Abstract
The hook-basal body (HBB) is a key intermediate structure in the flagellar assembly pathway in Salmonella typhimurium. The FlgM protein inhibits the flagellum-specific transcription factor ς28 in the absence of the intact HBB structure and is secreted out of the cell following HBB completion. The flk gene encodes a positive regulator of the activity of FlgM at an assembly step just prior to HBB completion: at the point of assembly of the P- and L-rings. FlgM inhibition of ς28-dependent class 3 flagellar gene transcription was relieved in P- and L-ring assembly mutants (flgA, flgH, and flgI) by introduction of a null mutation in the flk gene (J. E. Karlinsey et al., J. Bacteriol. 179:2389–2400, 1997). In P- and L-ring mutant strains, recessive mutations in flk resulted in a reduction in intracellular FlgM levels to those seen in wild-type (Fla+) strains. The reduction in intracellular FlgM levels by mutations in the flk gene was concomitant with a 10-fold increase in transcription of the flgMN operon compared to that of the isogenic flk+ strain, while transcription of the flgAMN operon was unaffected. This was true for both direct measurement of the flgAMN and flgMN mRNA transcripts by RNase T2 protection assays and for lac operon fusions to either the flgAMN or flgMN promoter. Loss of Flk did not allow secretion of FlgM through basal-body structures lacking the P- and L-rings. Intracellular FlgM was stable to proteolysis, and turnover occured primarily after export out of the cell. Loss of Flk did not result in increased FlgM turnover in either P- or L-ring mutant strains. With lacZ translational fusions to flgM, a null mutation in flk resulted in a significant reduction of flgM-lacZ mRNA translation, expressed from the class 3 flgMN promoter, in P- and L-ring mutant strains. No reduction in either flgAMN or flgMN mRNA stability was measured in the absence of Flk in Fla+, ring mutant, or HBB deletion strains. We conclude that the reduction in the intracellular FlgM levels by mutation in the flk gene is only at the level of flgM mRNA translation.
Salmonella typhimurium can propel itself in a liquid environment by the rotation of 6 to 12 individual flagellar structures that are located peritrichously on the cell surface. The signal transduction pathway of the chemosensory system allows for the biased movement of an individual bacterium across a chemical gradient, a process called chemotaxis (reviewed in references 2 and 43). The flagella can be signaled to rotate in a clockwise manner, which causes the bacterium to tumble and change direction, or if this signal is suppressed, they will rotate in a counterclockwise manner, which propels the bacterium in a forward direction. The flagella are biased toward counterclockwise rotation when approaching an attractant or moving away from a repellent and are biased toward clockwise rotation when moving away from an attractant or toward a repellent.
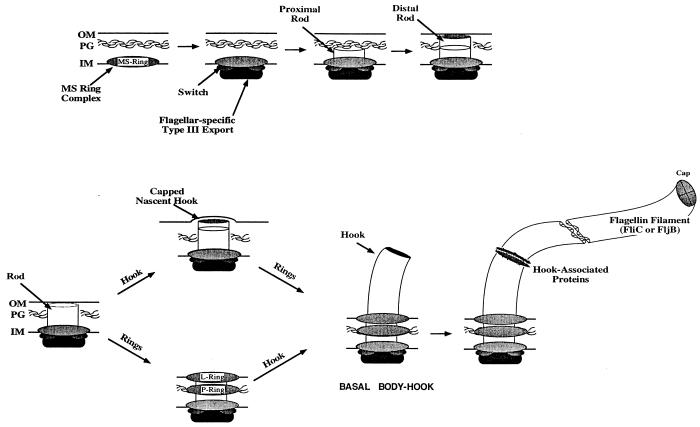
The flagellum is generally broken down into three main structural components: (i) the basal body, (ii) the hook, and (iii) the long external filament (for recent reviews of flagellar structure and assembly, see references 1 and 33). Several stages of flagellar assembly are diagrammed in Fig. 1. The basal body traverses from the cytoplasm to the outside of the cell. Assembly begins with formation of the C- and MS-rings at the cytoplasmic base of the basal body. The C-ring extends into the cytoplasm and includes a type III secretion machinery and proteins that control the direction of flagellar rotation in response to the chemotactic signal transduction system. The C-ring is attached to the MS-ring, which is embedded within the cytoplasmic membrane. The next structure to be assembled is the rod that extends from the MS-ring to the lipopolysaccharide (LPS) layer. This is followed by P- and L-ring assembly in the peptidoglycan and LPS layers, respectively, which must occur before an external hook structure can polymerize. Assembly of extracytoplasmic components of the hook-basal body (HBB) requires the type III secretion system, whereas P- and L-ring subunits are exported out of the cytoplasm via the signal peptide-dependent general secretory pathway. Initiation of hook assembly can occur prior to P- and L-ring assembly, but elongation is blocked by the peptidoglycan and LPS layers (21). The hook is thought to act as a universal joint between the rotating part of the basal body and the long external filament. Hook completion is determined by the fliK gene product, which somehow signals the type III export apparatus to stop the export of hook proteins and initiate export of the flagellin subunits and associated proteins (13, 25, 47). The flgK and flgL genes encode the hook-associated proteins, which form small subunit rings at the hook-filament junction (17). The fliD gene encodes a flagellar cap protein located at the tip of the flagellum (18). Flagellin is encoded by alternatively expressed fliC and fliB genes encoding antigenically distinct flagellin subunits (44). Flagellin subunits polymerize first between the cap and the FlgL ring and then between the cap and the tip of the growing filament.
FIG. 1.
Morphological pathway of flagellar assembly. MS-ring assembly in the inner membrane (IM) is followed by assembly of the switch complex and the flagellum-specific type III export apparatus. The rod components require the type III system to be exported across the inner membrane and into the periplasmic space, where they are assembled into a structure that penetrates the peptidoglycan layer (PG). The P- and L-rings (peptidoglycan and lipopolysaccharide, respectively) are assembled independently of hook initiation; however, hook elongation through the outer membrane (OM) requires the assembled P- and L-rings. After ring assembly, the hook is completed and the biosynthesis of the HBB intermediate structure is finished. At this point the class 3 flagellar proteins, including FlgM, are secreted. The hook-associated proteins and Cap are added to the end of the hook, followed by flagellin polymerization initially between the hook-associated proteins and Cap and then between the Cap and the elongating filament.
Regulation of the 50-plus genes in the flagellar and chemotaxis regulon occurs in coordination with flagellar assembly. The expression of these genes is organized into a regulatory hierarchy of three major classes: class 1, class 2, and class 3 (22). Class 1 genes, flhD and flhC, are cotranscribed and represent the top of the flagellar transcriptional hierarchy. They are transcribed from a ς70-dependent promoter, which is affected by a large number of global regulatory signals (28, 30, 31). The FlhD and FlhC proteins are required for expression of the rest of the genes in the regulon. These proteins form a heteromultimeric transcriptional activator complex which directs ς70-dependent transcription of promoters for class 2 genes (30). Class 2 genes encode proteins required for the structure and assembly of the HBB intermediate flagellar structure and the FlgM and ς28 regulatory proteins. Class 3 genes include ς28-dependent promoters and encode proteins required late in flagellar assembly, including the flgK, flgL, and fliD genes, and the filament genes fliC and fliB. In addition, genes required for chemotactic signal transduction are also class 3 genes.
This initial simplicity was complicated by the finding that multiple promoters transcribe most of the flagellar genes. The ς28 protein is an alternative transcription factor that interacts with RNA polymerase to direct transcription specifically from class 3 promoters but will also transcribe class 2 flagellar genes (23, 37). Several of the class 2 gene promoter regions have been shown to be transcribed by either ς28 holoenzyme or ς70 holoenzyme directed by the FlhDC complex (32). In vivo, all class 2 operons were shown to be dependent on either the flhDC operon or the ς28 structural gene, fliA, for expression (23). These results suggest that initiation of flagellar biosynthesis is completely dependent on FlhDC but that continued HBB gene expression results from FlhDC-dependent and/or ς28-dependent class 2 transcription. The class 3 structural genes are of two types; the flgK, flgL, and fliD genes are transcribed from both a class 2 promoter and a ς28-dependent class 3 promoter (26). The remaining class 3 structural genes are exclusively transcribed by ς28-dependent promoters (23).
The class 3 genes were originally defined by their dependence on a functional HBB structure for their expression (22). This ability to couple class 3 gene expression to HBB completion is accomplished by the action of the FlgM protein on ς28-dependent transcription (8, 9). FlgM is an anti-ς28 factor (38). When the HBB structure is incomplete or defective, FlgM interacts directly with ς28 to inhibit its activity as a transcription factor. FlgM can sense that the HBB structure is complete and export competent for external class 3 flagellar proteins by itself being a class 3 exported protein (16, 24). When the HBB structure is defective, FlgM inhibits ς28-dependent transcription and is not found in the external growth medium. In a strain that makes the HBB structure, FlgM is found in the external growth medium and ς28-dependent transcription occurs.
Recently, a novel flagellar regulatory gene, flk, was described as a gene whose product can sense the completion of the flagellar P- and L-rings of the basal body, which occurs just prior to hook assembly (20). The flgA and flgI genes are required for P-ring assembly, where the flgI gene encodes the structural component, and the flgH gene encodes the structural component for the L-ring (33). Like all genes involved in HBB assembly, mutations in any of the flgA, flgH, and flgI genes result in FlgM-dependent inhibition of ς28 activity (8). Unlike the other HBB genes, however, the negative regulatory effect of mutations in the flgA, flgH, or flgI gene can be suppressed by recessive insertion mutations in either flgM or flk (20). Insertions in flgM, but not those in flk, will suppress the negative regulatory effect caused by the loss of the remaining HBB genes (8, 20). In this paper, we provide evidence that the flk gene plays a role in the translational control of flgM gene expression.
MATERIALS AND METHODS
Strains.
The bacterial strains used in this study and their origins are listed in Table 1.
TABLE 1.
