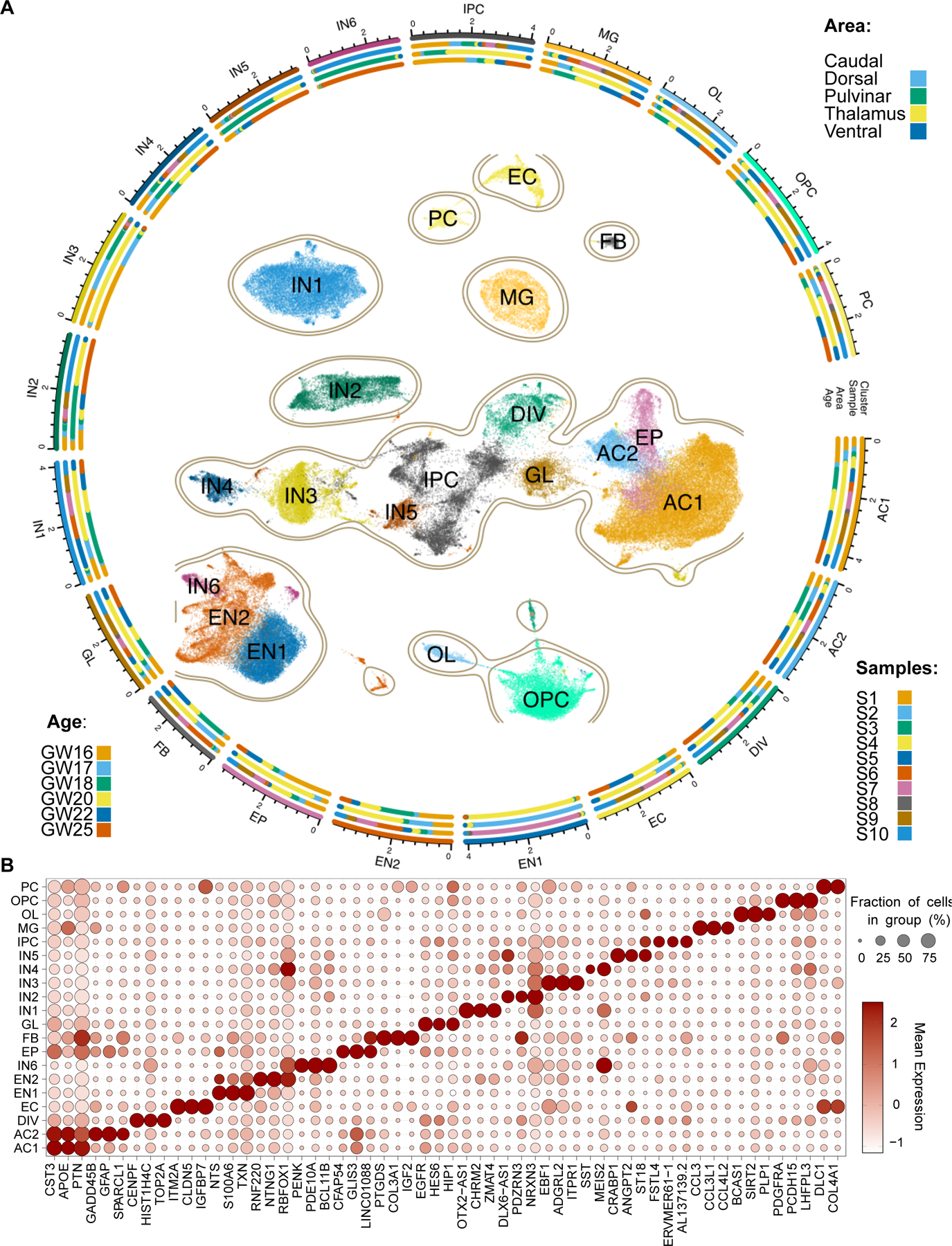
A. Constellation plot representing cellular diversity in second trimester human thalamus. PC - pericytes, OPC - oligodendrocyte precursor cells, OL - oligodendrocytes, MG - microglia, IPC - intermediate progenitor cells, IN1-6 - Inhibitory GABAergic neurons 1–6, GL - glial progenitor cells, FB - fibroblasts, EP - ependymal cells, EN1-2 - excitatory glutamatergic neurons 1–2, EC - endothelial cells, DIV - dividing cells, AC1-2 - astrocytes 1–2. B. Dot plot represents enrichment of select marker gene expression across cell types in the second trimester human thalamus.
Neural and glial progenitors were identified based on their expression of HES1 (fig. S3D). However, unlike in first trimester, we did not detect OLIG3+ clusters associated with thalamic progenitor cells (fig. S3D), in line with the early window of neurogenesis observed in mouse (57–59). However, we identified MKI67+ dividing cells (DIV) and NEUROD1/LHX1+ IPCs (IPC) that were enriched for MEIS1 and PAX3, markers of pretectal progenitors (Fig. 2B and fig. S3D) (5). This observation suggests ongoing neurogenesis in the pretectum, consistent with prior findings in the diencephalon (58). We identified two molecularly distinct astrocyte populations, AC1 and AC2, based on their expression of known astrocyte markers AQP4 and GJA1 (60–63) (fig. S3D). These populations are differentiated by LGR6 (AC1), a marker recently found to be enriched in thalamic astrocytes in the adult brain (64), and SPARCL1 (AC2) (Fig. 2B) (65). Ependymal cells (EP) exhibited similar transcriptomic profiles to astrocytes but were distinguished based on expression of FOXJ1 (fig. S3D) (66). We additionally observed a glial population (GL) that expresses EGFR, ASCL1, OLIG1, and OLIG2, but not PDGFRA, which is a signature of multipotent glial progenitor cells that may produce both astrocytes and oligodendrocytes (Fig. 2B) (67, 68). In contrast, OPCs and oligodendrocytes (OL) were distinguished from GL based on the expression of canonical markers PDGFRA and MBP, respectively (Fig. 2B) (69, 70). These results suggest that the end of neurogenesis and the onset of gliogenesis in the human thalamus occurs by GW16, which represents the earliest sample in our second trimester dataset.
