Abstract
The Covid-19 infection outbreak led to a global epidemic, and although several vaccines have been developed, the appearance of mutations has allowed the virus to evade the immune response. Added to this is the existing risk of the appearance of new emerging viruses. Therefore, it is necessary to explore novel antiviral therapies. Here, we investigate the potential in vitro of plant extracts to modulate cellular stress and inhibit murine hepatitis virus (MHV)-A59 replication. L929 cells were treated with P2Et (Caesalpinia spinosa) and Anamu SC (Petiveria alliacea) plant extracts during infection and virus production, ROS (reactive oxygen species), UPR (unfolded protein response), and autophagy were assessed. P2Et inhibited virus replication and attenuated both ROS production and UPR activation induced during infection. In contrast, the sustained presence of Anamu SC during viral adsorption and replication was required to inhibit viral infection, tending to induce pro-oxidant effects, and increasing UPR gene expression. Notably, the loss of the PERK protein resulted in a slight decrease in virus yield, suggesting a potential involvement of this UPR pathway during replication. Intriguingly, both extracts either maintained or increased the calreticulin surface exposure induced during infection. In conclusion, our findings highlight the development of antiviral natural plant extracts that differentially modulate cellular stress.
Keywords: Natural products, Coronavirus, Antiviral activity, Autophagy, Endoplasmic reticulum stress, Calreticulin
1. Introduction
Coronaviruses are classified into the Coronaviridae family belonging to Ribovira, order Nidovirales, and suborder Coronavirinae. Coronaviruses are a highly diverse family of positive-sense single-stranded RNA (+ssRNA) enveloped viruses, and the Orthocoronavirinae subfamily consists of four genera: alpha, beta, gamma, and delta [1]. Betacoronaviruses genus includes SARS-CoV, MERS, and SARS-CoV-2 viruses, among others that affect human health, and the prototype mouse hepatitis virus (MHV) [2]. The SARS-CoV-2 virus appeared in late 2019 in Wuhan, China. It is responsible for the COVID-19 pandemic, which has had a huge global impact. The higher rates of contagiousness during COVID-19 infection and lethality increased the search for new therapeutic strategies [3].
The cell stress response plays a significant role during viral adsorption and replication in RNA viruses. The induction of oxidative stress and antioxidant response deprivation plays a significant role during viral adsorption and replication in RNA viruses allowing infection maintenance through homeostatic disbalance [4]. Usually, during the induction of oxidative stress two main mechanisms are activated in the cells to remove the protein damage and help to restore cellular homeostasis, autophagy, and unfolded protein response (UPR) [5,6]. Autophagy may eventually enable viral assembly during replication of RNA viruses such as influenza [7,8]. In this sense, different reports tried to elucidate whether coronaviruses induce autophagy for their replication [9]. Some authors suggest that increased autophagy in cells infected with MHV is necessary to promote the viral replication cycle [10]. However, more recent evidence suggests an inhibitory role of the autophagic process by betacoronaviruses [7]. SARS-CoV and MERS-CoV protease PLP2 block the autophagosome and lysosome fusion, suppressing the autophagic flux [11]. MERS-CoV depends on the degradation of beclin-1 protein induced by the SPK2 ubiquitin ligase to block the autophagy and promote viral replication, thus, generating a possible therapy target that promotes an autophagic flux enough to overpass this block and reduce viral replication [12]. Similarly, recent studies showed that betacoronavirus infection disrupts the delicate balance of endoplasmic reticulum (ER) homeostasis, leading to ER stress and the unfolded protein response (UPR) activation to counteract the disturbance in ER function. The UPR is a complex signaling network encompassing three branches: PERK, IRE1, and ATF6. This intricate interplay between betacoronaviruses and the ER stress response is now recognized as a critical factor influencing viral replication, pathogenesis, and the host immune response [13].
On the other hand, it has been proposed that polyphenols and other natural compounds with antioxidant activities can be used to develop new therapies against COVID-19 [14]. Polyphenols can interfere with the adsorption and penetration of the virus in the host cells. For example, luteolin and quercetin inhibit the entry of the SARS-CoV-2 virus into Vero E6 cells, associated, at least for luteolin, with its ability to bind protein S of the virus with high affinity. In addition, polyphenols could also disrupt viral replication. There is evidence that epigallocatechin gallate and quercetin inhibit the SARS-CoV-2 viral protease 3CLpro, which is needed to process the viral polyprotein; meanwhile, resveratrol inhibits the viral RNA-dependent RNA polymerase (RdRp) in MERS infection [14]. The study of natural extracts has also shown that different natural compounds can promote autophagy and regulate the REDOX balance in several cell lines. These findings suggest the role of natural compounds in interfering with viral infection processes; however, there is no clear evidence in this sense [15].
We obtained two natural extracts from Petiveria alliacea L. and Caesalpinia spinosa (Molina) Kuntze plants. We have previously shown that the standardized extract called P2Et from C. spinosa (Molina) Kuntze contains various compounds, a diversity of polyphenols such as gallic acid and ethyl gallate, with antioxidant properties over cancer cells [16]. Recently, we showed that the P2Et extract promotes apoptosis in different ways in tumor cells depending on the induction of ERs and autophagy via PERK and Beclin-1, respectively [17,18]. In contrast, Anamu SC extract (obtained from P. alliacea L.) presents some sulfur compounds, sterols, triterpenes, and flavonoids as its main compounds. Anamu SC could induce oxidative stress because it interferes with glycolysis and mitochondrial respiration in cancer cells [19]. Furthermore, we investigated the antiviral properties of P2Et and Anamu SC extracts against SARS-CoV-2, the causative agent of COVID-19. In vitro experiments using Vero E6 cells revealed a significant inhibition of viral replication by both extracts. Subsequently, a phase II multicentric randomized, double-blind clinical trial was conducted to assess the impact of P2Et treatment on clinical outcomes and immunological response in COVID-19 patients [20].
Considering the activities of the extracts related to processes such as modulation of oxidative stress, ERs, and autophagy induction, this study aimed to evaluate the possible inhibitory effects of P2Et and Anamu SC extracts on viral infection caused by coronaviruses. Our results showed that the extracts present activities in a differential way where P2Et inhibits viral replication, being antioxidant and decreasing the UPR; meanwhile Anamu-SC decrease virus infection and was slightly pro-oxidant, increasing the UPR.
2. Material and methods
2.1. Plant extract preparations and ultra-performance liquid chromatography analysis
The Colombian Environmental Ministry agreement authorized the Access to Genetic Resources and Derived Products No. 220/2018 (RGE 0287-6) to use plant material for research. Caesalpinia spinosa (Molina) Kuntze plant pods were collected in Villa de Leyva, Boyacá, Colombia, and identified by Luis Carlos Jimenez from the Colombian National Herbarium (voucher specimen number COL 523714). P2Et, an ethanolic Caesalpinia spinosa (Molina) Kuntze extract, was produced under good manufacturing practice (GMP); it was standardized and chemically characterized as previously described [16]. In each assay, dried P2Et extract was diluted in 95 % ethanol, obtaining a 25 mg/mL fresh solution. For Anamu SC extract, plant material was identified by Antonio Luis Mejia from the Colombian National Herbarium, voucher number COL 333406. Anamu SC extract was produced from leaves of Petiveria alliacea L., as we described before [21]. The extract was obtained through supercritical fluids in La Corporación Universitaria Lasallista at 60 °C, 400 bar, 30 kg/h, and ethyl acetate 15 % like cosolvent. The stock solution of the extracts was prepared by reconstitution in 95 % ethanol at a concentration of 25 mg/mL and stored at 4 °C.
