Two cancer histotypes diverge from a common cell of origin epigenetically locked in different cell states, highlighting the importance of considering cell state to better understand the cell of origin of cancer.
Abstract
Clear cell ovarian carcinoma (CCOC) and endometrioid ovarian carcinoma (ENOC) are ovarian carcinoma histotypes, which are both thought to arise from ectopic endometrial (or endometrial-like) cells through an endometriosis intermediate. How the same cell type of origin gives rise to two morphologically and biologically different histotypes has been perplexing, particularly given that recurrent genetic mutations are common to both and present in nonmalignant precursors. We used RNA transcription analysis to show that the expression profiles of CCOC and ENOC resemble those of normal endometrium at secretory and proliferative phases of the menstrual cycle, respectively. DNA methylation at the promoter of the estrogen receptor (ER) gene (ESR1) was enriched in CCOC, which could potentially lock the cells in the secretory state. Compared with normal secretory-type endometrium, CCOC was further defined by increased expression of cysteine and glutathione synthesis pathway genes and downregulation of the iron antiporter, suggesting iron addiction and highlighting ferroptosis as a potential therapeutic target. Overall, these findings suggest that while CCOC and ENOC arise from the same cell type, these histotypes likely originate from different cell states. This “cell state of origin” model may help to explain the presence of histologic and molecular cancer subtypes arising in other organs.
Significance:
Two cancer histotypes diverge from a common cell of origin epigenetically locked in different cell states, highlighting the importance of considering cell state to better understand the cell of origin of cancer.
Introduction
Cell-of-origin and genetic mutations are often considered the most important determinants in the initiation and shaping of the final molecular and phenotypic landscape of cancer cells (1, 2). This framework, however, is insufficient to explain the two ovarian carcinoma histotypes, which share the same cell of origin and common genetic mutations, yet present striking differences in cellular phenotype and clinical behavior.
Epithelial ovarian cancer, or ovarian carcinoma, has historically been treated as one disease entity. In recent years, it has become clear that different histotypes of ovarian carcinoma have distinct etiologies as well as genetic and epigenetic underpinnings of the disease (3–6). High-grade serous ovarian carcinoma (HGSOC) is the most common histotype (∼75%), and one of the first cancers to be comprehensively characterized by The Cancer Genome Atlas (TCGA) Project. Other histotypes are not included in TCGA and remain poorly understood. Clear cell ovarian carcinoma (CCOC) accounts for approximately 5% to 12% of ovarian carcinomas cases. It is generally unresponsive to chemotherapy and has a worse prognosis than HGSOC when discovered at late stages (7). Notably, CCOC is more common in East Asian women (8), and accounts for as much as 30% of ovarian carcinoma in the Japanese population. Endometrioid ovarian carcinoma (ENOC) accounts for an additional 5% to 10% of epithelial ovarian carcinomas. Other histotypes, such as mucinous ovarian carcinoma (MOC) and low-grade serous carcinoma (LGSOC) are less common and comprise approximately 3% and 5% to 8% of all ovarian carcinomas, respectively (9). Despite similarity in nomenclature, LGSOCs exhibit distinct clinical behavior and molecular profiles compared with HGSOC tumors. Primary MOCs, which develop from benign and borderline tumors at the ovary (10), are often confused with metastases originating from mucus-secreting cells that line the gastrointestinal tract, endocervix, and other organ sites (9).
The origin of ovarian cancer has been the subject of intense debate for over two decades (3). It was only recently that most researchers agreed on a unique feature of ovarian carcinoma: most ovarian carcinoma histotypes arise from cells that are not native to the ovary (11). HGSOC likely originates from the fallopian tube epithelium (FTE; ref. 12), whereas ENOC and CCOC are thought to arise from ovarian endometriotic cysts, particularly atypical endometriosis (13). Endometriosis is a chronic disease that affects approximately 10% of uterus-bearing individuals of reproductive age in the United States (14–16), characterized by the presence of endometrium-like tissue outside of the uterine cavity, elevated systemic inflammation, and a diversity of clinical symptoms. Endometriotic tissue thickens and bleeds in response to changes in hormone levels during the menstrual cycle, similar to the eutopic endometrium.
The two endometriosis-associated histotypes display distinct cellular phenotypes and clinical behaviors, particularly with CCOC being chemoresistant and ENOC associated with better prognosis (7). Interestingly, ENOC and CCOC have very similar mutation profiles (17, 18). In contrast to the observation of near-universal, early-occurring TP53 mutations in HGSOC, these two histotypes instead share frequent somatic mutations affecting PIK3CA and the chromatin regulator ARID1A (17, 18). Importantly, these mutations are also frequently present in nonmalignant endometriotic lesions (19–22), suggesting that they may not be directly responsible for malignant transformation, let alone for histotype divergence. Previous studies on epigenomic and transcriptomic profiles of FTE, including ENOC and CCOC, focused primarily on clustering and molecular subtype identification, prognostic markers, or comparison to HGSOC (23–25). Little work has been done to understand the divergence between ENOC and CCOC, with only a single study suggesting that CCOC arises from a particular cell type located in the endometrium (26), a theory that has garnered some controversy (27).
These histotypes show important differences in gene expression, which have provided some insight into their biology. The most well-known difference between histotypes is the universal overexpression of hepatocyte nuclear factor-1β (HNF1B) in clear cell tumors (28, 29). Germline variants of HNF1B are associated with susceptibility to ovarian cancer histotypes (5), with opposing effects for CCOC and ENOC, the protective allele for ENOC is risk-conferring for CCOC, and vice versa. The “clear cell” phenotype observed in CCOC is associated with the accumulation of intracellular glycogen. HNF1B regulates multiple genes in the glycolytic and glucose metabolism pathways and is linked to increased glucose uptake and lactate secretion (30). However, there remains a lack of understanding of this apparent miswiring in CCOC.
The poor prognosis and lack of effective treatment options for advanced stage CCOC make it a research priority (31, 32). With RNA transcription and DNA methylation analysis of CCOC and ENOC tumors, we observe that they transcriptionally resemble two menstrual cycle states of normal endometrium. We propose that while these histotypes originate from the same cell type, they arise from different cell states (here defined as transitory transcriptional program in the same cell type). Furthermore, the histotypes appear to be epigenetically locked into the different menstrual cycle states through epigenetic control of estrogen signaling. In addition, by comparing different histotypes to their corresponding cell states of origin, we dissected cancer-specific pathways and processes that may offer therapeutic opportunities for these histotypes, particularly CCOC.
Materials and Methods
Patient samples and preparation
Ovarian carcinoma samples were selected from the UBC/VGH Gynecological Tissue Bank. Patients were recruited with written informed consent for prospective molecular analysis. Representative formalin-fixed paraffin-embedded (FFPE) slides from each case were subjected to a centralized pathology review to confirm the histotype. Frozen tissue specimens were also reviewed to ensure minimal cellularity for analysis. DNA and/or total RNA were extracted from frozen tissue sections (10–40 10 μm sections depending on tissue face size) using Qiagen QIAamp DNA or RNA Blood and Tissue Kits (Qiagen), following the manufacturer's protocols.
For normal-appearing uterine tissue, FFPE biopsies from women between the ages of 21 to 44, which had undergone biopsy for abnormal uterine bleeding were categorized into menstrual cycle phase (proliferative or secretory) by histomorphology (33). We excluded specimens with any visible pathology, coexisting malignancy, or known somatic alterations (33).
DNA methylation profiling
Three major histotypes of epithelial ovarian cancer were examined using the Infinium HumanMethylation450 BeadChip (HM450 array), including 60 HGSOC, 48 CCOC, and 19 ENOC samples. Bisulfite conversion was performed on 1 μg of genomic DNA from each sample using the EZ-96 DNA Methylation Kit (Zymo Research) according to the manufacturer's instructions. We assessed the amount of bisulfite-converted DNA and completeness of bisulfite conversion using a panel of MethyLight-based quality control (QC) reactions, as described previously (34). Bisulfite-converted DNA was whole-genome amplified and enzymatically fragmented prior to hybridization to the arrays. These samples were processed in the same facility using the same protocol as TCGA samples.
Transcriptome profiling
cDNA libraries for 28 CCOC and 29 ENOC were prepared using a strand‐specific RNA‐Seq Sample Preparation Kit (stranded, polyA+) from Illumina. Data were generated from paired-end sequencing at Canada's Michael Smith Genome Sciences Centre on Illumina platforms: HiSeq 2500 using V3 or V4 chemistry and paired‐end 125 base reads targeting 200 million paired-end reads per sample.
DNA methylation data processing and sample quality control
Raw IDAT files were processed using the R package SeSAMe (35) with background correction, nonlinear dye bias correction, and nondetection masking (any data point not significantly different from the background was replaced with NA). Probes with design issues were masked (35). DNA methylation β values, ranging from 0 to 1 (with “0” indicating fully unmethylated and “1” fully methylated), were calculated as the quantitative percentage of methylated signals over both methylated and unmethylated signals.
