Abstract
Legionella pneumophila, the causative organism of Legionnaires’ pneumonia, is spread by aerosolization from man-made reservoirs, e.g., water cooling towers and air conditioning ducts, whose nutrient-poor conditions are conducive to entrance into stationary phase. Exposure to starvation conditions is known to induce several virulence traits in L. pneumophila. Since catalase-peroxidases have been extremely useful markers of the stationary-phase response in many bacterial species and may be an avenue for identifying virulence genes in L. pneumophila, an investigation of these enzymes was initiated. L. pneumophila was shown to contain two bifunctional catalase-peroxidases and to lack monofunctional catalase and peroxidase. The gene encoding the KatB catalase-peroxidase was cloned and sequenced, and lacZ fusion and null mutant strains were constructed. Null mutants in katB are delayed in the infection and lysis of cultured macrophage-like cell lines. KatB is similar to the KatG catalase-peroxidase of Escherichia coli in its 20-fold induction during exponential growth and in playing a role in resistance to hydrogen peroxide. Analysis of the changes in katB expression and in the total catalase and peroxidase activity during growth indicates that the 8- to 10-fold induction of peroxidase activity that occurs in stationary phase is attributable to KatA, the second L. pneumophila catalase-peroxidase.
Legionella pneumophila is an intracellular parasite and the causative organism of Legionnaires’ pneumonia, a disease spread by aerosolization of the pathogen from environmental reservoirs. The reservoirs from which L. pneumophila is most frequently aerosolized are nutrient sparse: showerheads, respirators, air conditioning ducts and water cooling towers (10, 29, 30). These epidemiological considerations indicate that L. pneumophila must survive starvation conditions between periods of replication in a suitable host. It has recently been shown that several L. pneumophila virulence traits, including cytotoxicity, infectivity, and sodium sensitivity, are absent from exponentially growing cultures but are expressed in response to starvation. These observations led to the model where the stationary phase is a necessary prerequisite to the acquisition of virulence by legionella and not merely a stress state to be endured between rounds of intracellular multiplication (5). Stationary-phase gene expression has been associated with acquisition of virulence traits in other bacterial pathogens, e.g., Salmonella species, Pseudomonas aeruginosa, and Yersinia enterocolitica (2, 8, 18, 23). Therefore, genes in stationary-phase pathways of L. pneumophila may be tools for identifying genes in pathways leading to virulence.
In many bacterial species, genes controlling the antioxidant response play important roles in the stationary phase. The Escherichia coli KatE catalase is a hallmark of the stationary phase and is part of the cross-resistance to stress that accompanies starvation. E. coli katG, encoding a catalase-peroxidase, and xth, encoding the DNA repair enzyme exonuclease III, are other antioxidant enzymes that are expressed in the stationary phase under the control of RpoS, the stationary-phase sigma factor (14, 19, 21). We showed that the stationary-phase viability of a katG null mutant in Caulobacter crescentus is reduced by 6 orders of magnitude compared to that of the wild type, which is indicative of the importance of that catalase-peroxidase in stationary-phase survival (32).
Little is known about the L. pneumophila stationary-phase response. We began an investigation of catalase-peroxidases in L. pneumophila because in other bacterial species these genes have been extremely useful tools for studying the stationary-phase response. We report here that L. pneumophila contains two bifunctional catalase-peroxidases and no monofunctional catalases or peroxidases. We cloned the katB gene encoding one of the catalase-peroxidases, constructed lacZ fusion and null strains, and identified a role for katB in H2O2 resistance of the free-living organism and in infection of human macrophage lines.
MATERIALS AND METHODS
Media and growth conditions.
