Summary
Background
Escherichia coli sequence type 131 (ST131), specifically its fluoroquinolone-resistant H30R clade (ST131-H30R), is a global multidrug-resistant pathogen. The gut microbiome's role in ST131-H30R intestinal carriage is undefined.
Methods
Veterans and their household members underwent longitudinal fecal swab surveillance for ST131 in 2014–2018. The fecal microbiome was characterized by 16S rRNA qPCR and sequencing. We evaluated associations between ST131-H30R carriage and gut microbiome at baseline by random forest models to identify the most informative gut bacterial phyla and genera attributes for ST131 and ST131-H30R carriage status. Next, we assessed longitudinal associations between fecal microbiome and ST131-H30R carriage using a mixed-effects logistic regression with longitudinal measures.
Findings
Of the 519 participants, 78 were carriers of ST131, among whom 49 had ST131-H30R. At the baseline timepoint, H30R-positive participants had higher proportional abundances of Actinobacteria phylum (mean: 4.9% vs. 3.1%) than ST131-negative participants. H30R-positive participants also had higher abundances of Collinsella (mean: 2.3% vs. 1.1%) and lower abundances of Alistipes (mean: 2.1% vs. 2.6%) than ST131-negative participants. In the longitudinal analysis, Collinsella abundance correlated positively with ST131-H30R carriage status and negatively with the loss of ST131-H30R. Conversely, Alistipes corresponded with the loss and persistent absence of ST131-H30R even in the presence of a household exposure.
Interpretation
Abundances of specific fecal bacteria correlated with ST131-H30R carriage, persistence, and loss, suggesting their potential as targets for microbiome-based strategies to reduce carriage of ST131-H30R, a significant risk factor for invasive infections.
Funding
This work was supported in part by National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R21AI117654 and UM1AI104681 and the Office of Research and Development, Department of Veterans Affairs. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Keywords: Gut microbiome, Escherichia coli, ST131, H30R, Alistipes, Collinsella
Research in context.
Evidence before this study
Escherichia coli sequence type 131 and its fluoroquinolone-resistant H30R subclone lineage (ST131-H30R) is a leading cause of antimicrobial resistant E. coli infections globally. ST131-H30R is commonly carried in the gut as an opportunistic pathogen before subsequent infection of the colonized host. Gut microbiome determinants of ST131-H30R carriage are unclear and have not been evaluated in longitudinal studies.
Added value of this study
Using a large cross-sectional study, we used random forest models to identify bacteria associated with ST131-H30R carriage. Participants with ST131-H30R exhibited higher proportional abundance of Actinobacteria and Collinsella. Conversely, Alistipes was more common in participants without ST131-H30R carriage. These gut microbiome patterns were confirmed in an extensive longitudinal study of individuals and their household members, where Collinsella was present more frequently among persistent ST131-H30R carriers, while Alistipes increased in conjunction with loss of ST131-H30R carriage.
Implications of all the available evidence
Our data indicated that multiple gut bacteria corresponded with E. coli ST131-H30R gut carriage, persistence, and loss. Collinsella has been previously linked with gut inflammation and permeability, whereas Alistipes has been linked with anti-inflammatory effects and has been reported as inversely correlated with Escherichia carriage. This study supports these findings using longitudinal analyses, and extends them specifically to ST131-H30R, showing higher Collinsella abundance alongside persistent ST131-H30R carriage, and a significant increase in Alistipes abundance in conjunction with persistent loss of ST131-H30R. These gut microbiome taxa (Alistipes, Collinsella), or the pathways they represent, may provide targets for therapeutic interventions through which to reduce or prevent ST131-H30R carriage, thereby decreasing transmission and infections from this critical multidrug-resistant opportunistic pathogen.
Introduction
Escherichia coli (E. coli) sequence type 131 (ST131), especially its recently emerged H30R subclone and the two main clonal subsets within H30R—H30R1 and H30Rx—cause a substantial portion of antimicrobial-resistant infections in the U.S. and worldwide.1 Both H30R1 and H30Rx are characterized by fluoroquinolone resistance. Additionally, many H30R strains—especially within H30Rx—are co-resistant to multiple other antimicrobial agents, most notably advanced beta-lactams, due to plasmidic or chromosomal extended-spectrum beta-lactamases and, increasingly, carbapenemases.
Gut colonization by ST131-H30R significantly increases the risk for subsequent infection.2 Individuals can become colonized by antibiotic-resistant E. coli via horizontal transmission, such as through exposures from international travel,3 within households,4 or in the healthcare setting.5 A key feature of ST131-H30R—including the H30R1 and H30Rx subsets—that underlies its pandemic emergence is its ability to stably colonize the human gastrointestinal tract,2,6 which is mediated, at least in part through bacterial factors such as FimH,7,8 fluoroquinolone resistance,9 and accessory traits such as iron uptake systems and protectins.6 However, not all individuals exposed to known carriers of ST131-H30R become colonized.10,11
Studies of probiotics have found that certain gut microbiome features are associated with failed probiotic engraftment, suggesting that microbiome composition can determine whether colonization occurs after exposure to a new bacterium.12 This potential microbiome-mediated resistance to gut bacterial colonization may extend to opportunistic pathogens such as ST131-H30R, which if true could lead to probiotic solutions to protect against ST131-H30R colonization. Previous studies, including a longitudinal study of 27 Dutch nursing home residents,13 have examined the association of gut microbiome composition and host colonization by fluoroquinolone-resistant E. coli14 or multidrug-resistant organisms.15 However, the influence of gut microbiome on gut colonization by the major pandemic subclone ST131-H30R is unknown.
