Abstract
Objective
BRCA1/2 status is a key to personalized therapy for invasive breast cancer patients. This study aimed to explore the association between ultrasound radiomics features and germline BRCA1/2 mutation in patients with invasive breast cancer.
Materials and methods
In this retrospective study, 100 lesions in 92 BRCA1/2-mutated patients and 390 lesions in 357 non-BRCA1/2-mutated patients were included and randomly assigned as training and validation datasets in a ratio of 7:3. Gray-scale ultrasound images of the largest plane of the lesions were used for feature extraction. Maximum relevance minimum redundancy (mRMR) algorithm and multivariate logistic least absolute shrinkage and selection operator (LASSO) regression were used to select features. The multivariate logistic regression method was used to construct predictive models based on clinicopathological factors, radiomics features, or a combination of them.
Results
In the clinical model, age at first diagnosis, family history of BRCA1/2-related malignancies, HER2 status, and Ki-67 level were found to be independent predictors for BRCA1/2 mutation. In the radiomics model, 10 significant features were selected from the 1032 radiomics features extracted from US images. The AUCs of the radiomics model were not inferior to those of the clinical model in both training dataset [0.712 (95% CI, 0.647–0.776) vs 0.768 (95% CI, 0.704–0.835); p = 0.429] and validation dataset [0.705 (95% CI, 0.597–0.808) vs 0.723 (95% CI, 0.625–0.828); p = 0.820]. The AUCs of the nomogram model combining clinical and radiomics features were 0.804 (95% CI, 0.748–0.861) in the training dataset and 0.811 (95% CI, 0.724–0.894) in the validation dataset, which were proved significantly higher than those of the clinical model alone by DeLong’s test (p = 0.041; p = 0.007). To be noted, the negative predictive values (NPVs) of the nomogram model reached a favorable 0.93 in both datasets.
Conclusion
This machine nomogram model combining ultrasound-based radiomics and clinical features exhibited a promising performance in identifying germline BRCA1/2 mutation in patients with invasive breast cancer and may help avoid unnecessary gene tests in clinical practice.
Keywords: BRCA mutation, Invasive breast cancer, Ultrasound, Radiomics, Nomogram
1. Introduction
1.1. Background
Breast cancer has become the most commonly diagnosed malignancy and the leading cause of cancer-related mortality in women worldwide [1]. Pathogenic mutations of breast cancer susceptibility genes 1 and 2 (BRCA1/2), two distinct tumor suppressor genes playing an integral part in DNA damage repair by homologous recombination [2], are present in about 3%–5% of unselected breast cancer patients in China and worldwide [3,4]. The cumulative risk of breast cancer in a lifetime is 38%–90% for pathogenic BRCA1 mutation carriers and 37%–69% for BRCA2 mutation carriers [[5], [6], [7]]. Breast cancer patients carrying pathogenic BRCA1/2 mutations have a higher incidence of a contralateral or a new ipsilateral disease [8] and have different choices in chemotherapy and targeted therapy. For BRCA1/2 carriers, platinum agents, rather than taxanes, are recommended for patients with advanced breast cancer [9]. Furthermore, olaparib, a poly ADP-ribose polymerase (PARP) inhibitor targeting the DNA damage response in BRCA1/2-mutated tumors, also expanded the options for BRCA1/2-mutated patients with HER-2 negative or advanced breast cancer [10]. Therefore, determining BRCA1/2 status in patients with breast cancer is of great help in predicting prognosis and making treatment decisions in clinical practice.
However, due to the complexity and heterogeneity of BRCA1/2, the detection of pathogenic BRCA1/2 mutations is highly dependent on next-generation sequencing technology [11], which is expensive, time-consuming, and of low availability. Because of these limitations, guidelines only recommend genetic testing for patients who meet certain criteria and, therefore, bear a relatively high risk of carrying pathogenic or likely pathogenic (P/LP) BRCA1/2 mutations [12]. The criteria proposed by the National Comprehensive Cancer Network (NCCN) are mainly comprised of clinicopathological characteristics, including diagnosis at a young age, triple-negative breast cancer (TNBC), a second breast cancer, personal or family history of other BRCA1/2-related malignancies, etc [12]. However, the NCCN criteria showed limited power in identifying BRCA1/2 mutations. In a retrospective cohort study involving 4196 breast cancer patients, the BRCA1/2 mutation rate was 9.4% in patients who met the NCCN criteria, while 7.9% in those who did not [13]. It was estimated that merely 30% of breast cancer patients with BRCA1/2 mutations had been identified in the United States [13], indicating that underdiagnosis of BRCA1/2 mutations in clinical practice is far from negligible. A more efficient method is needed to recognize BRCA1/2 mutation carriers.
Ultrasound (US) is widely used to screen and evaluate breast lesions. Compared to other imaging techniques, e.g., MRI or mammogram, US offers the advantages of real-time display and radiation-free nature, as well as being convenient and affordable. US-based radiomics also has been attracting attention in recent years. In radiomics studies, high-throughput quantitative features are extracted from digital medical images [14]. The features could be used to identify tumor information beyond the recognition of naked eye, hence carrying the potential for reflecting biological behaviors and characteristics at the genetic or molecular level [15,16]. For breast cancer, several radiomics models based on US images have been constructed and proved useful in distinguishing benign and malignant tumors, identifying lymph node metastasis, classifying molecular subtypes, evaluating Ki-67 levels, and predicting neoadjuvant treatment response, etc [[17], [18], [19], [20]]. These indicated that US-based radiomics analysis carries a great potential for identifying BRCA1/2 mutation in breast cancer patients.
To the best of our knowledge, this is the first study to explore the association between BRCA1/2 status and US radiomics features of breast cancer lesions. The present study aimed to explore whether radiomics features of gray-scale US images could be utilized to identify BRCA1/2 mutation, and further develop a model combining radiomics features and clinical factors to assist identifying BRCA1/2-mutated patients with invasive breast cancer.