The UPLC analysis was conducted using an Acquity H Class UPLC system from Waters® equipped with an el Acquity photodiode array (PDA) detector, quaternary pump, degasser, and autosampler. Empower® 3 software was employed for data processing. A BEH Shield C18 1.7 μm 2.1 mm × 150 mm Waters® column was utilized, maintained at a temperature of 35 °C ± 1. The elution process involved a gradient of acetonitrile (solvent A) and 0.1 % formic acid (solvent B), with a flow rate of 0.37 mL min^(−1) and an injection volume of 3 μL. A wavelength of 274 nm was employed for detection, while the UV spectrum was monitored within a range of 200–400 nm. Peaks were characterized by comparing their retention times and UV spectra with reference standards. The concentration of the P2Et extract was determined to be 2000 μg/mL. The standard solutions and the extract were dissolved in methanol and water (1:1 v/v). Individual compounds were quantified using an eleven-point linear regression curve (r^2 > 0.997). All standard solutions were analyzed in triplicate, and the average peak areas were calculated. The analysis showed the presence of marker compounds ethyl gallate (78.97 ± 1.49 μg/mL), gallic acid (59.82 ± 0.72 μg/mL), and methyl gallate (20.1 ± 0.62 μg/mL) according to previous batch analyses (Supplementary Fig. 1a) [16]. It is important to note that all experiments were conducted using the same batch of P2Et.
For the Anamu SC, a BEH Shield C18 1.7 μm 2.1 mm × 100 mm Waters® column was employed, maintained at a temperature of 25 °C ± 1. The elution gradient consisted of acetonitrile (solvent A) and 0.1 % formic acid (solvent B), with a flow rate of 0.15 mL min^ (−1) and an injection volume of 3 μL. Detection was performed at a wavelength of 274 nm, and the UV spectrum was monitored 200–400 nm. The UPLC-PDA chromatographic profile of the Anamu SC extract revealed the identification of myricetin and dibenzyl disulfide (Supplementary Fig. 1B).
As previously reported, the cytotoxic activity for P2Et and Anamu SC was evaluated using MTT assay [22]. The P2Et and Anamu SC were dissolved in ethanol, and this vehicle was used as a negative control. The IC50 (50 % inhibition of cell growth) value was calculated using a nonlinear regression log (inhibitor) versus response-variable slope graph in GraphPad Prism (GraphPad Prism Software, Inc.).
2.2. Cell culture, virus production and plaque assay
Virus stocks of mouse hepatitis virus A59 strain (MHV-A59, VR-764) and their producer cell lines NCTC-1469 (CCL-9.1) were obtained from the American Type Culture Collection (Manassas, VA, USA). NCTC-1469 cells were used for propagation of MHV-A59, and cell cultures were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 4.5 g/l d-glucose, two mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin and 10 % horse serum (GIBCO Laboratories, Grand Island, N.Y.). Propagation of virus stocks was performed using permissive cells cultured on T75 flaks and infected with the multiplicity of infection (MOI) of 1.0 or 0.1 for 1 h at 37 °C of virus suspension. MHV-A59 infected cells were frozen in their culture flasks after 48h. Then, cells were subjected to three freeze-thaw cycles to allow the release of the virus. The contents of the flasks were centrifuged at 3.000g for 10 min to separate the virus from cell debris. The supernatant was filtered through 0.20 μm, tittered on L929 cells, and freezer at −70 °C. For titration, L929 cells, kindly provided by Dr. Carlos Guerrero (Universidad Nacional de Colombia), were seeded in 12-well plates at a concentration of 1.2 × 105 per well and cultured overnight. The supernatant virus was assayed following serial dilution in DMEM after removing the culture medium. For each 10-fold dilution up to 10-5, each well was inoculated with 200 μl of the virus suspension. Following adsorption at room temperature for 60 min, plates were overlaid with DMEM plus 2 % FBS containing 0.6 % agarose. Plaques were visualized in the L929 monolayers at 48 h post-infection with 0.5 % crystal violet following fixation with 20 % formalin for 30 min. Finally, plaque-forming units (PFU) per milliliter were calculated.
Melanoma B16–F10 cell line (American Type Culture Collection, Manassas, VA, USA) was cultured in RPMI-1640 (Gibco, Life Technologies, NY, USA). Obtaining the CRISPR clones (PERK KO, beclin K.O., and XBP-1 KO) was previously described [17,18]. The medium was supplemented with heat-inactivated fetal calf serum (10 %), 2 mmol/L l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 25 mmol/L Hepes buffer (Gibco, Life Technologies, NY, USA). Cells were incubated in a humidified environment at 37 °C and 5 % CO2 and grown until 75 % confluence. Cells were collected using 1x trypsin/EDTA (ethylenediamine-tetra-acetic acid) 0.25 % phenol red (Gibco, Life Technologies, NY, USA). B16–F10 cells and clones were seeded in 12-well plates at a concentration of 1.5 × 105 per well and cultured overnight until confluence, and the infection process was carried out as previously mentioned.
2.3. Viral inhibition assays
L929 cells were seeded in 12-well plates at a concentration of 1.2 × 105 per well and cultured overnight. Cells were pretreated for 2 h with the extracts or controls, washed twice with PBS 1X, and inoculated with 200 μl of the virus suspension to 0.1, 0.01, or 0.001 MOI in DMEM, 1 h at room temperature (RT) in agitation. The virus inoculum was removed, and alternatively, cells were treated again or not with extracts for 6, 12, or 24h. Supernatants were collected for plaque assays. The cells were collected to evaluate proteins by Western blot and ROS assays by flow cytometry.
2.4. ROS evaluation
The generation of intracellular reactive oxygen species (ROS) in L929 cells was performed using the 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) ROS-sensitive fluorescent probe (1 μM). L929 cells grown in 12-well until confluence, then infected with a MOI of 0.1, 0.01, or 0.001 of the MHV-A59 virus in the presence or absence of ½ of the IC50 (inhibitory concentration 50) calculated in viability assays of P2Et and Anamu SC (SC) extracts, or with chloroquine (CQ). Treatments were placed all the time, both during adsorption and viral replication. After 24 h of infection with MHV-A59, L929 cells were recovered, and 2.5 × 105 cells per condition were labeled with the H2DCFDA probe for 30 min at 37 °C, and before of analyzed by flow cytometry, propidium iodide was added to a final concentration 1 μg/mL. Then, cells were acquired using FACSAria IIU, and the results were subsequently analyzed using FlowJo 10.8.1 Software. The data are expressed as the ratio of fluorescence/fluorescence of the non-stimulated control. Each assay was performed in duplicate, and the results are expressed as the mean ± SD for 3 independent experiments.
2.5. UPR and autophagy analysis
The induction of UPR and autophagy during infection and treatments were analyzed. Thus, L929 was pretreated for 2h or not with P2Et and Anamu SC, washed twice with PBS 1X, and inoculated with 200 μl of the virus suspension to 0.1 MOI in DMEM, 1 h at RT in agitation. Then they were infected and cultured for 12 h. As a control, non-infected cells were used. Plant extracts were maintained during infection time. The protein and RNA were obtained to analyze UPR and autophagy gene expression by Western blot and reverse transcription-polymerase chain reaction (qPCR). Autophagy was evaluated by transfecting L929 cells with mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) protein that allows differentiation between autophagosomes and autolysosomes, detecting by confocal microscope yellow dots or red dots, respectively [23]. After transfection, L929 cells were infected or not in the presence of treatments.
2.6. Autophagy analysis by confocal microscopy
L929 cells were transfected with mRFP-GFP tandem fluorescent-tagged LC3 protein and cultured on slides covered with fibronectin. Then, the cells were infected or not with a MOI of 0.1 in the presence or not of the extracts P2Et and Anamu SC extracts or of CQ for 12 h. Nuclei cells were stained with 300 nM DAPI, then cells were fixed with 4 % PFA solution, and coverslips with cells were mounted on slides with ProLong® Antifade Reagent (Life Technologies). A z-stack of 640 × 640 pixels images was acquired with a laser scanning confocal microscope FV1000 (Olympus) using UPLSAPO 60×1.35 NA oil immersion objective. 405 nm, 488 nm, and 543 nm laser lines were used to excite DAPI, GFP, and RFP fluorescences at 10 us/pixel speeds. Images were acquired and analyzed using the Fluoview software of Olympus. Red and yellow puncta were counted.