Glutamatergic excitatory neuron clusters were defined based on shared expression of LHX9 and SLC17A6 (Fig. 2B and fig. S3D), and consisted of two clusters (EN1-2). EN1 was enriched for neurotensin (encoded by NTS), calcyclin (encoded by S100A6), and SOX2, whereas EN2 was enriched for RNF220, CRTAC1, and FOXP2 (Figs. 2B and 3B). EN1 and EN2 likely relate to the SOX2 and FOXP2-expressing subtypes recently observed in the embryonic mouse thalamus (5). Three classification schemes exist for thalamic glutamatergic neurons in the adult brain for which molecular markers have been identified. The core/matrix model classifies thalamic neurons based on whether they project to layer IV or layer I in the cortex, and in primates can be molecularly identified based on their expression of CALB1 or PVALB (71). The first order/higher order model relates to thalamic nuclei and their involvement in sensory relay pathways or transthalamic cortico-cortical communication (72), and their transcriptional signatures have been ascertained in mice (73). Finally, transcriptomic profiling of the different thalamocortical projection systems in mice revealed three major genetic profiles (primary, secondary, tertiary) found across thalamic pathways, and can be identified based on expression of TNNT1, NECAB1, or CALB2 (74). We sought to determine whether any of these classification schemes could be applied to the second trimester human brain based on gene expression. We first explored whether EN1 and EN2 could be associated with core or matrix identity based on expression of PVALB and CALB1, respectively. These genes were detected in only a sparse number of cells, suggesting that these genes turn on later in development (fig. S3D). We next sought to determine whether EN1 and EN2 were associated with transcriptional signatures of first order and higher order nuclei or with primary/secondary/tertiary profiles by performing gene signature scoring using the UCell package (75). We identified that EN1 was enriched for signatures of first order nuclei compared to EN2, but were not enriched for any of the primary/secondary/tertiary profiles (Fig. 3C). However, EN1 was enriched for CALB2, a marker for tertiary profile neurons that relate to matrix neurons in intralaminar nuclei (fig. S3D) (74). In contrast, EN2 was enriched for signatures of higher order nuclei and secondary profile neurons compared to EN1, including NECAB1 (Fig. 3C and fig. S3D). Secondary profile neurons relate to matrix neurons that are enriched in higher order nuclei (74). Collectively, our data suggests that the genetic programs underlying sensory and higher-order relays may begin to be established during prenatal development in humans, prior to the onset of sensory experience. EN1 may be enriched for neurons associated with first order nuclei, and with matrix neurons in intralaminar nuclei based on expression of CALB2. EN2 is associated with matrix neurons enriched in higher order nuclei.
Fig. 3. Spatially mapped thalamic glutamatergic neuron subtypes in human mid-gestation thalamus.
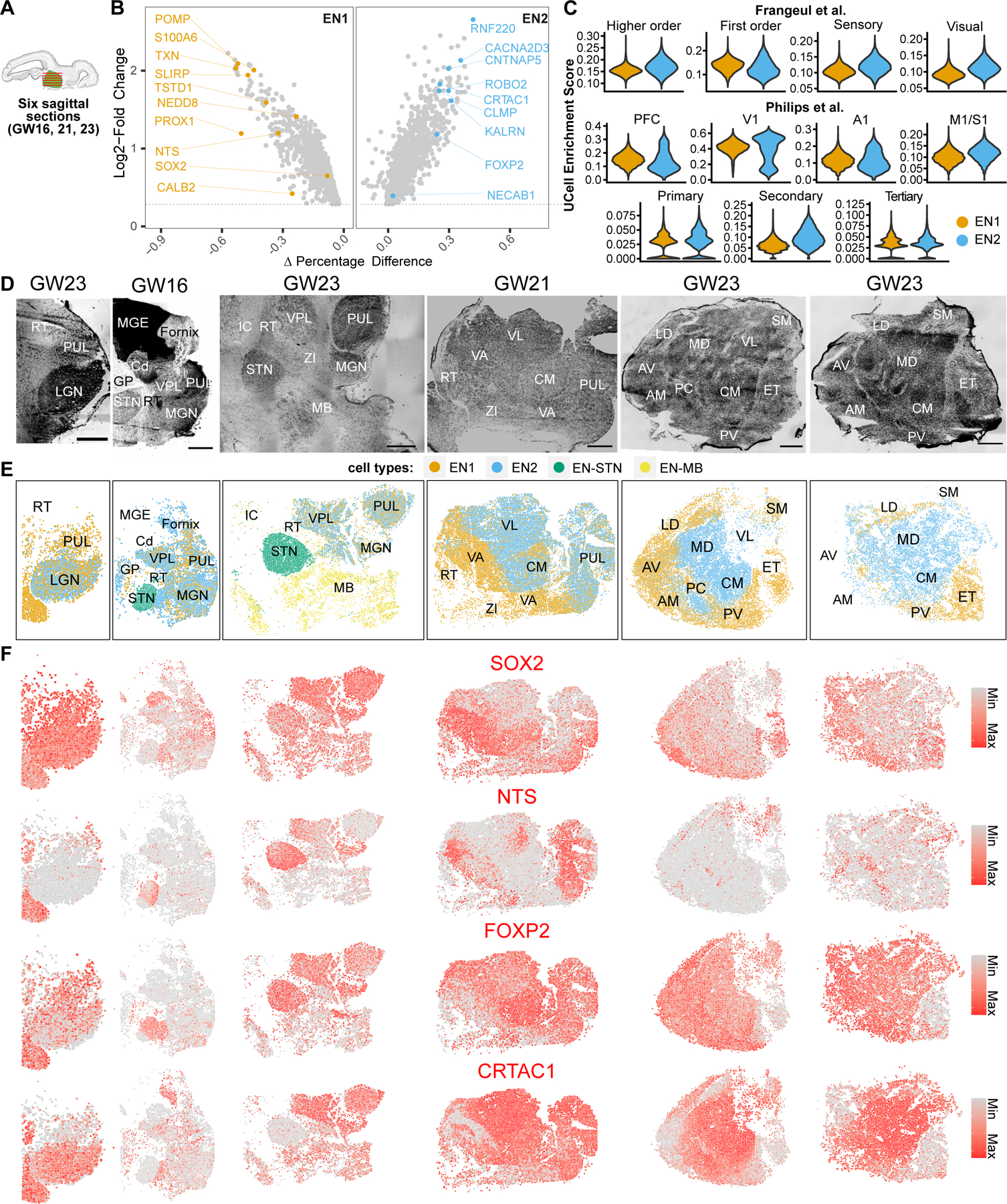
A. Eight sagittal sections total were collected from three biologically independent second trimester thalamus specimens (GW16, 21, 23). Six representative sections are shown here and the other two are shown in figs. S6 and S7. B. Marker genes enriched for glutamatergic subtypes EN1 and EN2. C. UCell enrichment scores for gene signatures relating to known classes of thalamus neurons from adult mouse studies calculated for glutamatergic neuron subtypes EN1 and EN2. PFC - prefrontal cortex, V1 - Visual cortex, A1 - Auditory cortex, M1/S1 Motor/Somatosensory cortex. D. Nissl stains of sagittal sections. Sections are in order from left (lateral- most) to right (medial-most). Scale bar: 2 mm. RT - Reticular Nucleus, PUL - Pulvinar, LGN - Lateral Geniculate Nucleus, MGE - Medial Ganglionic Eminence, Cd - Caudate, GP - Globus Pallidus, STN - Subthalamic Nucleus, VPL - Ventral Posterior Lateral Nucleus, MGN - Medial Geniculate Nucleus, MB - Midbrain, VA - Ventral Anterior Nucleus, VL - Ventral Lateral Nucleus, CM - Centromedian Nucleus, ZI - Zona Incerta, AV - Anteroventral Nucleus, AM - Anteromedial Nucleus, LD - Dorsolateral Nucleus, MD - Dorsomedial Nucleus, PC - Paracentral Nucleus, PV - Paraventricular Nucleus, SM - Stria Medullaris, ET - Epithalamus. E. Spatial clustering analysis of MERFISH transcriptomics data. EN1 and EN2 from spatial clustering analysis relate to EN1 and EN2 from scRNA-seq data based on gene expression profiles. EN-STN and EN-MB are distinct clusters not represented by our scRNA-seq, and relate to glutamatergic neurons in the subthalamic nucleus and midbrain, respectively. F. Spatial feature plots showing the expression of two example marker genes for EN1 (SOX2, NTS) and EN2 (FOXP2, CRTAC1) glutamatergic neuron subtypes in MERFISH datasets.