2. Methods and materials
2.1. Patient enrollment
This study was approved by the Institutional Review Board of Sun Yat-sen University Cancer Center, with the requirement for informed consent waived. Breast cancer patients who took the BRCA1/2 gene test from Nov 2017 to Dec 2021 in Sun Yat-sen University Cancer Center were investigated.
The inclusion criteria were (a) female patients with US-suspected breast masses pathologically diagnosed as invasive breast cancer; (b) patients who took the BRCA1/2 gene test by next-generation sequencing technology using blood samples. BRCA1/2 genetic testing was recommended by physicians when risk factors, e.g. diagnosis at a young age, were detected in patients and brought concerns about familial breast cancer. Genetic testing was also done when requested by patients who did not necessarily carry risk factors. Patients were included in either case.
The exclusion criteria were (a) patients carrying BRCA1/2 variants of uncertain significance, whose impact on protein function is unknown; (b) patients who underwent treatment or invasive biopsy before US examination; (c) non-invasive carcinoma, e.g., ductal carcinoma in situ; (d) missing images or images of low quality, e.g., massive lesions beyond the limited width of the US probe; (e) incomplete clinicopathologic information, e.g., for multiple lesions observed in a single patient, the ones without separately reported immunohistochemical staining results were excluded; (f) difficult to match US images and pathological reports. The flow chart of the study population is shown in Fig. 1.
Fig. 1.
Flow chart of patient enrollment. US, ultrasound; P/LP, pathogenic or likely pathogenic; gBRCA1/2, germline BRCA1/2.
2.2. Clinical information collection and US image acquisition
Medical records of the patients were thoroughly reviewed. The age at first diagnosis of breast malignancy, and personal or family history of any BRCA1/2-related malignancy, including breast, ovarian, pancreatic, and prostate cancers, were recorded.
All included lesions were diagnosed as invasive breast cancer with available immunohistochemical (IHC) staining results. Estrogen receptor (ER) or progesterone receptor (PR) negative were defined as <1% positive tumor cells, while positive as 1% or more positive tumor cells [21,22]. The cut-off value for Ki-67 was set to 20%. When Ki-67 ≥ 20%, it is marked as high expression level; otherwise, as low expression level [23]. Regarding human epidermal growth factor receptor 2 (HER2) status, 3+ was defined as HER2 positive, while 0 or 1+ as HER2 negative. When IHC results were scored as 2+, it should be further confirmed by fluorescence in situ hybridization (FISH) according to ASCO/CAP guidelines [24].
All included patients underwent US examination before receiving any treatment or invasive biopsy. The US examinations were performed by qualified radiologists using multiple ultrasound systems, including LOGIQ E9 and LOGIQ S8 (GE Healthcare, Chicago, IL, USA), MyLab Twice (Esaote, Genoa, Italy), Aloka Arietta 60 (Hitachi Healthcare Corporation, Tokyo, Japan), EPIQ 7 (Philips Healthcare, Best, the Netherlands), Acuson Sequoia and Acuson Juniper (Siemens Medical Solutions USA, Malvern, PA, USA), Aplio 400 (Toshiba Medical Systems, Tochigi, Japan), Resona 7T, I9S and DC-8 (Mindray Medical International Co., Ltd., Shenzhen, China), all with a 6–15L or 6–18L linear array transducer. US images were saved in the format of JPEG. US and pathological reports were compared carefully to match the images with the pathological results. For each lesion, the largest longitudinal image of satisfying quality was included in further analysis.
2.3. Region of interest (ROI) segmentation and feature extraction
Segmentation of the ROIs was manually performed by delineating the tumor areas on gray-scale images in NovoUltrasound Kit (NUK, version 1.5.0, GE Healthcare, Shanghai, China), a software for quantitative analysis of medical images, by two experienced radiologists (Fig. 2). Radiomics features included first-order, shape, and textural features including gray-level co-occurrence matrix (GLCM), gray-level run-length matrix (GLRLM), gray-level size zone matrix (GLSZM), neighbourhood gray-tone difference matrix (NGTDM), gray-level dependence matrix (GLDM) features. From each US image, 1032 radiomics features were extracted. Part of the features were obtained from the wavelet transform that uses two orthogonal filters, low-pass (L) and high-pass (H), to generate 4 subgroups of signals from a single image, including one approximation (LL) and three detailed features of different directions [horizontal (LH), vertical (HL) and diagonal (HH)], therefore providing multi-resolution time/spatial-frequency analysis for texture of medical images [25].
Fig. 2.
Workflow of construction of predictive models for gBRCA1/2 mutation in patients with invasive breast cancer. Details of each step are shown on both sides. US, ultrasound; gBRCA1/2, germline BRCA1/2.
Fifty US images were randomly chosen for further stability analysis. One radiologist segmented the ROIs twice with an interval of eight weeks to evaluate intra-observer agreements, and two radiologists segmented the ROIs independently to evaluate inter-observer agreements. The intra-class correlation coefficients (ICCs) were utilized to evaluate intra- and inter-observer agreements of the extracted features. Features with intra- or inter-observer ICCs less than 0.75 were considered of relatively low reproducibility and were removed from the following analysis.
2.4. Data preprocessing
The cohort was randomly assigned to either the training or the validation datasets in a ratio of 7:3. The training and the validation datasets were utilized to develop and evaluate the predictive models, respectively. The included radiomics features were standardized by Z-score standardization before analysis.
2.5. Feature selection and model construction
Logistic models were developed from the training dataset and validated by the validation dataset. Using age, menopause status, personal and family history of BRCA1/2-related malignancies, and pathological findings, the clinical model was developed. Features with a p-value < 0.05 in the univariate logistics regression analysis were further involved in multivariate logistics regression analysis, which was done in a backward stepwise manner. In each step, factors with a p-value > 0.05 were omitted, and those with a p-value < 0.05 in the final step were considered significant predictors of BRCA1/2 mutation.