SNP probes (‘rs’ probes) were used to examine potential sample swaps that could occur in genomic studies. No swaps were identified. DNA methylation β values for three MIR141/200C promoter probes (“cg12161331,” “cg18185189,” “cg19794481”) were examined to track mesenchymal content within each sample. MIR141/200C is considered a master regulator of epithelial/mesenchymal phenotype transition, and this process is controlled by its promoter methylation state (36). Methylation levels at these three probes were highly correlated with the mesenchymal content in the flow sorting results (37). Tumor samples with a mesenchymal content of >65% were removed, together with one CCOC sample with an ambiguous pathology report. In total, 54 HGSOC, 41 CCOC, and 18 ENOC were included for further methylation analysis.
Transcriptome data processing and sample quality control
In-house generated CCOC and ENOC RNA-seq data were combined with publicly available paired-end RNA-seq data for 84 HGSOC tumor samples, 20 normal endometrium, and 3 normal epithelial brushings of the fallopian tube. Each raw sequencing file (FASTQ format) was aligned to the human reference genome (GRCh37) using STAR version 2.7 (38) with default settings. Estimation of gene-level abundance was performed using RSEM version 1.3.1 (39). Raw read count output from RSEM was further batch-corrected using the R package sva (function ComBat_seq) and normalized using Reads Per Kilobase of transcript per million mapped reads (RPKM) with library size scale factors estimated using R package edgeR. Log2-transformed RPKM values were used to visualize downstream expression. Uniform Manifold Approximation and Projection (UMAP) was performed using the R package uwot. Two CCOC and two ENOC samples were removed because of clear grouping with different histotypes in the UMAP analysis and ambiguous pathology reports. Furthermore, three CCOC samples were excluded because of their high mesenchymal content, as assessed using the HM450 array. In total, 84 HGSOC, 23 CCOC, 27 ENOC, and all normal samples were included in further tumor transcriptomic analysis.
Validation of RNA-seq data for expression of key genes through menstrual cycle
Affymetrix Human Genome U133 Plus 2.0 Array data from GSE4888 (40) were downloaded in MINiML format. The data represented the output of the Gene Chip Operating System version 1.1 using Affymetrix default analysis settings and global scaling as the normalization method. Array probes corresponding to HNF1B and ESR1 were obtained from the hgu133plus2.db Bioconductor R package, specifically the ProbeAnnDbBimap class object, hgu133plus2ALIAS2PROBE. Patients with no menstrual phase information were excluded (n = 6). Next, to account for probe intensity differences, z scores were calculated for each probe (ESR1, n = 9; HNF1B, n = 2) using the remaining 22 samples. The z-scores were plotted using phase grouping.
DNA methylation analyses
Unsupervised hierarchical clustering was performed on the top variable CpG probes (N = 10,000, filtered by SDs) across all ovarian tumor samples measured on the HM450 array using the R function hclust. Differentially methylated regions (DMR) were calculated using the R package DMRcate (41), based on its default FDR cutoff of 0.05 and MIR200c methylation was used as a covariate to adjust for purity. The β cut-off was set at 0.2. The significant DMRs were divided into hypermethylated in CCOC and hypermethylated in ENOC whereafter a locus overlap analysis (LOLA) for enrichment of genomic features (LOLA) was done using the LOLA core database with the “ucsc_features” and “encode_tfbs” collections. The userUniverse parameter was specified as 200 bp, centered on all 450k array probes. ESR1 promoter methylation was visualized using the VisualizeGene function from SeSAMe. Silencing events for MLH1 were called using its corresponding promoter probe cg00893636 at a beta cut-off of 0.1. Probes hypermethylated in CCOC relative to ENOC and overlapping with the ERalpha tfbs dataset were extracted and plotted as a heatmap ordered by average methylation in the normal endometrium.
Transcriptomic analyses
Differential gene expression analysis was performed using the R package edgeR v3.34.1 (42), with batch-corrected read counts as input. Genes with less than three read counts in more than 80% of samples were filtered out prior to differential analysis to minimize multiple testing on minimally expressed features. Differentially expressed genes were defined on the basis of an FDR cutoff of 0.05, and an absolute fold change greater than 2. The top-ranked differentially expressed genes (DEG) were ordered on the basis of their fold changes after satisfying an FDR significance level of 0.05. Pathway enrichment analysis and visualization were performed using the R package clusterProfiler v4.0.5, with biological process ontology terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets (43). Upset plots were generated using UpSetR package v1.4.0. Enrichment networks were visualized using the cnetplot function in the R package enrichplot v1.12.3.
DNA methylation datasets for normal endometrium and fallopian tube samples
Additional DNA methylation data for normal endometrial and fallopian tube samples were downloaded from TCGA (44). Data on additional FTE samples were obtained from GSE65820 (45), and GSE81224 (46). Early- and mid-secretory methylation endometrial samples were obtained from the GSE90060 dataset (47).
IHC
IHC for HNF1B was performed on a Leica Bond platform using anti-HNF1B primary antibody (HPA002083 rabbit polyclonal; Millipore Sigma) at 1:200 using ER1 antigen retrieval (Leica) followed by polymer detection (29, 48, 49). Staining was interpreted using established standards (29, 48–50), where nuclear staining was visible and scored as negative/absent (complete absence or focal in <50% of epithelial cells) or positive/present (diffuse staining of >50% of epithelial cells). A positive control of ovarian clear cell carcinoma was used in each staining batch (29, 50).
Data availability
Transcriptome and methylation array data generated in this study were deposited in the NCBI Gene Expression Omnibus Series GSE226872 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE226872). All other raw data generated in this study are available upon request from the corresponding author. Publicly available data generated by others were used by the authors. Raw RNA-seq datasets of normal endometrium samples and normal epithelial brushings of the fallopian tube were downloaded from GSE132711 (51) and GSE114493 (52), respectively. Sequencing data for primary tumor tissues from HGSOC were downloaded from GSE102073 (52). Functional gene sets were downloaded from the Molecular Signatures Database (v7.2). Expression data shown in Fig. 3A were from GSE4888. The images presented in Fig. 3D were cropped from these images retrieved from the Human Protein Atlas with the links below, as orthogonal datasets to validate our discovery with two different antibodies: (1) Antibody 1 (CAB068192): 33-year-old female (Patient ID 2941) and (2) Antibody 2 (HPA002083): 27-year-old female (Patient ID 2004; https://www.proteinatlas.org/ENSG00000275410-HNF1B/tissue/endometrium#img).
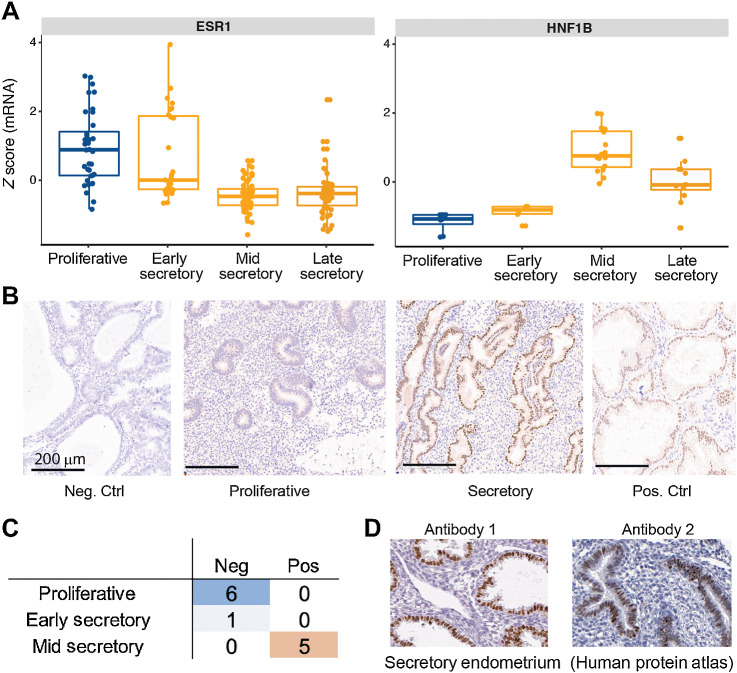
Figure 3.
Validation of HNF1B expression in normal mid-secretory endometrium. A, Expression z-scores averaged from all probes for ESR1 and HNF1B on an external microarray-based dataset (GSE4888), validating expression of HNF1B in mid-to-late secretory phase. B, IHC staining of endometrium at various menstrual cycle phases for HNF1B. Control specimens used are tissues from endometrioid endometrial carcinoma (negative) and clear cell carcinoma of the ovary (positive) that have been previously described as being negative or positive for HNF1B (respectively). These were rerun in the same experimental lot as endometrium samples. Note that stromal cells further serve as internal negative control in the CCOC control specimen as staining is restricted to nuclei of tumor epithelium. C, Tabulation of HNF1B IHC results from a panel of 12 normal endometrium samples shows segregation by menstrual cycle phase. D, HNF1B IHC results from Human Protein Atlas, with two different antibodies, for two normal endometrium samples at the secretory phase showing strong staining for HNF1B.