We also identified six clusters corresponding to GABAergic neurons based on the expression of GAD1 and SLC32A1 (fig. S3D). IN1 expressed OTX2 and SOX14 but were mostly negative for NKX2.2, a marker for prosomere 2-derived GABAergic neurons, suggesting that they represent midbrain-derived GABAergic neurons (fig. S3D) (53). These cells were additionally enriched for OTX2-AS1, CHRM2, ZMAT4, and LMO1 (Fig. 2B and fig. S3D). Because this GABAergic neuron subtype was not detected in our first trimester data, we suspect that this population likely represents a migratory stream that enters the thalamus after GW10. IN3 expressed EBF1, EBF3, ISLR2, and ESRRB, suggestive of pretectal identity (Fig. 2B and fig. S3D) (29, 76). IN4 expressed markers of thalamic reticular nucleus including SIX3, ISL1, and SST (Fig. 2B) (77). The remaining three populations (IN2, IN5, and IN6) expressed the pan-telencephalic marker FOXG1 (fig. S3D) (45). Previous anatomical studies have suggested that in humans, ganglionic eminence-derived GABAergic neurons migrate into thalamus (26, 27). FOXG1+ clusters could be distinguished based on differential enrichment for PDZRN3 in IN2, CRABP1, ANGPT2 and MAF in IN5, and PENK, RARB, and RXRG in IN6 (Fig. 2A and fig. S3D). IN5 is reminiscent of a recently described primate-specific CRABP1/MAF1 positive interneuron population derived from the medial ganglionic eminences (MGE) (28). In summary, we identified GABAergic populations of diencephalic, midbrain, and telencephalic origin, the latter two of which emerge during the second trimester of development in humans.
Spatial registration of thalamic neuron diversity
To investigate the spatial distribution of cell types in the developing human thalamus, we performed MERFISH on eight sagittal sections total collected from three biologically independent mid-gestation thalamus specimens using the Vizgen platform (Fig. 3A). These sections collectively captured the majority of thalamic nuclei as annotated by the Bayer and Altman atlas of the second trimester human brain (Fig. 3D) (78). Our gene panel was designed using cluster markers from second trimester sc/snRNA-seq data, which allowed us to visualize glutamatergic and GABAergic neurons, astrocytes, oligodendrocytes, microglia, endothelial cells, pericytes, and fibroblasts (Table S3, Figs. 3, 4, and 5).
Fig. 4. Spatially mapped GABAergic neuron subtypes across the human mid-gestation thalamus.
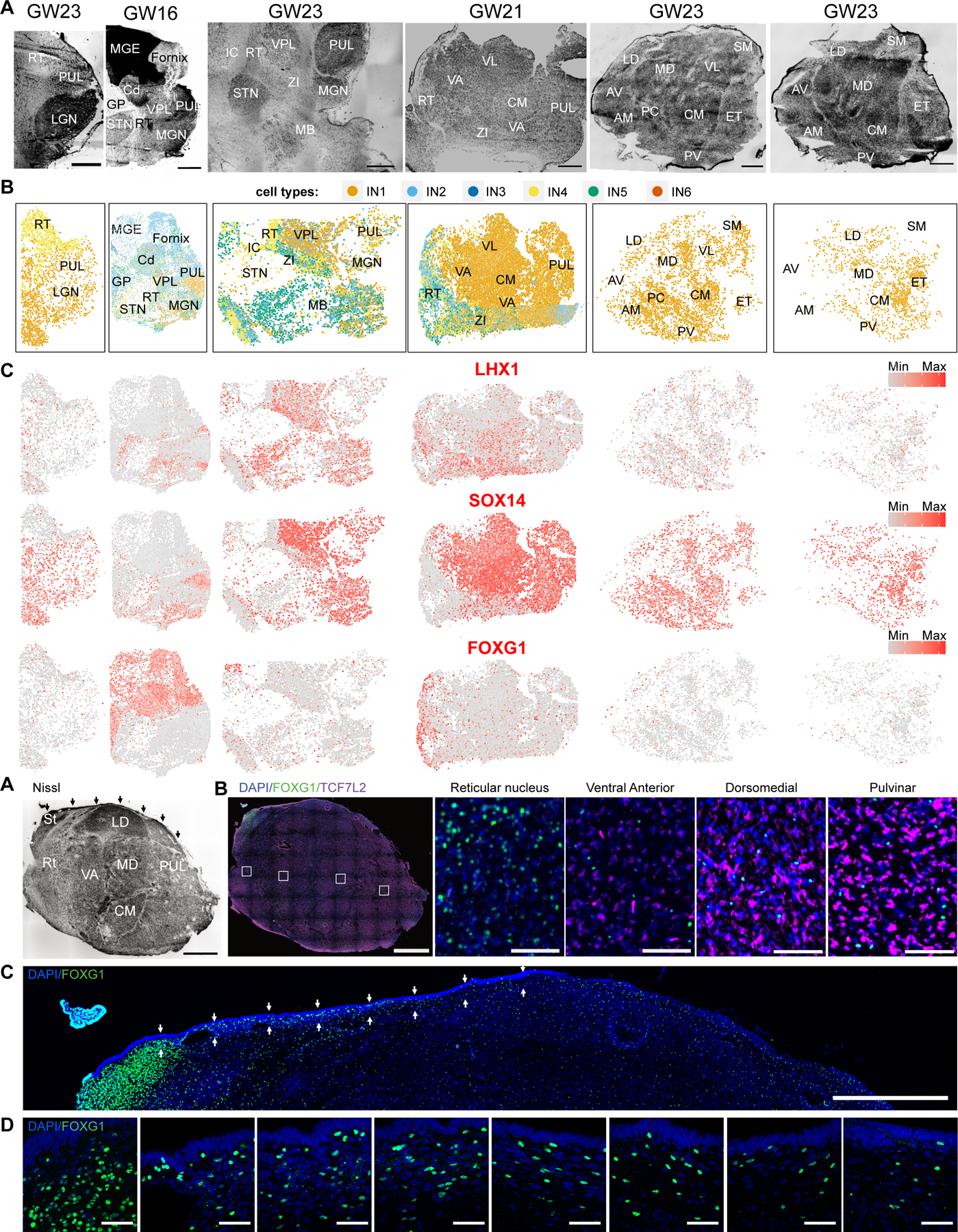
A. Nissl stains of sagittal sections. Sections are in order from left (lateral- most) to right (medial-most). Scale bar: 2 mm. RT - Reticular Nucleus, PUL - Pulvinar, LGN - Lateral Geniculate Nucleus, MGE - Medial Ganglionic Eminence, Cd - Caudate, GP - Globus Pallidus, STN - Subthalamic Nucleus, VPL - Ventral Posterior Lateral Nucleus, MGN - Medial Geniculate Nucleus, MB - Midbrain, VA - Ventral Anterior Nucleus, VL - Ventral Lateral Nucleus, CM - Centromedian Nucleus, ZI - Zona Incerta, AV - Anteroventral Nucleus, AM - Anteromedial Nucleus, LD - Dorsolateral Nucleus, MD - Dorsomedial Nucleus, PC - Paracentral Nucleus, PV - Paraventricular Nucleus, SM - Stria Medullaris, ET - Epithalamus. B. Spatial clustering analysis of MERFISH transcriptomics data depicting GABAergic neuron subtypes. C. Expression of GABAergic neuron