2.7. Western blot analysis
Cells were treated and infected as described above. Then the cells were lysed on ice for 20 min with RIPA buffer. A total of 20 μg of protein were separated on 15 % (for LC3 I/II) or 10 % polyacrylamide SDS-PAGE and transferred to PVDF membranes. The PVDF membranes were blocked in TBS-T supplemented with 5 % Bovine Serum Albumin (BSA) or 5 % Nonfat Milk nonfat for 1 h at room temperature CO2, following the manufacturer's instructions for the primary antibody. Primary antibodies anti-PERK (3192, Cell Signaling Technology), anti-p-PERK (3179, Cell Signaling Technology), anti-LC3 I/II (12741, Cell Signaling Technology), anti-β-actin (MAB8929, R&D systems), anti-Keap-1 (8047, Cell Signaling Technology), and anti-nucleoporin p62 clone 53 (610498, Becton–Dickinson) were incubated overnight. Appropriate secondary antibodies conjugated with horseradish peroxidase were used (Molecular Probes, Invitrogen) and detected using Super Signal West Pico chemiluminescent substrate (Pierce). Band protein images were obtained using iBright™ FL1500 Imaging System (Thermofisher).
2.8. Reverse transcription–polymerase chain reaction (qPCR)
Total RNA from treated cells with or without MHV infection or supernatants was extracted using TRIzol LS reagent (Life Technologies, Waltham, MA, USA) following the manufacturer's instructions. RNA quality and quantity were assessed with a NanoDrop spectrophotometer (NanoDrop Technologies). Complementary DNA (cDNA) was synthesized from total RNA with SuperScripts III Reverse Transcriptase, following the manufacturer's instructions (Life Technologies Corporation, Invitrogen, NY, USA). For real-time PCR reaction, 600 ng of cDNA, DNA Master Plus SYBR Green I (Roche Applied Science, IN, USA), and 250 nM forward and reverse primers were added in a total volume of 20 μL. Primer sequences are shown in Table 1. Reactions were performed in duplicates using QuantStudio 3 Real-Time PCR systems (ThermoFisher Scientific, Waltham, MA, USA). The relative expression of the genes associated with the UPR was normalized to the endogenous control gene GAPDH. Relative expression was calculated using the 2−ΔΔCT comparative method [24].
Table 1.
Primers used in the SYBR green real-time PCR analyses.
| Primer name | Sequence (5′ - 3′) | Annealing temperature (°C) |
|---|---|---|
| MHV_F | CAGATCCTTGATGATGGCGTAGT | 57 |
| MHV_R | AGAGTGTCCTATCCCGACTTTCTC | |
| CHOP_F | GTCCCTAGCTTGGCTGACAGA | 59 |
| CHOP_R | TGGAGAGCGAGGGCTTTG | |
| sXBP1_F | AAGAACACGCTTGGGAATGG | 56 |
| sXBP1_R | CTGCACCTGCTGCGGAC | |
| tXBP1_F | GACAGAGAGTCAAACTAACGTGG | 56 |
| tXBP1_R | GTCCAGCAGGCAAGAAGGT | |
| BiP_F | TCATCGGACGCACTTGGAA | 56 |
| BiP_R | CAACCACCTTGAATGGCAAGA | |
| EDEM_F | AAGCCCTCTGGAACTTGCG | 59 |
| EDEM_R | AACCCAATGGCCTGTCTGG | |
| GAPDH_F | TCAACAGCAACTCCCACTCTTCCA | 56 |
| GAPDH_R | ACCCTGTTGCTGTAGCCGTATTCA |
2.9. Statistical analysis
Statistical analysis of the significance between two groups was calculated using two-tailed unpaired and paired Student's t-test for most statistical analyses and a p value of <0.05 was considered statistically significant. The statistical test results are indicated in each figure: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.001. Statistical analyses were performed in GraphPad Prism version 8.0.
3. Results
3.1. Natural extracts P2Et and Anamu SC decrease virus infection by MHV-A59
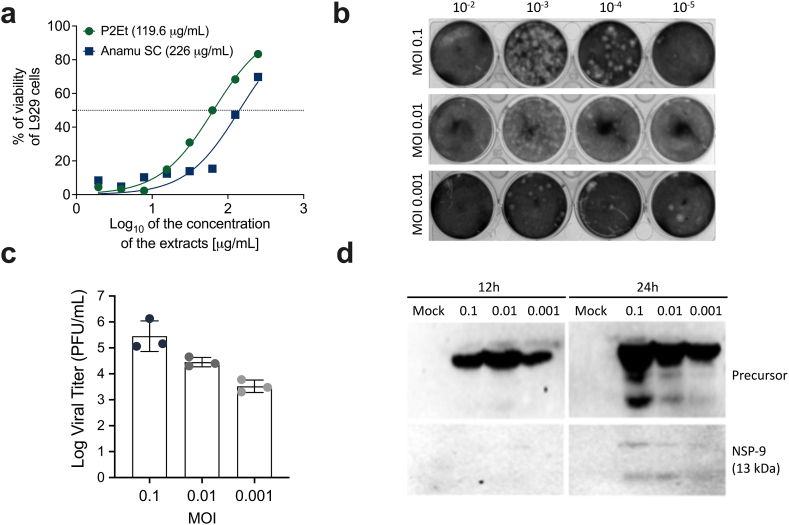
Initially, the viability of L929 fibroblasts was assessed to determine if the extracts alone had any impact. The IC50 was determined using an MTT assay. The IC50 values for Anamu SC and P2Et were found to be 226 ± 57.4 μg/mL and 119.6 ± 8.5 μg/mL, respectively (Fig. 1a). To ensure cell viability during the treatment with the extracts, half of the IC50 was used in the viral inhibition experiments. A lower concentration (the fifth part of the IC50) was included to determine if it similarly affected virus infection. Then, we evaluated the permissibility of murine fibroblast cell line L929 for MHV-A59 infection using three different MOI 0.01 viral particles per cell as described before [25], and one MOI higher (0.1) or one lower (0.001) than 0.01. After infection, supernatants were collected, and the virus yield was determined by plaque-forming units (PFU) assay on L929. An apparent dose-response effect was found (Fig. 1b and c). Furthermore, we evaluated viral proteins present in supernatants detecting NSP9 by Western blot. At 12 h post-infection, the large polyprotein precursor was detected. However, mature NSP9 was detected only 24 h post-infection (Fig. 1d).
Fig. 1.