E. coli DH5α was used for cloning. The liquid and solid media for culturing E. coli were Luria Bertani medium (31) with the following antibiotics at the indicated final concentrations: sodium ampicillin (100 μg/ml), kanamycin sulfate (50 μg/ml), gentamicin sulfate (5 μg/ml), and chloramphenicol (25 μg/ml). The culture media for L. pneumophila were AYE broth (17) and CYE plates (9) with the following antibiotic concentrations: kanamycin sulfate (25 μg/ml), gentamicin sulfate (5 or 10 μg/ml), and chloramphenicol (5 μg/ml). The temperature for culturing was 37°C. The parental L. pneumophila strain for genetic constructions was the wild-type strain JR32 (36) (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| L. pneumophila | ||
| JR32 | Homogeneous salt-sensitive isolate of AM511 (Philadelphia-1 Smr r−m+) | H. Shuman (36) |
| PB102 | JR32 katB+katB::lacZ (katB fusion strain) | This work |
| PB117 | JR32 katB::ΩCm (katB null mutant) | This work |
| E. coli | ||
| UM383 | HB101 katG17::Tn10(Tet)katE::Tn10(Kan) | P. Loewen |
| UM383(pMMB207αB::katB) | UM383 with a PCR-amplified katB gene in the Hind/Pst site of pMMB207αB | This work |
| Plasmids | ||
| pJBZ280 | lacZ with polylinker upstream for constructing translational fusions; ColE1 ori (Kanr) | M. R. K. Alley (3) |
| pNPTS138 | Derivative of pLITMUS 38 cloning vector with nptI, RK2 oriT, and Bacillus subtilis sacB; Kanr | 27 |
| pHP45 ΩCm | pBR322 derivative containing the Cmr gene with transcriptional and translational terminators and a polylinker at each end of the antibiotic cassette (Ampr) | 28 |
| pMMB207αB | RSF1010 derivative; IncQ lacIq Genr Ptac oriT; multiple cloning site α complementation | 36 |
| pMMB207αB::katB | pMMB207αB with Hind/Pst fragment containing PCR-amplified katB gene in Hind/Pst site | This work |
| pNPTS138 katB::ΩCm | pNPTS138 with katB null allele (katB::ΩCm) in BamHI site | This work |
Southern blotting.
Restriction digests of 4 μg of genomic DNA (32, 33) were electrophoresed overnight in 0.8% agarose and blotted by capillary transfer to nitrocellulose (BA85; Schleicher and Schuell). Hybridization was performed at 62°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate. Washing was performed at 55°C in 2× SSC–0.1% sodium dodecyl sulfate.
Cloning of L. pneumophila katB.
A BamHI genomic blot of wild-type L. pneumophila DNA was hybridized with an E. coli katG probe to identify 6.4- and 5.7-kb bands. The probe was a PCR fragment of E. coli katG from the third nucleotide of the ATG start codon through the codon for the penultimate amino acid, L725 (34). The 5.7-kb band was gel purified and ligated into the BamHI site of pUC12, producing a library of ≈600 Apr colonies, which were patched to grids. Blots of BamHI-digested plasmid DNA from progressively smaller pools were hybridized with the E. coli katG probe. By this approach a plasmid containing the 5′ end of L. pneumophila katB was isolated. To isolate an overlap containing the 3′ end, a 9-kbp fraction was gel purified from an EcoRI/HindIII genomic digest of strain JR32 and ligated into pUC12, producing a library which was screened as described above. In this cloning, the probe was a 2.1-kb HindIII/BamHI fragment containing the 5′ end of katB and some 5′ upstream sequence. The overlap fragment contained ≈7 kb beyond the BamHI site in the 5.7-kbp fragment initially cloned.
Construction of the chromosomal katB::lacZ translational fusion.
The 5.7-kb BamHI fragment containing the 5′ end of katB and upstream sequence was subcloned into the BamHI site of pJBZ280 (3) (Table 1) to create an in-frame translational fusion. E. coli colonies harboring the construct were blue on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates, indicating that the Legionella promoter was recognized in E. coli. This construct was transformed by electroporation into strain JR32, and Kanr transformants were selected. Three cycles were performed, in which cultures were grown overnight without kanamycin and then plated without kanamycin and isolated colonies were patched to identify Kanr strains. Southern blotting demonstrated that the fusion had integrated into the chromosome adjacent to katB. The fusion strain, PB102, was blue with X-Gal overlay (36). A L. pneumophila chromosomal integrant derived from pJBZ280 with katB in the wrong orientation with respect to lacZ was white with X-Gal and showed no significant LacZ activity in assays of liquid cultures.
Construction of L. pneumophila katB null mutant.
A katB null mutant was made by allelic exchange with sucrose counterselection (27, 32, 33, 36). The 5.7-kb BamHI fragment containing the 5′ end of katB in pUC12 was cleaved at the unique ApaI site. This site was filled in with Klenow fragment and blunt-end ligated with the ΩCm cassette excised from pHP45 ΩCm (28) with HindIII and blunted. The resulting 8.5-kb fragment containing the null allele was excised from pUC12 with BamHI and subcloned into the sucrase vector pNPTS138 (27). Wild-type L. pneumophila JR32 was transformed by electroporation with pNPTS138::katB::ΩCm. Individual Cmr transformants were streaked on CYE–chloramphenicol–2% sucrose to identify Cmr Sucr Kans strains (33, 36). Allelic-exchange mutants were identified by Southern blotting, catalase and peroxidase enzyme assays, and activity staining of gels.