subtype markers across the sections. D-E. Nissl (D) and Immunostaining (E) of FOXG1 and TCF7L2 on a lateral section in a sagittal section from a biologically independent GW19 thalamus. Arrows in Nissl stain highlight the corpus gangliothalamicus, which is a thin migratory stream from MGE to thalamus. Small dotted boxes are zoom-in images depicted on the right. Large dotted box is a zoom-in image depicted in Fig. 4F. Scale bar for Nissl and low magnification images: 2 mm. Scale bar for high magnification images: 50 µm. St: Stria terminalis. F. Zoom in of the dorsal region of the thalamus section depicted in (E) to highlight FOXG1 staining within and below the corpus gangliothalamicus (highlighted by the arrows). Scale bar: 2 mm. G. High magnification (63X) images of regions highlighted by arrows in Fig. 4F highlights FOXG1 staining within the corpus gangliothalamicus, in order from left to right. Scale bar: 50 µm.
Fig 5. Spatially mapped non-neural cell subtypes across the human mid-gestation thalamus.
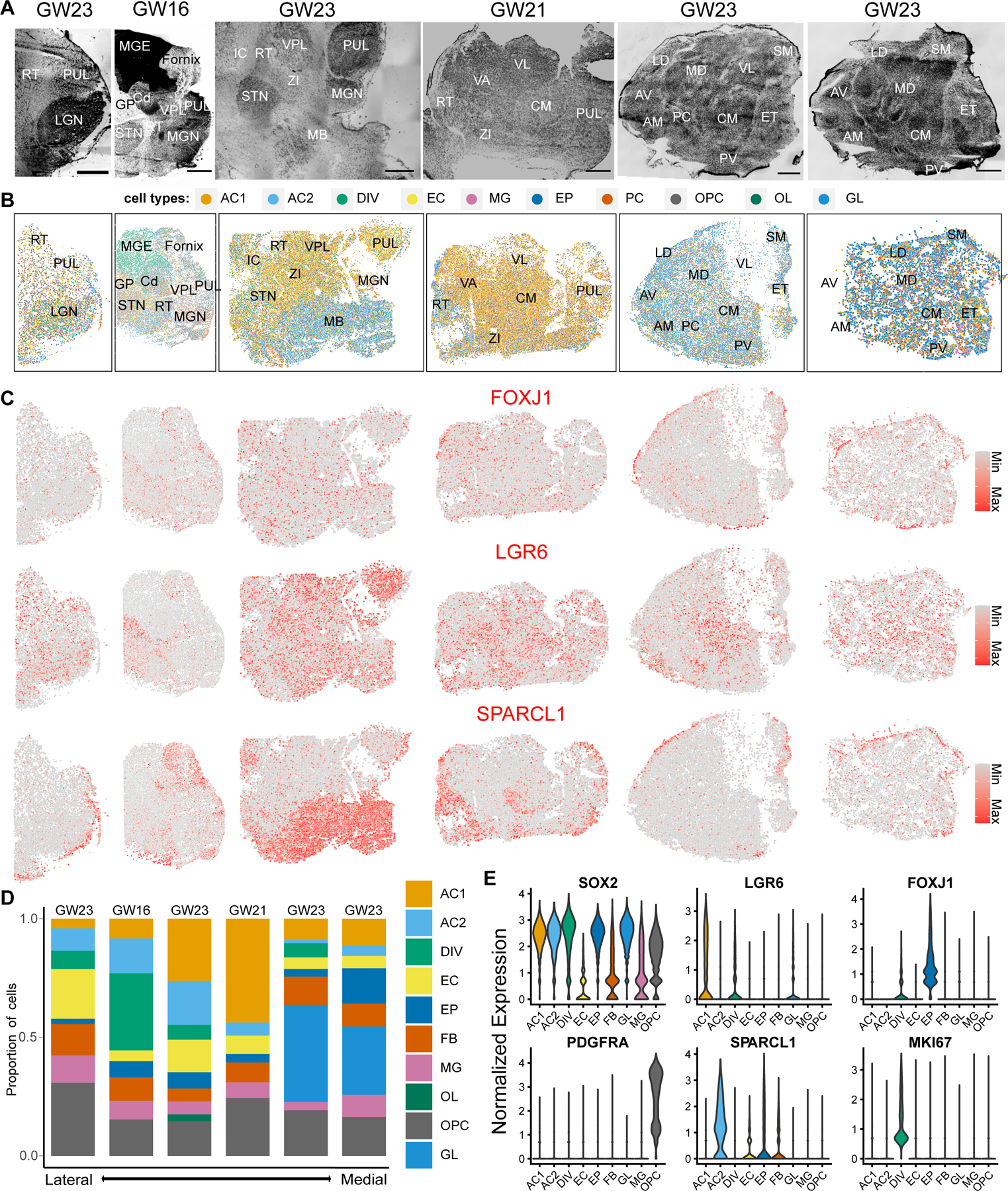
A. Nissl stains of sagittal sections. Sections are in order from left (lateral-most) to right (medial-most). Scale bar: 2 mm. RT - Reticular Nucleus, PUL - Pulvinar, LGN - Lateral Geniculate Nucleus, MGE - Medial Ganglionic Eminence, Cd - Caudate, GP - Globus Pallidus, STN - Subthalamic Nucleus, VPL - Ventral Posterior Lateral Nucleus, MGN - Medial Geniculate Nucleus, MB - Midbrain, VA - Ventral Anterior Nucleus, VL - Ventral Lateral Nucleus, CM - Centromedian Nucleus, ZI - Zona Incerta, AV - Anteroventral Nucleus, AM - Anteromedial Nucleus, LD - Dorsolateral Nucleus, MD - Dorsomedial Nucleus, PC - Paracentral Nucleus, PV - Paraventricular Nucleus, SM - Stria Medullaris, ET - Epithalamus. B. Spatial clustering analysis of MERFISH transcriptomics data depicting non-neuronal subtypes. AC1 and AC2 - astrocyte subtypes, DIV - dividing cells, EC - endothelial cells, EP - ependymal cells, FB - fibroblasts, MG - microglia, OL - oligodendrocytes, OPC - oligodendrocyte progenitor cells, GL - bipotent glial progenitor cells. C. Spatial feature plots depicting normalized expression of EP (FOXJ1), AC1 (LGR6), and AC2 (SPARCL1) markers. D. Relative proportion of non-neural cell types across lateral-to-medial sections. E. Violin plots depicting normalized expression of glial, astrocytes, ependymal, and oligodendrocyte precursor marker genes across the 2 lateral-most GW23 sections.
We first analyzed the distribution of glutamatergic clusters EN1 and EN2 (Fig. 3E; fig. S5B; fig. S7B; fig. S8; fig. S9). We observed an enrichment for EN1 neurons in rostral regions of the thalamus, with a greater abundance of these cells observed in AV/AM, LD, VA, PC, and PV (Fig. 3E). In contrast, EN2 exhibited a more caudal bias, with an enriched distribution in the MD, VL, and CM (Fig. 3E). Sensory nuclei such as LGN, MGN, and VPL contained similar abundance of both EN1 and EN2 populations (Fig. 3E and fig. S7B). Pulvinar, which represents an evolutionarily expanded thalamic nucleus (79), showed an overall enrichment for EN1 in the lateral-most section and an enrichment for EN2 in more medial sections (Fig. 3E). This difference within the pulvinar may relate to its functional organization, whereby lateral pulvinar is involved with the visual pathway