Based on radiomics features, another logistic model was developed. A maximum relevance minimum redundancy (mRMR) algorithm was used to rank the radiomics features and select a subset of features. The mRMR method aims to select features that correlate with the target class the most (high relevance) while being mutually exclusive (low redundancy) [26] and is widely used in the analysis of medical images. Then, using multivariate logistic least absolute shrinkage and selection operator (LASSO) regression, features were further narrowed down. Cross-validation was used to determine the optimal coefficients. The features remaining in LASSO regression were included in the radiomics model.
The radiomics scores were calculated and combined with significant clinical predictors to construct a comprehensive logistic model. A nomogram was developed according to the results. Fig. 2 shows the workflow of this study.
2.6. Statistical analysis
The data were expressed as number (%) or mean ± standard deviation. Model performances were evaluated by depicting receiver operating characteristic curves (ROC). Decision curve analyses (DCA) were performed to quantify the net benefits and assess the usefulness of the models in instructing clinical decisions. For each model, the optimal cut-off value was determined by maximizing the Youden index, which equals sensitivity + specificity - 1. Accuracy, sensitivity, specificity, positive predictive values (PPV), negative predictive values (NPV), and area under curve (AUC) values were then calculated; and the DeLong’s tests were used to compare the difference between AUCs. For all statistical analyses, Python v3.8.8 was used. A two-tailed p-value < 0.05 was considered significant.
3. Results
3.1. Patient characteristics
A total of 490 breast cancer lesions in 449 patients were included, among which, 100 (20.4%) lesions identified in 92 patients (20.5%) carried pathogenic or likely pathogenic (P/LP) BRCA1/2 mutations.
BRCA1/2-mutated and non-mutated cases showed differences in many aspects (Table 1). Patients with BRCA1/2 mutation tended to be firstly diagnosed with breast malignancy at a younger age (average 40.1 vs 43.1, p = 0.009) and had a higher ratio of having a family history of BRCA1/2-related malignancies (47.3% vs 15.5%, p < 0.001). In the analysis of IHC results, BRCA1/2-mutated lesions demonstrated a higher ratio of negative ER (42.0% vs 31.3%, p = 0.043), negative HER2 (96.0% vs 74.4%, p = 0.001) and, therefore, a higher ratio of TNBC (31.0% vs 18.5%, p < 0.001). Higher expression of Ki-67 was also shown in BRCA1/2-mutated lesions (96.0% vs 80.3%, p < 0.001).
Table 1.
Clinicopathologic Characteristics of BRCA1/2-mutated and non-BRCA1/2-mutated Patients and Lesions.
| Characteristics |
Non-gBRCA1/2-mutated |
gBRCA1/2-mutated |
P value |
|
|---|---|---|---|---|
| Number of patients | 357 (79.5%) | 92 (20.5%) | ||
| Age at first diagnosis/year | 43.1 ± 10.3 | 40.1 ± 8.6 | 0.009 | |
| Menopause | 90 (28.2%) | 16 (19.8%) | 0.131 | |
| Bilateral∗ | 24 (6.7%) | 12 (13.0%) | 0.054 | |
| Previous BC | 28 (4.5%) | 11 (8.8%) | 0.216 | |
| Family History† | 52 (15.5%) | 41 (47.3%) | <0.001 | |
| Number of lesions | 390 (79.6%) | 100 (20.4%) | ||
| Tumor Size/mm | 25.2 ± 12.3 | 25.1 ± 10.1 | 0.986 | |
| Side (Left) | 218 (55.9%) | 48 (48.0%) | 0.157 | |
| ER status | Positive | 268 (68.7%) | 58 (58.0%) | 0.043 |
| Negative | 122 (31.3%) | 42 (42.0%) | ||
| PR status | Positive | 237 (60.8%) | 53 (53.0%) | 0.158 |
| Negative | 153 (39.2%) | 47 (47.0%) | ||
| HER2 status | Positive | 100 (25.6%) | 10 (10.0%) | 0.001 |
| Negative | 290 (74.4%) | 90 (90.0%) | ||
| Ki-67 | ≥20% | 313 (80.3%) | 96 (96.0%) | <0.001 |
| <20% | 77 (19.7%) | 4 (4.0%) | ||
| Molecular subtype | Luminal A | 52 (13.3%) | 4 (4.0%) | <0.001 |
| Luminal B1 | 166 (42.6%) | 55 (55.0%) | ||
| Luminal B2 | 67 (17.2%) | 5 (5.0%) | ||
| TNBC | 72 (18.5%) | 31 (31.0%) | ||
| HER2+ HR- | 33 (8.5%) | 5 (5.0%) | ||
BC: breast cancer; ER: oestrogen receptor; PR: progesterone receptor; HR: hormone receptor; HER2: human epidermal growth factor receptor 2; TNBC: triple-negative breast cancer.
Including synchronous and metachronous breast cancer.
Family history of gBRCA1/2-related malignancy, including breast, ovarian, pancreatic and prostate cancer.
The cases enrolled in the study were randomly assigned in a ratio of 7:3 as the training and the validation datasets. The clinical characteristics of the patients and the pathological characteristics of the lesions were compared between the two datasets (Table 2). For patients, no statistically significant difference was found in terms of the age at diagnosis, menopause status, family history of BRCA1/2-related malignancies, bilateral lesions, and previous breast cancer history cancer. For breast lesions, there was also no statistically significant difference in tumor size, IHC results including ER, PR, HER2 status, and Ki-67. Similar distributions in molecular subtypes were also presented between the training and the validation datasets.
Table 2.