List of bacterial strains
| S. typhimurium strain | Genotype | Source or referencea |
|---|---|---|
| SJW198 | ΔflgH2136 fliBe,n,x hin(vh2) | S. Yamaguchi |
| SJW200 | ΔflgI2453 fla fliBe,n,x hin(vh2) | S. Yamaguchi |
| SJW203 | ΔflgHI958 fliBe,n,x hin(vh2) | S. Yamaguchi |
| SJW204 | ΔflgE1204 fliBe,n,x hin(vh2) | S. Yamaguchi |
| SJW1376 | ΔflgI1376 fliBe,n,x hin(vh2) | S. Yamaguchi |
| SJW1518 | Δfla-2157 (ΔflgG-L) fliBe,n,x hin(vh2) | S. Yamaguchi |
| SJW1525 | flgB2164 fliBe,n,x hin(vh2) | S. Yamaguchi |
| SJW1529 | ΔflgA1529 fliBe,n,x hin(vh2) | S. Yamaguchi |
| TH1479 | fliA5059::Tn10dTc | |
| TH2157 | flgB2164 fljB5001::MudJ | 8 |
| TH2164 | flgI2002 fljB5001::MudJ | 8 |
| TH2413 | flk-5206::Tn10dCm | 8 |
| TH2512 | flgA5210::Tn10dTc | 10 |
| TH2541 | flk-5212::Tn10dTc | 8 |
| TH2575 | flgA5211::MudA | 10 |
| TH2592 | fljB5001::MudJ | |
| TH2877 | flgM5207::MudA | 10 |
| TH3282 | flk-5212::Tn10dTc fljB5001::MudJ | |
| TH3303 | flgB2164 fljB5001::MudJ | |
| TH3441 | ΔflgA1529 fljB5001::MudJ | |
| TH3442 | ΔflgH2136 fljB5001::MudJ | |
| TH3443 | ΔflgI2453 fljB5001::MudJ | |
| TH3444 | ΔflgI1376 fljB5001::MudJ | |
| TH3445 | ΔflgHI958 fljB5001::MudJ | |
| TH3451 | ΔflgA1529 fljB5001::MudJ flk-5212::Tn10dTc | |
| TH3452 | ΔflgH2136 fljB5001::MudJ flk-5212::Tn10dTc | |
| TH3453 | ΔflgI2453 fljB5001::MudJ flk-5212::Tn10dTc | |
| TH3454 | ΔflgI1376 fljB5001::MudJ flk-5212::Tn10dTc | |
| TH3455 | ΔflgHI958 fljB5001::MudJ flk-5212::Tn10dTc | |
| TH3504 | fljB5001::MudCm flgB2164 | |
| TH3505 | fljB5001::MudCm | |
| TH3506 | fljB5001::MudCm flk-5212::Tn10dTc | |
| TH3507 | flgB2164 fljB5001::MudCm flk-5212::Tn10dTc | |
| TH3508 | ΔflgA1529 fljB5001::MudCm | |
| TH3513 | ΔflgA1529 fljB5001::MudCm flk-5212::Tn10dTc | |
| TH3558 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm | |
| TH3559 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm flgB2164 | |
| TH3560 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgA1529 | |
| TH3561 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgH2136 | |
| TH3562 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgI2453 | |
| TH3564 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgHI958 | |
| TH3565 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm flk-5212::Tn10dTc | |
| TH3566 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm flgB2164 flk-5212::Tn10dTc | |
| TH3567 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgA1529 flk-5212::Tn10dTc | |
| TH3568 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgH2136 flk-5212::Tn10dTc | |
| TH3569 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgI2453 flk-5212::Tn10dTc | |
| TH3571 | ataA::[P22 Δ(mnt-arc)::Kmr PfliA-lac-600]fljB5001::MudCm ΔflgHI958 flk-5212::Tn10dTc | |
| TH3590 | DUP1127[(purB1879)*MudA*(flgA5211)] | |
| TH3591 | DUP1127[(ΔflgA1529 purB1879)*MudA*(flgA5211)] | |
| TH3592 | DUP1127[(ΔflgG-L2157 purB1879)*MudA*(flgA2511 ΔflgG-L2157)] | |
| TH3593 | DUP1127[(purB1879)*MudA*(flgA5211)]flk-5206::Tn10dCm | |
| TH3594 | DUP1127[(ΔflgA1529 purB1879)*MudA*(flgA5211)]flk-5206::Tn10dCm | |
| TH3595 | DUP1127[(ΔflgG-L2157 purB1879)*MudA*(flgA5211 ΔflgG-L2157)]flk-5206::Tn10dCm | |
| TH3596 | DUP1113[(purB1879)*MudA*(flgM5207 flgA5210::Tn10dTc)] | |
| TH3597 | DUP1113[(ΔflgA1529 purB1879)*MudA*(flgM5207 flgA5210::Tn10dTc)] | |
| TH3598 | DUP1113[(ΔflgG-L2157 purB1879)*MudA*(flgM5207 flgA5210::Tn10dTc ΔflgG-L2157)] | |
| TH3599 | DUP1113[(purB1879)*MudA*(flgM5207 flgA5210::Tn10dTc)]flk-5206::Tn10dCm | |
| TH3600 | DUP1113[(ΔflgA1529 purB1879)*MudA*(flgM5207 flgA5210::Tn10dTc)]flk-5206::Tn10dCn | |
| TH3601 | DUP1113[(ΔflgG-L2157 purB1879)*MudA*(flgM5207 flgA5210::Tn10dTc ΔflgG-L2157)]flk-5206::Tn10dCm | |
| TH3745 | DEL[(flgA5211)*MudJ*(flgN5220)] DEL1103(tct-zfg-3516::Tn10dTc-fljB5001::MudJ) fla-2050 (ΔfliA-D) | |
| TH3907 | flgA5210::Tn10dTc flgM5223 flgM5208::MudK | |
| TH4023 | DUP1142[(purB1879)*MudB*(flgM5208 flgM5223 flgA5210::Tn10dTc)] | |
| TH4024 | DUP1142[(ΔflgA1529 purB1879)*MudB*(flgM5208 flgM5223 flgA5210::Tn10dTc)] | |
| TH4025 | DUP1142[(ΔflgG-L2157 purB1879)*MudB*(flgM5208 flgM5223 flgA5210::Tn10dTc ΔflgG-L2157)] | |
| TH4026 | DUP1142[(purB1879)*MudB*(flgM5208 flgM5223 flgA5210::Tn10dTc)]flk-5206::Tn10dCm | |
| TH4027 | DUP1142[(ΔflgA1529 purB1879)*MudB*(flgM5208 flgM5223 flgA5210::Tn10dTc)]flk-5206::Tn10dCm | |
| TH4028 | DUP1142[(ΔflgG-L2157 purB1879)*MudB*(flgM5208 flgM5223 flgA5210::Tn10dTc ΔflgG-L2157)]flk-5206::Tn10dCm | |
| TH4029 | DUP1142[(purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)] | |
| TH4030 | DUP1142[(ΔflgA1529 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)] | |
| TH4031 | DUP1142[(ΔflgG-L2157 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529 ΔflgG-L2157)] | |
| TH4032 | DUP1142[(purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)]flk-5206::Tn10dCm | |
| TH4033 | DUP1142[(ΔflgA1529 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)]flk-5206::Tn10dCm | |
| TH4034 | DUP1142[(ΔflgG-L2157 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529 ΔflgG-L2157)] flk-5206::Tn10dCm | |
| TH4041 | ΔflgA1529 flgM5223 flgM5208::MudK | |
| TH4045 | DUP1142[(purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)]fliA5050::Tn10dTc | |
| TH4046 | DUP1142[(ΔflgA1529 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)] fliA5050::Tn10dTc | |
| TH4047 | DUP1142[(ΔflgG-L2157 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529 ΔflgG-L2157)] fliA5050::Tn10dTc | |
| TH4048 | DUP1142[(purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)]fliA5050::Tn10dTc flk-5206::Tn10dCm | |
| TH4049 | DUP1142[(ΔflgA1529 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529)] fliA5050::Tn10dTc flk-5206::Tn10dCm | |
| TH4050 | DUP1142[(ΔflgG-L2157 purB1879)*MudB*(flgM5208 flgM5223 ΔflgA1529 ΔflgG-L2157)] fliA5050::Tn10dTc flk-5206::Tn10dCm | |
| TT10213 | purB1879::MudA | John Roth |
Unless indicated otherwise, all strains were constructed during the course of this work.
Media and standard genetic manipulations.
Media, growth conditions, transductional methods, and motility assays were performed as described previously (7, 8).
Plasmid constructions.
Plasmids p322KS and p322SK were constructed as described previously (20). The pGEM3Z vector was obtained from Promega Corp (Madison, Wis.). pJK282 served as a template for RNase T2 probes and was made as follows. A BspDI-HindIII 3,455-bp fragment from pMH71 (14) containing the flgAMN operon as well as the flgB and flgC genes was ligated into BspDI-HindIII-cut p322KS. pTX592 contained a 1.3-kbp insert obtained by PCR amplification with pJK286 (20) and primers 5′-GCCGGATCCTTCCCGTCCACGCA and 5′-TTAAAGCTTCGACGCGTTCCATATTAA. This insert contained the first 182 bp of the flgB coding region, the entire flgA gene, and the first 198 bp of the flgM coding region. The amplified PCR product was digested with BamHI and HindIII at sites that were engineered in the primers and were ligated to BamHI- and HindIII-cut pGEM3Z to give plasmid pTX592. Plasmids pTX593 and pTX594 were obtained by digesting the 1.3-kbp PCR products with BamHI and AvaI and HindIII and AvaI, respectively, and ligating the 777- and 511-bp fragments to the pGEM3Z vector cut with the same enzymes. pTX593 contained the first 182 bp of the flgB coding region, the intercistronic region of flgB and flgA, and the first 439 bp of the flgA coding region. pTX594 contained the last 221 bp of the flgA coding region, the intercistronic region of flgA and flgM, and the first 218 bp of the flgM coding region. pTX595 contained a 704-bp BamHI-to-HpaI fragment from pMS531 (42) cloned into the pGEM3Z vector. It contained 122 bp of the region upstream from fliA and the first 582 bp of the fliA coding region.
Construction of pJK188 for the pulse-chase experiments was done as follows. A BspHI-HindIII 400-bp fragment filled in with Klenow from pMC64 (5) containing the flgM gene was ligated into pGEX-2T (Amersham Pharmacia Biotech, Piscataway, N.J.) cut with SmaI. The N terminus of FlgM was fused in frame to the C terminus of glutathione S-transferase (GST) to create a 39-kDa fusion protein under the inducible Ptac promoter.
Tandem chromosomal duplications of flgM-lac operon fusions.
Duplications used to measure transcription separately from the flgM class 2 and class 3 promoters were constructed as described previously (15). Separately, a P22 lysate, grown on either the flgA5211::MudA or the flgM5207::MudA strain, was mixed with an equal titer of P22 lysate grown on the purB1879::MudA strain. The mixed lysates were used to transduce strain LT2 to MudA-encoded ampicillin resistance. Because of the large size of the MudA transposon (38 kbp) relative to the size of DNA packaged by P22 (42 kbp), inheritance of MudA by homologous recombination requires that two transduced fragments, each carrying part of the Mud transposon, enter a recipient cell to inherit the Mud insertion. Inheritance of MudA by homologous recombination involves three recombinational exchanges; one exchange joins the donor MudA fragments and two exchanges occur between the composite fragment and the recipient chromosome (15). Mixed lysates from either TH2575 (flgA::MudA) or TH2877 (flgM::MudA) and TT10213 (purB::MudA) were used to transduce LT2 to MudA-encoded ampicillin resistance. Duplication recombinants were distinguished from the parental recombinant types by the fact that they maintained wild-type copies of the purB and flgA genes and when cultures were grown in the absence of ampicillin, such that selection for MudA at the duplication join point was removed, they gave off Aps Lac− (white in the presence of X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside]) segregants. Loss of MudA is indicative of recombination between the duplicated homologies when selection for the MudA insertion at the join point is removed (15). Duplications were constructed in ΔflgA and ΔflgG to -L genetic backgrounds by transduction, selecting for MudA-encoded ampicillin resistance and screening for nonmotile transductants that kept the flg allele(s) of the recipient strain.