and medial pulvinar is involved with multi-sensory, associative, and higher order cognitive function (3, 80). The overall biased distribution of EN1 and EN2 neurons is consistent with the embryonic mouse thalamus (5). Epithalamus, which is a neighboring diencephalic region derived from prosomere 2 like the thalamus, contained glutamatergic neurons associated with EN1 signatures (Fig. 3E). This is in line with previously reported similarities in gene expression profiles between epithalamus and thalamus (29, 81). However, other neighboring regions, including the subthalamic nucleus and midbrain, contained glutamatergic neurons that clustered separately from EN1 and EN2 (EN-STN, EN-MB) (Fig. 3E).
We next analyzed the distribution of GABAergic neuron subtypes revealed by our MERFISH data (Fig. 4A; figs. S6A and S7C). Midbrain-derived OTX2/SOX14-positive, NKX2.2-negative cells (IN1) represented the most abundant population of GABAergic neurons in the thalamus, in line with recent descriptions in the mouse and marmoset (53) (Fig. 4, B and D; figs. S6B and S7C). In humans, these cells were distributed throughout the thalamus (Fig. 4, B and D; figs. S6B and S7C), unlike in mice where they are enriched in the LGN (53). GABAergic neurons derived from the prethalamus (IN4) and telencephalon-derived neurons (IN2, IN5) were enriched in the reticular nucleus and zona incerta, but also found sparsely distributed across the thalamic nuclei (Fig. 4, B and D; fig. S7C). Abundant IN2 population was also present in the MGE and basal ganglia structures that were visible in our sections such as caudate and globus pallidus, consistent with their telencephalic identity (Fig. 4B). IN5 was enriched in epithalamus (fig. S7). Finally, FOXG1-positive IN6 cluster was observed in the caudate but not the thalamus, suggesting that this sparse population may have been accidentally included in the sc/snRNASeq data generation due to inaccurate dissection (Fig. 4B). We also could not detect IN3, which we hypothesized were pretectal GABAergic neurons, likely due to lack of representation of pretectum in our spatial transcriptomics datasets.
We sought to validate the existence of telencephalon-derived cells in the thalamus by performing immunostaining against FOXG1 in a biologically-independent GW19 sample. Nissl staining revealed the proposed migratory stream between the MGE and the thalamus previously observed in humans, the corpus gangliothalamicus, which is defined as a thin stream of cells starting from the stria terminalis and lining the dorsal edge of the thalamus (Fig. 4D). We observed that cells within this migratory stream stained positive for FOXG1, confirming telencephalic identity (Fig. 4, F and G). Whereas previous reports suggest that these cells migrate predominantly to mediodorsal and pulvinar nuclei, we observe FOXG1+ cells sparsely throughout the thalamus, with enrichment in the reticular nucleus (Fig. 4, F and G; fig. S11). We additionally performed in situ hybridization against FOXG1 and CRABP1 across seven sections total collected from three biologically independent thalamus samples. We further confirmed the presence of FOXG1/CRABP1-expressing cells within the thalamus and epithalamus, and enrichment in the reticular nucleus, confirming our MERSCOPE results (fig. S11).