Our main study objective was to determine how the gut microbiome correlates with carriage, persistence, and loss of ST131-H30R. We identified and followed the members of households within which at least one individual carried fluoroquinolone-resistant E. coli and/or ST131. Using the combination of a cross-sectional study of 519 participants and a nested longitudinal household study of 141 participants, we determined how gut microbiome composition correlates with the temporal dynamics of ST131 carriage.
Methods
Ethics and sample collection
Fecal swabs from consenting subjects were collected according to a protocol approved by the University of Minnesota Institutional Review Board (1412M58401), Minneapolis VA Health Care System IRB (4490-A), and George Washington University IRB (121508). As described elsewhere,6,16 study subjects included military veterans under care at the Minneapolis Veterans Affairs Medical Center (MVAMC) and their household members. Veterans were recruited prospectively (2014 through 2018) by sending invitations for study participation to all newly discharged MVAMC inpatients and randomly selected outpatients. Veterans who agreed to participate were encouraged to refer all available adult and child household members and pets. Swabs were mailed to the research laboratory at room temperature in commercial transport medium, along with basic demographic information. Other clinical data were not obtained.
To assess persistent ST131 carriage, subjects whose initial fecal swab yielded fluoroquinolone-resistant E. coli and/or ST131 (as detected using the below-described methods) were offered serial fecal sampling, along with their household members. Serial sampling was done monthly for six months, then every three months, until the first of the following occurred: the end of the study period, the subject/household declined further follow-up, or on two consecutive sampling occasions no household member yielded the household's initial strain of interest.
Culture methods
In the research laboratory, fecal swabs provided by study participants were screened for fluoroquinolone-resistant E. coli by selective culture (using Tergitol-7 agar plates with and without ciprofloxacin 4 μg/mL), for overnight incubation at 37 °C. Individual colonies of presumptive E. coli—i.e., lactose-positive colonies with a characteristic E. coli morphology—were assessed for species identity using indole and citrate phenotype (indole-positive, citrate-negative).
Molecular characterization of E. coli isolates
We used standard protocols for detection of fluoroquinolone-resistant ST131-H30 isolates using highly sensitive and specific established PCR-based assays, as previously reported.6,16 In brief, up to 10 presumptive E. coli colonies per plate were screened for clonality by using random amplified polymorphic DNA (RAPD) analysis. Using duplicate boiled lysates for template DNA and relevant positive and negative controls, one representative colony per unique RAPD profile per sample was screened for ST131 by PCR and ST131 isolates were screened to identify the H30 and H30Rx subclones. Fluoroquinolone-resistant ST131-H30 isolates were classified as ST131-H30R, and H30R isolates that tested negative for H30Rx were classified as H30R1.16
ST131-H30R carriage pattern categorization
Based on their E. coli strain carriage pattern, we assigned participants to one of five categories. These included: 1) sustained positive (i.e., participants with ST131-H30R at all time points); 2) exposed, sustained negative (i.e., participants exposed to a ST131-H30R positive household member but who remained ST131-H30R negative); 3) sustained loss (i.e., participants with at least one ST131-H30R isolate, followed by at least two samplings without ST131-H30R, and who never re-acquired ST131-H30R); 4) unexposed negative (i.e., participants without ST131-H30R and not exposed to a household member with ST131-H30R); and 5) other (i.e., participants not falling into the above four categories).
Sample processing and microbiome characterization
DNA isolation and purification were performed using the collected fecal swabs. Total DNA was extracted from 200 μl of undiluted swab eluent using the MagAttract PowerMicrobiome DNA/RNA kit (Qiagen Inc., Valencia, CA, USA) and eluted into 100 μl of elution buffer and stored at −80 °C until analysis.
The prevalence and proportional abundance of specific bacterial taxa in each fecal swab were characterized by sequencing the 16S rRNA gene V3–V4 region as described in Fadrosh et al.17 using MiSeq Reagent Kit v3 (600-cycle) (Illumina Inc., San Diego, CA). A negative-extraction control (NEC) was included with each batch of extraction and sequenced in order to assess for cross-contamination. No-template controls (NTCs) and positive-template controls (PTCs) were included to assess for cross-contamination and verify PCR performance.
An in-house pipeline built with published tools was used for amplicon data processing. Briefly, primers were clipped by cutadapt v2.418 and resultant sequences quality-trimmed using Trimmomatic v0.39.19 then processed with DADA2 v1.1020 to identify amplicon sequence variants (ASVs). ASVs were queried using BLAST v.2.2.30+21 with cut off values of 99% nucleotide identity and 100% query coverage to remove bloomed ASVs22 (https://github.com/knightlab-analyses/bloom-analyses/blob/master/data/newbloom.all.fna). After removing bloom ASVs, the remaining ASVs were classified at 80% bootstrap confidence level using the Naïve Bayesian Classifier (v.2.12).23 Additional details can be found at https://github.com/araclab/mb_analysis.
Samples with under 1000 reads were excluded from analysis. After filtering out rare taxa (any genera accounting for <0.0025% of the entire dataset), the resultant sequencing data were used to calculate the prevalence and the proportional abundance of each taxon for each sample (i.e., Number of 16S rRNA gene sequences assigned to a taxon divided by the total number of 16S rRNA sequences). Data from this study is available at SRA project number PRJNA838578.