Results
Tumor sample quality control, purity, and composition
In bulk tumor-based studies, tumor purity and composition have a substantial impact on molecular readout due to mixed signals from tumor and stromal cells. To exclude any potential impact of tumor purity on our analysis and to capture the tumor microenvironment as an important feature of the tumors, we first used multiple orthogonal deconvolution methods to estimate tumor purity and stromal composition, including canonical marker genes for multiple cell types (Fig. 1). These included two DNA methylation-based methods and one RNA-seq–based method.
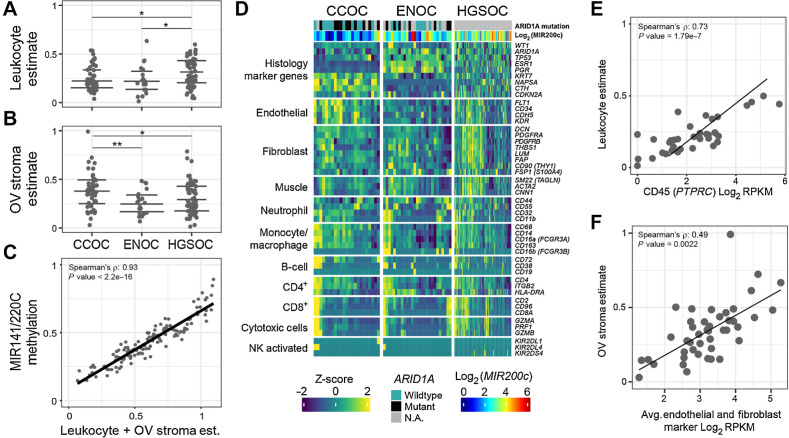
Figure 1.
Quality control of samples used in this study. A–C, Cellular composition and tumor purity estimation from DNA methylation with three orthogonal methods. A, Estimate of immune cell fraction with leukocyte-specific DNA methylation signature for the OV histotypes (see Materias and Methods). Y-axis indicates the estimated leukocyte proportion of each sample, with samples divided by subtype on the x-axis. Significance level: *, P ≤ 0.05; **, P ≤ 0.01. Horizontal lines denote the first quantile, median, and third quantile. B, Estimate of ovarian stroma-like component fraction (y-axis), with tissue-specific methylation signature (see Materials and Methods). C, Total mesenchymal fraction estimated by DNA methylation β values at the polycistronic MIR141/200C promoter (y-axis) is highly correlated with the sum (x-axis) of leukocyte fraction (A) and stroma fraction (B). D, RNA-seq shows histotype and cellular composition differences by expression of known marker genes for various cell populations. Color represents the Z-score of counts per million for marker genes (rows) across samples (columns). Samples are clustered within each histotype and genes are clustered within each marker category indicated on the left of the heatmap. Canonical histotype markers are plotted as controls (top). E, DNA methylation–based leukocyte estimates from A (y-axis) correlate to PTPRC mRNA expression for samples, with matching RNA-seq and methylation data. F, As in E, but with ovarian stroma estimate from DNA methylation (y-axis) and the RNA-seq stroma markers (x-axis).
The first method estimated the composition of immune cells and ovarian stroma in tumors with a cell type-specific methylation signature, as described previously (53, 54). The endometriosis-derived histotypes, CCOC and ENOC, had comparable levels of immune cell infiltration, both significantly lower than that of HGSOC (Wilcox tests, P < 0.05, Fig. 1A). CCOC was characterized by extensive stromal content, particularly when compared with ENOC (Wilcox test P = 0.007, Fig. 1B). Interestingly, ARID1A mutants had significantly lower tumor purity in CCOC (Wilcox test, P = 0.029; Supplementary Fig. S1A) but not in ENOC. The overall immune cell fraction did not differ according to ARID1A status (Supplementary Fig. S1B), whereas samples with mutations tended to have lower stroma content in both CCOC and ENOC (Supplementary Fig. S1C).
The second method used the DNA methylation fraction (measured by the beta value) at the polycistronic MIR141/200C promoter region. This promoter region is fully unmethylated in epithelial/carcinoma cells and fully methylated in mesenchymal stromal cells. Thus, the methylation level can be used as a direct surrogate for mesenchymal contamination in tumors of epithelial origin (37). This fraction should include two major nontumor components from Method 1. Indeed, the sum of leukocyte and stroma contents from Method 1 showed a high correlation with the total mesenchymal content assessed by Method 2 (Spearman ρ = 0.93, P < 2.2e−16, Fig. 1C), validating each other.
Next, we visualized the mRNA levels of key marker genes in the known cell populations (Fig. 1D). Canonical histotype markers were plotted as controls (top). Although these gene products are typically used as protein markers, the mRNA level often correlates with the final protein level and should show cell-type specificity. As expected, the mRNA levels of these marker genes showed clear segregation according to histotype. This validated the pathology-reviewed histotype labels of each sample. Consistent with the DNA methylation-based estimates, CCOC and ENOC were depleted in T cells compared with HGSOC (Supplementary Fig. S1D). There were no observed differences between subtypes for expression of CD4+, CD8+ or monocyte/macrophase markers (Supplementary Figs. S1E–S1G). In contrast, the stromal content in CCOC observed in DNAm-based estimates appeared to be attributable to endothelial cells. DNA methylation-based estimates again correlated well with the expression levels of marker genes (Fig. 1E and F).
On the basis of these analyses, we excluded three samples profiled using DNA methylation arrays with low purity (less than 35% tumor) from subsequent analyses (Supplementary Table S1).
Histotype differences reflect menstrual cycle differences
UMAP showed that tumors largely clustered by annotated histologies with a few exceptions (Supplementary Fig. S2). A sample with both gene expression and DNA methylation data were annotated as ENOC, but consistently showed clustering with CCOC for both data types (Supplementary Fig. S2). Pathology report review revealed documentation ambiguity in this sample and in three others; these samples were excluded from subsequent analyses (Supplementary Table S2).
To provide a reference point for the ovarian cancer datasets, we included transcriptomic data from two normal tissue types: FTE and endometrium (Supplementary Table S2). When the UMAP included these additional normal samples, the FTE samples clustered with HGSOC (Fig. 2A); this is expected, as FTE has been suggested as the cell of origin for HGSOC. The normal endometrium, however, unexpectedly split into two clusters, with one group of endometrial samples clustering with CCOC and the other cluster with ENOC (Fig. 2A). Examination of all covariates associated with normal endometrium revealed that this split correlated with the menstrual cycle phase of the endometrium: the mid-secretory endometrium (secE) clustered with CCOC, whereas the proliferative endometrium (proE) clustered with ENOC.
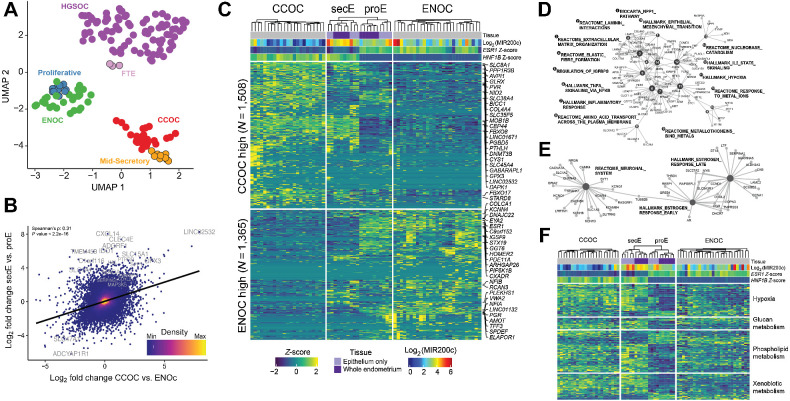
Figure 2.
CCOC and ENOC resemble different phases of normal cyclic endometrium based on gene expression profile. A, RNA-seq UMAP shows tumors clustering by histotype and with putative corresponding normal cell(s) of origin. Each dot represents a sample, with colors indicating sample type. B, Globally, CCOC–ENOC expression differences for all genes (log2-fold change, x-axis) are positively correlated with fold change between secretory and proliferative endometrium (y-axis). Top 20 DEGs by P value for secE versus proE are labeled. C, Significant DEGs between CCOC and ENOC also separate endometrium of different phases (FDR <0.05 and absolute fold change >2). Each row represents a DEG and each column a sample. Both rows and columns are clustered by Euclidian distance after first grouping by fold change sign (rows) as well as into CCOC, ENOC, and endometrium (columns). Note that secE and proE separate perfectly based on CCOC versus ENOC DEGs. Gene expression (Log2 RPKM) is row normalized into Z-scores, capped at ±2. The 25 most up- and downregulated genes by P value are labeled. D, Gene-concept network plot showing enriched molecular pathways for genes upregulated in both CCOC relative to ENOC and secE relative to proE. Numbered nodes represent pathways, with DEGs in that pathway connected to the corresponding node. The size of each node is scaled on the basis of the number of overlapped DEGs in that pathway. E, As in D, but for pathways enriched in genes overexpressed in ENOC relative to CCOC and proE relative to secE. F, CCOC and ENOC share key metabolic pathways with secE and proE, respectively. Color of the heatmap represents Z-score as in C for genes (rows) from four pathways significantly overrepresented in DEGs between secretory and proliferative endometrium.