Spatial registration of non-neuronal diversity in the thalamus
We finally assessed the distribution of non-neuronal cell types (Fig. 5B and fig. S7D). LGR6+ astrocytes (AC1) appeared to be the most abundant glial population in lateral sections whereas the bipotent glial progenitors (GL) were the most abundant in medial sections (Fig. 5, B–D), consistent with the lateral-to-medial maturation gradient that was previously observed in the context of neurogenesis in the rodent thalamus (58, 82). SPARCL1+ AC2 astrocytes were enriched in extrathalamic regions such as the midbrain and reticular nucleus, but were also present to a lesser degree in the thalamus (Fig. 5, B and C; fig. S7D). FOXJ1+ ependymal cells were enriched along the periphery of the thalamus and within the centromedian nucleus (Fig. 5, B and C; fig. S7D). Other cell types, including dividing cells, OPCs, oligodendrocytes, microglia, pericytes, fibroblasts were found distributed broadly throughout the thalamus (Fig. 5, B–D).
Thalamic nuclei exhibit differences in gene expression during the second trimester
We identified several genes enriched in anatomically-defined thalamic nuclei. For example, INHBA, TLL1, ITGA8, and KCTD8 were enriched in centromedian nucleus (fig. S14). Ventral anterior nucleus was highly enriched for PENK, GABRG1, TSHZ2, and CDH9 (fig. S14). Reticular nucleus showed enriched expression of CNTNAP4, EPHA5, PDE1C, and PVALB (fig. S14). Zona incerta was highly enriched for LHX5, PAX6, ISL1, and DLX2 (fig. S14). Given the spatial bias we observed for many neuron-enriched genes, we sought to determine whether thalamic nuclei can be molecularly distinguished in mid-gestation in the human brain. We designed a gene panel containing 140 genes based on top markers for neuronal cell types, and performed MERSCOPE on a sagittal section from GW21 human thalamus. Clustering analysis revealed segmentation of the thalamus into regions reminiscent of thalamic nuclei independently identified by Nissl staining (fig. S14). These results confirm that spatial variation of neuronal gene expression parcellates the thalamus into molecularly-defined nuclei as early as the second trimester.
Discussion
Together, our study identifies the molecular trajectories of neuronal differentiation in the developing human diencephalon and suggests three principles of neuronal differentiation and spatial organization. 1), human thalamic neuronal subtypes begin to emerge during the first trimester of development and start to organize into spatially distinct nuclei during the second trimester. 2) molecularly defined subtypes of glutamatergic neurons are shared across thalamic nuclei, and their composition can be used to distinguish first-order and higher-order thalamic nuclei. 3) we detect six molecular subtypes of GABAergic neurons that contribute to the developing diencephalon in humans. These include prethalamus-derived, telencephalon-derived, and midbrain-derived GABAergic neurons that together contribute substantially to the cellular composition of diencephalic subregions in humans. In particular, we find that midbrain- and telencephalon-derived GABAergic neurons appear in the developing thalamus as early as the second trimester of development.
Our study confirms previously hypothesized migration of telencephalon-derived GABAergic neurons into the thalamus, which may contribute to the expansion of this structure in humans (54, 55). These cells maintain expression of FOXG1 during the second trimester, and form a thin stream along the dorsal edge of the thalamus that coincides with the corpus gangliothalamicus, a migratory pathway between the MGE and the thalamus that was previously identified using Golgi and Nissl stains (54, 55, 83). In addition to the previously identified localization of these cells in mediodorsal and pulvinar nuclei, our results indicate that FOXG1-positive cells appear to migrate throughout the thalamus, with particular enrichment in the reticular nucleus. We also observe their presence in the epithalamus, suggesting a greater caudal dispersion of these cells across prosomere 2 of the diencephalon than previously appreciated. Finally, a subset of FOXG1-expressing cells expresses CRABP1, a gene involved in the retinoic acid signaling pathway. CRABP1-positive GABAergic neurons have been recently identified in studies of primate forebrain interneuron diversity (28, 32). Consistently, CRABP1-positive GABAergic neurons expressed LHX6 and MAF in our single cell sequencing data, suggesting medial ganglionic eminence origin, but did not express TAC3.
Together, our findings underscore the complexity of neurodevelopmental cell lineage relationships in primates and provide a cellular substrate that could contribute to the evolution of the human thalamus. Additional work is needed to further characterize the molecular diversity of neural stem and progenitor cells in the first trimester human diencephalon and molecular mechanisms underlying their fate specification. These include cells that give rise to the epithalamus and pretectum, which was not a focus of the current work. Furthermore, the ontogeny of cell types in the thalamus will need to be confirmed using lineage tracing or fate mapping tools in human and non-human primate model systems, our understanding of which largely comes from rodent studies. The developmental origins of GABAergic subtypes are of particular interest. The majority of GABAergic neurons in mouse thalamus are midbrain-derived and reside in the dorsal lateral geniculate nucleus, but the number of GABAergic neurons appears to be significantly increased in primates and are distributed throughout the thalamus. It would be interesting to confirm whether the most abundant GABAergic subtype, SOX14+ IN1, are truly midbrain-derived, and whether FOXG1+ telencephalon-derived subtypes originate primarily from MGE or also from the other ganglionic eminences. Furthermore, comparative studies are required to determine whether telencephalon-derived GABAergic neurons are specific to humans, as has been previously hypothesized (54, 55). Finally, future studies are required to determine whether GABAergic neuron subtypes in human thalamus exhibit enrichment in first or higher-order nuclei, and whether they exhibit differences in their physiological properties or patterns of connectivity, during later time points after which their migration has concluded. The thalamus has been increasingly implicated in neuropsychiatric disorders, including autism spectrum disorder, schizophrenia, bipolar disorder, and attention deficit/hyperactivity disorder (84–89). Studies of evolutionarily divergent cell types may provide inroads into better understanding human disease.
Supplementary Material
Acknowledgements:
We thank Matthew Schmitz and members of the Nowakowski lab for providing helpful feedback during manuscript preparation. Widefield imaging data for this study were acquired at the Center for Advanced Light Microscopy - Weill Innovation Center Microscopes. This publication was supported by and coordinated through BICCN. This publication is part of the Human Cell Atlas-www.humancellatlas.org/publications/
Funding:
This study was supported by NIH awards: Brain Initiative award UM1MH130991 (to TJN), NIMH award R01MH128364 (to TJN), NINDS award R01NS123263 (to TJN), R01NS123912 (to TJN), a Simons Foundation grant SF810018 (to TJN), a grant from the Esther A & Joseph Klingenstein Fund (to TJN), Shurl and Kay Curci Foundation (to TJN) and the Sontag Foundation (to TJN), and gifts from Schmidt Futures and the William K. Bowes Jr Foundation (to TJN). T.J.N. is a New York Stem Cell Foundation Robertson Neuroscience Investigator.