Enzymatic assays.
Peroxidase activity was assayed at pH 6.4, monitoring the oxidation of dianisidine at 460 nm (ɛM = 11.3 mM−1 cm−1 [6]). Catalase activity was assayed at pH 7.2, monitoring the decomposition of H2O2 at 240 nm (ɛM = 39.4 M−1 cm−1 [1]). One unit of activity equals 1 μmol of H2O2 decomposed per min. Catalase activity was visualized as clear zones in nondenaturing polyacrylamide gels via inhibition of diaminobenzidine oxidation after permeation of the gel with a mixture of horseradish peroxidase, H2O2, and diaminobenzidine. Peroxidase activity was visualized as brown zones by omitting horseradish peroxidase from the protocol (7, 13). β-Galactosidase was assayed with o-nitrophenyl-β-d-galactoside as the substrate; activity was expressed in Miller units (22).
Measurement of resistance to hydrogen peroxide.
Zone of inhibition tests were performed by using overnight cultures in AYE with the appropriate antibiotic (0.1 ml in 3 ml of 0.8% agar without nutrients on CYE plates without antibiotics). Whatman 3MM disks (7-mm diameter) received 10 μl of freshly diluted H2O2. The plates were incubated for 48 h.
Growth of L. pneumophila in cultured THP-1 macrophage cells.
THP-1 cells were maintained and prepared for infection as previously described (33). L. pneumophila cells from overnight cultures in AYE were diluted in RPMI 1640 supplemented with 10% fetal calf serum, 1% glutamine, and 20% normal human serum and then added to the THP-1 cells (see Fig. 4 for further details). Aliquots removed daily were plated on CYE plates without added antibiotics.
FIG. 4.
Growth of L. pneumophila in cultured macrophage cells. Adherent THP-1 cells (3 × 105 per well) were infected on day 0 with 1 × 103 to 5 × 103 cells of the wild-type strain JR32 (•) or the katB null strain PB117 (■) from overnight cultures in AYE medium. Error bars indicate standard deviations.
PCR methodology.
PCR was used to amplify a fragment of 2.7 kb, beginning 304 nucleotides (nt) upstream of the katB ATG translational start and ending 234 nt downstream of the katB translational stop. The PCR mixture contained 40 ng of HindIII-digested JR32 genomic DNA, 20 pmol each of 5′ and 3′ primers (Fig. 1), 2 mM each deoxynucleoside triphosphate dNTP and 2.5 U of Taq DNA polymerase in 100 μl. The amplification program was 1 cycle of 1 min at 94°C, 1 min at 55°C, and 2.5 min at 72°C; 25 cycles of 1 min at 94°C, 1 min at 55°C, and 4 min at 72°C; and a final cycle of 1 min at 94°C, 1 min at 55°C, and 5 min at 72°C. Following digestion with HindIII and PstI, the PCR fragment was ligated into pMMB207αB (Table 1).
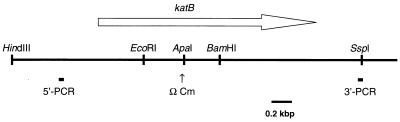
FIG. 1.
Restriction map of L. pneumophila katB region. The thick horizontal line depicts the region sequenced (3,865 nt). Every nucleotide reported was identified in two to five sequencing runs, each with a different sequencing primer. Beneath the line are indicated the locations of the 5′ (TGATGCAGGCCTGCAGTTCACCAGTCAGCAGAGCG) and 3′ (GGTTTACAAAGCTTAGATGAGACTTACAGAAACG) PCR primers (5′-PCR and 3′-PCR) and the site of insertion of the ΩCm cassette (ΩCm). The underlined regions are PstI and HindIII sites, respectively, that replaced 6 nt in the genomic L. pneumophila sequence. The arrow indicates the direction of transcription and the position of the katB open reading frame.
Nucleotide sequence accession number.
The GenBank accession number for the katB gene is AF078110.