Establishment of the viral infection model. (a) The IC50 for P2Et and Anamú SC was calculated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The results are expressed as the mean ± SD for 3 independent experiments. (b) Plaque assay images using infectious MHV-A59 viral particles at a multiplicity of infection (MOI) of 0.1, 0.01, and 0.001 to infect L929 cells, after 24 h of infection, supernatants were recovered and titrated using dilutions 10−2, 10−3, 10−4 and 10−5. (c) Mean virus supernatant titers (Log Plaque Forming Units (PFU)/mL) of plate assays. (d) Cells infected in this multiplicity of infection (MOI) were recovered at 12 or 24 h and the proteins were resolved in SDS-PAGE and after being transferred to polyvinylidene difluoride (PVDF) membranes they were tested with an anti-NSP9 antibody. The results are expressed as the mean ± SD for 3 independent experiments. The p values were calculated using Student's t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
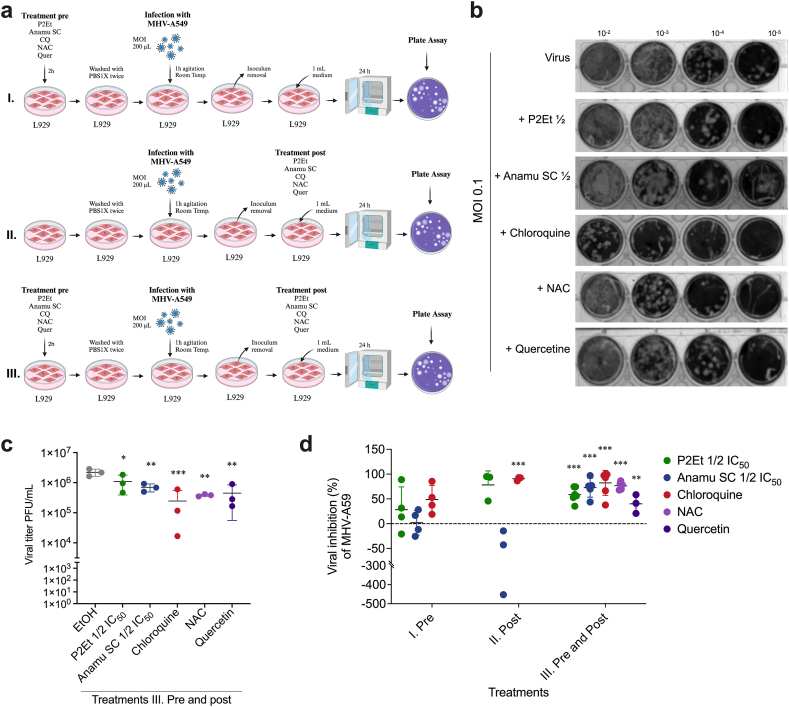
L929 cells were infected with the half or fifth part of the IC50 of each extract to determine whether P2Et or Anamu SC could inhibit viral infection. The extracts were added during the 2 h before starting the viral adsorption (treatment I. Pre), during the virus replication (treatment II. Post), or both, maintaining the extract before viral adsorption and virus replication (treatment III. Pre and post) (Fig. 2a). Culture supernatants were collected after infection and used to assess the number of infectious virus particles released. Using a MOI of 0.1, the inhibition of virus yield was observed under treatment with both P2Et and Anamu SC, mainly when the extracts were maintained all the time (Fig. 2b and c). Surprisingly, our findings revealed distinct effects of the extracts when administered at different stages of the viral life cycle. Although Anamu SC extract tended to increase virus production when administered solely during virus replication, it hinted at an ability to interfere with viral adsorption. In contrast, both P2Et and Anamu SC extracts, when administered during both adsorption and viral replication stages, effectively reduced the infection caused by the MHV-A59 virus in L929 cells. Therefore, in order to have a comparative approximation on the role of cellular stress in the antiviral activity associated with these natural extracts, it was decided to carry out the other analyzes maintaining the extracts both during adsorption and during viral replication. The antiviral effects of the extracts were further confirmed by comparison to the control molecules, chloroquine, N-acetylcysteine (NAC), and quercetin, which exhibited inhibition of virus yield (Fig. 2b and d). Supplementary Fig. 2 shows that when cells were treated with P2Et during virus infection at MOI of 1, 0.1, and 0.001, there is a tendency that the cycles required to amplify the N gene increase, reflecting that less viral genome is released in the supernatants. Similar results were observed with chloroquine while Anamu SC presented ambiguous results using 1/5 or 1/2 of IC50. These results highlight the potential of P2Et and Anamu SC extracts as therapeutic candidates against MHV-A59 infection.
Fig. 2.
P2Et and Anamu SC inhibit viral production. L929 cells were infected with a multiplicity of infection (MOI) of 0.1 of the MHV-A59 virus in the presence or absence of ½ of the IC50 of the P2Et and Anamu SC extracts, with chloroquine, quercetine, and N-acetylcysteine (NAC). Treatments were placed all the time both during adsorption and viral replication. In some experiments some treatments were applied only during viral adsorption or only during viral replication. After 24 h of infection the supernatants were recovered and tittered in a plaque assay using dilutions of 10−2, 10−3, 10−4 and 10−5. (a) Scheme that represents the different treatment conditions (I. Pre, II. Post or III. Pre and post infection). (b) Antiviral efficacy of the treatments in plaque assay. (b) Mean virus supernatant titers (Plaque Forming Units (PFU/mL)). (c) Percentages of viral inhibition of MHV-A59. In all cases, data are represented as the mean ± SD for 3 to 5 independent experiments. The p values were calculated using Student's t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
3.2. P2Et and Anamu SC modulate oxidative stress induced by MHV-A59
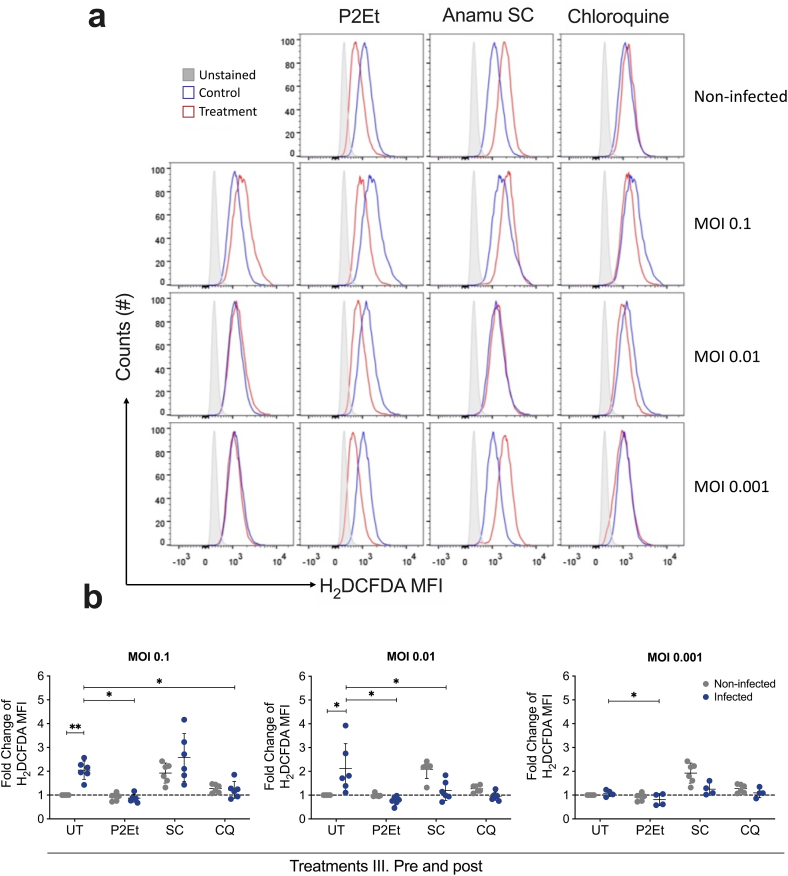
It has been proposed that compounds that modulate oxidative stress can affect viral adsorption and replication. Therefore, we wanted to evaluate whether treatment with natural extracts during coronavirus infection affected intracellular ROS levels contributing to the virus infection. For this, extracts were used before the adsorption and during virus replication (III. Pre y post). As expected, the viral infection of L929 fibroblasts with MHV increases ROS detection, depending on the number of infectious viruses used. In principle, the two extracts behaved differently on uninfected L929 cells where P2Et was antioxidant while Anamu SC was pro-oxidant. However, treatment with the P2Et extract decreased ROS induced by the viral infection in a MOI-dependent manner.
Interestingly, the effect of Anamu SC on infected cells depended on the level of oxidative stress generated by the virus. It maintained ROS induced by a viral MOI of 0.1 with a slight increase and behaved as an antioxidant with a virus MOI of 0.01 (Fig. 3a and b). Additionally, we also observed a decrease in the protein expression of Keap1 by Western blot, one protein involved with oxidative stress regulation, when L929 cells were infected with MHV, the virus interaction with Keap1 had been reported in several viral diseases as a mechanism used for viral replication after the induction of oxidative stress [26]. The effect over Keap1 was contra rested by P2Et, CQ, NAC, and Quer, showing a relevant role of oxidative stress in viral replication (Fig. 4b). On the other hand, the treatment with Anamu SC decrease Keap1 expression in uninfected and infected cells showing that the inhibitory role of Anamu SC could be independent of the oxidative stress induction. These results suggest that P2Et's antiviral activity is linked to oxidative stress reduction, whereas Anamu SC suppresses viral infection through a variety of mechanisms, including oxidative stress modulation. Nevertheless, the precise relationship between oxidative modulation and antiviral effect should be confirmed by combining the extracts with treatments, antioxidants, or pro-oxidants.
Fig. 3.
P2Et and Anamu SC extracts differentially modulate oxidative stress induced by viral infection. L929 cells were infected with a multiplicity of infection (MOI) of 0.1, 0.01 or 0.001 of the MHV-A59 virus in the presence or absence of ½ of the IC50 of the P2Et and Anamu SC (SC) extracts, or with chloroquine (CQ). Treatments were placed all the time both during adsorption and viral replication. After 24 h of infection with MHV-A59 the L929 cells were recovered and challenged with the H2DCFDA probe. Then, cells were acquired using FACSAria IIU, and the results were subsequently analyzed using FlowJo 10.8.1 Software. (a) Representative image of the mean fluorescence intensity (MFI) histograms of the probe. (b) Analysis of the data obtained from different independent experiments. UT (untreated cells). In all cases, data are represented as the mean ± SD for 3 independent experiments. The p values were calculated using Student's t-test. *p < 0.05; **p < 0.01.
