Abstract
Objective
This study aimed to determine the diagnostic accuracy of the antigen rapid diagnostic test (Ag-RDT) as a screening tool for SARS-CoV-2 infection compared to Quantitative reverse transcription polymerase chain reaction (qRT-PCR).
Methods
This study was conducted at six referral hospitals in Oromia Region, Ethiopia. One thousand seven hundred twenty-one patients who visited the hospitals for various medical conditions were tested with qRT-PCR and/or Ag-RDTs. Qualitative detection of SARS-CoV-2 antigen was performed using the Panbio™ COVID-19 Ag rapid test device.
Results
Compared with qRT-PCR, Ag-RDTs had a sensitivity of 33.3 % (95%CI: 30.9%–35.9 %) and a specificity of 99.3 % (95%CI: 98.8%–99.7 %) to detect active SARS-CoV-2 infection. The area under the receiver operator curve was 0.67 (95%CI: 0.63–0.69). The sensitivity of Ag-RDTs appeared high in patients with shortness of breath (73.3 %) and those presenting with all three symptoms – fever, cough, and dyspnea (71.4 %). In all instances, specificity was more than 98 %. The Ag-RDT positivity rate also correlated well with viral load: 51.7 % in cases with cycle threshold (Ct) < 25 (high viral load) and only 3.4 % when Ct > 25 (low viral load).
Conclusion
Although Ag-RDT for diagnosing SARS-CoV-2 is a good option as a point-of-care screening tool, it has a low sensitivity to detect active infections. Using Panbio™ COVID-19 Ag Rapid test for diagnostic and treatment decisions may lead to a false negative, resulting in patient misdiagnosis, ultimately contributing to disease spread and poor patient outcome.
Keywords: Panbio, Ag-RDT, qRT-PCR, COVID-19, Comparison, Ethiopia
1. Introduction
Despite the initially feared looming crisis, Africa's coronavirus disease 2019 (COVID-19) pandemic reportedly remained mild [1,2]. Due to inadequate monitoring and testing capability, the actual burden of the pandemic in most low-income nations was poorly understood [[3], [4], [5], [6], [7]]. However, a high seroprevalence of severe acute respiratory syndrome coronavirus type-2 (SARS-CoV-2) has been detected among healthcare workers (HCWs) in various African nations. For instance, SARS-CoV-2 seroprevalence of 41.2 % in the Democratic Republic of Congo [8], 45.1 % in Nigeria [9], and 60 % among blood donors in South Africa were reported by early 2021 [10]. Likewise, a serosurvey conducted in Ethiopia between August 2020 and April 2021 among healthcare workers and community participants revealed an escalation of SARS-CoV-2 seroprevalence from 10 % to nearly 60 % [5].
These seroepidemiological studies show that due to probably the dominance of the younger population and other factors, Africa, despite the high infection rate, did not face as many humanitarian crises as reported elsewhere [[10], [11], [12]]. Nonetheless, as the risk posed by the virus remains real, such settings must implement a system for early detection and treatment of cases, at least in hospitals.
qRT-PCR test remains the only diagnostic test approved globally as a confirmatory test for COVID-19, though it is assumed to miss about a third of cases. The turnaround time of the result using qRT-PCR is at least 24 h [6]. However, Ag-RDT performs well in individuals with high viral load during early infection and will be most reliable in settings where the incidence of SARS-CoV-2 infection is ≥ 5 %.
When there is no transmission or low transmission, the positive predictive value of Ag-RDTs will be low. In such a setting, qRT-PCR is preferable for first-line testing or confirmation of Ag-RDT positive results. The WHO recommends using Ag-RDT that meet minimum performance requirements of ≥80 % sensitivity and ≥97 % specificity, although Ag-RDTs are less sensitive than qRT-PCR, particularly in asymptomatic populations. Ag-RDTs should be reserved for use in symptomatic individuals meeting the case definition for COVID-19 and to test asymptomatic individuals at high risk of infection, including contacts and health workers, particularly in settings where qRT-PCR testing capacity is limited. Although multiple antigen and serological tests are now available, none is officially approved to confirm active infection. Besides, serologic studies remain the only option to identify the community's infection burden and assess outbreak dynamics [13].