Tandem chromosomal duplications of flgM-lacZ gene fusions.
Duplications used to measure translation of the flgM5208-lacZ gene fusion separately from either the flgM class 2 or class 3 promoters were constructed by first transducing strains TH3590, TH3591, TH3592, TH3593, TH3594, and TH3595 with a P22 transducing lysate grown on strain TH3907 (flgA5210::Tn10dTc flgM5223 flgM5208::MudK) selecting for Tcr. Recombination between the lac operon regions and the chromosomal regions clockwise to the site of the flgA5210::Tn10dTc insertion in both the donor and recipient DNA segments yielded Tcr, Apr, and Kms transductants in which the flgA5211::MudA duplication join point was converted to an flgM5208::MudB join point to yield strains TH4023, TH4024, TH4025, TH4026, TH4027, and TH4028, respectively. All of these constructs carry the polar flgA5210::Tn10dTc near the join point and the flgM5223 (L66S) allele in the FlgM-LacZ fusions. This means that the FlgM-LacZ fusion containing the flgM5223 (L66S) allele is transcribed and translated only from the class 3 flgMN promoter. Ampicillin was added to the growth medium to select for the duplication. When grown in the absence of ampicillin, the duplication strains segregated, yielding Aps, Lac−, and Tcs segregants, as expected.
To construct strains expressing the FlgM-LacZ fusion containing the flgM5223 (L66S) allele from just the class 2 flgAMN promoter, the flgA5210::Tn10dTc allele in strains TH4023 through TH4028, described above, was first removed by transducing these strains to tetracycline sensitivity (35) with a P22 transducing lysate grown on strain TH4041 (ΔflgA1529 flgM5223 flgM5208::MudK). A three-factor cross between the flgM5208::MudK, flgA5210::Tn10dTc, and ΔflgA1529 alleles revealed that the ΔflgA1529 allele lies between the flgM5208::MudK and flgA5210::Tn10dTc alleles (data not shown). This cross resulted in tetracycline-sensitive recombinants with a very high probability (>99%) of coinheriting the ΔflgA1529 allele. To eliminate background spontaneous tetracycline-sensitive (Tcs) revertants, phage lysates were mixed with cells at a high multiplicity of infection (i.e., 10) and were diluted 100- to 1,000-fold prior to being plated on Tcs selection medium (35). In this way, the number of Tcs transductants was 1,000-fold higher in frequency than that of spontaneous Tcs revertants on cell-only control plates. These crosses yielded strains TH4029 through TH4034, which were Tcs, Apr, and Kms. Finally, TH4029 through TH4034 were transduced to Tcr with a P22 transducing lysate on strain TH1479 (fliA5059::Tn10dTc). Introduction of the fliA::Tn10dTc allele prevents expression of the FlgM-LacZ fusion from the class 3 flgMN promoter (10). This yielded strains TH4045 through TH4050. In these strains, the FlgM-LacZ fusion carrying the flgM5223 (L66S) allele is expressed only from the class 2 flgAMN promoter.
β-Galactosidase assays.
β-Galactosidase assays were performed in triplicate on mid-log-phase cells as previously described (34). β-Galactosidase activities are expressed as nanomoles per minute per optical density unit at 650 nm.
Quantitative immunoblots.
Intracellular levels of FlgM protein were determined by quantitative immunoblotting (3) with the following modifications. Cell lysates were prepared from mid-log-phase cells. Cells were pelleted from 3 ml of culture, resuspended in 50 μl of 1× sample buffer (29), and heated for 3 min. Protein concentrations from dilutions of the prepared lysates were determined with a Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, Calif.). A total of 50 μg of each cell lysate was separated on 16.5% Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (40) and electrotransferred to Westran polyvinylidene difluoride membranes (Schleicher & Schuell, Keene, N.H.) in CAPS (3-[cyclohexylamino]-1-propane sulfonic acid) buffer (36). The blots were probed with anti-FlgM rabbit polyclonal antibodies and were immunostained by an alkaline phosphate-based reaction as described previously (16). Densitometric measurements of stained bands were accomplished by scanning the blots on a Agfa Arcus II Flatbed scanner by using Adobe Photoshop support software and then quantifying the image intensities with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Standard protein concentration curves with purified FlgM protein were performed and showed the linearity intensity of immunostaining versus the protein concentration to be between 1 and 25 ng.
Pulse-chase experiments.
Bacterial cells were grown in 1× E minimal medium supplemented with 0.2% glycerol and amino acid pools 6 to 11 without methionine (7) at 37°C to 40 Klett units. The cells were labeled for 1 min with [35S]methionine-cysteine protein labeling mix (New England Nuclear, Boston, Mass.) at a final concentration of 50 μCi/ml. Cold methionine was added at a final concentration of 0.5%, and duplicate 1-ml samples were taken at various time points. Trichloroacetic acid (TCA) was added to a final concentration of 5%, and the mixture was incubated on ice for 20 min. The TCA precipitate was centrifuged in an Eppendorf microcentrifuge at 16,000 × g for 5 min at 4°C and was then washed two times in 80% cold acetone and dried. The pellet was resuspended in 50 μl of 1× SDS-sample buffer (29) and boiled for 3 min. Equal counts per minute from the time course samples were added to 900 μl of radioimmunoprecipitation buffer (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS) (12) as well as a radiolabeled lysate from TH3746. Lysate from TH3746 has an N-terminal flgM fusion protein with glutathione S-transferase (39 kDa versus 10 kDa for native FlgM protein) and served as an internal control for the immunoprecipitation reaction. Anti-FlgM antibody (16) was added, and the mixture was incubated on ice for 1 h. A total of 50 μl of IgGSORB prepared as per the manufacturer’s instructions (The Enzyme Center, Malden, Mass.) was added, and the mixture was incubated on ice for 30 min. The immunoprecipitated material was washed three times with cold RIPA buffer and then resuspended in 25 μl of 1× SDS-sample buffer. The samples were electrophoresed on 10% Tricine SDS-PAGE gels (40), and the gels were fixed in 10% methanol–10% acetic acid and dried. The gels were analyzed with a PhosphorImager (Molecular Dynamics), and the band intensities were quantified (phosphorimager units [PU]) by using ImageQuant software (Molecular Dynamics). Relative levels of FlgM were calculated as (PU of FlgM band/PU of GST-FlgM band) × 100.
FlgM export assay.
Bacterial cells were grown in 25 ml of 1XE minimal medium supplemented with 0.4% glycerol and amino acid pools 6 to 11 (7) to 100 Klett units. The cells were pelleted at 17,000 × g for 30 min at 4°C, and the supernatant was saved and filtered through a 0.45-μm-pore-size cellulose acetate filter to remove any remaining bacterial cells. A 10-ml sample of filtered supernatant was filtered onto a prewetted BA85 0.2-μm-pore-size nitrocellulose filter (Schleicher & Schuell). The proteins were eluted by the addition of 50 μl of 1× SDS-sample buffer (29) and were heated at 65°C for 30 min. Samples were then analyzed by immunoblotting using anti-FlgM antibodies as described above.
RNA isolation and RNase T2 assays of chromosomal transcripts.
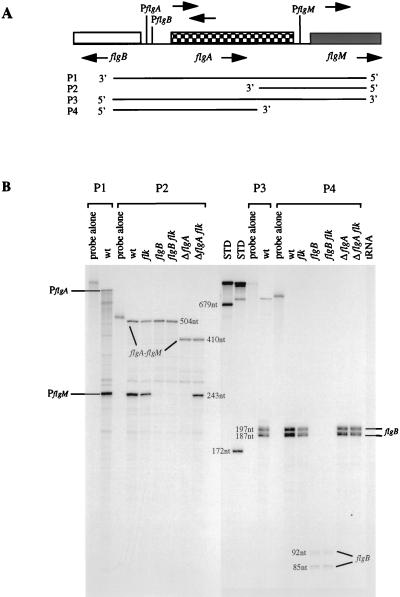
RNA was purified from cells grown exponentially (50 to 65 Klett units at 660 nm) at 37°C by adding portions of bacterial cultures directly to lysis solutions without intervening steps as described previously (46). RNase T2 protection assays of transcripts from the bacterial chromosomes were completed as described previously (46). RNA probes P1 to P4 covering parts of the flgB, flgA, and flgM regions are shown in Fig. 6 and were synthesized by using the following phage RNA polymerase and linearized plasmid templates: probe P1, SP6 and pTX592 with EcoRI; probe P2, SP6 and pTX594 with EcoRI; probe P3, T7 and pTX592 with HindIII; and probe P4, SP6 and pTX593 with EcoRI. An RNA probe for the detection of fliA transcript was synthesized by SP6 polymerase and pTX595 linearized with BamHI. A series of labeled RNA molecules of known lengths were used as size standards to determine the lengths of the protected probes (46). Each hybridization reaction mixture contained 40 μg of total RNA or tRNA as a negative control. The opposite strand of probe P2 and the fliA probe were also synthesized and used to test for DNA contamination or antisense transcription. No bands were detected with these probes. The radioactivity in the bands on the gels was measured with the InstantImager (Packard, Meriden, Conn.).
FIG. 6.
RNase T2 assay showing the effects of flk mutation on the amounts of transcripts in the flgB, flgA and flgM regions. (A) Structure and transcription of the flgB, flgA, and flgM regions in S. typhimurium. The figure, which is drawn to scale, is based on sequences in the GenBank database (accession no. D00498 for flgA and flgB and accession no. M74222 for flgM). The start sites of the promoters PflgB and PflgA were based on consensus sequences for promoters in class 2 flagellar operons (22). The start site of PflgM was mapped previously (9). Also indicated are the RNA probes P1 to P4 used in the RNase T2 protection assay. (B) Protected transcripts detected with RNA probes P1 to P4. Total RNA isolated from exponentially growing cells was hybridized with RNA probes P1 to P4 as described in Materials and Methods. The bands indicated correspond to transcription initiation from PflgA and PflgM, the flgA-flgM cotranscript, and transcripts in the flgB region. Lane 1, RNA probe P1; lane 2, protected transcripts obtained from hybridization of probe P1 with total RNA from TH3505 (wild type [wt]); lane 3, RNA probe P2; lanes 4 to 9, protected transcripts obtained from hybridization of probe P2 with total RNA obtained from TH3505 (wt), TH3506 (flk), TH3504 (flgB2164), TH3507 (flgB2164 flk), TH3508 (ΔflgA1529), and TH3515 (ΔflgA1529 flk), respectively; lanes 10 and 11, molecular weight standards [STD]; lane 12, RNA probe P3; lane 13, protected transcripts obtained from hybridization of probe P3 with total RNA from TH3505 (wt); lane 14, RNA probe P4; lanes 15 to 20, protected transcripts obtained from hybridization of probe P4 with total RNA obtained from TH3505 (wt), TH3506 (flk), TH3504 (flgB2164), TH3507 (flgB2164 flk), TH3508 (ΔflgA1529), and TH3513 (ΔflgA1529 flk), respectively. A total of 40 μg of total RNA was used in each hybridization. tRNA was used as a negative control (lane 21).