RESULTS
Cloning the katB catalase-peroxidase gene of L. pneumophila.
Genomic Southern blots were hybridized with probes derived from E. coli katE and katG, encoding a monofunctional catalase and bifunctional catalase-peroxidase, respectively. BamHI and PstI genomic digests each showed two bands with the katG probe and no significant hybridization with the katE probe.
Bands of 6.4 and 5.7 kbp were observed in the BamHI digest. The 5.7-kbp BamHI band was cloned by hybridization. The sequence at one end was highly homologous to that at the 5′ end of bacterial catalase-peroxidase genes and the amino-terminal sequences of the encoded enzymes. The nucleotide sequence of the entire catalase-peroxidase gene was determined by isolating a fragment containing the 3′ end (Fig. 1). This catalase-peroxidase gene was named katB. Sequences upstream and downstream of katB were not homologous to any sequences in the databases. The 6.4-kbp band has been shown to contain a second catalase-peroxidase gene, katA (4).
KatB catalase-peroxidase sequence and enzyme activity.
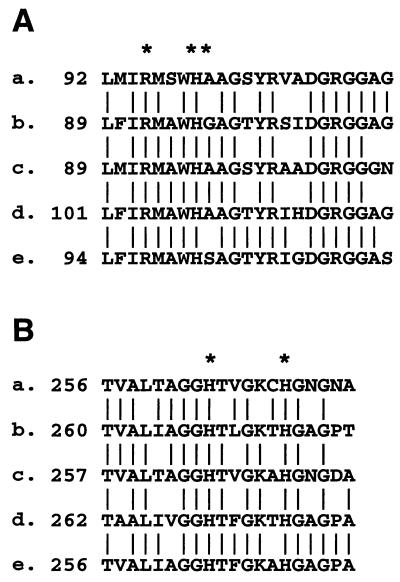
The katB open reading frame (ORF) encoded a protein of 721 amino acids highly homologous to bacterial catalase-peroxidases over its entire amino acid sequence: it was 65% identical to the catalase-peroxidase of Bacillus stearothermophilus and 57 to 60% identical to catalase-peroxidases of E. coli, Mycobacterium tuberculosis, and Rhodobacter capsulatus. In the absence of the three-dimensional structure data for catalase-peroxidases, putative heme ligands and active site residues have been identified by sequence homologies with the monofunctional cytochrome c peroxidase (CCP) from Saccharomyces cerevisiae (35). Residues forming the peroxide binding site on the distal side of the heme (Fig. 2A) and residues which bind the proximal side of the heme (Fig. 2B) in CCP are conserved in L. pneumophila KatB. In addition, the adjacent sequences were highly homologous with those in other catalase-peroxidases. L. pneumophila KatB showed no homologies to monofunctional catalases or to monofunctional peroxidases from fungi or bacteria that use nonheme cofactors, e.g., manganese peroxidase, NADH flavoprotein peroxidase, or the lignin glycoprotein peroxidases with bound calcium.
FIG. 2.
Conserved active site residues in bacterial catalase-peroxidases. The asterisks mark the conserved residues that are homologous to residues of known function in yeast CCP. L. pneumophila KatB (a), E. coli KatG; (b), R. capsulatus (c), M. tuberculosis ATCC 25618 (d), and B. stearothermophilus (e) sequences are shown. (A) Distal side of heme. Residues marked by asterisks correspond to those forming the binding site for peroxide in CCP (R-48, W-51, and H-52 in the CCP sequence). (B) Proximal side of heme. Residues marked by asterisks correspond to CCP residues which form the fifth coordination position of the heme (H-175) and a site of hydrogen bonding to a heme propionate (H-181).
The katB ORF plus 304 nt of upstream sequence and 234 nt of downstream sequence was amplified by PCR from L. pneumophila genomic DNA and cloned into the broad-host-range vector pMMB207αB, which is maintained in E. coli and L. pneumophila. This construct, pMMB207::katB, was transformed into strain UM383, an E. coli katE katG null mutant. Cell extracts of the transformant contained catalase and dianisidine peroxidase activities of 1.2 and 0.003 U/mg of cell protein, respectively. No detectable catalase or dianisidine peroxidase activity was found in extracts of untransformed strain UM383. These data indicate that L. pneumophila katB encodes a functional catalase-peroxidase which can be expressed in E. coli and that katB is not a pseudogene.
Construction of a katB null strain.