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability: Processed scRNA-seq are available on UCSC cell browser at http://cells.ucsc.edu/?ds=dev-thal. Processed MERFISH data are available on Dryad: https://doi.org/10.5061/dryad.98sf7m0q1
References and Notes
- 1.Guillery RW, Anatomical evidence concerning the role of the thalamus in corticocortical communication: a brief review. J Anat 187 ( Pt 3), 583–592 (1995). [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin MKL, Balaram P, Kaas JH, The evolution and functions of nuclei of the visual pulvinar in primates. J Comp Neurol 525, 3207–3226 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Homman-Ludiye J, Bourne JA, The medial pulvinar: function, origin and association with neurodevelopmental disorders. J Anat 235, 507–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molnar Z, Development and evolution of thalamocortical interactions. Eur J Morphol 38, 313–320 (2000). [PubMed] [Google Scholar]
- 5.Govek KW et al. Developmental trajectories of thalamic progenitors revealed by single-cell transcriptome profiling and Shh perturbation. Cell Rep 41, 111768 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaduri A et al. An atlas of cortical arealization identifies dynamic molecular signatures. Nature 598, 200–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron D et al. Single-Nuclei RNA Sequencing of 5 Regions of the Human Prenatal Brain Implicates Developing Neuron Populations in Genetic Risk for Schizophrenia. Biol Psychiatry 93, 157–166 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Berg M et al. FastCAR: Fast Correction for Ambient RNA to facilitate differential gene expression analysis in single-cell RNA-sequencing datasets. bioRxiv, 2022.2007.2019.500594 (2022). [DOI] [PMC free article] [PubMed]
- 9.Bais AS, Kostka D, scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 36, 1150–1158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muskovic W, Powell JE, DropletQC: improved identification of empty droplets and damaged cells in single-cell RNA-seq data. Genome Biol 22, 329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korsunsky I et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eze UC, Bhaduri A, Haeussler M, Nowakowski TJ, Kriegstein AR, Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat Neurosci 24, 584–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puelles L, Rubenstein JL, Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci 16, 472–479 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Walther C, Gruss P, Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113, 1435–1449 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Mastick GS, Andrews GL, Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci 17, 190–207 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Bluske KK, Dickel LK, Nakagawa Y, Basal progenitor cells in the embryonic mouse thalamus - their molecular characterization and the role of neurogenins and Pax6. Neural Dev 6, 35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vue TY et al. Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol 505, 73–91 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Zhou CJ, Pinson KI, Pleasure SJ, Severe defects in dorsal thalamic development in low-density lipoprotein receptor-related protein-6 mutants. J Neurosci 24, 7632–7639 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Ferre A, Navarro-Garberi M, Bueno C, Martinez S, Wnt signal specifies the intrathalamic limit and its organizer properties by regulating Shh induction in the alar plate. J Neurosci 33, 3967–3980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peukert D, Weber S, Lumsden A, Scholpp S, Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol 9, e1001218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sellers K, Zyka V, Lumsden AG, Delogu A, Transcriptional control of GABAergic neuronal subtype identity in the thalamus. Neural Dev 9, 14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho EA, Dressler GR, TCF-4 binds beta-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech Dev 77, 9–18 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Ferran JL, Sanchez-Arrones L, Sandoval JE, Puelles L, A model of early molecular regionalization in the chicken embryonic pretectum. J Comp Neurol 505, 379–403 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Bluske KK, Kawakami Y, Koyano-Nakagawa N, Nakagawa Y, Differential activity of Wnt/beta-catenin signaling in the embryonic mouse thalamus. Dev Dyn 238, 3297–3309 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan R, Graf M, Mason I, Lumsden A, Kiecker C, Complex and dynamic patterns of Wnt pathway gene expression in the developing chick forebrain. Neural Dev 4, 35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merchan P, Bardet SM, Puelles L, Ferran JL, Comparison of Pretectal Genoarchitectonic Pattern between Quail and Chicken Embryos. Front Neuroanat 5, 23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morona R, Ferran JL, Puelles L, Gonzalez A, Embryonic genoarchitecture of the pretectum in Xenopus laevis: a conserved pattern in tetrapods. J Comp Neurol 519, 1024–1050 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Guo Q, Li JYH, Defining developmental diversification of diencephalon neurons through single cell gene expression profiling. Development 146, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferran JL et al. Early pretectal gene expression pattern shows a conserved anteroposterior tripartition in mouse and chicken. Brain Res Bull 75, 295–298 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Kiecker C, Lumsden A, Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci 7, 1242–1249 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Jeong Y et al. Spatial and temporal requirements for sonic hedgehog in the regulation of thalamic interneuron identity. Development 138, 531–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabo NE, Zhao T, Zhou X, Alvarez-Bolado G, The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J Neurosci 29, 2453–2466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudmant PH et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis MY et al. The evolution and population diversity of human-specific segmental duplications. Nat Ecol Evol 1, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Florio M et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Heide M et al. Human-specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science 369, 546–550 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Xing L et al. Expression of human-specific ARHGAP11B in mice leads to neocortex expansion and increased memory flexibility. EMBO J 40, e107093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa Y, OĽeary DD, Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci 21, 2711–2725 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulfone A et al. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci 13, 3155–3172 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Guo Q, Li JY, Transcription factor Gbx2 acts cell-nonautonomously to regulate the formation of lineage-restriction boundaries of the thalamus. Development 136, 1317–1326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagutin OV et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev 17, 368–379 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobos I et al. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci 8, 1059–1068 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Jones EG, Rubenstein JL, Expression of regulatory genes during differentiation of thalamic nuclei in mouse and monkey. J Comp Neurol 477, 55–80 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Tao W, Lai E, Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron 8, 957–966 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto-Torii K et al. Differential activities of Sonic hedgehog mediated by Gli transcription factors define distinct neuronal subtypes in the dorsal thalamus. Mech Dev 120, 1097–1111 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Delogu A et al. Subcortical visual shell nuclei targeted by ipRGCs develop from a Sox14+-GABAergic progenitor and require Sox14 to regulate daily activity rhythms. Neuron 75, 648–662 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Virolainen SM, Achim K, Peltopuro P, Salminen M, Partanen J, Transcriptional regulatory mechanisms underlying the GABAergic neuron fate in different diencephalic prosomeres. Development 139, 3795–3805 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Jones E, The Thalamus, 2nd Edn Cambridge University Press. Cambridge.[Google Scholar], (2007). [Google Scholar]