Determination of mRNA stability.
The stability of the flgAMN and flgMN mRNA was determined by previously described methods (11, 45). Rifampin was added to exponentially grown cells to a final concentration of 500 mg/ml, and the RNA was purified at different time points following addition of rifampin. Transcripts expressed from the flgAMN and flgMN promoters were analyzed as described above except that the radioactivity in the bands was measured with a PhosphorImager (Molecular Dynamics). The flgAMN and flgMN mRNA half-lives were calculated as described previously (45).
RESULTS
Mutations in the HBB structure result in an increase in the intracellular levels of FlgM, and loss of Flk restores wild-type levels of FlgM in ring mutant strains.
Mutations in genes required for HBB formation result in the loss of class 3 gene expression (8). Because the negative regulator FlgM is believed to be exported in response to completion of the HBB structure, it was expected that a defective structure would lead to increased levels of FlgM protein in the cell. This model predicts that FlgM protein might accumulate in strains defective in HBB formation and result in the inhibition of ς28-dependent class 3 gene expression. In order to test if a defective HBB structure leads to an increase in the intracellular levels of FlgM, quantitative immunoblot assays were used to measure the intracellular levels of FlgM in a variety of flagellar mutant strains.
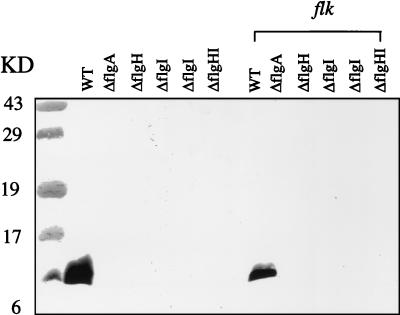
We performed quantitative immunoblot assays with anti-FlgM antibody on eight isogenic strains (Fig. 2) to measure the effects of rod (flgB), P-ring (flgA, flgI), and L-ring (flgH) mutations on the levels of FlgM in the cell. Each HBB mutation resulted in about a twofold increase in the levels of FlgM in the cell compared to that for the isogenic Fla+ strain. This result supports the original hypothesis that mutations in the HBB structure prevent the export of the FlgM negative regulator, which then accumulates in the cell to inhibit ς28-dependent class 3 gene transcription.
FIG. 2.
Effects of HBB mutations and Flk on intracellular levels of FlgM protein. A quantitative immunoblot was performed with anti-FlgM antibody on cell extracts prepared from exponentially growing cells of isogenic wild-type and flagellar mutant strains. WT, TH2592; flk, TH3282; flgB, TH2157; flgB flk, TH3303; flgA, TH3441; flgA flk, TH3451; flgH, TH3442; flgH flk, TH3452; flgI, TH3444; flgI flk, TH3454.
The effects of a null mutation in the flk gene on the levels of FlgM protein were determined in Fla+, ring, and rod mutant strains (Fig. 2). As mentioned above, intracellular levels of FlgM increased approximately twofold when export of FlgM was blocked by the presence of either a ring or a rod mutation compared to the levels of FlgM present in wild-type cells. Insertional inactivation of the flk gene had no effect on intracellular FlgM levels in either the wild-type or rod mutant strains, but intracellular levels of FlgM were reduced in the ring mutant strains in the presence of the flk null allele. This result demonstrates that inactivation of the flk locus in ring mutant strains leads to a reduction in the intracellular concentration of the FlgM anti-sigma factor. This could account for the observed expression of the fliB-lac fusion in P- and L-ring mutant strains when the flk gene is also disrupted (20).
FlgM turnover is unaffected by mutations in flk.
The reduction in the intracellular levels of FlgM protein by loss of Flk in the ring mutant strains would account for the increase in class 3 gene transcription. There are several mechanisms by which the loss of Flk could lead to reduced FlgM levels. Flk could affect FlgM stability, FlgM export, transcription of the flgM gene, flgM mRNA translation, flgM mRNA stability, FlgM anti-ς28 activity, or some other mechanism that affects FlgM protein levels.
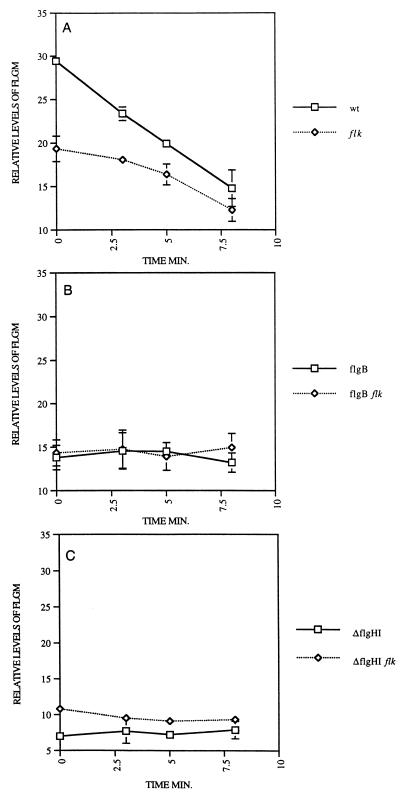
One mechanism to reduce FlgM protein levels in the cell by loss of Flk in a ring mutant strain would be to decrease FlgM protein stability. The possibility that the reduction of intracellular FlgM levels by inactivation of flk in ring mutant strains was due to an increase in FlgM protein degradation was tested by measuring the effect of inactivation of flk on FlgM half-life. FlgM turnover was measured by adding 35S-labeled methionine and cysteine to growing cells followed by a chase with unlabeled methionine and cysteine. At various times after introduction of label, both intracellular and secreted labeled FlgM were measured (see Materials and Methods) to determine if loss of Flk results in an increase in FlgM turnover in P- and L-ring mutant strains. We found that FlgM turnover occurred only in export-competent (Fla+) cells. (Note that we measured the combined precipitated FlgM from both inside and outside the cells.) In Fla+ cells, the measured half-life of FlgM was 7.3 min (Fig. 3A). This was increased to 11 min in the presence of a flk null allele. If FlgM export was prevented by a mutation in either an flgB or a ΔflgHI double mutant strain, then FlgM had no detectable turnover (Fig. 3B and C). (The flgB mutation is defective in the formation of the proximal rod portion of the basal body, while the ΔflgHI mutant strain is deleted for the P- and L-ring structural genes.) Loss of Flk did not result in increased FlgM turnover in the ΔflgHI strain even though loss of Flk did allow class 3 gene expression in the same ΔflgHI mutant background. These results suggest that the reduction in intracellular FlgM seen by loss of Flk in ring mutant strains does not result from an increase in FlgM turnover. They also suggest that FlgM is stable to proteolysis inside the cell but not in the spent growth medium.
FIG. 3.
Turnover of intracellular FlgM protein as measured by pulse-chase experiments with [35S]Met-Cys on exponentially growing cells of isogenic strains. (A) Intracellular turnover of labeled FlgM in a wild-type (wt) strain (half-life = 7.3 min) (TH2592) and an isogenic flk mutant strain (half-life = 11 min) (TH3282). (B) Intracellular turnover of labeled FlgM protein in isogenic flgB (rod-defective, TH2157) and flgB flk (TH3303) mutant strains. (C) Intracellular turnover of labeled FlgM protein in isogenic ΔflgHI (ring-defective, TH3445) and ΔflgHI flk (TH3455) mutant strains.
FlgM export is unaffected by mutations in flk.
The possibility that the reduction of intracellular FlgM levels by inactivation of flk in ring mutant strains was due to export of FlgM was assayed by measuring levels of exported FlgM in the external medium in a variety of flagellar mutant backgrounds. The results presented in Fig. 4 show that FlgM is present in the spent growth medium of a wild-type flagellar strain but not in that of isogenic ring mutant strains. Introduction of an insertion mutation in flk had no effect on FlgM export. In a wild-type Fla+ strain, FlgM is present in the external growth medium with or without a mutation in flk. In ring mutant strains no FlgM was detected in the spent growth medium even in the absence of a functional flk gene. These results do not support a model in which loss of Flk allows FlgM export in ring mutant strains.
FIG. 4.
Effect of Flk on the secretion of FlgM into the external growth medium. Immunoblot analysis was performed with anti-FlgM antibody on protein found in the spent growth medium of exponentially growing isogenic strains. WT, TH2592; flgA, TH3441; flgH, TH3442; flgI [left], TH3443; flgI [right], TH3444; flgHI, TH3445; WT flk, TH3282; flgA flk, TH3451; flgH flk, TH3452; flgI flk [left], TH3453; flgI flk [right], TH3454; flgHI flk, TH3455.
Effects of mutations in flk on expression of lac operon fusions to the flgM class 2 and class 3 promoters in wild-type and flagellar mutant strains.
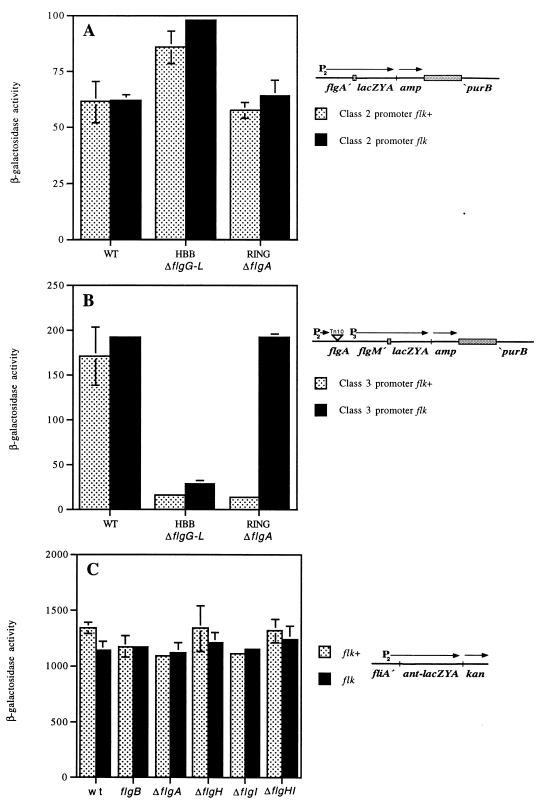
To measure the effect of Flk on expression from the flgAMN class 2 promoter and the flgMN class 3 promoter in response to flagellar structural mutations, tandem chromosomal duplications were constructed between MudA insertions in the purB and either the flgA or flgM gene by a previously described method (15). DNA sequence analysis revealed that the flgA5211::MudA and the flgM5207::MudA insertions used in these assays resulted from Mud transposition into the flgA gene just after codon 180 and into the flgM gene at codon 86 (CTC leu, 5′CT-Mud; however, insertion recreates a Leu codon at the fusion join point [10]). The duplication between MudA insertions in flgA and purB resulted in the lac operon being expressed from the class 2 promoter of the flgAMN operon (Fig. 5A). The duplication between MudA insertions in the flgM and purB genes resulted in the lac operon being expressed from the class 3 promoter of the flgMN operon (Fig. 5B). In this purB-flgM duplication, transcription from the upstream class 2 promoter is prevented by a polar flgA::Tn10dTc insertion mutation characterized previously (10).