Globally, CCOC–ENOC differences correlated with normal secE-proE differences (Spearman ρ = 0.31, P < 2.2e-16; Fig. 2B). Next, we identified DEGs between CCOC and ENOC, with absolute fold changes greater than 2 (N = 2,873; FDR < 0.05; Supplementary Table S3). When clustered on these genes, normal endometrium samples again clustered by menstrual cycle phase, with expression patterns in proE reflecting ENOC and secE reflecting CCOC (Fig. 2C). This included differential expression of estrogen receptor 1 (ESR1), progesterone receptor (PGR), and HNF1B.
Next, we identified genes upregulated in both CCOC relative to ENOC (Supplementary Table S3) and secE relative to proE (Supplementary Table S4) and tested them for enrichment of biological pathways using the MSigDB C2 collections (Fig. 2D). CCOC and secE shared upregulation of several hallmark pathways, such as epithelial to mesenchymal transition (Fig. 2D; node #14), hypoxia (node #11), inflammatory response (node #7), and extracellular matrix organization (node #3). ENOC- and proE-shared upregulated genes were involved in early and late estrogen responses (Fig. 2E). Expression across key pathways characteristic of CCOC/ENOC differences, such as hypoxia, glucan, phospholipid, and xenobiotic metabolism, also showed similar parallelism between the cancer and normal tissue subgroups (Fig. 2F), emphasizing that these known histotype differences can be at least partially explained by cell states corresponding to menstrual cycle phases and are not necessarily cancer-specific.
Validation of HNF1B expression in normal endometrium
Expression of HNF1B has been deemed as a central feature of CCOC cancer cells, but our data suggest that it is not cancer-specific, but rather tied to specific menstrual phases of normal endometrium. To validate this, we used a public microarray-based normal endometrium RNA expression dataset (40) with annotated menstrual cycle phase information. In this orthogonal external dataset, HNF1B showed menstrual cycle variation, with the highest HNF1B expression level in the mid-secretory phase (Fig. 3A). ESR1 is well established to have prominent expression in the glandular epithelium in the proliferative and early secretory phases (55, 56) and is included as a control. We validated HNF1B protein expression by IHC in the normal human endometrium, independently dated during pathology review (Fig. 3B; ref. 33). This confirmed strong mid-secretory expression, consistent with the mRNA results. Specifically, HNF1B protein expression was low in the proliferative and early secretory phases but became positive in the mid- and late-secretory phases (Fig. 3C; P = 0.002, Fisher exact test between proliferative and mid/late secretory). Finally, images from the human protein atlas of the two available HNF1B antibodies validated HNF1B expression in the normal secretory endometrium (proteinatlas.org; Fig. 3D; ref. 57).
Epigenetic mechanisms lock in cellular states
Transcriptional state per se is not heritable through cell division. Genetic or epigenetic alterations are required to propagate transcriptional states from parent to daughter cells during tumor cell proliferation. Therefore, to understand how initial cell states can be maintained during tumor initiation and progression, we examined DNA methylation in different histotypes. We identified 1,339 DMRs between CCOC and ENOC; 1,018 were hypermethylated in CCOC, and 321 were hypermethylated in ENOC (FDR <0.05; Supplementary Table S5).
With these DMRs, we used LOLA (58) to test for the enrichment of transcription factor binding sites (TFBS) separately in CCOC hypermethylated DMRs or ENOC hypermethylated DMRs (Supplementary Table S6). Sites of higher methylation in CCOC than in ENOC were enriched for binding sites of estrogen receptor (ER; ref. Fig. 4A) and other TF related to ER signaling, such as FOXA1/2 and GATA3 (59).
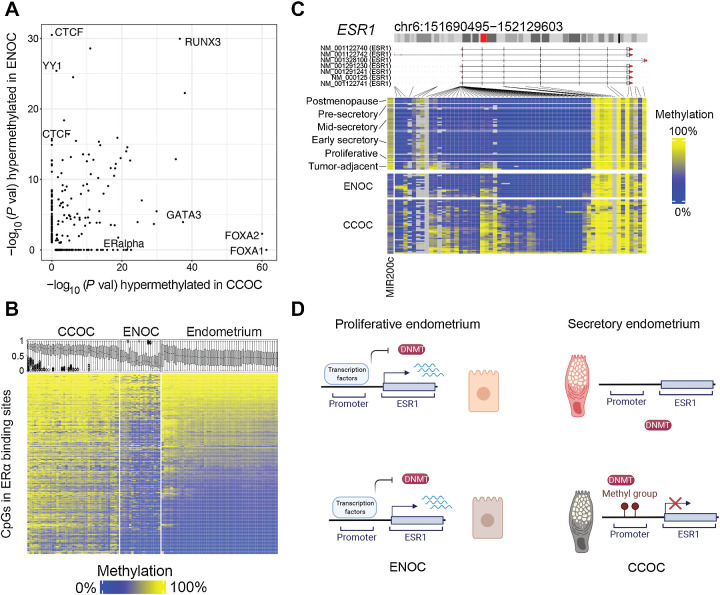
Figure 4.
Epigenetic differences between CCOC and ENOC reveal how cell state differences are propagated through mitosis. A, −Log10 (P value) for TFBS enrichment for probes hypermethylated in ENOC compared with CCOC (y-axis) and CCOC compared with ENOC (x-axis). Each dot represents a TFBS region set. Labels are shown for region sets related to chromatin architecture (enriched for hypermethylation in ENOC) and those related to estrogen signaling (enriched for hypermethylation in CCOC). B, Heatmap of probes (rows) overlapping ERα TFBSs sorted by average methylation in endometrium. Boxplots show the methylation distribution for these probes for samples (columns). CCOC shows gain of methylation at ERα binding sites compared with normal endometrium. C, In addition, the promoter of ERα’s encoding gene, ESR1, gains methylation in CCOC at most probes around the transcription start site; ESR1 transcription start site is unmethylated in normal endometrium. D, A model for ESR1 promoter methylation in normal endometrium and tumors. In normal endometrium, regardless of the phase, the ESR1 promoter is unmethylated, which allows for cyclic modulation by transcription factors through normal monthly cycling. In the cell of origin of CCOC, which is likely secretory endometrium-like, ESR1 is not expressed, and DNA methylation can accumulate stochastically and then become clonally expanded.
Indeed, ENOC had much lower methylation across regions identifiable as ER-binding sites (Fig. 4B), consistent with the overactivation of ER signaling in this histotype. In contrast, CCOC contained the majority of methylated ER-binding sites. In addition, the estrogen receptor (ESR1) gene was methylated across the CCOC samples (Fig. 4C). This promoter hypermethylation appeared to be cancer-specific, as the region was uniformly unmethylated in the normal endometrium of all phases, despite the modulation in transcription level through the menstrual cycle (some methylation in tumor-adjacent normal is presumably due to tumor contamination or field effects). This lack of methylation in the normal endometrium likely provides a permissive state that allows for maximum flexibility during normal cycling (Fig. 4D, top). Active expression repels the DNA methyltransferase (DNMT) machinery (60), whereas periods of low or no expression may allow for aberrant accumulation of methylation at the ESR1 promoter in CCOC or its progenitors. In this model (Fig. 4D, bottom), DNA methylation at both the ESR1 promoter likely inhibited the response to estrogen signaling in CCOC precursors and restricted cells to a secretory-like state. Extensive DNA methylation at ER binding sites suggests inactive ER-associated regulatory elements in CCOCs.
Transcriptomic comparison with corresponding normal tissue types for cancer-specific changes
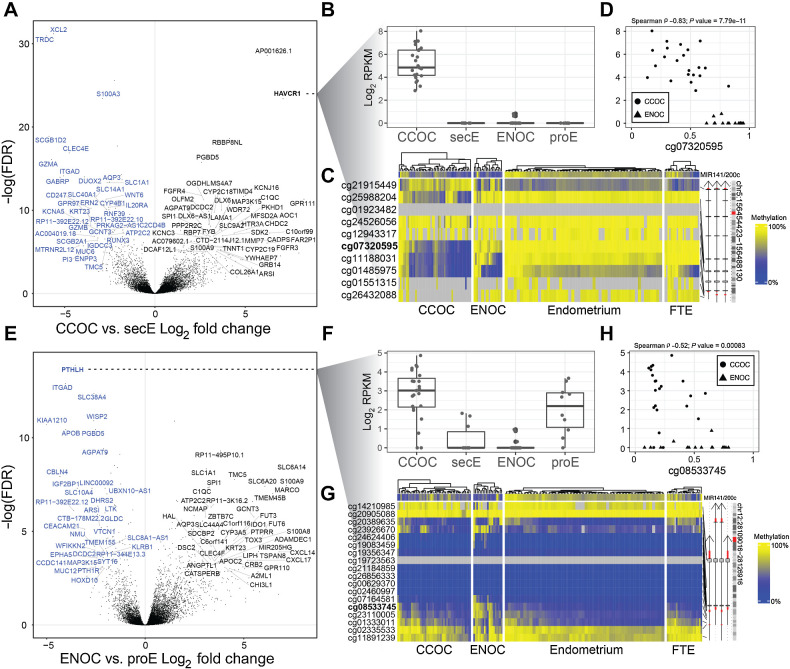
To isolate cancer-specific transcriptional changes, we compared each cancer type to the corresponding normal tissue and cell state that it most resembled: CCOC versus secE, and ENOC versus proE (Supplementary Tables S7 and S8). In this analysis, Hepatitis A virus receptor/kidney injury molecule 1 (HAVCR1) was the most overexpressed protein-coding gene in CCOC (Fig. 5A). HAVCR1 was not expressed in the normal endometrium in either phase, and its expression was limited or absent in ENOC (Fig. 5B). Likewise, the promoter region for HAVCR1 was unmethylated in CCOC compared with that in ENOC and other normal tissue types in the female reproductive tract (Fig. 5C), and the expression level was inversely correlated with DNA methylation levels (Fig. 5D). Residual methylation in CCOC appears to be attributable to the presence of noncarcinoma cells in the bulk tumor (as measured by MIR200C promoter methylation level in Fig. 5C) and increased as tumor purity decreased, suggesting consistent clonal loss of methylation of HAVCR1 across CCOC. Other top upregulated transcripts included LINC01671 (AP001626.1), RBBP8 N-terminal like (RBBP8NL), and laminin subunit alpha 1 (LAMA1), among many others. FGF receptor 3 (FGFR3) was also consistently upregulated in CCOC compared with secE, consistent with the dense stroma observed in CCOC.