Ethiopia started using Ag-RDTs for COVID-19 diagnosis as the point-of-care and for screening purposes, even though the test's sensitivity and specificity were not fully known or validated under specific conditions. If proven sensitive and specific, the antigen tests may help shorten the turnaround time by many hours to days and save meagre resources. Timely detection and isolation of cases and their contacts are crucial to help curtail this unprecedented pandemic [14]. Direct detection of SARS-CoV-2 viral proteins (antigens) in nasal swabs and other respiratory secretions using lateral flow immunoassays (also known as Ag-RDTs) offers a faster and less expensive method to test SARS-CoV-2 than qRT-PCR [14]. This strategy relies on robust, rapid, and easy-to-perform diagnostic tools that can be used to test large numbers of samples quickly. Hence, this study aimed to compare the Ag-RDTs with qRT-PCR among a sample of patients visiting the selected hospitals for various medical conditions in Ethiopia during the study period.
2. Methods and materials
2.1. Study setting
The study was conducted at six hospitals with the highest caseloads and catchment populations in four towns of Oromia Regional State. The region accounts for 32 % of Ethiopian land size with an area of 363399.8 Km2. Among 105 functional hospitals in the Oromia Region, 25 served as COVID-19 treatment centres in addition to the usual health services. There were 15 qRT-PCR COVID-19 testing centres and more than 60 Ag-RDT sites as point-of-care in selected health facilities during the pandemic in the Oromia Region. These six selected hospitals were referral and university teaching hospitals and were among the health facilities providing COVID-19 testing services by qRT-PCR and Ag-RDTs.
2.2. Study design and participants
A cross-sectional study was conducted among inpatients and outpatients visiting six selected hospitals during August 9–23, 2021. The participants were suspected COVID-19 cases, inpatients, pre-operative patients, and outpatients who visited the selected hospitals during the study period. The hospitals were Jimma University Specialized Hospital (Jimma Town), Wollega University Teaching and Referral Hospital and Nekemte General Hospital (Nekemte Town), Ambo University Teaching and Referral Hospital and Ambo General Hospital (Ambo Town), and Adama Hospital Medical College (Adama Town). All patients consented to give nasopharyngeal qRT-PCR swabs, and Ag-RDTs were recruited. Nasopharyngeal samples were collected and handled based on the national COVID-19 sample collection, processing, and testing guidelines.
2.3. Sample size calculation
Considering different assumptions (like high patient caseload and the number of confirmed COVID-19 cases), one thousand seven hundred twenty-one participants were selected and proportionally allocated to the selected hospitals accordingly (Jimma University Specialized Hospital 352, Wollega University Specialized Hospital and Nekemete general hospital 505, Ambo University Specialized Hospital and Ambo general hospital 468, and Adama Hospital Medical College 396).
2.4. Data collection procedures
Demographic and clinical characteristics of respondents were collected using an electronic data collection tool. We recruited twelve data collectors and supervisors (epidemiologists and public health emergency officers), sixteen laboratory professionals (for sample collection and handling), and twelve laboratory professionals (working at accredited COVID-19 testing centres for testing). Even though they all were professionals, they were trained in the specific data collection tools tailored to this study, sample collection, handling, and testing procedures. The training was also given on data collection software by an expert data manager. The English version of the survey tools was translated into local languages (Afaan Oromo and Amharic). The data collectors used the local language versions for data collection. The supervisors checked the work of data collectors daily using an online Open Data Kit (CSEntry) application software for the completeness of the survey tools and data quality. The data was sent to the central server, and data completeness was checked immediately by the research team. The laboratory samples were stored at the appropriate temperature by maintaining cold chain management at regional laboratories and hospitals.
2.5. Clinical specimens and SARS-CoV-2 tests
Two nasopharyngeal (NP) swabs were collected per participant for performing Ag-RDT and qRT-PCR tests in parallel.