Comparative gut microbiome analysis
We first used cross-sectional data to identify gut microbiome features that are associated with ST131-H30R carriage, then used longitudinal data to determine how the identified gut microbiome features correlate with ST131-H30R carriage and loss. Samples from pets were excluded from all analyses. Primary analyses focused on comparing ST131-negative participants to H30R-positive participants, given the clinical ambiguity of non-H30R ST131 strain carriage. Supplemental analyses included comparisons across four mutually exclusive carriage subcategories (ST131-negative, non-H30R ST131, ST131-H30R1, ST131-H30Rx).
The cross-sectional study included, for ST131 carriers, the first ST131-positive sample and, for ST131 non-carriers, the first sample overall, which by definition was ST131-negative. Alpha-diversity was measured using the Shannon diversity index. Among bacterial phyla with ≥10% prevalence (i.e., proportion of subjects positive), we identified phyla associated with ST131-H30R carriage using analyses as described below.
We next used longitudinal data to identify gut bacterial taxa associated with ST131-H30R carriage. For this, we considered three sets of candidate genera. The first set comprised genera that were members of phyla identified as being associated with ST131-H30R carriage in the cross-sectional study and also present in at least 25% of the overall study population. The second set comprised the 10 most-informative genera, as identified by using the mean decrease in Gini values in conjunction with the largest number of inclusions in trees in random forest models. The third set comprised two genera that we regarded a priori as being of interest: Bacteroides and Escherichia/Shigella. For the random forest model, we selected a breakpoint for the number of taxa to be included by identifying an elbow on a scree plot based on the Gini and margin values.
We used Chi-squared tests and Wilcoxon Rank–Sum tests to identify associations between ST131-H30R carriage and the prevalence and proportional abundance of phyla and genera. Correlations between ST131-H30R carriage and the proportional abundance of phyla and genera were evaluated among longitudinal samples using a mixed effects logistic regression model, allowing for repeated measures by individual. Statistical analyses were conducted in SAS, version 9.4, and R, version 3.3.1.
Role of the funding source
The funders had no role in study design, data collection, analysis, or interpretation, or any aspect pertinent to the study.
Results
Participant characteristics and ST131 carriage
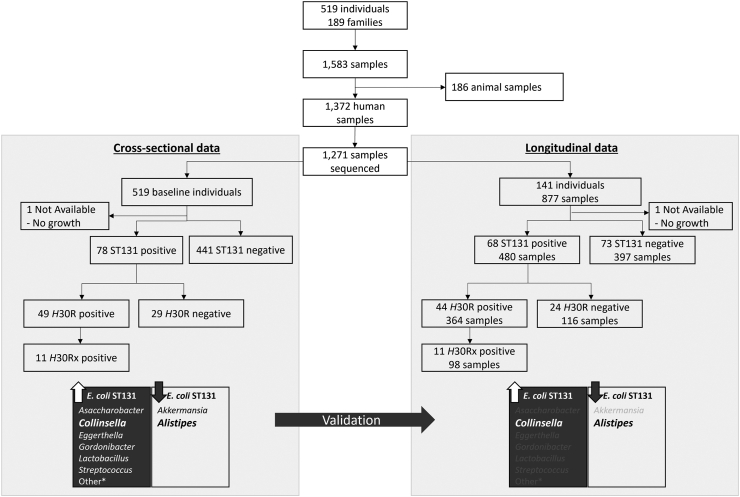
Of the 519 participants, 62.8% were veterans, 31.6% were a veteran's spouse, and 5.6% were a child or non-spouse adult in the household (Table 1). The initial fecal screening by conventional culture and PCR identified 78 (15.0%) participants as carriers of ST131 and/or fluoroquinolone-resistant E. coli. All members of households with an ST131 E. coli carrier were invited to undergo longitudinal surveillance; an average of 1.6 participants were enrolled per household. In total, we followed longitudinally 141 participants, including 44 of 49 H30R-positive participants (mean, 8.5 samples each [SD = 3.8]) and 73 of 441 ST131-negative participants (mean, 5.5 samples each [SD = 2.3]).
Table 1.
Demographic and other characteristics of participants.
| Baseline characteristics | No. (column %) |
|||
|---|---|---|---|---|
| Non-ST131 n = 441 | Any ST131 n = 78 | ST131 |
||
| Non-H30R n = 29 | H30R n = 49 | |||
| Participant | ||||
| Veteran (patient) | 279 (63.3%) | 47 (60.3%) | 18 (62.1%) | 29 (59.2%) |
| Child | 17 (3.9%) | 2 (2.6%) | 1 (3.5%) | 1 (2.0%) |
| Spouse | 136 (30.8%) | 28 (35.9%) | 10 (34.5%) | 18 (36.7%) |
| Other Adult | 9 (2.0%) | 1 (1.3%) | 0 (−) | 1 (2.0%) |
| Patient type | ||||
| Inpatient | 186 (42.2%) | 25 (32.1%) | 14 (48.3%) | 12 (24.5%) |
| Outpatient | 255 (57.8%) | 53 (68.0%) | 15 (51.7%) | 37 (75.5%) |
| Longitudinal visits | ||||
| ≥2 longitudinal visits | 84 (19.0%) | 57 (73.1%) | 22 (75.9%) | 36 (73.5%) |
Prevalence and dynamics of intestinal E. coli and ST131
According to genus-level sequencing data, at baseline 463 (89.2%) of the 519 study participants carried Escherichia. The proportional abundance of Escherichia at baseline ranged from 0% to 92.1% (mean 18.8%, SD 19.1) among the 519 participants.