An ΩCm cassette (26, 28), which interrupts transcription and translation, was cloned into the ApaI site within the Pro-280 codon of KatB. Exchange of wild-type katB for katB::ΩCm was accomplished by using sucrose counterselection and confirmed by Southern blotting (data not shown). A nonfunctional gene product was expected because residues essential for enzymatic function, i.e., a Trp proposed as the site of free radical formation and an Asp which stabilizes a histidine ligand of the heme (12), lie carboxyl terminal to the site in cytochrome c peroxidase, which is homologous to L. pneumophila Pro-280.
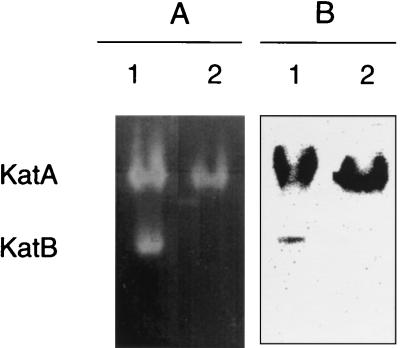
Cell extracts of wild-type and katB null strains were electrophoresed under nondenaturing conditions, and the acrylamide gels were stained for catalatic and diaminobenzidine peroxidatic activities. With the wild type (Fig. 3A and B, lanes 1), each enzyme stain visualized two bands. The coincidence of catalatic and peroxidatic staining for the two activities suggested that L. pneumophila expressed two catalase-peroxidases under the growth conditions used. With null strain PB117 katB::ΩCm, the band with greater mobility was absent (Fig. 3A and B, lanes 2). These data demonstrated that katB encoded the catalase-peroxidase with faster mobility and confirmed that strain PB117 was a functional katB null mutant. The slower catalase-peroxidase band was labeled KatA, as it is encoded by the second catalase-peroxidase gene implicated in genomic blots (4). Our data show that KatA and KatB account for the total catalatic and peroxidatic activity observed under the growth conditions used.
FIG. 3.
Zymogram analysis for catalatic and peroxidatic activities. Cell extracts of overnight AYE cultures (500 μg of protein per lane) were electrophoresed under nondenaturing conditions, and the gel was stained to visualize catalatic (A) or diaminobenzidine-peroxidase activity (B). The two catalase-peroxidases in L. pneumophila are indicated. Lanes 1, strain JR32 (wild type); lanes 2, strain PB117 katB::ΩCm (katB null).
Characterization of the katB null mutant. (i) Growth in media and sensitivity to hydrogen peroxide.
The katB null mutation had no effect on exponential growth. The doubling times of wild-type strain JR32 and null strain PB117 were identical in AYE broth: 170 ± 14 and 180 ± 12 min, respectively. Similarly, there was no difference in survival in the stationary phase. During 4 days in the stationary phase the titer of katB-null and wild-type L. pneumophila decreased similarly. This contrasted with the 104- to 106-fold decrease in survival of a L. pneumophila sodC null mutant (33).
Although the null mutant was no different from the wild type in growth and survival under normal aerobic culture conditions, it was more sensitive to an imposed H2O2 stress (Table 2). A second, independently isolated katB null strain showed an identical phenotype, consistent with the H2O2 phenotype being attributable to the katB null mutation and not to a spontaneous mutation elsewhere in the chromosome.
TABLE 2.
Resistance to hydrogen peroxidea
| Strainb | Diam of clearing (cm) at H2O2 concn (M) of:
|
|||
|---|---|---|---|---|
| 1 | 2 | 5 | 10 | |
| Wild type | 2.95 ± 0.05 | 3.4 | 4.15 ± 0.05 | 4.9 |
| katB null | 3.2 | 3.65 ± 0.05 | 4.4 | 5.13 ± 0.03 |
Overnight cultures in top agar; 10 μl of solution at the indicated H2O2 concentration were applied to the disk. Where no error is listed, the duplicate determinations gave identical numerical values.
Wild type, strain JR32; katB null, PB117 katB::ΩCm.
(ii) Growth in THP-1 macrophage line.