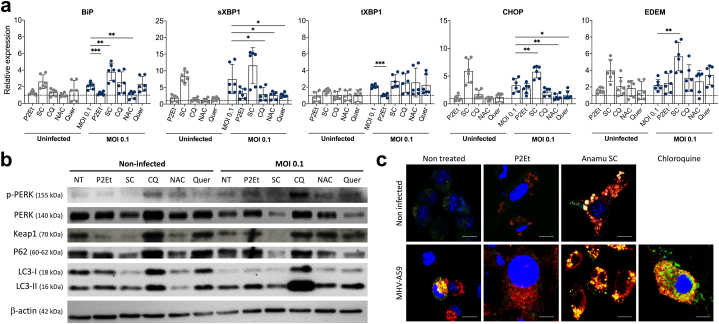
Fig. 4.
P2Et and Anamu SC differentially modulate the UPR response and autophagy in virus-infected cells. (a) L929 cells were infected with a multiplicity of infection (MOI) 0.1 of the MHV-A59 virus in the presence or absence of ½ of the IC50 of the P2Et and Anamu SC (SC) extracts, or with chloroquine (QC), N-acetylcysteine (NAC) and quercetin (Quer). After 12 h of infection the mRNA was recovered by trizol which was subsequently used to generate cDNA and then used to amplify with the appropriate primers of the BiP, CHOP, EDEM sXBP-1 and total XBP-1 genes. Expression levels were normalized and expression levels relative to cells treated with the mock negative control were calculated and shown. Each assay was performed in duplicate for 3 independent experiments. Gray date represented non-infected cells and blue date represented infected cells. (b) L929 cells treated with mock or infected with MOI of 0.1, 0.01 and 0.001 of MHV-A59 or treated with P2Et, Anamu SC (SC), or chloroquine (CQ) for 12 or 24 h were lysed and the proteins were used to determine the levels of LC3I and LC3-II, p62, PERK, p-PERK and keap-1 by Western blot. (c) L929 cells transfected with the LC3-GFP-RFP expression plasmid and seeded on fibronectin-coated slides. The cells were infected or not with a MOI of 0.1 in the presence or not of the extracts P2Et and Anamu SC extracts, or of CQ. Cells were visualized using the Olympus FV1000 confocal microscope. In all cases, data are represented as the mean ± SD. The p values were calculated using Student's t-test. *p < 0.05; **p < 0.01.
3.3. P2Et and Anamu SC modulate the UPR response and autophagy during MHV-A59 infection
Oxidative stress in cells can trigger stress response mechanisms such as the induction of the unfolded protein response (UPR) and autophagy. Coronaviruses have been proposed to have the ability to modulate both UPR and autophagy to promote viral replication [13]. P2Et extract has recently shown that it can regulate both mechanisms, UPR and autophagy [17,18]. Initially, to evaluate the participation of the UPR response in the antiviral mechanisms of our extracts, the expression of BiP, CHOP, EDEM, sXBP-1, and tXBP-1 genes was evaluated using the mRNA recovered from infected cells treated or not with P2Et, Anamu SC, and controls (NAC and quercetin) for 12 h (Fig. 4a). As expected, the single infection of the cells with MHV-A59 increased the expression of all UPR-related genes, particularly the generation of a splicing-induced form of XBP-1 (sXBP-1). Interestingly, P2Et does not induce ER stress on uninfected L929 cells. At the same time, in non-infected cells, Anamu SC increased the expression of all evaluated markers (Fig. 4a).
Additionally, when P2Et was used as a treatment during viral infection, the expression of BiP and tXBP-1 decreased markedly, similar to the results observed with the antioxidant NAC and the polyphenol quercetin downregulate some of the genes during infection. In contrast, Anamu SC clearly increased the expression of BiP, CHOP, and EDEM, with a tendency to increase sXBP-1 (Fig. 4a). On the other hand, EDEM increase has been associated with better antiviral response promoting cell survival, decreasing ER stress, and the secretion of viral particles [27,28]. Our results showed that Anamu SC antiviral activity would be involved with increased EDEM. Furthermore, we observed a significant increase in EDEM expression, approximately four-folds higher than uninfected and untreated cells, indicating a potential association between REDOX regulation, UPR, and viral infection. Western blot analysis revealed that MHV-A59 infection activates UPR via PERK phosphorylation, and both P2Et and chloroquine treatments further enhanced this effect (Fig. 4b). EDEM is involved in the quality control mechanism of the ER and plays a role in ER-associated degradation (ERAD) of misfolded proteins. While EDEM itself is not directly linked to autophagy, a functional relationship exists between ERAD and autophagy contributing to cell homeostasis [29].
In this study, the role of autophagy was assessed in cells infected with P2Et or treated with Anamu SC at 12 and 24 h. Both the infection and treatment increased the levels of the LC3-II form associated with autophagy induction, with chloroquine inhibiting autophagy and leading to an excessive accumulation of LC3-II (Fig. 4b). To analyze autophagic flux, cells were transfected with LC3-GFP-RFP plasmids and then infected with the virus. Confocal microscopy revealed the presence of yellow dots (indicating autophagosome accumulation) and red dots (representing autolysosomes). The virus slightly induced autophagosome accumulation, possibly as an escape mechanism from autolysosome degradation, while P2Et induced autophagic flux in both uninfected and infected cells, as previously reported [17].
Interestingly, the Anamu SC extract generated of large autophagosomes in infected and uninfected cells. However, a significant number of autolysosomes (indicated by red dots) were observed, indicating an active autophagic flux (Fig. 4c). Furthermore, Western blot results of cells treated with Anamu SC exhibited a decrease of p62 level, implying an enhanced autophagic flux. It is worth noting that previous studies have demonstrated that excessive protein accumulation can lead to the formation of protein aggregates independent of autophagy, and incorporation of LC3 during transient transfection [30,31]. In this context, the observed autophagic flux induced by Anamu SC may be associated with UPR activation. These findings suggest that both extracts possess the potential to modulate autophagy, possibly through distinct mechanisms. However, further investigation is warranted to elucidate these mechanisms in a more comprehensively.
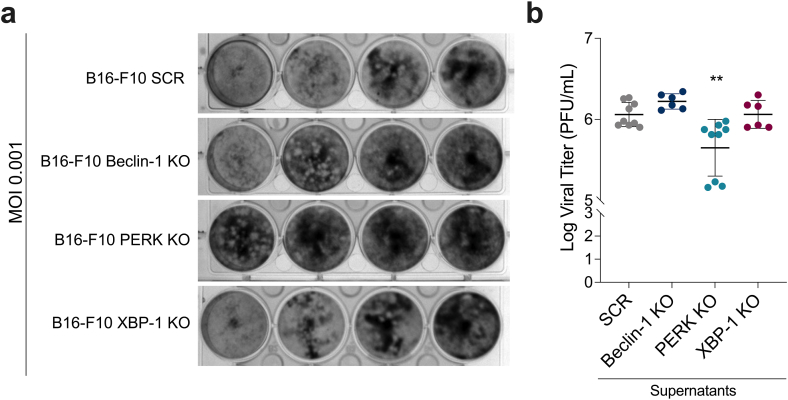
3.4. Loss of PERK decreases slightly virus yield after MHV-A59 infection
To investigate the involvement of UPR and autophagy role in MHV-A59 virus infection, we used B16–F10 melanoma cells that were genetically modified to lack expression of specific genes associated with these pathways (PERK, XBP-1 and Beclin-1) using the CRISPR/Cas-9 system [17,18]. We determined the viral yield by measuring PFU in the supernatants of B16–F10 cells infected with varying MOIs. Interestingly, B16–F10 cells exhibited increased permissiveness to MHV-A59 virus infection compared to control cells (Fig. 5a). This could be attributed to the higher expression of the murine virus receptor MHV-A59 in epithelial cells, as it belongs to the carcinoembryonic antigen family. Notably, the loss of the PERK gene resulted in a decrease in virus yield, suggesting that UPR mediated by PERK plays a role in virus replication. However, the absence of Beclin-1 and XBP-1 did not significantly impact virus yield (Fig. 5a and b), indicating that they may not directly influence viral replication. These findings provide valuable insights into the role of UPR, mainly via the PERK pathway, in mediating MHV-A59 virus replication.