Clinicopathologic characteristics of the patients and lesions enrolled.
| Characteristics |
Total |
Training |
Validation |
P value |
|
|---|---|---|---|---|---|
| Number of patients | 449 | 311 | 138 | ||
| BRCA1/2-mutated | 92 (20.5%) | 64 (20.6%) | 28 (20.3%) | 0.944 | |
| Age at first diagnosis/years | 42.5 ± 10.0 | 42.8 ± 10.0 | 41.9 ± 10.0 | 0.397 | |
| Menopause | 106 (23.6%) | 75 (24.1%) | 31 (22.4%) | 0.704 | |
| Bilateral∗ | 36 (8.0%) | 23 (7.4%) | 13 (9.4%) | 0.467 | |
| Previous BC | 39 (8.7%) | 26 (8.4%) | 13 (9.4%) | 0.714 | |
| Family history∗∗ | 93 (20.7%) | 66 (21.2%) | 27 (19.6%) | 0.690 | |
| Number of lesions | 490 | 343 | 147 | ||
| BRCA1/2-mutated | 100 (20.4%) | 70 (20.4%) | 30 (20.4%) | 1.000 | |
| Tumor Size/mm | 25.2 ± 11.9 | 24.9 ± 12.1 | 25.7 ± 11.4 | 0.506 | |
| Side (Left) | 266 (54.3%) | 183 (53.4%) | 83 (56.5%) | 0.528 | |
| ER status | Positive | 326 (66.5%) | 221 (64.4%) | 105 (71.4%) | 0.133 |
| Negative | 164 (33.5%) | 122 (35.6%) | 42 (28.6%) | ||
| PR status | Positive | 290 (59.2%) | 197 (57.4%) | 93 (63.3%) | 0.230 |
| Negative | 200 (40.8%) | 146 (42.6%) | 54 (36.7%) | ||
| HER2 status | Positive | 110 (22.4%) | 84 (24.5%) | 26 (17.7%) | 0.099 |
| Negative | 380 (77.6%) | 259 (75.5%) | 121 (82.3%) | ||
| Ki-67 | ≥20% | 409 (83.5%) | 289 (84.3%) | 120 (81.6%) | 0.475 |
| <20% | 81 (16.5%) | 54 (15.7%) | 27 (18.4%) | ||
| Molecular subtype | Luminal A | 56 (11.4%) | 37 (10.8%) | 19 (12.9%) | 0.177 |
| Luminal B1 | 221 (56.5%) | 147 (42.9%) | 74 (50.3%) | ||
| Luminal B2 | 72 (14.7%) | 54 (15.7%) | 18 (12.2%) | ||
| TNBC | 103 (21.0%) | 75 (21.9%) | 28 (19.0%) | ||
| HER2+ HR- | 38 (7.8%) | 30 (8.7%) | 8 (5.4%) | ||
BC: breast cancer; ER: oestrogen receptor; PR: progesterone receptor; HR: hormone receptor; HER2: human epidermal growth factor receptor 2; TNBC: triple-negative breast cancer.
Including synchronous and metachronous breast cancer.
Family history of BRCA1/2-related malignancy, including breast, ovarian, pancreatic and prostate cancer.
3.2. Clinical model construction and validation
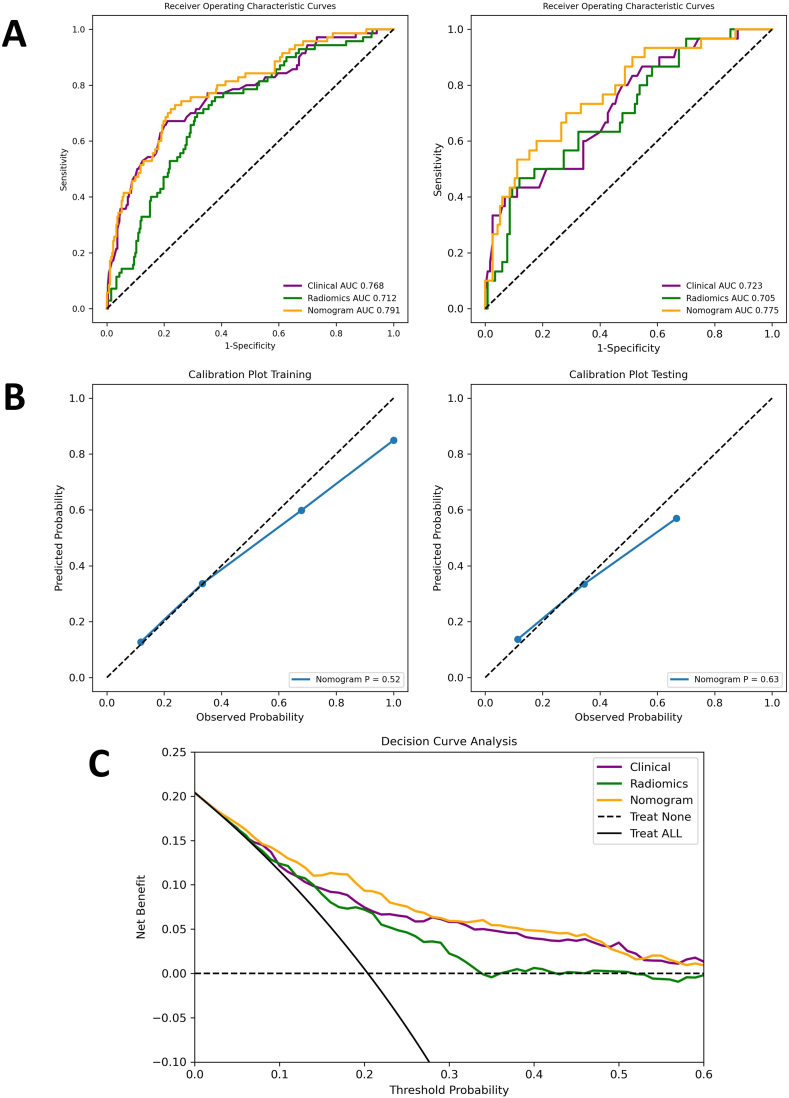
The clinicopathologic characteristics were analyzed using univariate and multivariate logistics regression (Table 3). Only factors with p-value < 0.05 were further included in multivariate analysis. According to the result, age at first diagnosis (p = 0.007), family history of BRCA1/2-related malignancies (p < 0.001), HER2 status (p = 0.003), and Ki-67 level (p = 0.015) were independent predictors for BRCA1/2 mutation. A four-factor clinical model for identifying BRCA1/2 mutation was constructed using the β coefficients summarized in Table 3. The AUCs in the training and the validation datasets were 0.768 (95% CI, 0.704–0.835) and 0.723 (95% CI, 0.625–0.828), respectively (Fig. 6A).