Figure 5.
Comparison with matched normal yields cancer-specific alterations. A, Volcano plot showing gene expression alterations between CCOC and secE. X-axis is log2-fold change and y-axis is negative log10-transformed FDR. Top ranked DEGs are labeled in black (upregulated in CCOC relative to secE) and blue (downregulated). B,HAVCR1 is expressed only in CCOC samples; lines indicate first quartile, median, and third quartile. C,HAVCR1 shows demethylation in three probes near the transcription start site in CCOC samples only. D, For samples with matched expression and methylation data, HAVCR1 expression corresponds to methylation at the transcription start site as illustrated by the probe cg07320595. E, As in A, but for ENOC vs. proE. F, Expression by sample group boxplots for PTHLH showing that this gene is downregulated in ENOC relative to proE but upregulated in CCOC relative to secE. These demonstrate that using a matched normal is critical because the merged signals from whole endometrium without regard to phase would obscure the change in ENOC relative to its putative cell state of origin. G, Methylation at the PTHLH gene shows some hypermethylation at the transcription start site in ENOC, but this does not fully explain the expression. H, As in D but for PTHLH. Athough expression is significantly associated with methylation at the transcription start site probe cg08533745, it does not seem to fully explain suppressed expression of PTHLH in ENOC samples.
Top downregulated genes in CCOC compared with secE include T Cell Receptor Delta Constant (TRDC), a surface marker for γδ T cells; chemokine (C motif) ligand 2 (CXL2), a chemokine expressed in activated cells; granzyme A (GZMA) and granzyme B (GZMB), characteristic genes of cytotoxic cells. We also examined the expression levels of key marker genes for immune cells and related cell types in CCOC, ENOC, and normal endometrium (Supplementary Fig. S3). The microenvironment of CCOC was similar to that of secE, both featuring an abundance of endothelial cells. However, consistent with the granzyme results, killer cell immunoglobulin-like receptor (KIR) genes (KIR2DL4, KIR2DL1, KIR2DS4, etc.) were significantly lower in CCOC than in secE (Supplementary Fig. S3). It appears that although CCOC largely resembles secE in terms of cell composition, its microenvironment is characterized by a lack of activated cytotoxic cells.
For the ENOC-proE comparison (Fig. 5E), the top up genes were enriched for solute carriers (SLC), including SLC6A14, which is responsible for non-polar amino acids, and SLC6A20, a proline transporter. Potassium channel genes (KCN) were also visibly enriched for the top expressed genes and resulted in a single biological process term enriched for ENOC-proE DEGs: chronic inflammatory response (P = 2.7e−5), and nine molecular function terms were enriched (FDR <0.05), including RAGE receptor binding, carboxylic acid transporter activity, Toll-like receptor binding, and long-chain fatty acid binding. The most downregulated gene in ENOC compared with proE was parathyroid hormone-like hormone (PTHLH), which encodes a parathyroid hormone-related protein (PTHrP). Interestingly, PTHLH was upregulated in CCOC (Fig. 5F). This downregulation of ENOC appeared to be mediated by DNA hypermethylation (Fig. 5G and H). Related to this, the receptor of PTHrPs, PTH1R was also 10 times lower in ENOC than in CCOC (Supplementary Table S3).
Notably, only a very small subset of genes was consistently up- or downregulated (39 and 8 genes, respectively) in both histotypes compared with their normal counterparts (Supplementary Fig. S4). Most of these common genes appeared to be associated with cell type differences in the microenvironment (e.g., GZMA), instead of tumor cell-specific changes. Taken together, the oncogenic pathways are likely divergent in these two histotypes.
Pathway enrichment for CCOC–ENOC contrasted with secE–proE comparisons
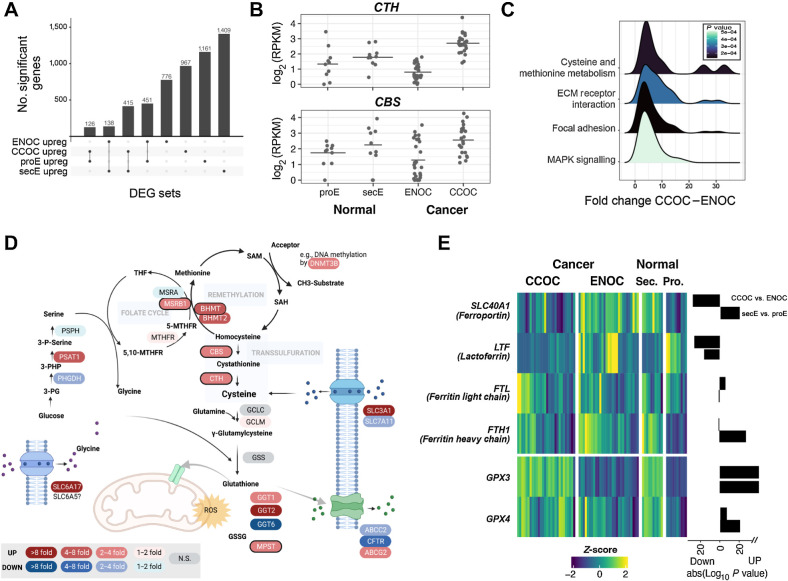
We reasoned that dissecting the differences between CCOC and ENOC into those shared with the normal secE–proE difference, and those that were unique to CCOC and ENOC, will help better delineate key molecular drivers of tumorigenesis for both histotypes. Genes upregulated in CCOC relative to ENOC (fold change >2, FDR <0.05) were three times as likely to overlap with those upregulated in secE, compared with those upregulated in proE. Similarly, genes upregulated in ENOC were three times more likely to overlap with those upregulated in proE (Fig. 6A).
Figure 6.
CCOC–ENOC differences beyond proEM–secEM differences highlight alterations in cysteine/methionine biogenesis and iron metabolism. A, Overlap of genes upregulated in ENOC (relative to CCOC), CCOC (relative to ENOC), proEM (relative to secEM), and secEM (relative to proEM) showing the shared gene sets used for enrichment testing. B,CTH and CBS are two highly differentially expressed genes between ENOC and CCOC, whereas not different between two phases of normal endometrium. C, Distribution of fold changes for genes in the four significantly enriched KEGG pathways upregulated in CCOC. D, Cysteine synthesis pathway with the fold change between CCOC and ENOC indicated for each expressed gene. E, Heatmap of iron and ferroptosis-related genes showing Z-score and absolute log10 (P value) for CCOC vs. ENOC and secEM vs. proEM, with direction indicated as a barplot on the right of the heatmap.
In addition to those shared with secE, 967 genes were upregulated exclusively in CCOC. These genes were enriched for only two biological process terms: homocysteine metabolism and regulation of anatomic structure. In particular, cystathionine gamma-lyase (CTH) was upregulated 3.9 times (FDR = 5.1e−12) and cystathionine β-synthase (CBS) was upregulated 2.2 times (Fig. 6B; FDR = 0.0045) compared with ENOC (Supplementary Table S3). Both CTH and CBS are key genes involved in cysteine synthesis via homocysteine transulfuration. Interestingly, in the CCOC–ENOC comparison without considering how secE and proE compared, cysteine and methionine metabolism was the top enriched KEGG pathway (Fig. 6C; Supplementary Table S9). Given these two results indicating the importance of cysteine synthesis, we explored the expression behavior of all genes involved in cysteine/glutathione metabolism and homeostasis (Fig. 6D). Strikingly, 22 of 24 genes in these pathways exhibited a significant difference between CCOC and ENOC (Supplementary Table S10), with many genes mirroring the difference between secE and proE (Supplementary Fig. S5). The cysteine transporter SLC3A1 (rBAT) was also nine times upregulated in CCOC compared with ENOC (FDR = 1.8E−16; Supplementary Fig. S5). The increased cysteine influx and biogenesis of cysteine converged, highlighting the importance of cysteine in CCOC. Interestingly, the other cysteine transporter, SLC7A11 (xCT), was downregulated by over two-fold in CCOC (FC = 0.41, P = 9E−7). This downregulation was significant, considering that SLC7A11 expression was 4-fold higher in secE than in proE cells. Unlike rBAT, xCT takes up cysteine via a 1:1 exchange of glutamate, which might not be favored by cells if glutamate is needed.