According to PCR testing, 78 (15.0%) of the 519 participants had ST131 in one or more samples, including 49 (9.2%) with H30R, among which 38 (7.1%) were H30R1 and 11 (2.1%) were H30Rx (Fig. 1). Of the 141 participants with longitudinal data, 25 (18%) were persistently H30R-positive (i.e., sustained positives), 11 (8%) were persistently H30R-negative despite household exposure (i.e., exposed, sustained negatives), 14 (10%) were H30R-positive, then persistently H30R-negative (i.e., sustained loss), 62 (44%) were H30R-negative in the absence of household exposure (unexposed negatives), and 27 (19%) exhibited other patterns.
Fig. 1.
Participant flow for inclusion into cross-sectional and longitudinal analyses, for the ST131, H30, and H30R categories. Flowchart for the cross-sectional and longitudinal data, including characterization of the number of families, individuals, samples, and molecular characterization of E. coli isolates. High-level associations between taxa and E. coli ST131 carriage are provided at the bottom, with cross-sectional association findings being validated in a subset of individuals with available longitudinal data.
Cross-sectional analysis for gut microbiome characteristics associated with ST131-H30R carriage
To identify gut bacterial taxa associated with carriage of ST131-H30R, we first performed a cross-sectional analysis involving all 519 participants, focusing initially on phyla, then genera. Alpha diversity was similar across all ST131 sub-categories (non-H30R, H30R, and H30Rx; p = 0.95), but was significantly lower when comparing ST131-negative individuals compared with ST131-positive individuals (p = 0.012, Supplemental Figure S4).
At the phylum level, ST131-H30R carriage was associated with both a higher proportional abundance of Actinobacteria and, conversely, a lower proportional abundance of Verrucomicrobia. Specifically, H30R-positive participants had a significantly higher proportional abundance of Actinobacteria (mean 4.9%, SD 4.1), as compared to ST131-negative participants (mean 3.1%, SD 4.6) (p = 0.002) (Table 2). Additionally, H30Rx-positive participants had the highest abundance of Actinobacteria (mean 6.7%, SD 4.0), whereas participants with no detectable Escherichia had the lowest abundance (mean 1.3%, SD 1.6) (Supplemental Table S1, Supplemental Figure S1). By contrast, H30R-positive participants had a significantly lower proportional abundance of Verrucomicrobia (mean 0.3%, SD 1.0) than did ST131-negative participants (mean 1.3%, SD 3.4) (p = 0.04).
Table 2.
Prevalence and mean proportional abundance of genera in cross-sectional analysis.
| Phylum or genus | Random foresta | Prevalenceb, column percent |
p value |
Proportional abundancec, mean (SD) |
p value |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. non-ST131 |
B. any ST131 |
C. ST131, non-H30R |
D. ST131, H30R |
A vs B | A v D | A. non-ST131 |
B. any ST131 |
C. ST131, non-H30R |
D. ST131, H30R |
A vs B | A v D | ||
| N = 441 | N = 78 | N = 29 | N = 49 | N = 441 | N = 78 | N = 29 | N = 49 | ||||||
| Actinobacteria | 94 | 96 | 100 | 94 | 0.51 | 0.90 | 3.1 (4.6) | 4.7 (3.9) | 4.3 (3.6) | 4.9 (4.1) | <0.001 | 0.001 | |
| Asaccharobacterd | X | 25 | 46 | 48 | 45 | <0.001 | 0.004 | 0.1 (0.1) | 0.1 (0.2) | 0.1 (0.2) | 0.1 (0.2) | <0.001 | 0.008 |
| Bifidobacterium | 62 | 71 | 72 | 69 | 0.15 | 0.30 | 0.9 (2.4) | 0.8 (1.2) | 0.8 (1.2) | 0.7 (1.3) | 0.32 | 0.78 | |
| Collinsella | X | 67 | 82 | 90 | 78 | 0.007 | 0.12 | 1.1 (2.5) | 2.1 (2.5) | 1.9 (2.3) | 2.3 (2.7) | <0.001 | 0.001 |
| Eggerthella | X | 47 | 62 | 72 | 55 | 0.02 | 0.31 | 0.2 (0.4) | 0.6 (1.1) | 0.7 (1.0) | 0.6 (1.2) | 0.001 | 0.18 |
| Gardnerella | X | 7 | 26 | 38 | 18 | <0.001 | 0.006 | 0.0 (0.3) | 0.2 (1.0) | 0.1 (0.3) | 0.3 (1.3) | <0.001 | 0.004 |
| Gordonibacter | 32 | 54 | 55 | 53 | <0.001 | 0.003 | 0.3 (1.0) | 0.5 (1.2) | 0.5 (1.2) | 0.5 (1.2) | <0.001 | 0.01 | |
| Bacteroidetes | 100 | 100 | 100 | 100 | 1.00 | 1.00 | 30.5 (15.7) | 27.6 (13.0) | 24.5 (13.0) | 29.4 (12.8) | 0.20 | 0.78 | |