Fig. 5.
Loss of PERK decreases viral production in B16 cells. B16 SCR or KO cells for PERK, XBP-1 or beclin-1 were infected with the MHV-A59 virus at a multiplicity of infection (MOI) of 0.01. After 24 h the supernatants were collected and the viral production was calculated through the plaque formation assay using dilutions of 10−2, 10−3, 10−4 and 10−5. (a) Representative images in plaque assay. (b) Mean virus supernatant titers (Log 10 Plaque Forming Units (PFU/mL)). Data are represented as the mean ± SD for 3 independent experiments (triplicate for SCR and PERK KO and duplicate for Beclin and XBP-1). The p values were calculated using Student's t-test. **p < 0.01; ****p < 0.0001.
3.5. P2Et and Anamu SC support the generation of DAMPs induced during viral infection by MHV-A59
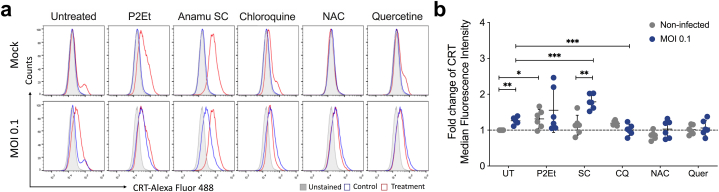
While the impact of viral infection on the generation of damage-associated molecular patterns (DAMPs), such as calreticulin exposure on the cell surface, remains largely unexplored, it has been speculated that DAMP accumulation could serve as potent signals to enhance the antigen-specific immune response [32]. Calreticulin (CRT) exposure is crucial in immunogenic cell death, regulated by UPR proteins. Some studies have indicated that viruses possess proteins capable of disrupting pathways associated with CRT exposure on the membrane of infected cells, thereby dampening the antiviral immune response [33,34]. Considering the role of the UPR in MHV-A59 infection with our previous findings demonstrating the ability of the P2Et extract to modulate this signaling pathway [18], we aimed to investigate whether the modulation of oxidative stress using plant extracts could also contribute to the activation of the immune system. Thus, L929 cells were initially infected with an MOI of 0.1 or left uninfected. Subsequently, the cells were treated with P2Et and Anamu SC extracts, chloroquine, NAC, or quercetin. Flow cytometry analysis was performed after 24 h to assess calreticulin levels on the cell surface. Our findings revealed that virus infection led to a modest increase in calreticulin surface expression. Interestingly, treatment of uninfected cells with P2Et, Anamu SC, or chloroquine tended to enhance calreticulin levels on the cell surface, while antioxidant treatments showed no effect, as expected. Interestingly, only the Anamu SC extract increased calreticulin levels during the infection, probably associated with the UPR increase (Fig. 6). These results showed that Anamu SC, unlike other treatments, increases calreticulin surface exposure and maintains the exposure after the infection.
Fig. 6.
P2Et and Anamu SC extracts maintain the generation of DAMPs induced by infection with the MHV-A59 virus in L929 cells. L929 cells were infected with a multiplicity of infection (MOI) of 0.1 of the MHV-A59 virus in the presence or absence of ½ of the IC50 of the P2Et and Anamu SC extracts, or with chloroquine (CQ), N-acetylcysteine (NAC), and quercetin (Quer). After 24 h of infection the cells were recovered and labeled with antibodies to visualize the surface expression of calreticulin (CRT). Then, cells were acquired using FACSAria IIU, and the results were subsequently analyzed using FlowJo 10.8.1 Software. (a). Representative histograms of mean fluorescence intensity (MFI) of CRT. (b) Fold change of MFI of CRT. Data are represented as the mean ± SD for 3 independent experiments. The p values were calculated using Student's t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
4. Discussion
This study's primary goal was to evaluate the P2Et and Anamu SC extracts' inhibitory effects on viral adsorption and/or replication. The findings demonstrated that whilst P2Et has a more noticeable effect on viral reproduction, Anamu SC mainly seems to inhibit viral adsorption. (Fig. 2). These findings are consistent with prior studies illustrating the antiviral properties of plant-derived natural compounds, which operate through various mechanisms. For instance, delphinidin and epigallocatechin-3-gallate (EGCG) directly interfere with the attachment of the hepatitis C virus (HCV) to host cells, thereby preventing viral entry but not replication [35]. Other polyphenols, such as isoquercetin, prevent clathrin-mediated internalization by the Zika virus [36]. Also, Polygonum cuspidatum, a bioactive botanical material, has been shown to alter dengue virus infectivity, possibly by interacting with structural components on the surface of viral particles such as with the E envelope glycoprotein, which is responsible for cell adhesion and viral entry [37,38]. Consequently, resveratrol an essential antioxidant representant secreted by plants, allows the retention of HMGB1 (high mobility group box 1) at the nuclear level, and that, consequently, can bind to the region of the interferon-stimulated gene promoter, contributing to decreased viral replication. Thus, our findings, combined with the existing literature, suggest that natural compounds, including those present in P2Et and Anamu SC extracts, can exert antiviral effects through diverse mechanisms that target different stages of the viral lifecycle.
Prior research has demonstrated how viral infections disturb the redox balance, creating a pro-oxidant milieu that promotes virus reproduction and heightens pathogenicity [[39], [40], [41]]. Our goal was to investigate how the extracts affect oxidative stress during virus infection. Anamu SC extract increased ROS in uninfected cells, and its impact on oxidative stress depended on the viral load. In contrast, P2Et extract consistently maintained its antioxidant activity regardless of the virus load. Further analysis identified sulfur compounds, sterols, triterpenes, and flavonoids, known as REDOX and UPR regulators, in the Anamu SC extract [42]. In viral infections, NRF2 is activated both directly and indirectly. In the case of Marburgvirus (MARV), a hemorrhagic fever virus of the Filoviridae family, when it binds to KEAP1, the viral protein VP24 releases and activates NRF2 directly. In HIV infection, a human retrovirus belonging to the lentivirus family, increased ROS levels produced by infected macrophages or glial cells trigger NRF2 activation via the Tat transcription factor. These findings suggest that viruses like HIV and MARV exploit the cellular antioxidant response for their benefit during replication [26]. In the context of our study, the differential effects on intracellular ROS levels during MHV-A59 cell infection and treatment with natural extracts may reflect variations in their antiviral capacity, highlighting the delicate balance between antioxidant and pro-oxidant environments in viral infection. P2Et, primarily composed of polyphenols, consistently exerted its antioxidant effect regardless of the viral dose. The induced cellular stress during viral infection, exacerbated by Anamu SC extract, could explain the increased virus replication observed when the extract was used solely during virus replication. The scientific literature has documented on multiple occasions the antiviral effect induced by antioxidant mechanisms as increases the concentration of enzyme antioxidants. However, it is important to note that research on the prooxidant effect as an antiviral strategy has received limited attention to date. These findings emphasize the importance of investigating the interplay between oxidative stress, viral infection, and the modulation of cellular redox pathways.
In this context, alterations in ROS levels are accompanied by protein synthesis damage during viral infections, leading to the induction of the UPR that aims to restore ER homeostasis by regulating protein folding, trafficking, and degradation. Recent studies have highlighted the induction of ER stress during coronavirus infections activating all three UPR branches (IRE1, PERK, and ATF6) [13]. As expected, cells infected with coronavirus strain MHV-A59 upregulated UPR-related genes, particularly IRE-1a/XBP-1. Similar effects have been observed in other viral models like dengue virus, where UPR response and autophagy induction aid viral replication [43]. Notably, the use of P2Et therapy during viral infection reduced the expression of BiP and tXBP-1, akin to the effects of antioxidants NAC and quercetin. The downregulation of UPR-related genes by P2Et indicates its potential in alleviating ER stress caused by viral infection, which is necessary for viral protein synthesis. This highlights the potential of plant-derived antioxidants in coronavirus prevention. On the other hand, Anamu SC treatment during viral infection increased the expression of BiP, CHOP, and EDEM, suggesting a stronger antiviral response, reduced ER stress, and inhibited viral particle secretion. This increase in EDEM expression indicates a possible link between REDOX regulation, UPR, and viral infection.