FIG. 5.
Effects of Flk on flgM gene expression from both the class 2 flgA promoter and class 3 flgM promoter and the effect of Flk on expression of the fliA gene. (A) Expression of an flgA-lac operon fusion from the class 2 flgA promoter in isogenic wild-type and flagellar mutant strains. WT, TH3590, a duplication strain carrying an flgA-lac fusion and an intact flgAMN operon; WT flk, TH3593 (TH3590 flk-5206::Tn10dCm), the identical strain defective in flk; ΔflgG-L flk+, TH3592 (same as TH3590 except both duplicated regions carry the ΔflgG-L2157 allele), a deletion of basal-body genes; ΔflgG-L flk, TH3595 (TH3592 flk-5206::Tn10dCm); ΔflgA flk+, TH3591 (same as TH3590 except the flgAMN operon carries a nonpolar internal deletion of the flgA gene); ΔflgA flk, TH3594 (TH3591 flk-5206::Tn10dCm). (B) Expression of an flgM-lac operon fusion from the class 3 flgM promoter in isogenic wild-type and flagellar mutant strains. For the results from all the strains presented in this panel, expression of the intact flgMN operon from the flgA promoter is prevented by the introduction of a polar upstream flgA::Tn10dTc insertion. WT, TH3596, a duplication strain carrying an flgM-lac fusion and an intact flgMN operon; WT flk, TH3599 (TH3596 flk-5206::Tn10dCm), the identical strain defective in flk; ΔflgG-L flk+, TH3598 (same as TH3596 except both duplicated regions carry the ΔflgG-L2157 allele), a deletion of basal-body genes; ΔflgG-L flk, TH3601 (TH3598 flk-5206::Tn10dCm); ΔflgA flk+, TH3597 (same as TH3590 except the flgAMN operon carries a nonpolar internal deletion of the flgA gene); ΔflgA flk, TH3600 (TH3597 flk-5206::Tn10dCm). (C) Expression of the lac operon from an isolated fliA promoter which includes DNA from −588 to +19 relative to the transcriptional start site from the fliA gene in the isogenic wild-type, the flagellar rod (flgB), and the different flagellar ring (flgA, flgH, and flgI) mutant strains with and without a mutation in the flk locus. WT, TH3558; WT flk, TH3565; flgB flk+, TH3559; flgB flk, TH3566; flgA flk+, TH3560; flgA flk, TH3567; flgH flk+, TH3561; flgH flk, TH3568; flgI flk+, TH3562; flgI flk, TH3569; flgHI flk+, TH3564; flgHI flk, TH3571.
Expression of lac from the flgAMN class 2 promoter was increased about 40% in the presence of an HBB deletion compared to the level of expression in the wild-type strain. In a ring mutant strain, transcription from the flgAMN class 2 promoter was unaffected (Fig. 5A). Introduction of an insertion mutation in the flk locus had no significant effect on the expression of the flgAMN class 2 promoter in any of the three backgrounds tested (Fig. 5A).
Expression of lac from the flgMN class 3 promoter was reduced 10-fold in the presence of either an HBB deletion mutation or a ring mutation (Fig. 5B). Introduction of an insertion mutation in the flk locus had no effect on expression of the flgMN class 3 promoter in the wild-type strain and increased its expression only slightly (less than twofold) in the HBB deletion mutant strain; however, in the ring mutant background, loss of flk by insertional inactivation restored expression from the class 3 flgMN promoter to levels seen in the isogenic wild-type strain (Fig. 5B). This result does not support the model that the reduction in the levels of FlgM protein seen in a ring flk double mutant compared to the levels in the isogenic ring mutant flk+ strain is due to a reduction in transcription of the flgM gene. In fact, expression of flgM was increased in the ring mutant strain by introduction of the flk null allele, and the class 3 flgMN promoter is regulated like any other class 3 promoter in response to effects of HBB mutations and flk.
Effect of mutations in flk on expression of lac operon fusions to the fliA promoter region in wild-type and flagellar mutant strains.
We examined the effect of Flk on expression of the fliA promoter in a rod mutant and the different ring mutant strains (Fig. 5C). It was possible that the increased expression from class 3 promoters in ring mutant strains by introduction of an flk null allele could be due to a reduction of FlgM levels or an increase in FliA (ς28) levels by increasing transcription from the fliA promoter. Introduction of the fliA gene on a multicopy plasmid overcomes FlgM inhibition of class 3 gene expression (16). A defective bacteriophage P22 vector carrying the lac operon transcribed from the fliA promoter region from −588 to +19 relative to the translational start site was constructed (5a). This phage was lysogenized into a variety of flagellar mutant strains with and without a functional flk gene, and the effect of Flk on fliA transcription was determined. Expression of lac from the fliA promoter was unaffected by a mutation in the rod structural gene, flgB, or any of the genes needed for P- or L-ring formation. Introduction of an insertion in the flk gene had no effect on expression of the fliA promoter in any of the strains tested (Fig. 5C). Thus, the increased expression from class 3 promoters in ring mutant strains by introduction of a flk null allele does not coincide with a reduction in transcription of the flgM gene or an increase in transcription of the fliA gene.
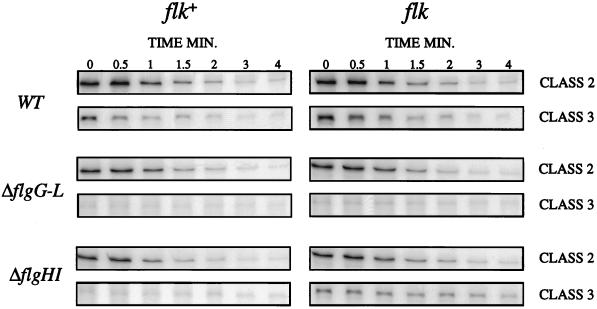
Effects of Flk on flgM mRNA synthesis.
The effects of the flk mutation on class 2 and class 3 transcription of flgM were also examined at the level of mRNA synthesis by using RNase T2 protection assays. Levels of mRNA transcripts from the class 2 and class 3 flgM promoters were measured directly in wild-type, rod (flgB), and ring (flgA) mutant strains with and without a functional flk+ gene (Table 2; Fig. 6). The ring mutant used in these experiments was the ΔflgA allele (ΔflgA1529) previously shown to have no effect on transcription or translation of the downstream flgM gene (10). Protected transcripts were detected with RNA probes P1 to P4 shown in Fig. 6A. As controls, the levels of class 2 transcripts from the flgB to -L operon and the fliAZY operon (probe not shown) were also measured.
TABLE 2.
Effects of flk mutation on the amounts of transcripts from class 2 and class 3 promoters in parent, flgB2164, and ΔflgA1529 strainsa
| Transcript | Transcript amount (cpm)b
|
|||||
|---|---|---|---|---|---|---|
| TH3505 (parent) | TH3506 (flk) | TH3504 (flgB) | TH3507 (flgB flk) | TH3508 (ΔflgA) | TH3513 (ΔflgA flk) | |
| flgA-flgMc | 274 | 197 | 256 | 266 | 251 | 261 |
| PflgM | 818 | 413 | 36 | 34 | 33 | 477 |
| PflgBd | 912 | 551 | 162 | 153 | 760 | 719 |
| PfliA | 840 | 499 | 413 | 440 | 416 | 563 |
Class 2 promoters include PflgA, which produces flgA-flgM cotranscript, and PflgB and PfliA (22). Class 3 promoters include PflgM (10).
Total RNA was isolated from cells grown exponentially in Luria-Bertani medium at 37°C and was hybridized with RNA probes in RNase T2 protection assays. flgA-flgM and PflgM, PflgB, and PfliA transcripts were quantitated from hybridized transcripts with probes P2 and P4 (Fig. 5) and the probe for PfliA (Materials and Methods), respectively. Hybridized RNA fragments were analyzed and quantitated as described in Materials and Methods. cpm, counts per minute.
The protected flgA-flgM cotranscripts in ΔflgA1529 mutants were 410 nt in length compared to 504 nt in the flgA+ parent.
Quantitation of PflgB included the sum of the 197- and 187-nt transcripts in the flgB+ parent and the 85- and 92-nt transcripts in the flgB2164 mutant.
We performed RNase T2 assays in six isogenic strains (TH3505, parent; TH3506, flk; TH3504, flgB2164; TH3507, flgB2164 flk; TH3508, ΔflgA1529; TH3513, ΔflgA1529 flk) (Table 2) to measure the effects of flk mutation on the amounts of transcripts from class 2 (PflgA, PflgB, and PfliA) and class 3 promoters (PflgM) in parent, non-ring (rod) mutant (flgB), and ring mutant (ΔflgA) strains. The transcription start site from the class 3 PflgM promoter has been previously mapped (10) and a predicted 234-nucleotide (nt) protected transcript (Fig. 6B) from this promoter was detected by the overlapping P1 (lane 1) and P2 (lane 2) probes. The amount of PflgM transcript decreased twofold in the flk mutant compared to that in the parent, TH3505 (Table 2). Mutation in flgB or flgA in the flk+ strain caused a dramatic (23- or 25-fold, respectively) decrease in PflgM transcript amounts (Table 2), consistent with the β-galactosidase activities of operon fusion strains. However, flk mutation reversed this decrease in the amount of PflgM transcript in the flgA mutant but not in the flgB mutant. The PflgM transcript amount was 14-fold higher in the flgA flk double mutant than the amount in the flgA mutant but remained low in the flgB flk double mutant, as in the flgB mutant.
In contrast to their effects on class 3 promoters, flgB, flgA, and flk mutations did not cause large changes in the amounts of transcript from class 2 promoters (Table 2). Transcripts initiated from PflgA gave rise to an flgA-flgM cotranscript detected as the predicted 504-nt full-length protected transcripts with probe 2 and as 410-nt transcripts in ΔflgA1529 mutants (Fig. 6B). Amounts of flgA-flgM transcript decreased slightly in the flk mutant and were similar in flgB, flgB flk, flgA, and flgA flk mutant strains compared to that in the parent strain. Transcripts initiated from PflgB were detected as 197- and 187-nt protected fragments with probes P3 and P4 in flgB+ strains. The 5′ ends of the flgB transcript were mapped to 27 and 37 nt downstream of the predicted PflgB transcript start site based on consensus sequences for class 2 promoters (22). It is possible that the detected transcripts were processed transcripts from the predicted promoter. The amounts of flgB transcript quantitated as the sum of the 197- and 187-nt species were slightly lower in the flk mutant than in the parent but were similar in flgA and flgA flk strains. The protected flgB transcripts in flgB2164 mutants were detected as shorter (92 and 85 nt) and less abundant species than the native flgB transcripts. The flk mutation had no effect on the amount of the mutant flgB transcript. Protected transcripts initiated from PfliA were detected by the fliA probe as a predicted 630-nt species (data not shown). The flk mutation caused a slight decrease of PfliA transcript in the wild-type flagellar background but did not change the transcript amounts in flgB and flgA mutant strains. These results are consistent with the genetic results obtained for the lac operon fusions presented above.