| Alistipes | X | 88 | 82 | 90 | 78 | 0.15 | 0.04 | 2.6 (3.8) | 1.8 (2.9) | 1.2 (1.4) | 2.1 (3.5) | 0.01 | 0.08 |
| Bacteroides | 100 | 100 | 100 | 100 | 0.55 | 0.64 | 23.9 (14.7) | 22.2 (11.8) | 20.4 (11.3) | 23.3 (12.1) | 0.52 | 0.78 | |
| Firmicutes | 100 | 100 | 100 | 100 | 0.55 | 0.64 | 39.7 (18.3) | 40.9 (14.3) | 39.2 (16.5) | 41.9 (12.9) | 0.35 | 0.22 | |
| Enterococcus | X | 43 | 53 | 55 | 51 | 0.10 | 0.26 | 0.8 (3.1) | 2.8 (8.0) | 4.5 (10.5) | 1.9 (6.0) | 0.03 | 0.17 |
| Lactobacillus | X | 33 | 65 | 62 | 67 | <0.001 | <0.001 | 0.3 (1.2) | 1.3 (3.3) | 1.0 (3.0) | 1.5 (3.5) | <0.001 | <0.001 |
| Streptococcus | X | 68 | 88 | 97 | 84 | <0.001 | 0.02 | 1.2 (3.9) | 2.0 (4.5) | 1.5 (2.0) | 2.3 (5.4) | <0.001 | 0.001 |
| Weissella | X | 2 | 9 | 14 | 6 | 0.002 | 0.11 | 0.0 (0.3) | 0.0 (0.2) | 0.0 (0.1) | 0.0 (0.2) | 0.003 | 0.10 |
| Fusobacteria | 13 | 14 | 14 | 14 | 0.78 | 0.79 | 0.2 (1.2) | 0.1 (0.8) | 0.1 (0.2) | 0.2 (1.0) | 0.87 | 0.85 | |
| Lentisphaerae | 10 | 5 | 0 | 8 | 0.16 | 0.65 | 0.0 (0.1) | 0.0 (0.0) | – | 0.0 (0.0) | 0.14 | 0.61 | |
| Proteobacteria | 98 | 100 | 100 | 100 | 0.20 | 0.31 | 24.8 (20.4) | 25.5 (17.5) | 31.3 (18.6) | 22.0 (16.0) | 0.35 | 0.74 | |
| Escherichia | 88 | 99 | 100 | 98 | 0.003 | 0.03 | 18.7 (19.6) | 19.5 (16.0) | 22.8 (16.8) | 17.6 (15.4) | 0.07 | 0.37 | |
| Oxalobacterd | X | 4 | 12 | 3 | 16 | 0.004 | <0.001 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.004 | <0.001 |
| Synergistetes | 13 | 13 | 0 | 20 | 0.98 | 0.15 | 0.1 (0.3) | 0.4 (3.0) | – | 0.6 (3.7) | 0.95 | 0.16 | |
| Verrucomicrobia | 52 | 38 | 34 | 41 | 0.048 | 0.22 | 1.3 (3.4) | 0.3 (0.9) | 0.2 (0.8) | 0.3 (1.0) | 0.003 | 0.04 | |
| Akkermansia | 52 | 38 | 34 | 41 | 0.03 | 0.14 | 1.3 (3.4) | 0.3 (0.9) | 0.2 (0.8) | 0.3 (1.0) | 0.005 | 0.02 | |
Bold denotes p < 0.05.
X indicates the taxon was selected as one of the top-10 most informative genera for predicting ST131 carriage using the random forest model.
Percent of participants in a group with a given taxon.
Mean proportional contribution of each taxon to each participant's gut microbiome (proportional abundance for a given sample was calculated as the number of reads of the given taxon divided by the total number of reads in that sample).
A genetic near neighbor of the taxon.
By contrast with Actinobacteria and Verrucomicrobia, the phyla Bacteroidetes, Firmicutes, and Proteobacteria occurred in nearly all participants, and did not differ significantly in prevalence or abundance by ST131 status or ST131 subclone carriage (not shown). Firmicutes was the most proportionally abundant of these phyla (mean 39.9%, SD 17.8), followed by Bacteroidetes (mean 30.1%, SD 15.4) and Proteobacteria (mean 24.9%, SD 20.0).
Cross-sectional analysis for genera and species associated with ST131-H30R carriage
At the genus level, although multiple genera were positively associated with H30R carriage, only Alistipes was negatively associated with H30R carriage. Specifically, of the genera examined, Gardnerella, Gordonibacter, Lactobacillus, Streptococcus, and a phylogenetic near neighbor of Asacharobacter all exhibited a higher prevalence and greater proportional abundances in association with carriage of ST131 and H30R, and Collinsella, Eggerthella, and Weissella exhibited similar association with carriage of ST131 (but not H30R).
By contrast, Alistipes and Akkermansia were negatively associated with H30R carriage. Specifically, as compared with H30R-positive participants, ST131-negative participants had a higher prevalence or proportional abundance of Alistipes (p = 0.04 and p = 0.08, respectively), and a higher proportional abundance of Akkermansia (p = 0.02), although a similar prevalence of Akkermansia (p = 0.14). Species-level associations are provided in supplemental analyses for Alistipes, along with the three most-prevalent genera associated with ST131 carriage (Collinsella, Lactobacillus, Streptococcus) (Supplemental Table S3).