Our mechanistic analyses found that silencing PERK, but not XBP-1 or Beclin-1, slightly decreased viral production during MHV-A59 infection, suggesting that induction of the UPR pathway by this pathway is essential to promote viral replication. On the other hand, we have recently shown that both the PERK genes of the UPR pathway and the beclin-1 protein genes of autophagy favour the exposure of DAMPs associated with immunogenic cell death (ICD) induced by the treatment of tumour cells with the P2Et extract [17,18]. Although the association of ICD induction during viral infection has yet to be widely described, the concept has begun to develop using viruses with oncolytic potential [44]. However, it was found that Anamu SC of L929 fibroblasts with MHV-A59 increased calreticulin exposure on the plasma membrane. This fact is consistent with the findings of UPR induction during MHV-A59 infection since the involvement of the PERK pathway in calreticulin exposure on the cell surface is established [45]. Interestingly, both P2Et and Anamu SC maintain calreticulin exposure induced by a viral infection in contrast to CQ, quercetin, or NAC treatments. The relevance of calreticulin exposure in the induction of the immune response during coronavirus infection should be studied further.
In a study mirroring observations in the MHV-A59 mouse model, both P2Et extract and Anamu SC were found to inhibit COVID-19 viral replication in Vero E6 cells from patient isolates. A Phase II clinical trial in COVID-19 patients treated with P2Et demonstrated a 2.7 day earlier hospital discharge compared to the placebo group, accompanied by a significant reduction in pro-inflammatory cytokines [20]. These findings suggest that these natural extracts, particularly P2Et, can effectively treat human coronavirus infections. However, further research is needed to understand the underlying mechanisms and the impact of these extracts on virus adsorption versus replication.
Our research suggested that these extracts have various mechanisms to combat coronaviruses. P2Et mainly hinders viral replication by reducing ROS production and ER stress, while Anamu SC extract increases ROS production and may aid protein degradation through the unfolded protein response (UPR). Anamu SC extract also enhances calreticulin exposure during virus-cell infection. The differing effects on intracellular ROS levels during MHV-A59 infection and treatment with these extracts highlight the delicate balance between antioxidants and pro-oxidants in viral infections. This underscores the importance of understanding how viral infections, cellular stress responses, and redox pathways interact. These distinct mechanisms of action provide a basis for further exploration of P2Et and Anamu SC plant extracts as potential therapies against coronaviruses.
The use of plants as a source of antiviral chemicals is noteworthy due to its quantity of chemical components, such as polyphenols, alkaloids, and terpenoids, as well as its sustainability when compared to the chemical production of pharmaceuticals. This diversity increases the number of therapeutic alternatives accessible and raises the likelihood of discovering useful chemicals, which may lead to more study and clinical trials. The complexities inherent in the interaction between plants and viruses, as well as the deepening understanding of the underlying mechanisms by which P2Et and Anamu SC inhibit viral replication, represent key challenges in our research that must be addressed to strengthen the knowledge base and maximize the applicability of our findings. The apparent differential modulation of oxidative stress by these two extracts with antiviral capacity represents a possible target for antiviral therapy; however, this association should be further evaluated mechanistically.
Availability of data and materials
The data supporting the findings of this study are available upon reasonable request from the corresponding author.
Competing interests/COI statement
S.F. and C.U. are inventors of a granted patent related to P2Et. The rest of the authors declares no competing interests.
Funding
Funding was provided by the Ministerio de Ciencias (MINCIENCIAS) in the Mincienciaton grant (Contract 378-2020, Code 1203101576633). Aditionally, KP, CA, CC and CU disclosed receipt of the following financial support from Ministerio de Ciencia, tecnología e Innovación, Ministerio de Educación Nacional, Ministerio de Industria, Comercio y Turismo e ICETEX, 2a Convocatoria Ecosistema científico - Colombia Científica 792–2017, Programa “Generación de alternativas terapéuticas en cáncer a partir de plantas a través de procesos de investigación y desarrollo traslacional, articulados en sistemas de valor sostenibles ambiental y económicamente” (Contrato no. FP44842-221-2018). We thank the Vicerrectoria de Investigaciones at the Pontificia Universidad Javeriana for the financial support to publish this research (Proposal ID00010641).
CRediT authorship contribution statement
Karol Prieto: Formal analysis, Investigation, Writing - original draft. Cindy Arévalo: Formal analysis, Investigation, Methodology, Writing - original draft, Writing - review & editing. Paola Lasso: Data curation, Investigation, Writing - original draft, Writing - review & editing. Carolina Carlosama: Formal analysis, Investigation. Claudia Urueña: Conceptualization, Formal analysis, Investigation. Susana Fiorentino: Conceptualization, Project administration, Resources, Writing - original draft. Alfonso Barreto: Conceptualization, Formal analysis, Funding acquisition, Investigation, Supervision, Validation, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Alfonso Barreto reports provided by Pontifical Xavierian University. Alfonso Barreto reports a relationship with Pontifical Xavierian University that includes: employment. Susana Fiorentino has patent #Patent related to P2Et licensed to US20230190848A1.
Acknowledgments
The authors would like to thank Pontificia Universidad Javeriana and the Colombian Environmental Ministry for allowing the use of genetic resources through the Contract of Access to Genetic Resources No. 220 of 2018 for Colombian plant material. The authors express gratitude to Laura Rojas for the UPLC analyzes of the extracts.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23403.
Contributor Information
Cindy Arévalo, Email: cindy.arevalo@javeriana.edu.co.
Paola Lasso, Email: plasso@javeriana.edu.co.
Carolina Carlosama, Email: carolina_carlosamap@javeriana.edu.co.
Claudia Urueña, Email: curuena@javeriana.edu.co.
Susana Fiorentino, Email: susana.fiorentino@javeriana.edu.co.