Table 3.
Univariate and multivariate logistic regression of clinical and pathological factors in the training dataset.
| Characteristics | Univariate |
Multivariate |
|||
|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | β coefficient | P value | |
| Age | 0.959 (0.932–0.986) | 0.004 | 0.957 (0.927–0.988) | −0.044 | 0.007 |
| Menopause | 0.698 (0.366–1.330) | 0.274 | |||
| Bilateral BC | 1.344 (0.601–3.006) | 0.471 | |||
| Previous BC | 1.473 (0.626–3.466) | 0.375 | |||
| Family History∗ | 5.207 (2.910–9.318) | <0.001 | 4.941 (2.670–9.143) | 1.598 | <0.001 |
| ER positive | 0.730 (0.427–1.250) | 0.252 | |||
| PR positive | 0.792 (0.467–1.342) | 0.386 | |||
| HER2 positive | 0.335 (0.153–0.731) | 0.006 | 0.280 (0.122–0.639) | −1.274 | 0.003 |
| Ki-67 ≥ 20% | 5.130 (1.551–16.962) | 0.007 | 4.609 (1.343–15.831) | 1.528 | 0.015 |
| TNBC | 1.738 (0.962–3.139) | 0.067 | |||
BC: breast cancer; ER: oestrogen receptor; PR: progesterone receptor; HER2: human epidermal growth factor receptor 2; TNBC: triple-negative breast cancer.
Family history of BRCA1/2-related malignancy, including breast, ovarian, pancreatic and prostate cancer.
Fig. 6.
The performance of the clinical, the radiomics and the nomogram models. (A) Receiver operating characteristic (ROC) curves in the training (left) and the validation datasets (right). (B) Calibration curves of the nomogram model in the training (left) and the validation datasets (right). (C) The decision curve analysis (DCA) figure of the three models in the whole cohort.
3.3. Radiomics model construction and validation
From each gray-scale US image, 1032 features were extracted. Fifty randomly selected lesions were used to evaluate the stability of radiomics features. 744 features, in which both intra- and inter-observer ICC were found >0.75, were considered stable and included in the following analysis.
Using the mRMR algorithm, a subset of ten features was acquired and further analyzed using LASSO regression. An optimal Lambda of 8.1113 was generated for the LASSO regression, and all the ten radiomics features remained with non-zero coefficients (Fig. 3A and B). A multivariate logistics regression model was developed, and the coefficients of selected features were summarized in Fig. 3C. The Pearson correlation coefficients between each pair of features were also calculated (Fig. 4). The AUCs of the radiomics model in the training and the validation datasets were 0.712 (95% CI, 0.647–0.776) and 0.705 (95% CI, 0.597–0.808), respectively (Fig. 6A).
Fig. 3.
Figures of logistic LASSO regression. (A) Cross-validation plot for the penalty term. (B) LASSO path plot of the model in the training dataset. (C) A non-zero coefficient profile plot of the ten selected radiomics features.
Fig. 4.
Pearson correlation coefficient heatmap of ten selected radiomics features for predicting gBRCA1/2 mutation. The shade of the color represents the correlation degree. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Combined nomogram model construction and validation
The radiomics scores of the US images were calculated according to the radiomics model. Through another round of multivariate logistics regression, a model combining radiomics scores and clinical factors was constructed, and a nomogram was developed (Fig. 5). In the nomogram model, age at first diagnosis (p = 0.008), family history of BRCA1/2-related malignancies (p < 0.001), HER2 status (p = 0.003), expression of Ki-67 (p = 0.016), and the radiomics score (p < 0.001) were independent predictive factors for BRCA1/2 mutation (Table 4).
Fig. 5.
The nomogram model combining a radiomics score and four clinical factors.
Table 4.
Multivariate logistic regression analysis of clinical factors and the radiomics score.
| Characteristics | OR (95% CI) | β coefficient | P value |
|---|---|---|---|
| Age | 0.957 (0.926–0.989) | −0.044 | 0.008 |
| Family History∗ | 4.336 (2.270–8.281) | 1.598 | <0.001 |
| HER2 positive | 0.272 (0.117–0.634) | −1.274 | 0.003 |
| Ki-67 ≥ 20% | 4.609 (1.343–15.831) | 1.528 | 0.016 |
| Radiomics score | 1.666 (1.302–2.154) | 0.511 | <0.001 |
HER2: human epidermal growth factor receptor 2.
Family history of BRCA1/2-related malignancy, including breast, ovarian, pancreatic and prostate cancer.
For comparison between the clinical and the radiomics models, no significant differences were detected in the training (p = 0.429) or the validation dataset (p = 0.820) (Table 5). The AUCs of the nomogram model were significantly higher than those of the clinical model in both the training (0.804 vs 0.768, p = 0.041) and the validation datasets (0.811 vs 0.723, p = 0.007) (Table 5), indicating improved diagnostic efficiency brought by radiomics features.
Table 5.
Performance of Different Models in the Training and Validation Datasets of gBRCA1/2 Mutation.
| Dataset | Model | AUC (95% CI) | ACC | Se | Sp | PPV | NPV |
|---|---|---|---|---|---|---|---|
| Training | Clinical | 0.768 (0.704–0.835) | 0.764 | 0.671 | 0.788 | 0.448 | 0.903 |
| Radiomics | 0.712 (0.647–0.776)∗ | 0.688 | 0.700 | 0.685 | 0.363 | 0.899 | |
| Nomogram | 0.804 (0.748–0.861)† | 0.732 | 0.800 | 0.714 | 0.418 | 0.933 | |
| Validation | Clinical | 0.723 (0.625–0.828) | 0.626 | 0.600 | 0.658 | 0.273 | 0.837 |
| Radiomics | 0.705 (0.597–0.808)∗ | 0.639 | 0.633 | 0.641 | 0.311 | 0.872 | |
| Nomogram | 0.811 (0.724–0.894)† | 0.721 | 0.800 | 0.701 | 0.407 | 0.932 |
ACC: accuracy; Sp: specificity; Se: sensitivity; PPV: positive predictive value; NPV: negative predictive value.