Infection of cultured macrophage-like cell lines has been the model for L. pneumophila infection of pulmonary macrophages in Legionnaires’ disease (30, 33, 36). L. pneumophila fails to replicate in most tissue culture media. Increases in the L. pneumophila titer in the medium are therefore attributable to release of the bacterium following invasion, intracellular replication, and lysis of a macrophage host. Wild-type L. pneumophila infected the THP-1 macrophage-like line as previously observed by us and others (33, 36) (Fig. 4). For katB null mutant PB117, the time course of the infection was reproducibly delayed by 2 days compared to that of the wild type. Further studies are necessary to discern if the delay is due to differences in entry, intracellular growth, or lysis. A similar delay was observed when cultured HL-60 macrophage-like cells were infected with PB117 katB::ΩCm.
An independently isolated katB null mutant exhibited an identical delay in infection of THP-1. This supports the contention that the above-mentioned infection phenotype is attributable to inactivation of katB and not to a spontaneous mutation in another gene.
Construction and characterization of a katB::lacZ fusion strain.
A translational fusion in a Kanr ColE1 plasmid was constructed with 4.6 kbp of upstream sequence and 1.1 kbp from the 5′ end of the katB ORF. Capitalizing on the generally poor maintenance of ColE1 plasmids in L. pneumophila, a cointegrate was isolated by selecting for Kanr transformants of wild-type L. pneumophila and then screening for Kanr after repeated culturing in the absence of kanamycin. The translational fusion strain, PB102, was blue by X-Gal overlay on CYE plates. Southern blotting (data not shown) confirmed that the fusion integrated adjacent to chromosomal katB. This strategy appears not to have been used previously with L. pneumophila and may be of general use in the construction of chromosomal fusions.
Exponential cultures of strain PB102 in AYE (optical density at 600 nanometers, 0.4 to 1.0) were treated with single additions of H2O2 to 15 or 60 μM. No change in LacZ activity was observed from 0.5 to 3 h after H2O2 challenge. These results suggested that the KatB catalase-peroxidase of L. pneumophila is relatively inert to hydrogen peroxide induction. In contrast, katG catalase-peroxidase was induced 20-fold when exponential cultures of C. crescentus were treated with the same range of H2O2 concentrations (32).
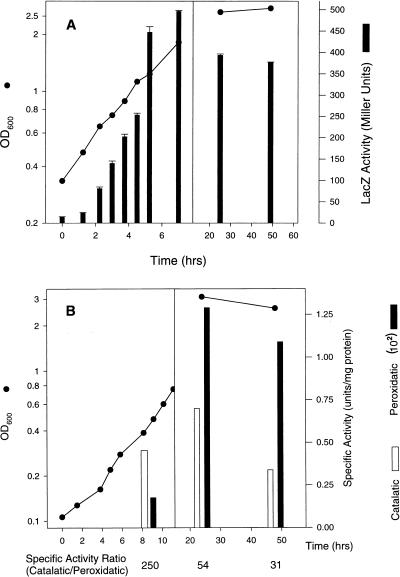
Expression of the katB::lacZ fusion increased about 30-fold during exponential growth and then decreased by about 25% in the stationary phase (Fig. 5A). A similar induction was seen for E. coli katG and was attributed to an increase in the number of respiratory centers during exponential growth, leading to increased H2O2 production per cell (11). Catalatic and peroxidatic activity during the transition from exponential to stationary phase was measured for wild-type L. pneumophila (Fig. 5B). Peroxidase activity in wild-type L. pneumophila increases about six- to eightfold in the stationary phase, in agreement with data in the literature (24).
FIG. 5.
Expression of catalatic and peroxidatic activities during growth. (A) Growth stage induction of the katB::lacZ fusion in the chromosomal fusion strain PB102. (B) Catalatic and dianisidine peroxidase specific activities of wild-type strain JR32 in mid-exponential and stationary phases and activity ratio (below abscissa). Error bars indicate standard deviations. OD600, optical density at 600 nm.
DISCUSSION
Studies of L. pneumophila virulence and intracellular multiplication have outpaced studies of its physiology and metabolism. Recent observations linking stationary-phase gene expression with the acquisition of L. pneumophila virulence traits (5) suggest that the study of stationary-phase physiology is an avenue for identifying virulence genes. We initiated studies of catalase-peroxidases in L. pneumophila because such enzymes are frequently linked with the stationary-phase stress response. The most recent studies of catalases and peroxidases in L. pneumophila, over a decade ago, reported a single bifunctional catalase-peroxidase, based on chromatographic separation of activities in a cell extract (24, 25). In the present study, we established that in fact two catalase-peroxidases are present, KatA and KatB. The discrepancy may be due to cochromatography of the two in the prior study or to strain differences, although both laboratories used the Philadelphia-1 strain or a derivative of it.