Effects of Flk on flgM mRNA stability.
Another mechanism by which Flk could reduce FlgM levels in P- and L-ring mutant strains could be through an effect on flgM mRNA stability. Given that the class 3 flgMN mRNA transcript increases 10-fold in an flk ring mutant compared to that in the ring mutant strain alone, any reduction in flgM mRNA stability might be expected to occur on this transcript, although it is possible that both the flgAMN and the flgMN transcripts are affected. The stabilities of the class 2 flgAMN and class 3 flgMN transcripts were determined in wild-type (Fla+), HBB deletion (ΔflgG to -L), and ring mutant backgrounds in the presence and absence of an flk null allele (Fig. 7 and Table 3). There was a slight increase in the mRNA stability (50 to 60%) of the class 2 transcript in the HBB deletion (ΔflgG to -L) strain and no effect of the ring mutation on the stability of the class 2 transcript. There were 1.5- and 2-fold increases in the half-lives of the class 3 transcript in the HBB deletion mutant and ring mutant backgrounds, respectively, compared to that of the Fla+ background. Finally, there was no effect of flk on the class 2 transcript in the different backgrounds and a less than twofold increase in stability in the ring mutant background. There was a further slight increase (less than twofold) in the stability of the class 3 transcript in the ring mutant background when Flk was removed, but removal of Flk did not significantly affect the stability of the class 3 transcript in the HBB deletion background. Thus, there is no evidence suggesting that the reduction of intracellular FlgM levels in ring mutant backgrounds is due to a decrease in either class 2 or class 3 flgM mRNA stability. If anything, there was a three- to fourfold increase in the stability of the class 3 transcript in the ring flk double mutant strain compared to that of the Fla+ (flk+/−) strains.
FIG. 7.
Stability of flgM mRNA transcripts. Class 2 flgAMN and class 3 flgMN mRNA stability assays were performed on flagellar wild-type (WT: flk+, LT2; flk, TH2413), HBB deletion (ΔflgG-L flk+, TH3586; ΔflgG-L flk, TH3886), and ring mutant (ΔflgHI flk+, TH3852; ΔflgHI flk, TH3888) strains in the presence and absence of a functional flk gene. The results of RNAase T2 protection assays on RNA samples, taken at the different time points as indicated following addition of rifampin, are shown (see Materials and Methods). The probe used in these assays was the P2 probe depicted in Fig. 6A, which hybridizes to both class 2 flgAMN and class 3 flgMN transcripts.
TABLE 3.
Effects of flk mutation on the half-lives of mRNA transcripts from class 2 flgAMN and class 3 flgMN promoters in wild-type, ring mutant (ΔflgHI), and HBB deletion (ΔflgG to -L) strains
| Flagellar genotype | mRNA half-lifea (min)
|
|||
|---|---|---|---|---|
| Class 2 transcript
|
Class 3 transcript
|
|||
| flk+ | flk | flk+ | flk | |
| Fla+ | 0.45 ± 0.05 | 0.53 ± 0.2 | 0.98 ± 0.2 | 0.63 ± 0.3 |
| ΔflgHI | 0.43 ± 0.01 | 0.54 ± 0.1 | 2.2 ± 0.8 | 3.7 ± 0.2 |
| ΔflgG to -L | 0.74 ± 0.2 | 0.8 ± 0.2 | 1.5 ± 0.2 | 1.8 ± 0.1 |
Half-lives were determined by linear regression analysis, as described in Materials and Methods, for two independent experimental trials, the results from one of which are shown in Fig. 7.
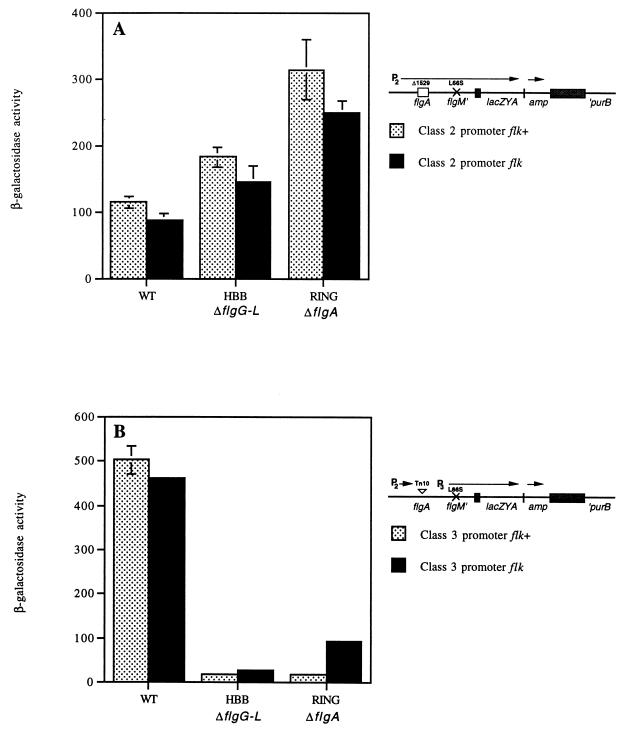
Characterization of a LacZ translational fusion to FlgM.
We have shown that the loss of flk results in a reduction in intracellular FlgM levels in P- and L-ring mutant strains (Fig. 2). Reduction in FlgM levels leads to increased transcription of the class 3 promoter, including the class 3 promoter for the flgM gene itself (20) (Table 2 and Fig. 5 and 6). FlgM turnover, FlgM secretion, flgM transcription, and flgM mRNA stability were not found to account for the observed reduction in FlgM levels (Fig. 3, 4, and 7 and Table 3). Another possibility is that translation of the class 2 and/or the class 3 flgM mRNA transcripts might be reduced by loss of flk in the ring mutant strains. We tested this possibility with a lacZ translational fusion to flgM and used β-galactosidase assays to examine the effects of flk on flgM translation. We previously reported the isolation of a MudK insertion in the flgM gene, flgM5208::MudK, that resulted in a translational fusion of lacZ to flgM (10). DNA sequence analysis revealed that the Mud transposed into the flgM gene at codon 86 (CTC Leu, 5′CT-MudK) (10). This is the exact same site of insertion for the flgM5207::MudA insertion used to characterize the effect of flk on flgM transcription from the class 3 promoter presented above (Fig. 5).
Effects of mutations in flk on expression of flgM-lacZ gene fusions expressed from the flgM class 2 and class 3 promoters in wild-type and flagellar mutant strains.
We decided to compare expression of β-galactosidase with the flgM-lacZ gene fusion (where β-galactosidase levels are dependent on both transcription and translation of flgM) to expression of β-galactosidase with the flgM-lac operon fusion (where β-galactosidase levels are dependent only on transcription of flgM). This is feasible only if the putative translational regulatory signals are 5′ to the site of insertion of the Mud transposon at amino acid 86 of FlgM. To measure the effect of Flk on translation of FlgM-LacZ expressed from the class 2 and class 3 promoters in response to flagellar structural mutations, tandem chromosomal duplications were constructed as described in Materials and Methods. The duplications resulted in two tandem chromosomal copies of the flgM region. One copy contains a wild-type flgM gene. The second copy of the flgM chromosomal region contains the flgM-lacZ gene fusion expressed from either the class 2 promoter of the flgAMN operon or the class 3 promoter of the flgMN operon. In all cases the fusion protein carried the L66S substitution mutation. This is a base substitution mutation at codon 66 in FlgM that results in substitution of leucine by serine at this position (6). The L66S mutation renders FlgM defective in binding to ς28 (6). This mutation was used in these assays so that changes in the level of the FlgM-LacZ protein fusion would not have the capacity to feedback-regulate its own expression at the class 3 promoter. In the cases where the FlgM-LacZ fusion was expressed from the class 2 promoter, an internal, nonpolar deletion of flgA, ΔflgA1529, was included which when duplicated for the flgA1529 allele renders the cell defective in P-ring assembly.
Effects of mutations in flk on expression of lac operon fusions to the flgM class 2 and class 3 promoters in wild-type and flagellar mutant strains.
Expression of flgM-lacZ from the flgAMN class 2 promoter increased less than twofold in the presence of an HBB deletion and increased about 2.5-fold in the ring mutant background compared to the level in the isogenic wild-type strain (Fig. 8A). Introduction of an insertion mutation in the flk locus showed only a slight reduction of about 20% in the expression of the FlgM-LacZ fusion from the flgAMN class 2 promoter in all of the three backgrounds tested (Fig. 8A).
FIG. 8.
Effects of Flk on flgM-lacZ expression from the class 2 flgAMN and class 3 flgMN promoters. (A) Expression of the flgM5208-lacZ gene fusion from the class 2 flgA promoter in isogenic wild-type and flagellar mutant strains. All strains harbor tandem, chromosomal duplications of the flgM through purB genes with the flgM5208-lacZ gene fusion located at the join point of the duplicated region. WT, TH4045; WT flk, TH4048; HBB ΔflgG-L flk+, TH4047 (same as TH4045 except both duplicated regions carry the ΔflgG-L2157 allele), HBB ΔflgG-L flk, TH4050 (TH4047 flk-5206::Tn10dCm); RING ΔflgA flk+, TH4046 (same as TH4045 except both the flgAM-lacZYA and flgAMN operons carry a nonpolar internal deletion of the flgA gene); RING ΔflgA flk, TH4049 (TH4046 flk-5206::Tn10dCm). All six strains carry the fliA5059::Tn10dTc insertion, which prevents expression of flgM and flgM-lacZ from the class 3 flgMN promoter (10). (B) Expression of the flgM5208-lac operon fusion from the class 3 flgM promoter in isogenic wild-type and flagellar mutant strains. All strains harbor tandem, chromosomal duplications of the flgM through purB genes with the flgM5208-lacZ gene fusion located at the join point of the duplicated region. WT, TH4023; WT flk, TH4026; HBB ΔflgG-L flk+, TH4025 (same as TH4023 except both duplicated regions carry the ΔflgG-L2157 allele); HBB ΔflgG-L flk, TH4028 (TH4025 flk-5206::Tn10dCm); RING ΔflgA flk+, TH4024 (same as TH4023 except that the flgAMN operon carries a nonpolar internal deletion of the flgA gene); RING ΔflgA flk, TH4027 (TH4024 flk-5206::Tn10dCm). All six strains carry the flgA5210::Tn10dTc insertion, which prevents expression of flgM-lacZ from the class 2 flgAMN promoter (10).