Longitudinal associations between genera and H30R carriage
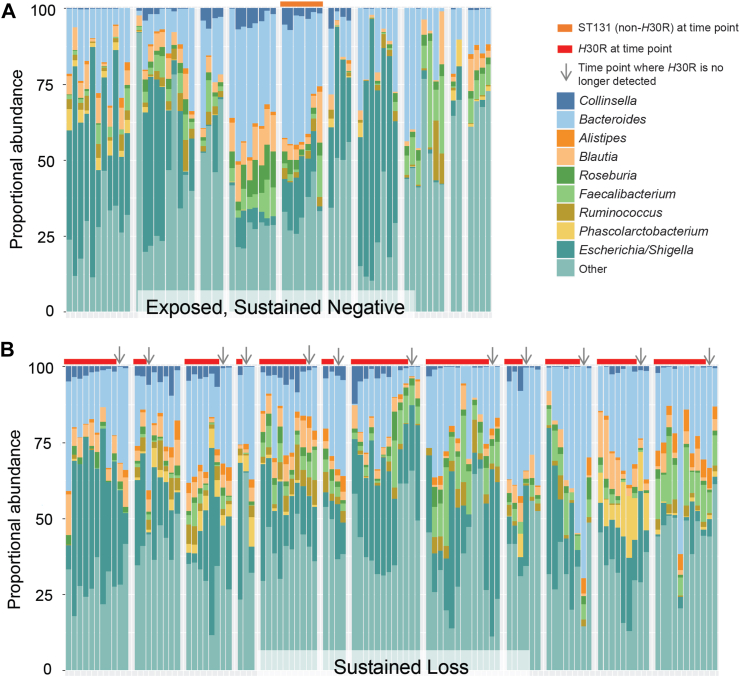
We next performed longitudinal analyses of the gut microbiome, limited to the genus level, from 25 participants with either of two clinically relevant H30R colonization patterns: exposed, sustained negative (participants with a H30R-positive household member who remained H30R-negative for the duration of the study) and sustained loss (H30R-positive individuals who became H30R-negative during the study). On average, participants contributed 7.6 visits.
Findings from the cross-sectional analysis were corroborated in the longitudinal analysis by using a mixed-effects logistic regression model that controlled for intra-individual variation over time and included all genera of interest from Table 2. Alistipes was the only genus significantly associated with absence of H30R (p = 0.005), whereas Collinsella was the only genus associated with presence of H30R (p = 0.035) (Supplemental Table S2). No other taxa displayed significant associations with H30R carriage. Among participants with sustained loss of H30R, Collinsella decreased in median proportional abundance with loss of H30R, from 1.5% to 0.5% (Fig. 2). Conversely, median proportional abundance of Alistipes increased from 0.8% to 2.0% after loss of H30R. Collinsella and Alistipes median proportional abundance were both low (0.2% and 0.4%, respectively) for participants who were exposed to a H30R-positive family member but remained ST131-negative.
Fig. 2.
Gut microbiota genera proportional abundance among exposed, sustained H30-negative (A) and sustained H30R loss participants (B). Each group of adjoined columns represents a sequential (from left to right) time series of samples from an individual participant. Columns represent the proportional abundance of genera at each time point. Colored horizontal bars above the columns represent ST131 (non-H30R, orange) and ST131-H30R (red) status over time for each participant. Arrows represent the time point where H30R is no longer detected for participants with sustained loss, which is generally accompanied by loss of Collinsella.
Discussion
In this study of veterans and their household members, we found that specific phyla and genera in the gut microbiome were consistently associated with ST131 and H30R carriage. In cross-sectional comparisons, the phylum Actinobacteria and certain common genera, including Lactobacillus, Streptococcus, and Collinsella (a member of Actinobacteria), were associated with presence of ST131 and H30R. In longitudinal analyses, the genus Collinsella again was associated with H30R-positivity, whereas the genus Alistipes was associated with absence and loss of ST131 and H30R. These findings identify potential opportunities for influencing gut colonization by ST131 and its H30R subclone.
The H30R subclone of E. coli, which has emerged as an important multidrug-resistant pathogen, causes a significant portion of antimicrobial-resistant infections in the U.S. and worldwide.1,24 Although H30R is known to be an excellent gut colonizer,6 the gut microbiome features associated with acquisition, persistence, and loss of H30R carriage are poorly understood. To our knowledge, this study provides the first description of the bacterial phyla and genera associated with H30R gastrointestinal carriage. If the observed associations represent a causal effect of specific microbiota on H30R colonization, interventions that shift those taxa could have important public health benefits.
Collinsella is a member of the phylum Actinobacteria which was associated with presence of ST131 and H30R in both the cross-sectional and longitudinal analyses (specifically, C. aerofaciens; Supplemental Table S3). Collinsella may be an important health-relevant constituent of the gut microbiome.25 Presence of Collinsella has been associated with diverse diseases, including irritable bowel syndrome, gestational diabetes mellitus, and atherosclerosis,26, 27, 28 whereas increased abundance of Collinsella aerofaciens has been associated with rheumatoid arthritis.29 Low dietary fiber intake by overweight pregnant women may allow overgrowth of Collinsella, have pro-inflammatory effects, and confer increased susceptibility to impaired glucose tolerance.30 Collinsella aerofaciens has also been linked with increased gut permeability and inflammation, specifically with production of IL-17A, along with high levels of alpha-aminoadipic acid and asparagine.29 Conceivably, the associations we observed of Collinsella with ST131 and H30R may be due to Collinsella-mediated modulation of the host inflammatory and immune systems in an ST131-favoring manner. If so, this would make Collinsella a potential intervention target. Alternatively, these associations may reflect nutritional or other exposures that jointly favor Collinsella, ST131, and H30R. Identification of such causal factors could have even broader health benefits. Evaluating causal mechanisms may require co-culture models, metagenomic, or meta-transcriptomic approaches.