, p > 0.05 in DeLong’s test, compared to the clinical model.
, p < 0.05 in DeLong’s test, compared to the clinical model.
To further evaluate the performance of the nomogram model, the calibration curves were drawn and the predicted BRCA1/2 status closely approximated the truth in both datasets (Fig. 6B). The decision curve analysis (DCA) curves also showed that the inclusion of radiomics features enhanced the clinical benefits for patients (Fig. 6C). The sensitivity, specificity, accuracy, PPVs, and NPVs of the above-mentioned models were also summarized in Table 5. The nomogram model achieved NPVs of 0.93 and sensitivity of 0.80 in both datasets.
4. Discussion
BRCA1/2 status is a key to personalized therapy for invasive breast cancer patients [27]. This study aims to identify BRCA1/2 mutation in invasive breast cancer cases by using both ultrasound (US) images and clinicopathological information. The main findings were as follows: (1) Ten US radiomics features of breast cancer lesions were found to be associated with BRCA1/2 status. (2) A nomogram model combining a radiomics score and four clinicopathological factors showed significant predictive power for BRCA1/2-mutated cases with AUCs of 0.80 and NPVs of 0.93 in the datasets.
Traditional imaging characteristics were found to have limited association with BRCA1/2 mutation status in breast cancer patients. Some studies reported that BRCA1/2-mutated patients were prone to have lower levels of breast parenchymal enhancement in MRI [28,29], while different opinions existed that no significant differences were shown in either morphological or kinetic features between MRI data of BRCA1/2 carriers and non-carriers [30]. For breast US images, multiple studies demonstrated no significant differences in tumor size, orientation, shape, margin, echo pattern, and posterior features according to BRCA1/2 status, whether in TNBC patients or unselected breast cancer patients [[31], [32], [33]].
Radiomics analysis has shown its potential in identifying tumor characteristics at the genetic level. Previous studies mainly focused on radiomics features of breast MR, which were found to be associated with gene mutations, e.g., HER-2, TP53, PI3K, to different degrees [[34], [35], [36], [37]]. Vasileiou and colleagues [38] utilized 41 breast MRI data to construct a radiomics model for predicting BRCA1/2 mutation and achieved an AUC of 0.865. However, the small sample size indicated inevitable overfitting and relatively low credibility.
Recently, a small number of studies have indicated that US radiomics is a useful tool to predict gene mutation in tumors. Wang and colleagues [39] utilized US radiomics features of 138 papillary thyroid cancer to predict BRAFV600E mutation and achieved an AUC of 0.938. However, similar studies that included gray-scale images alone yielded limited AUCs of 0.651–0.685 [40,41]. For breast cancer, in a study utilizing gray-scale US images of 312 cases to predict PIK3CA mutation, several machine learning and deep learning models were constructed and showed relatively strong predictive power (AUC 0.741–0.775) [42].
In the present study, US radiomics features demonstrated favorable discrimination of BRCA1/2 mutation. The radiomics model based on 10 features significantly elevated the performance of the nomogram model in both datasets, indicating that US radiomic analysis could provide a new perspective and act as an effective supplement to clinical information. Among the 10 features, six were obtained from the wavelet transform, which provided multi-resolution time/spatial-frequency analysis for texture of the lesions [25]. Half of the features were higher-order features, including 2 GLSZM, 2 GLCM and 1 NGTDM features, which evaluate the relationships between multiple pixels and demonstrate the characteristic patterns of gray-level distribution. Four features were first-order features describing simple pixel intensity characteristics. Only one shape feature, “original_shape2D_Elongation”, was included in the radiomics model, which indicated that a less eccentric shape may be associated with non-BRCA1/2-mutated lesions.
To be noted, with negative predictive values (NPVs) higher than 0.93 in this study, the nomogram model has great potential in avoiding unnecessary gene tests in clinical practice. When patients with breast cancer were considered as low-risk for BRCA1/2 mutation by the nomogram model, choosing not to take genetic testing might bring more benefits, e.g., reducing medical costs and mental stress. Notably, a BRCA1/2 mutation rate of about 20% was shown in this cohort, higher than the previously reported rate of cohorts that met the NCCN criteria (3.2%–12.0%) [43], which might be explained by the high proportion of positive family history (20.7%, 93/449) in this study. The relatively balanced ratio fortunately created an ideal condition for model construction. However, given that BRCA1/2 mutation rate in unselected breast cancer patients was merely 3%–5% [3,4], the nomogram model needs further validation or improvement in cohorts with lower mutation rates.
Another interesting finding is that Ki-67 level was firstly described as an independent predictor of BRCA1/2 mutation (OR 4.609, p = 0.015) in breast cancer patients in this study. Although Ki-67 was not mentioned in the NCCN criteria [12] and often neglected in BRCA1/2-related studies, higher Ki-67 levels in BRCA1/2-mutated breast cancer patients have been reported in multiple studies [29,[44], [45], [46]].
Despite several promising findings, the present study still has some limitations. Firstly, despite a relatively large sample size, the retrospective nature and single-center source inevitably brought selection bias. Multicenter studies would be desirable in the future. Secondly, the images were acquired from multiple US systems and under different acquisition conditions. In this case, radiomics predictors might be more robust, but radiomics analysis became much more challenging. Another limitation of this study is that only one gray-scale US image of the largest longitudinal plane of each lesion was involved, which might not completely reflect the heterogeneous characteristics of breast cancer. For future research, multi-modal US images such as shear-wave elastography and multiple planes of the lesions might be taken into consideration and help achieve better performance.