Expression of FlgM-LacZ from the flgMN class 3 promoter was reduced 25-fold in the presence of either an HBB deletion mutation or a ring mutation (Fig. 8B). This is presumably due to the FlgM-mediated inhibition of transcription from the class 3 promoter in an HBB mutant strain (Fig. 5B). Introduction of an insertion mutation in the flk locus had no effect on flgM expression in the wild-type strain and had a slight increase (less than twofold) in the HBB deletion mutant strain. There was a fivefold increase in expression in the ring mutant background (Fig. 8B). In these assays with the flgM-lacZ gene fusion, we did not observe restoration of β-galactosidase activity to wild-type levels by loss of flk as seen in the flgM-lac operon fusion constructs expressed from the class 3 promoter in the ring mutant background (Fig. 5B). Thus, by comparing the effect of Flk on expression of the flgM-lacZ translational fusion (Fig. 8) to the effect of Flk on expression of the flgM-lac transcriptional fusion (Fig. 5), we conclude that loss of Flk leads to a reduction in the translation of flgM expressed from the class 3 transcript in the ring mutant background but not in either the wild-type or the HBB deletion background.
DISCUSSION
Loss of Flk reduces FlgM levels in ring mutant strains.
The flagellar regulon is a regulatory network of over 50 genes in which expression of these genes is tightly coupled to the assembly of the flagellar organelle. The FlgM negative regulator is responsible for keeping expression of ς28-dependent promoters off until the HBB structure is complete. A novel genetic locus, flk, was discovered which allowed class 3 gene expression in ring mutant strains. Results presented in this report demonstrate that restoration of class 3 gene expression in ring mutant strains by loss of Flk is the result of reduced FlgM levels in the cell. This work examined many possible mechanisms in which loss of Flk could result in a reduction in FlgM levels in the cell. This work demonstrates that the Flk effect on class 3 transcription in ring mutant strains is not due to any of the following: (i) a reduction in transcription of the flgM gene, (ii) a reduction in flgM mRNA levels, (iii) a reduction in flgM mRNA stability, (iv) a decrease in FlgM protein stability, or (v) the ability to export FlgM through a “ringless” HBB structure. Transcription of flgM from the class 3 promoter increased about 15-fold when flk was mutated, suggesting that the effect of Flk on FlgM levels is posttranscriptional. Indeed flgM translation was reduced when a null mutation in flk was introduced into a ring mutant background (discussed below).
FlgM accumulates in export-deficient cells despite a reduction in flgM transcription.
The hypothesis that FlgM could sense the completion of the HBB structure by being a substrate for secretion through the completed structure was supported by the finding that intracellular levels of FlgM increase by about 2-fold in HBB mutant strains despite a 10-fold decrease in FlgM transcription from the class 3 promoter. We had shown previously that about 80% of flgM transcription during exponential growth comes from the class 3 promoter. We have determined that in HBB mutant strains, FlgM was stable and its turnover was dependent on the presence of a functional HBB structure, suggesting that turnover of FlgM in wild-type strains during exponential growth is dependent on a gene product produced or activated following HBB completion or that FlgM turnover occurs only on extracellular FlgM.
In examining the effect of Flk on flgM gene transcription, we saw that loss of Flk in ring mutant strains resulted in the simultaneous 10-fold increase in transcription from the flgM class 3 promoter and a return of intracellular FlgM down to wild-type levels. How does increased flgM transcription correlate with decreased FlgM protein levels? The answer has to do with the feedback regulation that FlgM has on its own structural gene expressed from the class 3 promoter in response to completion of the HBB structure. FlgM responds to the completed HBB structure by itself being a substrate for export through it into the external medium. Because FlgM is autoregulatory, the rate at which FlgM protein is produced (transcription-translation) and the rate at which FlgM is exported through the structure will both determine and depend on how much FlgM accumulates in the cell. FlgM inhibits ς28-dependent transcription, and 80% of flgM gene transcription is from a ς28-dependent, class 3 promoter. When the HBB is defective, FlgM protein accumulates and, as was shown in this work, it is a very stable intracellular protein. Accumulation of FlgM inhibits ς28-dependent transcription, including that directed by the class 3 promoter for the flgM gene. We have presented both genetic data, by individually assaying both the flgM class 2 (PflgA) and class 3 (PflgM) promoters fused to the lac operon (Fig. 5), and molecular data, by measuring the individual transcripts expressed from the PflgA and PflgM promoters (Fig. 6 and Table 2), to support this model.
Flk couples flgM translation to ring assembly.
The observation that the large amount of flgM-containing transcript from the class 3 promoter in a ring flk double mutant strain does not result in an increase in intracellular FlgM protein levels (Fig. 2 and 3) suggested that FlgM translation was reduced when flk was defective.
The effect of flk on flgM translation was tested with an FlgM-LacZ protein fusion in which the anti-ς28 activity was rendered defective by mutation (L66S). As a result of comparing the transcription of an flgM-lac operon fusion to the translation of an flgM-lacZ gene fusion in wild-type, HBB deletion, and ring mutant strains with and without a mutation in flk (Fig. 5 and 8), we can make the following conclusions. The effect of the HBB deletion resulted in a 50% increase in transcription of lacZ from the flgAMN class 2 promoter, while no increase in transcription was observed in the ring mutant background (Fig. 5A). The expression of the flgM-lacZ gene fusion also showed a 50% increase in the HBB deletion background (Fig. 7A), which can be attributed to the 50% increase in the HBB deletion background (Fig. 7A), which can be attributed to the 50% increase in transcription from the class 2 promoter (compare Fig. 5A to 8A). In the ring mutant background there was about a threefold increase in translation (Fig. 8A), and since there was no increase in transcription in the ring mutant strain (Fig. 5A), we conclude that there is a threefold increase in flgM translation in a ring mutant strain. We also conclude that there is differential transcriptional and translational regulation in different basal-body mutant strains. This implies that the coupling of flagellar gene expression to flagellar assembly occurs at assembly stages prior to HBB completion.
If we now compare the transcription and translation of flgM from the class 3 promoter, the effects are striking. As expected, the level of class 3 transcription is lower in either the HBB deletion or the ring mutant background (Fig. 5B). Loss of flk restores transcription of flgM class 3 transcription in the ring mutant background, but not in the HBB deletion background, to levels seen for the wild-type strain (Fig. 5B). This result is similar to what we observed previously for transcription from the class 3 fliB flagellin gene promoter (20). Finally, if we look at expression of the flgM-lacZ gene (translational) fusion, it is up 50% in the HBB mutant background (Fig. 8B), which can be accounted for by a 50% increase in transcription (compare Fig. 5B and 8B). In the ring mutant background, expression of the flgM-lacZ gene fusion is up when flk is mutated, but the level of β-galactosidase activity is only 20% of that measured in the Fla+ strains, whereas the amount of β-galactosidase activity from the flgM-lac operon fusion in the ring mutant background was at 100% of that measured in the Fla+ strains when flk was mutated (compare Fig. 5B and 8B). This result suggests that the translation of the flgM-lacZ gene fusion is reduced by the loss of Flk in the ring mutant strain. This accounts for the regulatory effect seen on flagellar class 3 gene expression in ring mutant strains. The results are in agreement with a model, presented in Fig. 9, that the Flk effects on FlgM protein levels are due, at least partly, to a reduction in the translation of the class 3 flgM mRNA. However, because the class 3 flgM mRNA is not transcribed in the ring mutant strains, there must be some effect of Flk on flgM expressed from the class 2 transcript. We observed a 20% reduction in flgM translation from the class 2 transcript in the ring mutant background when flk was mutated, but we saw the same effect in the Fla+ and HBB deletion backgrounds as well. It may be that our error limits prevent us from detecting the small, but significant, reduction in flgM translation from the class 2 transcript when flk is mutated compared to the levels of flgM translation in the Fla+ and HBB deletion backgrounds, and it is only translation of flgM from the class 3 transcript that is easily distinguished in the different backgrounds.
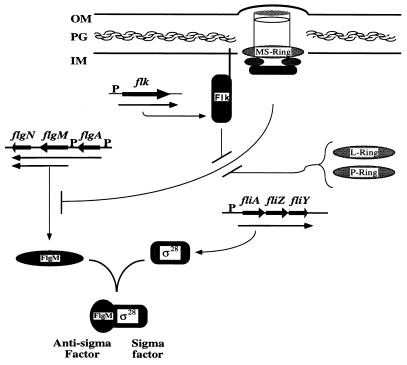
FIG. 9.
Model diagramming the role of the Flk protein in the regulation of flgM mRNA translation. The ringless basal body is depicted transversing the inner membrane (IM) and peptidoglycan (PG) layers but is unable to penetrate the outer membrane-LPS (OM) layer. Flk is depicted to interact with the ringless basal body through its C-terminal hydrophobic 18-amino-acid tail in the inner membrane. Prior to P- and L-ring assembly, the ringless basal-body structure either directly or indirectly inhibits flgM mRNA translation. This effect requires a functional hook gene (flgE), so either the ringless basal body contains a nascent hook structure or hook subunits act separately in the cytoplasm. A reduction in flgM mRNA translation and subsequent FlgM protein levels in the cell would relieve inhibition of ς28 and allow class 3 flagellar gene expression. The negative regulatory effect of the ringless basal-body structure on flgM mRNA translation is inhibited either by the incorporation of the P- and L-rings into the basal body structure or by the action of the Flk protein. The genes are named above the chromosomal open reading frames (thickest arrows). mRNA transcripts are depicted below the genes, and the arrows indicate the direction of transcription. The arrow below each transcript indicates translation to the indicated protein product.
Homologies between Flk and RNA binding proteins of E. coli.
An enhanced search of the protein database revealed homology between the so-called “S1 domain” of RNA binding proteins of E. coli (4) and a region of the deduced Flk protein sequence (amino acids 190 through 212 of S. typhimurium Flk and amino acids 192 through 214 of E. coli Flk). This could imply a direct interaction between Flk and the flgM mRNA.
FlgM, whose native structure is unfolded, is protected from intracellular degradation.
While it is not unusual for proteins to be stable to turnover during exponential growth, the stability of intracellular FlgM seemed surprising given the recent discovery that the FlgM protein exists as an unfolded polypeptide (6). How does an unfolded protein stay resistant to protein degradation within the cell? One possible answer to this question is that intracellular FlgM stays bound by other proteins. The C-terminal half of FlgM contains the anti-ς28 domain (6, 19), while the N-terminal portion is essential for FlgM export through the HBB structure (19). If binding of other proteins is necessary for FlgM stability, then two predictions can be made. First, in the absence of FlgM export, something may bind the N terminus of FlgM in the cell to prevent its degradation, while the ς28 protein binds the C terminus. If the level of FlgM exceeds that of ς28, something may bind the C terminus of FlgM to protect it from degradation. Furthermore, only insertions in the flgM gene relieve class 3 gene expression in strains defective in HBB genes, except in the case of ring mutants, which can be suppressed by insertions in flk as well as in flgM. This suggests that if binding proteins essential for intracellular FlgM stability exist, they may be either essential to cell viability or essential for class 3 gene expression. Otherwise, selections that yielded flgM mutants would have yielded mutations in other genes as well. A second prediction is that the ς28 protein may be required for FlgM stability unless the anti-ς28 domain of FlgM binds other proteins as well as ς28.