In addition, γ-glutamyl transpeptidase (GGT) cleaves extracellular glutathione such that cells have access to more cysteine, and the expression level of GGT is directly related to cisplatin treatment in prostate cancer (61). Interestingly, GGT1 and GGT2 were upregulated in CCOC and secE, whereas GGT6 was significantly downregulated (Supplementary Table S10). The glycine importer SLC6A17 was also highly upregulated in a few CCOC samples, whereas the glutathione exporter CFTR was downregulated (Supplementary Table S10), likely indicating a dependence on increased intracellular glutathione levels. Indeed, GLRX (Glutaredoxin) was the 11th most differentially expressed gene between CCOC and ENOC (FDR = 1.5E−33; fold change = 5.5; Supplementary Table S3).
Closely related to this observation, CCOC and ENOC also exhibited striking differences in iron storage and transportation (Fig. 6E). The iron antiporter ferroportin (SLC40A1) was 5-fold downregulated (P < 0.0001) in CCOC, despite secE expressing far more SLC40A1 than proE. This apparent “switch” highlights the importance of shutting down iron outflow for CCOC. In addition, lactoferrin (LTF) was 14-fold downregulated in CCOC compared with ENOC. Transferrin (TF) was 1.8-fold downregulated (unadjusted P = 0.04), and ferritin light chain (FTL) was 1.65-fold higher (unadjusted P = 0.003) in CCOC. These suggest iron addiction in CCOC. Human glutathione peroxidase 4 (GPX4), which prevents cells from entering ferroptosis with iron-induced ROS, was highly expressed in CCOC and secE together with the closely related GPX3, consistent with a high-iron state in CCOC.
Discussion
The study of cells-of-origin is an important aspect of cancer research (2). Traditionally, cell type has been the focus of research for the cell-of-origin for a particular cancer type. It is believed that cells-of-origin and genetic mutations jointly shape the characteristics of the cancer cells (1). However, in the case of ENOC and CCOC, which share the same cell-of-origin and have overlapping mutational profiles, how they yield phenotypically different cancer entities is an intriguing question.
Our results suggest that the same cell type in different cell states—endometrial or endometriotic progenitor/stem cells in proliferative and mid-to-late secretory phases—are likely associated with different transformation paths towards ENOC and CCOC, respectively. This offers a potential explanation for the presence of molecular and/or histologic subtypes of cancers arising in many different organs (1), in which the progenitor cell may have been “locked” at different cellular states that the particular cell lineage can adopt. These cells share the same functional type, and similar epigenetic profiles, but upon response to external signals—such as hormones—can adopt a different transcriptional state, reversible upon signal withdrawal. Deposition of epigenetic marks, such as DNA methylation, can be influenced by the current transcriptional state. Active transcription repels the DNA methylation machinery, whereas the latter can deposit the methyl mark to promoters of genes that are not expressed (60), subsequently “locks in” the unexpressed state, providing mitotically heritable variation for selection during clonal evolution. In the case of CCOC, the lack of ESR1 transcription in the secretory state permits stochastic deposition of DNA methylation at this promoter. This DNA methylation gain persists through mitotic division and prevents transcriptional changes in response to estrogen. Frequent clonal loss of the chromatin remodeler ARID1A in CCOC/ENOC may also reflect an oncogenic advantage for the cell-of-origin to not respond to such extrinsic signals and somehow stay “locked in” to existing cell states (62). We discuss this model in a cancer type that arise from the female reproductive tract, which exhibits exceptional plasticity, with both monthly modulation and remodeling during pregnancy. Nonetheless, cells in other tissues can also adopt various states under normal conditions, and subtypes arising in these other organ sites can be explained by a difference in initial cell state upon transformation, which is subsequently maintained through mitotic divisions by epigenetic mechanisms. For example, breast carcinoma histotypes may arise through a similar mechanism.
The cell state difference associated with the menstrual cycle offers a plausible explanation for the many known differences between CCOC and ENOC. The most well-known characteristics of CCOC that differentiate it from ENOC are (i) hobnail appearance, (ii) glycogen-filled cytoplasm, (iii) HNF1B expression, and (iv) resistance to chemotherapy. Accordingly, (i) the hobnail appearance is often seen as part of the Arias-Stella reaction in secretory (and gestational) endometrium (63). (ii) Intracellular glycogen concentration is also known to be low in proliferative endometrium and increases by over 10-fold by the early secretory phase (64). (iii) Our study validated the CCOC diagnostic biomarker HNF1B expression, both RNA and protein, in the mid-secretory endometrium and showed it to be absent in the proliferative endometrium. (iv) Resistance to chemotherapy may be explained, in part, by upregulated xenobiotic metabolism in the CCOC and secretory endometrium. On the other hand, the similarity between ENOC and proliferative endometrium also makes immediate sense—“endometrioid” literally means endometrium-like, and cancer represents a heightened proliferative state. Consistent with the biological similarities between ENOC and proE, progesterone treatment, which induces exit from the proliferation phase, can reduce the survival of primary cultures of endometrioid ovarian cancer (65). Hormone receptor positivity (66) and hormone responsiveness are well recognized in ENOC, and targeting is not uncommon. Nonetheless, 20% of ENOC are ER-negative (66), likely representing a further change from this base state.
The separation of these normal cell features from cancer-specific alterations is helpful to better define how cancer develops and point to true cancer-specific changes. HNF1B has been recognized as the most important CCOC marker, but here we show it is also expressed in the normal secretory endometrium. Instead, our analyses highlighted clonal loss of HAVCR1 promoter DNA methylation as a potential driver. Germline alterations in HAVCR1 are common in early-onset clear cell renal cell carcinoma (ccRCC), and elevated expression promotes angiogenesis via IL6 (67, 68). The IL6/STAT3/HIF1A axis has been identified as a key pathway in CCOC (69). In our data, IL6 RNA expression was 14.7 times higher on average in CCOC than in ENOC (Supplementary Table S3). The mechanism of HAVCR1 overexpression in ccRCC has been elusive, with only gene amplification examined and excluded (67). Our results suggest that DNA demethylation is a potential mechanism for the high expression observed in CCOC, and could also be responsible for HAVCR1 overexpression observed in other clear cell tumors. In addition, PTHLH was the top downregulated gene for ENOC. PTHLH was identified in a study of humoral hypercalcemia of malignancy (HHM), a paraneoplastic syndrome in which elevated levels of PTHrP lead to increased osteoclastic bone resorption and serum calcium levels. High expression of PTHLH in CCOC has been reported previously (70) and implicated in other cancer types (71). However, its promoter hypermethylation and associated transcriptional downregulation have not yet been reported, as in the case of ENOC. Parathyroid hormone 1 receptor (PTH1R) was also highly downregulated in ENOC. These suggest that HHM may be a CCOC-specific phenomenon, and the contrast between CCOC and ENOC with regard to this pathway warrants further exploration.
Our analysis also highlights a key therapeutic vulnerability of CCOC. On the basis of transcriptional analysis, this histotype demonstrates an apparent dependence on cysteine and iron. Endometriotic cysts contain chocolate-colored fluids from the menstruation-like blood. ENOC and CCOC appear to adopt different approaches to address the abundance of iron in the microenvironment. Although ENOC appears to keep iron out of the cells, likely with E2-driven intracellular iron efflux (72), we hypothesize that CCOC accumulates iron and likely relies on cysteine to counteract the high intracellular iron, based on gene expression profiles. Consistent with this hypothesis, a recent study showed that a subset of stromal cells with elevated expression of iron export proteins donate iron to the associated CCOCs (73). Increased iron content in cancer cells is associated with resistance to chemotherapy, a known feature of CCOC, and creates an attractive therapeutic target. A recent study (74) screened four clear cell ovarian cancer cell lines and showed that cysteine inhibition leads to ferroptosis in these cells. Another recent study (75) showed that cysteine deprivation resulted in cell death via oxidative stress and iron-sulfur cluster biogenesis deficits. Both these studies were done in cell lines only, but validated the key pathways discovered in our analysis based on primary human tumors. Our analysis provides transcriptomic explanations for these experimental results, and further underscores the importance of cysteine and iron metabolism in targeting CCOC, for which better therapeutic options are sorely needed. This dependency on cysteine and ferroptosis appears to be central to clear cell carcinomas across tissue types (76).