We cloned the gene for L. pneumophila katB catalase-peroxidase, encoding a typical bifunctional catalase-peroxidase, in polypeptide length and active site homologies with CCP. Functionally, KatB is similar to E. coli KatG catalase-peroxidase. The 30-fold induction of katB during exponential growth can be reasonably attributed to increased cellular production of H2O2 resulting from increased respiration and increased leakage of electrons to O2, as proposed for E. coli katG (11). Although L. pneumophila and E. coli catalase-peroxidases both play roles in resistance to H2O2, only E. coli katG is induced by H2O2. If katB is induced during exponential growth, why isn’t it induced by H2O2 addition? During exponential growth in E. coli, katG levels correlate with the rate of H2O2 production but not with the absolute level of H2O2 (11). If a similar mechanism operates in L. pneumophila, then a bolus addition of H2O2 may create a rate of increase in H2O2 concentration that is a poor mimic of physiological H2O2 production. In complex media, the doubling time of L. pneumophila is 3 h, compared to 30 min or less for that of E. coli. Consequently, respiration and respiratory generation of H2O2 during exponential growth are likely to occur at a lower rate in L. pneumophila than in E. coli, and katB induction may be tuned to smaller H2O2 gradients.
We and others have observed an 8- to 10-fold induction of peroxidatic activity in L. pneumophila stationary phase (24). Here we showed that this induction is largely, if not entirely, due to KatA, because expression of katB is reduced in the stationary phase relative to its maximum during exponential growth (Fig. 5A). We also showed that during growth the ratio of catalatic activity to peroxidatic activity changes substantially, from ≈250 in the exponential phase to ≈30 to 50 in the stationary phase (Fig. 5B). We directly determined the catalatic:peroxidatic activity ratio for KatB as ≈400 by expressing katB in a catalase and peroxidase mutant of E. coli. Therefore, the catalatic:peroxidatic activity ratio for KatA must be <30 to 50, as it is the major but not the exclusive catalase-peroxidase in the stationary phase (Fig. 3). A survey of the catalatic:dianisidine peroxidase activity ratios for purified bacterial catalase-peroxidases shows values ranging from 1,800 (R. capsulatus [16]) to 250 (Klebsiella pneumoniae [15]) to ≈100 (M. tuberculosis and E. coli [6, 20]). A ratio of less than 30, inferred for KatA, would be uncommonly low and indicative of an enzyme with a propensity towards peroxidatic versus catalatic activity.
In sum, our studies are the first molecular genetic investigation of catalase-peroxidases in L. pneumophila. We demonstrated a role for the KatB catalase-peroxidase in defense against H2O2 and in infection of macrophages. We also identified the KatA catalase-peroxidase as responsible for the stationary-phase increase in peroxidase activity and as a potentially useful marker for the stationary-phase gene expression that precedes virulence. In addition, KatA is predicted to have an uncommonly high peroxidatic activity relative to catalatic activity. Cloning of L. pneumophila katA and construction and characterization of katA fusion and null strains are in progress.
ACKNOWLEDGMENTS
This work was supported by grant MCB-9513706 to H.M.S. from the National Science Foundation.
We thank Peter Loewen for E. coli UM383, Howard Shuman for plasmid pMMB207αB and for HL-60 cells, Yves Brun for pJBz280 and pNPT5138, and Michelle Swanson for communication of results prior to publication. Gregory St. John and Paul S. Rava provided expert technical assistance.
REFERENCES
- 1.Aebi H. Catalase in vitro. Methods Enzymol. 1984;108:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 2.Albus A M, Pesci E C, Runyen-Janecky L J, West S E, Iglewski B H. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alley M R K, Gomes S L, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–342. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay, P., and H. Steinman. 1998. Unpublished data.