- 50.Jager P et al. Tectal-derived interneurons contribute to phasic and tonic inhibition in the visual thalamus. Nat Commun 7, 13579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun MM, Etheridge A, Bernard A, Robertson CP, Roelink H, Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development 130, 5579–5587 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Foroni L et al. The rhombotin gene family encode related LIM-domain proteins whose differing expression suggests multiple roles in mouse development. J Mol Biol 226, 747–761 (1992). [DOI] [PubMed] [Google Scholar]
- 53.Jager P et al. Dual midbrain and forebrain origins of thalamic inhibitory interneurons. Elife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letinic K, Rakic P, Telencephalic origin of human thalamic GABAergic neurons. Nat Neurosci 4, 931–936 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Rakic P, Sidman RL, Telencephalic origin of pulvinar neurons in the fetal human brain. Z Anat Entwicklungsgesch 129, 53–82 (1969). [DOI] [PubMed] [Google Scholar]
- 56.Bayer SA, Altman J, The human brain during the early first trimester (CRC Press, 2007). [Google Scholar]
- 57.Wong SZH et al. In vivo clonal analysis reveals spatiotemporal regulation of thalamic nucleogenesis. PLoS Biol 16, e2005211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Angevine JB Jr., Time of neuron origin in the diencephalon of the mouse. An autoradiographic study. J Comp Neurol 139, 129–187 (1970). [DOI] [PubMed] [Google Scholar]
- 59.Shi W et al. Ontogenetic establishment of order-specific nuclear organization in the mammalian thalamus. Nat Neurosci 20, 516–528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almad AA et al. Connexin 43 in astrocytes contributes to motor neuron toxicity in amyotrophic lateral sclerosis. Glia 64, 1154–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frigeri A et al. Localization of MIWC and GLIP water channel homologs in neuromuscular, epithelial and glandular tissues. J Cell Sci 108 ( Pt 9), 2993–3002 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Nielsen S et al. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J Neurosci 17, 171–180 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rochefort N, Quenech'du N, Ezan P, Giaume C, Milleret C, Postnatal development of GFAP, connexin43 and connexin30 in cat visual cortex. Brain Res Dev Brain Res 160, 252–264 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Bakken TE et al. Single-cell and single-nucleus RNA-seq uncovers shared and distinct axes of variation in dorsal LGN neurons in mice, non-human primates, and humans. Elife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kucukdereli H et al. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci U S A 108, E440–449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meletis K et al. Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol 6, e182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X et al. Decoding Cortical Glial Cell Development. Neurosci Bull 37, 440–460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang L, Li Z, Liu G, Li X, Yang Z, Developmental Origins of Human Cortical Oligodendrocytes and Astrocytes. Neurosci Bull 38, 47–68 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pringle NP, Mudhar HS, Collarini EJ, Richardson WD, PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development 115, 535–551 (1992). [DOI] [PubMed] [Google Scholar]
- 70.Zecevic N, Andjelkovic A, Matthieu J, Tosic M, Myelin basic protein immunoreactivity in the human embryonic CNS. Brain Res Dev Brain Res 105, 97–108 (1998). [PubMed] [Google Scholar]
- 71.Jones EG, The thalamic matrix and thalamocortical synchrony. Trends Neurosci 24, 595–601 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Sherman SM, Guillery RW, Distinct functions for direct and transthalamic corticocortical connections. J Neurophysiol 106, 1068–1077 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Frangeul L et al. A cross-modal genetic framework for the development and plasticity of sensory pathways. Nature 538, 96–98 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Phillips JW et al. A repeated molecular architecture across thalamic pathways. Nat Neurosci 22, 1925–1935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andreatta M, Carmona SJ, UCell: Robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J 19, 3796–3798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mallika C, Guo Q, Li JY, Gbx2 is essential for maintaining thalamic neuron identity and repressing habenular characters in the developing thalamus. Dev Biol 407, 26–39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y et al. Distinct subnetworks of the thalamic reticular nucleus. Nature 583, 819–824 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bayer SA, Altman J, The human brain during the second trimester (CRC Press, 2005). [Google Scholar]
- 79.Chalfin BP, Cheung DT, Muniz JA, de Lima Silveira LC, Finlay BL, Scaling of neuron number and volume of the pulvinar complex in New World primates: comparisons with humans, other primates, and mammals. J Comp Neurol 504, 265–274 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Kaas JH, Collins CE, The primate visual system (CRC Press, 2003). [Google Scholar]
- 81.Nagalski A et al. Molecular anatomy of the thalamic complex and the underlying transcription factors. Brain Struct Funct 221, 2493–2510 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Altman J, Bayer SA, Development of the rat thalamus: I. Mosaic organization of the thalamic neuroepithelium. J Comp Neurol 275, 346–377 (1988). [DOI] [PubMed] [Google Scholar]
- 83.Letinic K, Kostovic I, Transient fetal structure, the gangliothalamic body, connects telencephalic germinal zone with all thalamic regions in the developing human brain. J Comp Neurol 384, 373–395 (1997). [DOI] [PubMed] [Google Scholar]
- 84.Nair A et al. Impaired thalamocortical connectivity in autism spectrum disorder: a study of functional and anatomical connectivity. Brain 136, 1942–1955 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Norra C et al. High-frequency somatosensory thalamocortical oscillations and psychopathology in schizophrenia. Neuropsychobiology 49, 71–80 (2004). [DOI] [PubMed] [Google Scholar]
- 86.Roy DS et al. Anterior thalamic dysfunction underlies cognitive deficits in a subset of neuropsychiatric disease models. Neuron 109, 2590–2603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sukhodolsky DG, Leckman JF, Rothenberger A, Scahill L, The role of abnormal neural oscillations in the pathophysiology of co-occurring Tourette syndrome and attention-deficit/hyperactivity disorder. Eur Child Adolesc. Psychiatry 16, 51–59 (2007). [DOI] [PubMed] [Google Scholar]
- 88.Tomasi D, Volkow ND, Reduced local and increased long-range functional connectivity of the thalamus in autism spectrum disorder. Cereb. Cortex 29, 573–585 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moreau CA et al. Atlas of functional connectivity relationships across rare and common genetic variants, traits, and psychiatric conditions. medRxiv 2021.05.21.21257604 (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