5. Conclusion
This study constructed and validated a nomogram model combining clinical factors and US-based radiomics features to predict BRCA1/2 mutation in patients with invasive breast cancer. The nomogram model demonstrated favorable performance and may reduce underdiagnosis of BRCA1/2 mutated cases and avoid unnecessary gene tests in clinical practice. However, future prospective multicenter studies are required for further validation or improvement.
Declarations
Ethics statement
This study was approved by the Institutional Review Board of Sun Yat-sen University Cancer Center (B2021-453-01), with the requirement for informed consent waived. The publishment of the images included in this paper has earned the patients’ consent.
Data statement
In order to protect privacy of the patients, clinical and ultrasound data will not be open to public. The de-identified data can be obtained from the authors, Ruohan Guo (guorh@sysucc.org.cn) and Yiwen Yu (Yuyw@sysucc.org.cn), upon reasonable request.
Funding
This study received no funding.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Zhao W., Wiese C., Kwon Y., Hromas R., Sung P. The BRCA tumor suppressor network in chromosome damage repair by homologous recombination. Annu. Rev. Biochem. 2019;88:221–245. doi: 10.1146/annurev-biochem-013118-111058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun J., Meng H., Yao L., Lv M., Bai J., Zhang J., et al. Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients. Clin. Cancer Res. 2017;23:6113–6119. doi: 10.1158/1078-0432.CCR-16-3227. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong N., Ryder S., Forbes C., Ross J., Quek R.G. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin. Epidemiol. 2019;11:543–561. doi: 10.2147/CLEP.S206949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.A., Mooij T.M., Roos-Blom M.J., et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 6.Yao L., Sun J., Zhang J., He Y., Ouyang T., Li J., et al. Breast cancer risk in Chinese women with BRCA1 or BRCA2 mutations. Breast Cancer Res. Treat. 2016;156:441–445. doi: 10.1007/s10549-016-3766-3. [DOI] [PubMed] [Google Scholar]
- 7.Risch H.A., McLaughlin J.R., Cole D.E., Rosen B., Bradley L., Fan I., et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J. Natl. Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 8.Haffty B.G., Harrold E., Khan A.J., Pathare P., Smith T.E., Turner B.C., et al. Outcome of conservatively managed early-onset breast cancer by BRCA1/2 status. Lancet (London, England) 2002;359:1471–1477. doi: 10.1016/S0140-6736(02)08434-9. [DOI] [PubMed] [Google Scholar]
- 9.Yip C.H., Newman L.A. American Society of Clinical Oncology, American Society for Radiation Oncology, and Society of Surgical Oncology Guideline for Management of Hereditary Breast Cancer. JAMA Surg. 2021;156:284–285. doi: 10.1001/jamasurg.2020.6254. [DOI] [PubMed] [Google Scholar]
- 10.NCCN Clinical Practice Guidelines in Oncology: Breast Cancer, Version 1 NCCN National Comprehensive Cancer Network. 2023. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf
- 11.Hirotsu Y., Nakagomi H., Sakamoto I., Amemiya K., Mochizuki H., Omata M. Detection of BRCA1 and BRCA2 germline mutations in Japanese population using next-generation sequencing. Mol. Genet. Genom. Med. 2015;3:121–129. doi: 10.1002/mgg3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly M.B., Pal T., Berry M.P., Buys S.S., Dickson P., Domchek S.M., et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. : JNCCN. 2021;19:77–102. doi: 10.6004/jnccn.2021.0001. [DOI] [PubMed] [Google Scholar]
- 13.Beitsch P.D., Whitworth P.W., Hughes K., Patel R., Rosen B., Compagnoni G., et al. Underdiagnosis of hereditary breast cancer: are genetic testing guidelines a tool or an obstacle? J. Clin. Oncol. 2019;37:453–460. doi: 10.1200/JCO.18.01631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinker K., Chin J., Melsaether A.N., Morris E.A., Moy L. Precision medicine and radiogenomics in breast cancer: new approaches toward diagnosis and treatment. Radiology. 2018;287:732–747. doi: 10.1148/radiol.2018172171. [DOI] [PubMed] [Google Scholar]
- 17.Gu J., Jiang T. Ultrasound radiomics in personalized breast management: current status and future prospects. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.963612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X., Yao Z., Huang Y., Yu Y., Wang Y., Liu Y., et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat. Commun. 2020;11:1236. doi: 10.1038/s41467-020-15027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J., Fang Q., Yao J., Ge L., Hu L., Wang Z., et al. Integration of ultrasound radiomics features and clinical factors: a nomogram model for identifying the Ki-67 status in patients with breast carcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.979358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu J., Tong T., He C., Xu M., Yang X., Tian J., et al. Deep learning radiomics of ultrasonography can predict response to neoadjuvant chemotherapy in breast cancer at an early stage of treatment: a prospective study. Eur. Radiol. 2022;32:2099–2109. doi: 10.1007/s00330-021-08293-y. [DOI] [PubMed] [Google Scholar]
- 21.Fujii T., Kogawa T., Dong W., Sahin A.A., Moulder S., Litton J.K., et al. Revisiting the definition of estrogen receptor positivity in HER2-negative primary breast cancer. Ann. Oncol. 2017;28:2420–2428. doi: 10.1093/annonc/mdx397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison K.H., Hammond M.E.H., Dowsett M., McKernin S.E., Carey L.A., Fitzgibbons P.L., et al. Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J. Clin. Oncol. 2020;38:1346–1366. doi: 10.1200/JCO.19.02309. [DOI] [PubMed] [Google Scholar]
- 23.Goldhirsch A., Winer E.P., Coates A.S., Gelber R.D., Piccart-Gebhart M., Thürlimann B., et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018;36(20):2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 25.Abbasian Ardakani A., Bureau N.J., Ciaccio E.J., Acharya U.R. Interpretation of radiomics features-A pictorial review. Comput. Methods Progr. Biomed. 2022;215 doi: 10.1016/j.cmpb.2021.106609. [DOI] [PubMed] [Google Scholar]