By contrast with Collinsella, the genus Alistipes was negatively associated with H30R prevalence and ST131 proportional abundance. It has contrastingly been implicated as a risk factor for certain conditions, such as hypertension,31 but protective against others, including atrial fibrillation, cardiovascular disease,32, 33, 34 ulcerative colitis,35 and liver fibrosis.36 Studies have linked Alistipes with anti-inflammatory effects in the gut.37,38 These anti-inflammatory effects may be mediated by production of the short-chain fatty acids propionate and acetate,39,40 or through37,38 other immuno-modulatory mechanisms, leading to reduced inflammation in the gut and other organ systems.41,42 As with Collinsella, hypothesis testing regarding possible inflammation-mediated mechanisms for the observed relationship between Alistipes and H30R may be worthy of future consideration given the tremendous public health importance of H30R colonization.
Interestingly, multiple gut microbiome studies have observed an inverse correlation between Alistipes and both Escherichia and Streptococcus,32,34,42 Conceivably, reductions in Alistipes may contribute independently to both disease pathogenesis and ST131 and H30R carriage. Further evaluations of interactions between these key taxa may have implications for inflammation and disease and may elucidate potential targets for reductions in ST131-H30R colonization such as targeted probiotics and diet modifications.
The study had limitations. First, the limited demographic and clinical data precluded adjustment for potentially important exposures such as antibiotic use, diet, recent travel, and probiotic use; absence of these important factors in the available data limit our ability to infer causal associations. Second, fecal swabs underwent extended room temperature exposure between collection and processing. To address this, we used bioinformatics methods to reduce distortions from potential in vitro blooms and limited our analyses to prevalence and proportional abundance, which are less affected by overgrowth of specific taxa. The consistency of findings between prevalence and proportional abundance measures, and also between cross-sectional and longitudinal analyses, support validity of the findings with regards to both sensitivity of detecting ST131-H30R isolates and also conclusions regarding Alistipes, Collinsella, and other taxa of interest. Third, the study population was restricted temporally, geographically, and socio-culturally; other populations might yield different results. Fourth, fecal swabs may not accurately reflect the total gut microbiota. Fifth, while species-level data for the most prevalent genera associated with ST131-H30R were evaluated in supplemental cross-sectional analyses (Supplemental Table S3), we did not apply the longitudinal model due to concerns of sample size and overfitting with the large number of species. Sixth, the resolution of 16S rRNA gene amplicon sequencing may be limited to the genus level due to high similarity in the 16S rRNA gene for some closely related species. Lastly, due to sample size limitations, our primary analyses focused on comparing ST131 to no ST131 carriage, rather than across all possible subclone categories. Different E. coli subclones may exhibit differing niche adaptive abilities,43 setting the stage for niche-specific competition and behavior.44
In summary, gastrointestinal carriage of ST131, and specifically H30R, a leading antimicrobial-resistant pathogen, is associated with presence and abundance of Collinsella and certain other genera, whereas absence and loss of H30R are associated with Alistipes. If these associations involving the gut microbiota represent causal pathways that reduce ST131 carriage, bacteria negatively associated with ST131 carriage may be useful directly as a probiotic species, or indirectly via a prebiotic or other dietary manipulations that may promote their growth and thereby protect against ST131 colonization. Conversely, bacteria positively associated with ST131 carriage may be potential targets for vaccines or narrow-spectrum therapeutics such as bacteriophages that aim to mitigate an important public health threat.
Contributors
JRJ, LBP, CML conceived and initiated the study. JRJ, BJK, CC curated the data. BJK, KR, DEP, MA, CML conducted the statistical analysis. JRJ, CC supervised the study. JRJ, LBP, CML, DEP, MA drafted and revised the manuscript. All authors read and approved the final manuscript.
Data sharing statement
The dataset supporting the conclusions of this article is available at SRA project number PRJNA838578 Any additional relevant data are available upon request.
Declaration of interests
J. R. J. has received grants or consultancies from Achaogen/Cipla, Allergan, Janssen/Crucell, Melinta, Merck, Shionogi, Syntiron, and Tetraphase, and has patent application for tests to detect specific E. coli strains. All other authors report no actual or potential conflicts.
Acknowledgements
This work was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [1R21AI117654-01 to LBP and UM1AI104681 to JRJ], and the Office of Research and Development, Department of Veterans Affairs [1I01 CX000920-01 and 2I01CX000920-04 to JRJ]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veteran Affairs.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104909.