The ringless basal body is necessary for translational inhibition of flgM expression.
What is the mechanism by which ring completion is coupled to translation of flgM, and what is the role of Flk in this mechanism? All of the HBB genes except the ring genes, flgA, flgH, and flgI, are required to be functional for the Flk effect on class 3 gene expression to be seen. This suggests that it is the ringless HBB structure that is directly or indirectly responsible for reduction in FlgM protein levels in the absence of Flk (Fig. 9). If any one of the other 23 known HBB genes is mutated, then loss of Flk does not exhibit the large effect on class 3 gene expression. We propose that Flk is involved in detecting completion of the L- and P-rings for the purpose of initiating some class 3 gene derepression prior to completion of the L- and P-rings for the purpose of initiating some class 3 gene dererpression prior to completion of the HBB structure. Otherwise, class 3 gene derepression must wait until the hook is complete and FlgM is exported out of the cell. This would result in a lag time between hook completion and the transcription, translation, and assembly of flagellin filament protein. The following scenario is hypothesized for when flagellin is expressed only after FlgM export occurs: the hook is completed, FlgM is exported, transcription of the flagellin filament structural genes (fliC or fliB) is initiated, and flagellin mRNA is transcribed and translated into protein subunits ready for export and polymerization at the hook-filament junction. Thus, there would be a time loss between hook completion and filament polymerization, potentially conferring a competitive disadvantage. In the presence of Flk, the following scenario is hypothesized. The rings are completed, and at this moment two things happen simultaneously: (i) hook elongation begins and (ii) Flk, sensing ring completion and/or hook elongation, directly or indirectly inhibits translation of flgM mRNA while production of ς28 protein continues. The ς28/FlgM ratio increases and allows some transcription of flagellin subunits while the hook is elongating. Once hook biosynthesis is complete, FlgM is exported and flagellin transcription is fully derepressed, but enough flagellin has already been synthesized to initiate its export and polymerization without a lapse in time for flagellar assembly. Flk is predicted to eliminate the potential time loss from hook completion and FlgM export to flagellin expression, leading to more efficient biosynthesis of the flagellum and a greater survival advantage over a cell less efficient in flagellar assembly.
The above model also allows a role for Flk in detecting ringless HBB structures in wild-type cells. There are two distinct secretion pathways utilized in the assembly of the HBB. The type III flagellum-specific secretion system is required to direct components assembled beyond the cytoplasmic membrane into the hollow core of the growing structure to the site of assembly for each component. The rod components are assembled first, followed by the hook scaffolding protein FlgD and then the hook (21, 39). The P- and L-ring subunits are secreted into the periplasm by the general signal-sequence-dependent secretory pathway (14, 41). Thus, it is possible that ringless HBB structures could form while awaiting secretion of ring components into the periplasm. It is the presence of this structure that results in a reduction in FlgM levels in ring flk double mutant strains. Flk may be there to prevent this structure from reducing flgM translation until the rings form and HBB completion is imminent. This suggests that there may be conditions where ring assembly is not efficient or does not occur, leading to accumulation of the ringless HBB structures. Flk may be present to prevent any premature flagellin expression under such conditions.
The flk and rflH mutants may represent alleles of the same gene.
A new regulatory gene of the flagellar regulon, rflH, was recently described as having a negative regulatory role in FlgM export (27). Because the chromosomal location of the rflH locus is placed at a location similar to the flk locus, it is possible that the rflH mutations are alleles of the flk gene. The FliK protein determines hook length. (Note that FliK and Flk are not the same protein.) Null mutations in the fliK gene result in what is known as the polyhook phenotype, in which the transition from export of hook subunits to export of class 3-exported proteins (FlgM, FlgK, FlgL, FliD, FliC, and FljB) does not occur. As a result, null alleles of the fliK gene are nonmotile. Motile revertants of fliK null alleles map to the flhB locus, which is part of the type III flagellum-specific export apparatus (13, 25). This realization led to the model that the FliK protein measured hook length and that, when hook length reached about 55 nm, the FliK protein was able to signal the export machinery, at FlhB, to stop export of hook subunits and begin export of class 3 exported proteins. Thus, in a strain carrying a flhB suppressor of the fliK null allele, FlgM is exported and class 3 genes are derepressed in the absence of the fliK gene. However, if the hook gene is mutated in the same background, FlgM is not exported and class 3 transcription is repressed (27). Selection for class 3 gene expression in an flgE flhB (fliK bypass mutant) strain yielded mutations in rflH that allowed FlgM export in this strain. A model was proposed that export is controlled by a double-locked gate where RflH and FlhB act as locks to independently inhibit export of class 3 proteins and that these locks are released by a hook completion signal (sensed directly by RflH and via FliK to FlhB), resulting in class 3 protien export. If flk and rflH are the same locus, then this protein has a dual role in flagellar assembly at the level of ring completion (and FlgM translation) and at the level of preventing class 3 protein export in the absence of the hook structure. We suggest that Flk does not sense hook completion as proposed but that it senses the translation and/or export and/or polymerization of hook following ring completion. This model is consistent with the phenotypes of both the rflH and flk alleles reported.
ACKNOWLEDGMENTS
This work was supported by grant MCB-9318890 from the National Science Foundation and grants GM56141 and GM37561 from the NIH to K.T.H. and M.E.W., respectively. K.T.H. is a recipient of a Faculty Research Award from the American Cancer Society.
REFERENCES
- 1.Aizawa S-I. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;20:1–4. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 2.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 3.Blake M S. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 4.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 5.Chadsey M S, Karlinsey J E, Hughes K T. The flagellar anti-sigma factor, FlgM, actively dissociates Salmonella typhimurium RNA polymerase holoenzyme. 1998. Genes Dev., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Chilcott, G. S., and K. T. Hughes. Unpublished results.
- 6.Daughdrill G W, Chadsey M S, Karlinsey J E, Hughes K T, Dahlquist F W. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target ς28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 7.Davis R W, Botstein D, Roth J R. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 8.Gillen K L, Hughes K T. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J Bacteriol. 1991;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillen K L, Hughes K T. Molecular cloning of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillen K L, Hughes K T. Transcription from two promoters and autoregulation contribute to the control of expression of the Salmonella typhimurium flagellar regulatory gene flgM. J Bacteriol. 1993;175:7006–7015. doi: 10.1128/jb.175.21.7006-7015.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goluszko P, Moseley S L, Truong L D, Kaul A, Williford J R, Selvarangan R, Nowicki S, Nowicki B. Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae. J Clin Investig. 1997;99:1662–1672. doi: 10.1172/JCI119329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 13.Hirano T, Yamaguchi S, Oosawa K, Aizawa S-I. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homma M, Ohnishi K, Iino T, Macnab R M. Identification of flagellar hook and basal body gene products (FlaFV, FlaFVI, FlaFVII, and FlaFVIII) in Salmonella typhimurium. J Bacteriol. 1987;169:3617–3624. doi: 10.1128/jb.169.8.3617-3624.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes K T, Roth J R. Directed formation of deletions and duplications using Mu d(Ap Lac) Genetics. 1985;109:263–282. doi: 10.1093/genetics/109.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda T, Homma M, Iino T, Asakura S, Kamiya R. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 1987;169:1168–1173. doi: 10.1128/jb.169.3.1168-1173.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda T, Oosawa K, Hotani H. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J Mol Biol. 1996;259:679–686. doi: 10.1006/jmbi.1996.0349. [DOI] [PubMed] [Google Scholar]
- 19.Iyoda S, Kutsukake K. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol Gen Genet. 1995;249:417–424. doi: 10.1007/BF00287103. [DOI] [PubMed] [Google Scholar]
- 20.Karlinsey J E, Pease A J, Winkler M E, Bailey J L, Hughes K T. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J Bacteriol. 1997;179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 22.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon in Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutsukake K, Iino T. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J Bacteriol. 1994;176:3598–3605. doi: 10.1128/jb.176.12.3598-3605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 25.Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol. 1994;176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutsukake K, Ide N. Transcriptional analysis of the flgK and fliD operons of Salmonella typhimurium which encode flagellar hook-associated proteins. Mol Gen Genet. 1995;247:275–281. doi: 10.1007/BF00293195. [DOI] [PubMed] [Google Scholar]
- 27.Kutsukake K. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J Bacteriol. 1997;179:1268–1273. doi: 10.1128/jb.179.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutsukake K. Autogenous and global control of the flagellar master operon, flhDC, in Salmonella typhimurium. Mol Gen Genet. 1997;254:440–448. doi: 10.1007/s004380050437. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K, Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Matsumura P. The C-terminal region of the alpha subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J Bacteriol. 1995;177:5186–5188. doi: 10.1128/jb.177.17.5186-5188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Matsumura P. Differential regulation of multiple overlapping promoters in flagellar Class II operons in Escherichia coli. Mol Microbiol. 1996;21:613–620. doi: 10.1111/j.1365-2958.1996.tb02569.x. [DOI] [PubMed] [Google Scholar]
- 33.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 34.Maloy S R. Experimental techniques in bacterial genetics. Boston, Mass: Jones and Bartlett Publishers; 1990. [Google Scholar]
- 35.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 37.Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 38.Ohnishi K, Kutsukake K, Suzuki H, Iino T. A novel transcriptional regulatory mechanism in the flagellar regulon of Salmonella typhimurium: an anti-sigma factor inhibits the activity of the flagellum-specific sigma factor ς28. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- 39.Ohnishi K, Ohto Y, Aizawa S-I, Macnab R M, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schägger H, Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 41.Schoenhals G J, Macnab R M. Physiological and biochemical analyses of FlgH, a lipoprotein forming the outer membrane L ring of the flagellar basal body of Salmonella typhimurium. J Bacteriol. 1996;178:4200–4207. doi: 10.1128/jb.178.14.4200-4207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starnbach M N, Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992;6:459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- 43.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 44.Stocker B A D. Measurement of the rate of mutation of flagellar antigenic phase in Salmonella typhimurium. J Hyg. 1949;47:398–413. doi: 10.1017/s002217240001473x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsui H C T, Feng G, Winkler M E. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsui H C T, Pease A J, Koehler T M, Winkler M E. Detection and quantitation of RNA transcribed from bacterial chromosomes. Methods Mol Genet. 1994;3:179–204. [Google Scholar]
- 47.Williams A W, Yamaguchi S, Togashi F, Aizawa S-I, Kawagishi I, Macnab R M. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]