- 26.Peng H., Long F., Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 27.Tung N.M., Boughey J.C., Pierce L.J., Robson M.E., Bedrosian I., Dietz J.R., et al. Management of hereditary breast cancer: American Society of Clinical Oncology, American Society for Radiation Oncology, and Society of Surgical Oncology Guideline. J. Clin. Oncol. 2020;38:2080–2106. doi: 10.1200/JCO.20.00299. [DOI] [PubMed] [Google Scholar]
- 28.Grubstein A., Rapson Y., Benzaquen O., Rozenblatt S., Gadiel I., Atar E., et al. Comparison of background parenchymal enhancement and fibroglandular density at breast magnetic resonance imaging between BRCA gene mutation carriers and non-carriers. Clin. Imag. 2018;51:347–351. doi: 10.1016/j.clinimag.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 29.You C., Xiao Q., Zhu X., Sun Y., Di G., Liu G., et al. The clinicopathological and MRI features of patients with BRCA1/2 mutations in familial breast cancer. Gland Surg. 2021;10:262–272. doi: 10.21037/gs-20-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noh J.M., Han B.K., Choi D.H., Rhee S.J., Cho E.Y., Huh S.J., et al. Association between BRCA mutation status, pathological findings, and magnetic resonance imaging features in patients with breast cancer at risk for the mutation. J. Breast Cancer. 2013;16:308–314. doi: 10.4048/jbc.2013.16.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesurolle B., Kadoch L., El-Khoury M., Lisbona A., Dendukuri N., Foulkes W.D. Sonographic features of breast carcinoma presenting as masses in BRCA gene mutation carriers. J. Ultrasound Med. 2007;26:817–824. doi: 10.7863/jum.2007.26.6.817. [DOI] [PubMed] [Google Scholar]
- 32.Vijayaraghavan G.R., Kona M., Maheswaran A., Kandil D.H., Toke M.K., Vedantham S. Ultrasound imaging morphology is associated with biological behavior in invasive ductal carcinoma of the breast. J. Clin. Imag. Sci. 2021;11:48. doi: 10.25259/JCIS_60_2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karbasian N., Sohrabi S., Omofoye T.S., Le-Petross H., Arun B.K., Albarracin C.T., et al. Imaging features of triple negative breast cancer and the effect of BRCA mutations. Curr. Probl. Diagn. Radiol. 2021;50:303–307. doi: 10.1067/j.cpradiol.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Greca Saint-Esteven A., Vuong D., Tschanz F., van Timmeren J.E., Dal Bello R., Waller V., et al. Systematic review on the association of radiomics with tumor biological endpoints. Cancers. 2021;13 doi: 10.3390/cancers13123015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin P., Liu W.K., Li X., Wan D., Qin H., Li Q., et al. MRI-based radiogenomics analysis for predicting genetic alterations in oncogenic signalling pathways in invasive breast carcinoma. Clin. Radiol. 2020;75:561. doi: 10.1016/j.crad.2020.02.011. e1-.e11. [DOI] [PubMed] [Google Scholar]
- 36.Sun K., Zhu H., Chai W., Yan F. Multimodality MRI radiomics analysis of TP53 mutations in triple negative breast cancer. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1153261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun K., Zhu H., Chai W., Yan F. TP53 mutation estimation based on MRI radiomics analysis for breast cancer. J. Magn. Reson. Imag.: JMRI. 2023;57:1095–1103. doi: 10.1002/jmri.28323. [DOI] [PubMed] [Google Scholar]
- 38.Vasileiou G., Costa M.J., Long C., Wetzler I.R., Hoyer J., Kraus C., et al. Breast MRI texture analysis for prediction of BRCA-associated genetic risk. BMC Med. Imag. 2020;20:86. doi: 10.1186/s12880-020-00483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y.G., Xu F.J., Agyekum E.A., Xiang H., Wang Y.D., Zhang J., et al. Radiomic model for determining the value of elasticity and grayscale ultrasound diagnoses for predicting BRAF(V600E) mutations in papillary thyroid carcinoma. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.872153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon M.R., Shin J.H., Park H., Cho H., Hahn S.Y., Park K.W. Radiomics study of thyroid ultrasound for predicting BRAF mutation in papillary thyroid carcinoma: preliminary results. AJNR Am. J. Neuroradiol. 2020;41:700–705. doi: 10.3174/ajnr.A6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang J., Jiang S., Ma J., Xi X., Li H., Wang L., et al. Nomogram based on radiomics analysis of ultrasound images can improve preoperative BRAF mutation diagnosis for papillary thyroid microcarcinoma. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.915135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen W.Q., Guo Y., Ru W.E., Li C., Zhang G.C., Liao N., et al. Using an improved residual Network to identify PIK3CA mutation status in breast cancer on ultrasound image. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.850515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cropper C., Woodson A., Arun B., Barcenas C., Litton J., Noblin S., et al. Evaluating the NCCN clinical criteria for recommending BRCA1 and BRCA2 genetic testing in patients with breast cancer. J. Natl. Compr. Cancer Netw.: JNCCN. 2017;15:797–803. doi: 10.6004/jnccn.2017.0107. [DOI] [PubMed] [Google Scholar]
- 44.Stella S., Vitale S.R., Martorana F., Massimino M., Pavone G., Lanzafame K., et al. Mutational analysis of BRCA1 and BRCA2 genes in breast cancer patients from eastern sicily. Cancer Manag. Res. 2022;14:1341–1352. doi: 10.2147/CMAR.S348529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang G.T., Shi J.X., Hu X., Zhang C.H., Shan L., Song C.G., et al. The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: screening of 2,991 patients and 1,043 controls by next-generation sequencing. Int. J. Cancer. 2017;141:129–142. doi: 10.1002/ijc.30692. [DOI] [PubMed] [Google Scholar]
- 46.Ji G., Bao L., Yao Q., Zhang J., Zhu X., Bai Q., et al. Germline and tumor BRCA1/2 pathogenic variants in Chinese triple-negative breast carcinomas. J. Cancer Res. Clin. Oncol. 2021;147:2935–2944. doi: 10.1007/s00432-021-03696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]