Finally, CCOC and ENOC both resembled their corresponding normal tissues (secE and proE, respectively) not only in transcriptional states, but also in cellular composition of the microenvironment. For example, we showed CCOC and secE were both rich in endothelial cells, compared with ENOC and proE. CCOC and secE only appeared to differ in terms of cellular composition was the lack of activated cytotoxic cells, such as NK cells (Supplementary Fig. S3). The mechanism for NK inactivation in these tumors may be worth exploring. Other aspects of the tumor microenvironment and how they might affect the fate choice are also interesting questions. In particular, stroma is a potent regulator of the epithelial state, particularly in the female reproductive tract. The type and amount of stroma associated with endometriotic cells in establishing the lesion may affect the fate of epithelial cells. Because ENOC and CCOC display important differences in genes involved in iron homeostasis, the abundance of iron in the lesion may also contribute to fate decisions. The type of endometriosis (e.g., deep infiltrating, endometrioma, superficial; ref. 77) might also affect the histotype choice. Although not examined in the current study, these are interesting leads for further studies.
Supplementary Material
Figures S1-S5
Supplementary Tables S1-10
Acknowledgments
This research was supported by an NCI grant (R37CA230748) to H. Shen. K. Heinze was funded through a research scholarship by the Deutsche Forschungsgesellschaft (HE 8699/1–1).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
K. Heinze reports grants from Deutsche Forschungsgesellschaft during the conduct of the study. A. Leonova reports personal fees from Aima Laboratories outside the submitted work. C.L. Pearce reports grants from NIH and DoD during the conduct of the study and personal fees from Ovarian Cancer Research Alliance outside the submitted work. M.S. Anglesio reports grants from Michael Smith Health Research BC and NIH during the conduct of the study. H. Shen reports other support from FOXO Biotechnologies and AnchorDx outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
I. Beddows: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. H. Fan: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft. K. Heinze: Validation, methodology, writing–original draft, writing–review and editing. B.K. Johnson: Methodology, writing–original draft. A. Leonova: Formal analysis, validation, investigation, writing–original draft. J. Senz: Validation, investigation, methodology, writing–original draft. S. Djirackor: Investigation, writing– draft. K.R. Cho: Investigation, writing–original draft, writing–review and editing. C.L. Pearce: Investigation, writing–original draft, writing–review and editing. D.G. Huntsman: Supervision, investigation, writing–original draft. M.S. Anglesio: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing. H. Shen: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018;173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Visvader JE. Cells of origin in cancer. Nature 2011;469:314–22. [DOI] [PubMed] [Google Scholar]
- 3. Karnezis AN, Cho KR, Gilks CB, Pearce CL, Huntsman DG. The disparate origins of ovarian cancers: pathogenesis and prevention strategies. Nat Rev Cancer 2017;17:65–74. [DOI] [PubMed] [Google Scholar]
- 4. Kobel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 2008;5:e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shen H, Fridley BL, Song H, Lawrenson K, Cunningham JM, Ramus SJ, et al. Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun 2013;4:1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shih Ie M, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol 2004;164:1511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Landrum LM, Java J, Mathews CA, Lanneau GS Jr, Copeland LJ, Armstrong DK, et al. Prognostic factors for stage III epithelial ovarian cancer treated with intraperitoneal chemotherapy: a gynecologic oncology group study. Gynecol Oncol 2013;130:12–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee AW, Navajas EE, Liu L. Clear differences in ovarian cancer incidence and trends by ethnicity among Asian Americans. Cancer Epidemiol 2019;61:142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Craig O, Nigam A, Dall GV, Gorringe K. Rare epithelial ovarian cancers: low grade serous and mucinous carcinomas. Cold Spring Harb Perspect Med 2023;13:a038190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheasley D, Wakefield MJ, Ryland GL, Allan PE, Alsop K, Amarasinghe KC, et al. The molecular origin and taxonomy of mucinous ovarian carcinoma. Nat Commun 2019;10:3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubeau L, Drapkin R. Coming into focus: the nonovarian origins of ovarian cancer. Ann Oncol 2013;24Suppl 8(Suppl 8):viii28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dubeau L. The cell of origin of ovarian epithelial tumours. Lancet Oncol 2008;9:1191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anglesio MS, Bashashati A, Wang YK, Senz J, Ha G, Yang W, et al. Multifocal endometriotic lesions associated with cancer are clonal and carry a high mutation burden. J Pathol 2015;236:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saunders PTK, Horne AW. Endometriosis: etiology, pathobiology, and therapeutic prospects. Cell 2021;184:2807–24. [DOI] [PubMed] [Google Scholar]
- 15. Taylor HS, Kotlyar AM, Flores VA. Endometriosis is a chronic systemic disease: clinical challenges and novel innovations. Lancet 2021;397:839–52. [DOI] [PubMed] [Google Scholar]
- 16. Zondervan KT, Becker CM, . MSA Endometriosis. N Engl J Med 2020;382:1244–56. [DOI] [PubMed] [Google Scholar]
- 17. Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010;330:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010;363:1532–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anglesio MS, Papadopoulos N, Ayhan A, Nazeran TM, Noe M, Horlings HM, et al. Cancer-associated mutations in endometriosis without cancer. N Engl J Med 2017;376:1835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lac V, Verhoef L, Aguirre-Hernandez R, Nazeran TM, Tessier-Cloutier B, Praetorius T, et al. Iatrogenic endometriosis harbors somatic cancer-driver mutations. Hum Reprod 2019;34:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Praetorius TH, Leonova A, Lac V, Senz J, Tessier-Cloutier B, Nazeran TM, et al. Molecular analysis suggests oligoclonality and metastasis of endometriosis lesions across anatomically defined subtypes. Fertil Steril 2022;118:524–34. [DOI] [PubMed] [Google Scholar]
- 22. Suda K, Nakaoka H, Yoshihara K, Ishiguro T, Tamura R, Mori Y, et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep 2018;24:1777–89. [DOI] [PubMed] [Google Scholar]
- 23. Bartlett TE, Jones A, Goode EL, Fridley BL, Cunningham JM, Berns EM, et al. Intra-Gene DNA methylation variability is a clinically independent prognostic marker in women's cancers. PLoS One 2015;10:e0143178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bodelon C, Killian JK, Sampson JN, Anderson WF, Matsuno R, Brinton LA, et al. Molecular classification of epithelial ovarian cancer based on methylation profiling: evidence for survival heterogeneity. Clin Cancer Res 2019;25:5937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cunningham JM, Winham SJ, Wang C, Weiglt B, Fu Z, Armasu SM, et al. DNA methylation profiles of ovarian clear cell carcinoma. Cancer Epidemiol Biomarkers Prev 2022;31:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cochrane DR, Tessier-Cloutier B, Lawrence KM, Nazeran T, Karnezis AN, Salamanca C, et al. Clear cell and endometrioid carcinomas: are their differences attributable to distinct cells of origin? J Pathol 2017;243:26–36. [DOI] [PubMed] [Google Scholar]
- 27. Kolin DL, Dinulescu DM, Crum CP. Origin of clear cell carcinoma: nature or nurture? J Pathol 2018;244:131–4. [DOI] [PubMed] [Google Scholar]
- 28. Tsuchiya A, Sakamoto M, Yasuda J, Chuma M, Ohta T, Ohki M, et al. Expression profiling in ovarian clear cell carcinoma: identification of hepatocyte nuclear factor-1 beta as a molecular marker and a possible molecular target for therapy of ovarian clear cell carcinoma. Am J Pathol 2003;163:2503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kobel M, Kalloger SE, Lee S, Duggan MA, Kelemen LE, Prentice L, et al. Biomarker-based ovarian carcinoma typing: a histologic investigation in the ovarian tumor tissue analysis consortium. Cancer Epidemiol Biomarkers Prev 2013;22:1677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gounaris I, Brenton JD. Molecular pathogenesis of ovarian clear cell carcinoma. Future Oncol 2015;11:1389–405. [DOI] [PubMed] [Google Scholar]
- 31. Anglesio MS, Carey MS, Kobel M, Mackay H, Huntsman DG, Vancouver Ovarian Clear Cell Symposium S. Clear cell carcinoma of the ovary: a report from the first ovarian clear cell symposium, June 24th, 2010. Gynecol Oncol 2011;121:407–15. [DOI] [PubMed] [Google Scholar]
- 32. Karlan BY, Alvarez RD, StraussJF 3rd. Evolving paradigms in research and care in ovarian cancers. Obstet Gynecol 2016;128:771–4. [DOI] [PubMed] [Google Scholar]
- 33. Lac V, Nazeran TM, Tessier-Cloutier B, Aguirre-Hernandez R, Albert A, Lum A, et al. Oncogenic mutations in histologically normal endometrium: the new normal? J Pathol 2019;249:173–81. [DOI] [PubMed] [Google Scholar]
- 34. Campan M, Weisenberger DJ, Trinh B, Laird PW. MethyLight. Methods Mol Biol 2009;507:325–37. [DOI] [PubMed] [Google Scholar]