- 5.Byrne B, Swanson M S. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claiborne A, Fridovich I. Purification of the o-dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J Biol Chem. 1979;254:4245–4252. [PubMed] [Google Scholar]
- 7.Clare D A, Duong M N, Darr D, Archibald F, Fridovich I. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal Biochem. 1984;140:532–537. doi: 10.1016/0003-2697(84)90204-5. [DOI] [PubMed] [Google Scholar]
- 8.El-Gedaily A, Paesold G, Krause M. Expression profile and subcellular location of the plasmid-encoded virulence (Spv) proteins in wild-type Salmonella dublin. Infect Immun. 1997;65:3406–3411. doi: 10.1128/iai.65.8.3406-3411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feeley J C, Gibson R J, Gorman G W, Langford N C, Rasheed J K, Mackel D C, Baine W B. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields B S. The Molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodin D B, McRee D E. The Asp-His-Fe triad of cytochrome c peroxidase controls the reduction potential, electronic structure, and coupling of the tryptophan free radical to the heme. Biochemistry. 1993;32:3313–3324. [PubMed] [Google Scholar]
- 13.Gregory E M, Fridovich I. Visualization of catalase on acrylamide gels. Anal Biochem. 1974;58:57–62. doi: 10.1016/0003-2697(74)90440-0. [DOI] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecther M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 15.Hochman A, Goldberg I. Purification and characterization of a catalase-peroxidase and a typical catalase from the bacterium Klebsiella pneumoniae. Biochim Biophys Acta. 1991;1077:299–307. doi: 10.1016/0167-4838(91)90544-a. [DOI] [PubMed] [Google Scholar]
- 16.Hochman A, Shemesh A. Purification and characterization of a catalase-peroxidase from the photosynthetic bacterium Rhodopseudomonas capsulata. J Biol Chem. 1987;262:6871–6876. [PubMed] [Google Scholar]
- 17.Horwitz M A, Silverstein S C. Intracellular multiplication of Legionnaires’ disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J Clin Invest. 1983;71:15–26. doi: 10.1172/JCI110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iriarte M, Stainier I, Cornelis G R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995;63:1840–1847. doi: 10.1128/iai.63.5.1840-1847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins D E, Schultz J E, Matin A. Starvation-induced cross-protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1988;170:3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnsson K, Froland W A, Schultz P G. Overexpression, purification, and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J Biol Chem. 1997;272:2834–2840. doi: 10.1074/jbc.272.5.2834. [DOI] [PubMed] [Google Scholar]
- 21.Loewen P. Probing the structure of catalase HPII of Escherichia coli—a review. Gene. 1996;179:39–44. doi: 10.1016/s0378-1119(96)00321-6. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 23.Nickerson C A, Curtiss R., III Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pine L, Hoffman P S, Malcolm G B, Benson R F, Franzus M J. Role of keto acids and reduced-oxygen-scavenging enzymes in the growth of Legionella species. J Clin Microbiol. 1986;23:33–42. doi: 10.1128/jcm.23.1.33-42.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pine L, Hoffman P S, Malcolm G B, Benson R F, Keen M G. Determination of catalase, peroxidase, and superoxide dismutase within the genus Legionella. J Clin Microbiol. 1984;20:421–429. doi: 10.1128/jcm.20.3.421-429.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;20:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 27.Reisenauer A, Mohr C D, Shapiro L. Regulation of a heat shock ς32 homolog in Caulobacter crescentus. J Bacteriol. 1996;178:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remy F, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 29.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington, D.C: ASM Press; 1994. [Google Scholar]
- 30.Shuman H A, Purcell M, Segal G, Hales L, Wiater L A. Intracellular multiplication of Legionella pneumophila: human pathogen or accidental tourist? Curr Top Microbiol Immunol. 1998;225:99–112. doi: 10.1007/978-3-642-80451-9_6. [DOI] [PubMed] [Google Scholar]
- 31.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 32.Steinman H M, Fareed F, Weinstein L. Catalase-peroxidase of Caulobacter crescentus: Function and role in stationary-phase survival. J Bacteriol. 1997;179:6831–6836. doi: 10.1128/jb.179.21.6831-6836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St. John G, Steinman H M. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triggs-Raine B L, Doble B W, Mulvey M R, Sorby P A, Loewen P C. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J Bacteriol. 1988;170:4415–4419. doi: 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welinder K G. Bacterial catalase-peroxidases are gene duplicated members of the plant peroxidase superfamily. Biochim Biophys Acta. 1991;1080:215–220. doi: 10.1016/0167-4838(91)90004-j. [DOI] [PubMed] [Google Scholar]
- 36.Wiater L A, Sadosky A B, Shuman H A. Mutagenesis of Legionella pneumophila using Tn903dIIlacZ: identification of a growth-phase-regulated pigmentation gene. Mol Microbiol. 1994;11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]