Appendix A. Supplementary data
References
- 1.Johnson J.R., Porter S., Thuras P., Castanheira M. The pandemic H30 subclone of sequence type 131 (ST131) as the leading cause of multidrug-resistant Escherichia coli infections in the United States (2011-2012) Open Forum Infect Dis. 2017;4:ofx089. doi: 10.1093/ofid/ofx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tchesnokova V.L., Rechkina E., Chan D., et al. Pandemic uropathogenic fluoroquinolone-resistant Escherichia coli have enhanced ability to persist in the gut and cause bacteriuria in healthy women. Clin Infect Dis. 2020;70:937–939. doi: 10.1093/cid/ciz547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langelier C., Graves M., Kalantar K., et al. Microbiome and antimicrobial resistance gene dynamics in international travelers. Emerg Infect Dis. 2019;25:1380–1383. doi: 10.3201/eid2507.181492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valverde A., Grill F., Coque T.M., et al. High rate of intestinal colonization with extended-spectrum-β-lactamase-producing organisms in household contacts of infected community patients. J Clin Microbiol. 2008;46:2796–2799. doi: 10.1128/JCM.01008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilty M., Betsch B.Y., Bögli-Stuber K., et al. Transmission dynamics of extended-spectrum β-lactamase–producing enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis. 2012;55:967–975. doi: 10.1093/cid/cis581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson J.R., Clabots C., Porter S.B., Bender T., Johnston B.D., Thuras P. Intestinal persistence of colonizing Escherichia coli strains, especially ST131- H 30, in relation to bacterial and host factors. J Infect Dis. 2022 doi: 10.1093/infdis/jiab638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokurenko E.V., Chesnokova V., Dykhuizen D.E., et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connell I., Agace W., Klemm P., Schembri M., Mărild S., Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson J.R., Urban C., Weissman S.J., et al. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and bla CTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother. 2012;56:2364–2370. doi: 10.1128/AAC.05824-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson J.R., Davis G., Clabots C., et al. Household clustering of Escherichia coli sequence type 131 clinical and fecal isolates according to whole genome sequence analysis. Open Forum Infect Dis. 2016;3 doi: 10.1093/ofid/ofw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres E., López-Cerero L., Morales I., Navarro M.D., Rodríguez-Baño J., Pascual A. Prevalence and transmission dynamics of Escherichia coli ST131 among contacts of infected community and hospitalized patients. Clin Microbiol Infection. 2018;24:618–623. doi: 10.1016/j.cmi.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Zmora N., Zilberman-Schapira G., Suez J., et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–1405.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 13.Ducarmon Q.R., Terveer E.M., Nooij S., et al. Microbiota-associated risk factors for asymptomatic gut colonisation with multi-drug-resistant organisms in a Dutch nursing home. Genome Med. 2021;13:54. doi: 10.1186/s13073-021-00869-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liss M.A., Leach R.J., Rourke E., et al. Microbiome diversity in carriers of fluoroquinolone resistant Escherichia coli. Investig Clin Urol. 2019;60:75. doi: 10.4111/icu.2019.60.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araos R., Battaglia T., Ugalde J.A., Rojas-Herrera M., Blaser M.J., D'Agata E.M.C. Fecal microbiome characteristics and the resistome associated with acquisition of multidrug-resistant organisms among elderly subjects. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohamed M., Clabots C., Porter S.B., Thuras P., Johnson J.R. Isolation and characterization of Escherichia coli sequence type 131 and other antimicrobial-resistant gram-negative Bacilli from clinical stool samples from veterans. Antimicrob Agents Chemother. 2016;60:4638–4645. doi: 10.1128/AAC.00383-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fadrosh D.W., Ma B., Gajer P., et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- 19.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Amir A., McDonald D., Navas-Molina J.A., et al. Correcting for microbial blooms in fecal samples during room-temperature shipping. mSystems. 2017;2 doi: 10.1128/mSystems.00199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolas-Chanoine M.-H., Bertrand X., Madec J.-Y. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tourlousse D.M., Sakamoto M., Miura T., et al. Complete genome sequence of Collinsella aerofaciens JCM 10188 T. Microbiol Resour Announc. 2020;9 doi: 10.1128/MRA.00134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kassinen A., Krogius-Kurikka L., Mäkivuokko H., et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson F.H., Fåk F., Nookaew I., et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrocino I., Ponzo V., Gambino R., et al. Changes in the gut microbiota composition during pregnancy in patients with gestational diabetes mellitus (GDM) Sci Rep. 2018;8 doi: 10.1038/s41598-018-30735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J., Wright K., Davis J.M., et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Arango L.F., Barrett H.L., Wilkinson S.A., et al. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microb. 2018;9:189–201. doi: 10.1080/19490976.2017.1406584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S., Goel R., Kumar A., et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci. 2018;132:701–718. doi: 10.1042/CS20180087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jie Z., Xia H., Zhong S.-L., et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo K., Li J., Li K., et al. Disordered gut microbiota and alterations in metabolic patterns are associated with atrial fibrillation. GigaScience. 2019;8 doi: 10.1093/gigascience/giz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui X., Ye L., Li J., et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci Rep. 2018;8:635. doi: 10.1038/s41598-017-18756-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dziarski R., Park S.Y., Kashyap D.R., Dowd S.E., Gupta D. Pglyrp-regulated gut microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campion D., Giovo I., Ponzo P., Saracco G.M., Balzola F., Alessandria C. Dietary approach and gut microbiota modulation for chronic hepatic encephalopathy in cirrhosis. World J Hepatol. 2019;11:489–512. doi: 10.4254/wjh.v11.i6.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andoh A. Physiological role of gut microbiota for maintaining human health. Digestion. 2016;93:176–181. doi: 10.1159/000444066. [DOI] [PubMed] [Google Scholar]
- 38.Arpaia N., Campbell C., Fan X., et al. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature. 2013;504:451. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polansky O., Sekelova Z., Faldynova M., Sebkova A., Sisak F., Rychlik I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol. 2016;82:1569–1576. doi: 10.1128/AEM.03473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliphant K., Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. 2019;7:91. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker B.J., Wearsch P.A., Veloo A.C.M., Rodriguez-Palacios A. The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang W., Wu N., Wang X., et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meador J.P., Caldwell M.E., Cohen P.S., Conway T. Escherichia coli pathotypes occupy distinct niches in the mouse intestine. Infect Immun. 2014;82:1931–1938. doi: 10.1128/IAI.01435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter S.B., Johnston B.D., Kisiela D., Clabots C., Sokurenko E.V., Johnson J.R. Bacteriophage cocktail and microcin-producing probiotic Escherichia coli protect mice against gut colonization with multidrug-resistant Escherichia coli sequence type 131. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.887799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.