- 35. Zhou W, Triche TJ Jr, Laird PW, Shen H. SeSAMe: reducing artifactual detection of DNA methylation by infinium BeadChips in genomic deletions. Nucleic Acids Res 2018;46:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cherniack AD, Shen H, Walter V, Stewart C, Murray BA, Bowlby R, et al. Integrated molecular characterization of uterine carcinosarcoma. Cancer Cell 2017;31:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. George JW, Fan H, Johnson B, Carpenter TJ, Foy KK, Chatterjee A, et al. Integrated epigenome, exome, and transcriptome analyses reveal molecular subtypes and homeotic transformation in uterine fibroids. Cell Rep 2019;29:4069–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Betran AP, Ye J, Moller AB, Souza JP, Zhang J. Trends and projections of caesarean section rates: global and regional estimates. BMJ Glob Health 2021;6:e005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 2006;147:1097–121. [DOI] [PubMed] [Google Scholar]
- 41. Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, VL R, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015;521:489–94. [DOI] [PubMed] [Google Scholar]
- 46. Klinkebiel D, Zhang W, Akers SN, Odunsi K, Karpf AR. DNA methylome analyses implicate fallopian tube epithelia as the origin for high-grade serous ovarian cancer. Mol Cancer Res 2016;14:787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kukushkina V, Modhukur V, Suhorutsenko M, Peters M, Magi R, Rahmioglu N, et al. DNA methylation changes in endometrium and correlation with gene expression during the transition from pre-receptive to receptive phase. Sci Rep 2017;7:3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen W, Husain A, Nelson GS, Rambau PF, Liu S, Lee CH, et al. Immunohistochemical profiling of endometrial serous carcinoma. Int J Gynecol Pathol 2017;36:128–39. [DOI] [PubMed] [Google Scholar]
- 49. Hoang LN, McConechy MK, Meng B, McIntyre JB, Ewanowich C, Gilks CB, et al. Targeted mutation analysis of endometrial clear cell carcinoma. Histopathology 2015;66:664–74. [DOI] [PubMed] [Google Scholar]
- 50. Kalloger SE, Kobel M, Leung S, Mehl E, Gao D, Marcon KM, et al. Calculator for ovarian carcinoma subtype prediction. Mod Pathol 2011;24:512–21. [DOI] [PubMed] [Google Scholar]
- 51. Chi RA, Wang T, Adams N, Wu SP, Young SL, Spencer TE, et al. Human endometrial transcriptome and progesterone receptor cistrome reveal important pathways and epithelial regulators. J Clin Endocrinol Metab 2020;105:e1419–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ducie J, Dao F, Considine M, Olvera N, Shaw PA, Kurman RJ, et al. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat Commun 2017;8:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fan H, Atiya HI, Wang Y, Pisanic TR, Wang TH, Shih IM, et al. Epigenomic reprogramming toward mesenchymal-epithelial transition in ovarian-cancer-associated mesenchymal stem cells drives metastasis. Cell Rep 2020;33:108473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lessey BA, Killam AP, Metzger DA, Haney AF, Greene GL, McCarty KS Jr. Immunohistochemical analysis of human uterine estrogen and progesterone receptors throughout the menstrual cycle. J Clin Endocrinol Metab 1988;67:334–40. [DOI] [PubMed] [Google Scholar]
- 56. Lessey BA, Palomino WA, Apparao KB, Young SL, Lininger RA. Estrogen receptor-alpha (ER-alpha) and defects in uterine receptivity in women. Reprod Biol Endocrinol 2006;4 Suppl 1(Suppl 1):S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 58. Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and bioconductor. Bioinformatics 2016;32:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 2013;23:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 2015;517:640–4. [DOI] [PubMed] [Google Scholar]
- 61. Hanigan MH, Gallagher BC, Townsend DM, Gabarra V. Gamma-glutamyl transpeptidase accelerates tumor growth and increases the resistance of tumors to cisplatin in vivo. Carcinogenesis 1999;20:553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen H. Subclonal mutations in epigenetic regulators bring fitness in times of stress. Cancer Cell 2023;41:7–9. [DOI] [PubMed] [Google Scholar]
- 63. Dhingra N, Punia RS, Radotra A, Mohan H. Arias-Stella reaction in upper genital tract in pregnant and non-pregnant women: a study of 120 randomly selected cases. Arch Gynecol Obstet 2007;276:47–52. [DOI] [PubMed] [Google Scholar]
- 64. Milwidsky A, Palti Z, Gutman A. Glycogen metabolism of the human endometrium. J Clin Endocrinol Metab 1980;51:765–70. [DOI] [PubMed] [Google Scholar]
- 65. Pedernera E, Gomora MJ, Morales-Vasquez F, Perez-Montiel D, Mendez C. Progesterone reduces cell survival in primary cultures of endometrioid ovarian cancer. J Ovarian Res 2019;12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sieh W, Kobel M, Longacre TA, Bowtell DD, deFazio A, Goodman MT, et al. Hormone-receptor expression and ovarian cancer survival: an ovarian tumor tissue analysis consortium study. Lancet Oncol 2013;14:853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cuadros T, Trilla E, Sarro E, Vila MR, Vilardell J, de Torres I, et al. HAVCR/KIM-1 activates the IL-6/STAT-3 pathway in clear cell renal cell carcinoma and determines tumor progression and patient outcome. Cancer Res 2014;74:1416–28. [DOI] [PubMed] [Google Scholar]
- 68. Cuadros T, Trilla E, Vila MR, de Torres I, Vilardell J, Messaoud NB, et al. Hepatitis a virus cellular receptor 1/kidney injury molecule-1 is a susceptibility gene for clear cell renal cell carcinoma and hepatitis a virus cellular receptor/kidney injury molecule-1 ectodomain shedding a predictive biomarker of tumour progression. Eur J Cancer 2013;49:2034–47. [DOI] [PubMed] [Google Scholar]
- 69. Anglesio MS, George J, Kulbe H, Friedlander M, Rischin D, Lemech C, et al. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res 2011;17:2538–48. [DOI] [PubMed] [Google Scholar]
- 70. Yao M, Murakami T, Shioi K, Mizuno N, Ito H, Kondo K, et al. Tumor signatures of PTHLH overexpression, high serum calcium, and poor prognosis were observed exclusively in clear cell but not non clear cell renal carcinomas. Cancer Med 2014;3:845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li J, Karaplis AC, Huang DC, Siegel PM, Camirand A, Yang XF, et al. PTHrP drives breast tumor initiation, progression, and metastasis in mice and is a potential therapy target. J Clin Invest 2011;121:4655–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang Q, Jian J, Katz S, Abramson SB, Huang X. 17beta-Estradiol inhibits iron hormone hepcidin through an estrogen responsive element half-site. Endocrinology 2012;153:3170–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Atiya HI, Frisbie L, Goldfeld E, Orellana T, Donnellan N, Modugno F, et al. Endometriosis-associated mesenchymal stem cells support ovarian clear cell carcinoma through iron regulation. Cancer Res 2022;82:4680–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang WH, Huang Z, Wu J, Ding CC, Murphy SK, Chi JT. A TAZ-ANGPTL4-NOX2 axis regulates ferroptotic cell death and chemoresistance in epithelial ovarian cancer. Mol Cancer Res 2020;18:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Novera W, Lee ZW, Nin DS, Dai MZ, Binte Idres S, Wu H, et al. Cysteine deprivation targets ovarian clear cell carcinoma via oxidative stress and iron-sulfur cluster biogenesis deficit. Antioxid Redox Signal 2020;33:1191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun 2019;10:1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang Y, Nicholes K, Shih IM. The origin and pathogenesis of endometriosis. Annu Rev Pathol 2020;15:71–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S5
Supplementary Tables S1-10
Data Availability Statement
Transcriptome and methylation array data generated in this study were deposited in the NCBI Gene Expression Omnibus Series GSE226872 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE226872). All other raw data generated in this study are available upon request from the corresponding author. Publicly available data generated by others were used by the authors. Raw RNA-seq datasets of normal endometrium samples and normal epithelial brushings of the fallopian tube were downloaded from GSE132711 (51) and GSE114493 (52), respectively. Sequencing data for primary tumor tissues from HGSOC were downloaded from GSE102073 (52). Functional gene sets were downloaded from the Molecular Signatures Database (v7.2). Expression data shown in Fig. 3A were from GSE4888. The images presented in Fig. 3D were cropped from these images retrieved from the Human Protein Atlas with the links below, as orthogonal datasets to validate our discovery with two different antibodies: (1) Antibody 1 (CAB068192): 33-year-old female (Patient ID 2941) and (2) Antibody 2 (HPA002083): 27-year-old female (Patient ID 2004; https://www.proteinatlas.org/ENSG00000275410-HNF1B/tissue/endometrium#img).
Figure 3.
Validation of HNF1B expression in normal mid-secretory endometrium. A, Expression z-scores averaged from all probes for ESR1 and HNF1B on an external microarray-based dataset (GSE4888), validating expression of HNF1B in mid-to-late secretory phase. B, IHC staining of endometrium at various menstrual cycle phases for HNF1B. Control specimens used are tissues from endometrioid endometrial carcinoma (negative) and clear cell carcinoma of the ovary (positive) that have been previously described as being negative or positive for HNF1B (respectively). These were rerun in the same experimental lot as endometrium samples. Note that stromal cells further serve as internal negative control in the CCOC control specimen as staining is restricted to nuclei of tumor epithelium. C, Tabulation of HNF1B IHC results from a panel of 12 normal endometrium samples shows segregation by menstrual cycle phase. D, HNF1B IHC results from Human Protein Atlas, with two different antibodies, for two normal endometrium samples at the secretory phase showing strong staining for HNF1B.