Alfonso Barreto, Email: alfonso.barreto@javeriana.edu.co.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coley S.E., Lavi E., Sawicki S.G., Fu L., Schelle B., Karl N., Siddell S.G., Thiel V. Recombinant mouse hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. J. Virol. 2005;79:3097–3106. doi: 10.1128/JVI.79.5.3097-3106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/s0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C. Therapeutic modulation of virus-induced oxidative stress via the nrf2-dependent antioxidative pathway. Oxid. Med. Cell. Longev. 2018;2018 doi: 10.1155/2018/6208067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattarai K.R., Riaz T.A., Kim H.R., Chae H.J. The aftermath of the interplay between the endoplasmic reticulum stress response and redox signaling. Exp. Mol. Med. 2021;53:151–167. doi: 10.1038/s12276-021-00560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foo J., Bellot G., Pervaiz S., Alonso S. Mitochondria-mediated oxidative stress during viral infection. Trends Microbiol. 2022;30:679–692. doi: 10.1016/j.tim.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Zhou A., Zhang W.H., Dong X., Liu M.Y., Chen H.B., Tang B. The battle for autophagy between host and influenza A virus. Virulence. 2022;13:46–59. doi: 10.1080/21505594.2021.2014680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y., Bowman J.W., Jung J.U. Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 2018;16:340–353. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Wang K., Xing Y., Tu J., Yang X., Zhao Q., Li K., Chen Z. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014;5:912–927. doi: 10.1007/s13238-014-0104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mosbauer K., Zellner A., et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019;10:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue M., Feng L. The role of unfolded protein response in coronavirus infection and its implications for drug design. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.808593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paraiso I.L., Revel J.S., Stevens J.F. Potential use of polyphenols in the battle against COVID-19. Curr. Opin. Food Sci. 2020;32:149–155. doi: 10.1016/j.cofs.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limanaqi F., Busceti C.L., Biagioni F., Lazzeri G., Forte M., Schiavon S., Sciarretta S., Frati G., Fornai F. Cell clearing systems as targets of polyphenols in viral infections: potential implications for COVID-19 pathogenesis. Antioxidants. 2020;9 doi: 10.3390/antiox9111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandoval T.A., Urueña C.P., Llano M., Gomez-Cadena A., Hernandez J.F., Sequeda L.G., Loaiza A.E., Barreto A., Li S., Fiorentino S. Standardized extract from Caesalpinia spinosa is cytotoxic over cancer stem cells and enhance anticancer activity of doxorubicin. Am. J. Chin. Med. 2016;44:1693–1717. doi: 10.1142/S0192415X16500956. [DOI] [PubMed] [Google Scholar]
- 17.Prieto K., Lozano M.P., Uruena C., Almeciga-Diaz C.J., Fiorentino S., Barreto A. The delay in cell death caused by the induction of autophagy by P2Et extract is essential for the generation of immunogenic signals in melanoma cells. Apoptosis. 2020;25:875–888. doi: 10.1007/s10495-020-01643-z. [DOI] [PubMed] [Google Scholar]
- 18.Prieto K., Cao Y., Mohamed E., Trillo-Tinoco J., Sierra R.A., Urueña C., Sandoval T.A., Fiorentino S., Rodriguez P.C., Barreto A. Polyphenol-rich extract induces apoptosis with immunogenic markers in melanoma cells through the ER stress-associated kinase PERK. Cell Death Discov. 2019;5:134. doi: 10.1038/s41420-019-0214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernández J.F., Urueña C.P., Sandoval T.A., Cifuentes M.C., Formentini L., Cuezva J.M., Fiorentino S. A cytotoxic Petiveria alliacea dry extract induces ATP depletion and decreases β-F1-ATPase expression in breast cancer cells and promotes survival in tumor-bearing mice. Revista Brasileira de Farmacognosia. 2017;27:306–314. [Google Scholar]
- 20.Urueña C., Ballesteros-Ramírez R., Gomez Cadena A., Barreto A., Prieto K., Quijano S., Aschner P., Martínez C., Zapata-Cardona M.I., El Ahanidi H. Randomized double-blind clinical study in patients with COVID-19 to evaluate the safety and efficacy of a phytomedicine (P2Et) Front. Med. 2022;9 doi: 10.3389/fmed.2022.991873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahmani M., Shirzad H., Shahinfard N., Sheivandi L., Rafieian-Kopaei M. Cancer phytotherapy: recent views on the role of antioxidant and angiogenesis activities. In J Evid Based Complementary Altern Med. 2017;22:299–309. doi: 10.1177/2156587215625157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaneda D.M., Pombo L.M., Uruena C.P., Hernandez J.F., Fiorentino S. A gallotannin-rich fraction from Caesalpinia spinosa (Molina) Kuntze displays cytotoxic activity and raises sensitivity to doxorubicin in a leukemia cell line. BMC Compl. Alternative Med. 2012;12:38. doi: 10.1186/1472-6882-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Cadena A., Uruena C., Prieto K., Martinez-Usatorre A., Donda A., Barreto A., Romero P., Fiorentino S. Immune-system-dependent anti-tumor activity of a plant-derived polyphenol rich fraction in a melanoma mouse model. Cell Death Dis. 2016;7:e2243. doi: 10.1038/cddis.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmittgen T.D., Zakrajsek B.A., Mills A.G., Gorn V., Singer M.J., Reed M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 25.Schickli J.H., Zelus B.D., Wentworth D.E., Sawicki S.G., Holmes K.V. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 1997;71:9499–9507. doi: 10.1128/jvi.71.12.9499-9507.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herengt A., Thyrsted J., Holm C.K. NRF2 in viral infection. Antioxidants. 2021;10 doi: 10.3390/antiox10091491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perera N., Miller J.L., Zitzmann N. The role of the unfolded protein response in dengue virus pathogenesis. Cell Microbiol. 2017;19 doi: 10.1111/cmi.12734. [DOI] [PubMed] [Google Scholar]
- 28.Lazar C., Macovei A., Petrescu S., Branza-Nichita N. Activation of ERAD pathway by human hepatitis B virus modulates viral and subviral particle production. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiritoiu M., Chiritoiu G.N., Munteanu C.V.A., Pastrama F., Ivessa N.E., Petrescu S.M. EDEM1 drives misfolded protein degradation via ERAD and exploits ER-phagy as back-up mechanism when ERAD is impaired. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21103468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuma A., Matsui M., Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 31.Wang L., Chen M., Yang J., Zhang Z. LC3 fluorescent puncta in autophagosomes or in protein aggregates can be distinguished by FRAP analysis in living cells. Autophagy. 2013;9:756–769. doi: 10.4161/auto.23814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galluzzi L., Kepp O., Morselli E., Vitale I., Senovilla L., Pinti M., Zitvogel L., Kroemer G. Viral strategies for the evasion of immunogenic cell death. J. Intern. Med. 2010;267:526–542. doi: 10.1111/j.1365-2796.2010.02223.x. [DOI] [PubMed] [Google Scholar]
- 33.Kepp O., Senovilla L., Galluzzi L., Panaretakis T., Tesniere A., Schlemmer F., Madeo F., Zitvogel L., Kroemer G. Viral subversion of immunogenic cell death. Cell Cycle. 2009;8:860–869. doi: 10.4161/cc.8.6.7939. [DOI] [PubMed] [Google Scholar]
- 34.Garg A.D., Krysko D.V., Vandenabeele P., Agostinis P. The emergence of phox-ER stress induced immunogenic apoptosis. OncoImmunology. 2012;1:786–788. doi: 10.4161/onci.19750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calland N., Sahuc M.E., Belouzard S., Pène V., Bonnafous P., Mesalam A.A., Deloison G., Descamps V., Sahpaz S., Wychowski C., et al. Polyphenols inhibit hepatitis C virus entry by a new mechanism of action. J. Virol. 2015;89:10053–10063. doi: 10.1128/jvi.01473-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chojnacka K., Skrzypczak D., Izydorczyk G., Mikula K., Szopa D., Witek-Krowiak A. Antiviral properties of polyphenols from plants. Foods. 2021;10 doi: 10.3390/foods10102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo Y.-T., Liu C.-H., Li J.-W., Lin C.-J., Jassey A., Wu H.-N., Perng G.C., Yen M.-H., Lin L.-T. Identification of the phytobioactive Polygonum cuspidatum as an antiviral source for restricting dengue virus entry. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-71849-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zainal N., Chang C.-P., Cheng Y.-L., Wu Y.-W., Anderson R., Wan S.-W., Chen C.-L., Ho T.-S., AbuBakar S., Lin Y.-S. Resveratrol treatment reveals a novel role for HMGB1 in regulation of the type 1 interferon response in dengue virus infection. Sci. Rep. 2017;7:1–12. doi: 10.1038/srep42998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz K.B. Oxidative stress during viral infection: a review. Free Radic. Biol. Med. 1996;21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- 40.Checconi P., De Angelis M., Marcocci M.E., Fraternale A., Magnani M., Palamara A.T., Nencioni L. Redox-modulating agents in the treatment of viral infections. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21114084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khomich O.A., Kochetkov S.N., Bartosch B., Ivanov A.V. Redox biology of respiratory viral infections. Viruses. 2018;10 doi: 10.3390/v10080392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballesteros-Ramírez R., Aldana E., Herrera M.V., Urueña C., Rojas L.Y., Echeverri L.F., Costa G.M., Quijano S., Fiorentino S. Preferential activity of Petiveria alliacea extract on primary myeloid leukemic blast. Evid Based Complement Alternat Med. 2020;2020 doi: 10.1155/2020/4736206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y.R., Kuo S.H., Lin C.Y., Fu P.J., Lin Y.S., Yeh T.M., Liu H.S. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci. Rep. 2018;8:489. doi: 10.1038/s41598-017-18909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabiljo J., Laengle J., Bergmann M. From threat to cure: understanding of virus-induced cell death leads to highly immunogenic oncolytic influenza viruses. Cell Death Discov. 2020;6:48. doi: 10.1038/s41420-020-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panaretakis T., Kepp O., Brockmeier U., Tesniere A., Bjorklund A.C., Chapman D.C., Durchschlag M., Joza N., Pierron G., van Endert P., et al. Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death. The EMBO journal. 2009;28:578–590. doi: 10.1038/emboj.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available upon reasonable request from the corresponding author.