Ag-RDT: The test was performed by using the Panbio™ COVID-19 Ag Rapid Test. The NP swab was immersed into an extraction tube containing the buffer fluid from individuals who fulfilled the inclusion criteria. The specimens were collected at selected health facilities by trained laboratory technicians, and the test was performed following the manufacturer's instructions. The specimens were tested immediately after collection, and results were read at 15 min at the point of care. All the procedures complied with personal safety guidelines, including the use of personal protective equipment. The manufacturer reported a specificity of the Panbio COVID-19 Ag of 99.8 % (95%CI 98.65%–100 %) and an overall sensitivity of 98.1 % (95%CI: 93.2%–99.8 %). It was also reported that sensitivity is 100 % (95%CI: 92.3%–100 %) within the first three days of symptom onset and 96.6 % (95%CI: 88.1%–99.6) 4–7 days after symptom onset [15] [14,[16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26]].
qRT-PCR: The test was performed by using the Abbot Real-Time SARS CoV-2 Assay. The NP swab was mixed with 2 mL of Viral Transport Media (VTM), consisting of Hanks' balanced salt, 0.4 % fetal bovine serum, antibiotic, and antifungal agents, and transported to the test laboratory within 48 h of specimen collection. Samples were transported and stored at 2–8 °C at the testing laboratories (Adama Public Health Reference and Referral Laboratory and Nekemete Public Health Reference Referral Laboratory) for processing and testing. All specimens were processed in biosafety level-2 (BSL-2) facilities with full personal protective equipment. All the tests were performed according to the manufacturer's guidelines and the Ethiopian Public Health Institute recommendations. Ct-value was retrieved from the QRT-PCR machine, and the performance of QRT-PCR was interpreted based on Ct-value. According to the manufacturer's Clinical Performance Evaluation, the Positive Percent Agreement (PPA) between the two assays was 95.9 % (95%CL: 86.0%–99.5 %), and the Negative Percent Agreement (NPA) was 100 % (95%CL: 93.5%–100.0 %) [27].
2.6. Data management and analysis
Basic demographic, epidemiological, and clinical information were collected online through CSentry CSPro DataEntry 7.2.1, stored at the central server. They were exported to Microsoft Excel for data cleaning and then to Stata, version 14.0, for analysis. Receiver Operating Curve (ROC) and Kohen's kappa analysis were performed, and positive percent agreement, overall agreement, sensitivity, specificity, positive predictive value, and negative predictive value were calculated. The positivity rate was calculated by both tests with patients with different clinical scenarios. The result was summarized with absolute and relative (percentage) frequencies with point and interval (95 % confidence intervals, CI) estimates. A p-value less than 0.05 was considered statistically significant. All statistical computations were performed with the statistical software Stata version 14.0.
2.7. Ethical considerations
Ethical approval was received from the Jimma University Institute of Health (Ref No-JUIR/IRB/333/23) and the Oromia Health Bureau Ethical Review Board. Written informed consent was obtained from all participants before the data collection. Data collectors adhered to the standard COVID-19 prevention recommendations during the interview, including using face masks and physical distancing. Confidentiality of the collected data was ensured through anonymity. Participants with positive PCR and/or Ag-RDT results were transferred to treatment centres or followed at Home-based and Isolation care according to the COVID-19 National Case Management and Isolation Protocol.
3. Results
Overall, 1721 participants were involved in the study. Almost all (n = 1713, 99.5 %) were tested with Ag-RDTs as point-of-care testing, while only 1368 (79.5 %) were tested with qRT-PCR. In total, 1364 participants tested by qRT-PCR and Ag-RDTs were included in the final analysis for a head-to-head comparison. The rest were excluded from laboratory tests and final analysis due to the poor quality of the specimens and labeling errors.
3.1. Background characteristics of study participants
Among the participants, 52 % (708/1364 were females with a median age of 32 (IQR: 24–50) years. Participants with no formal education accounted for 31 % (426/1364), and 23.7 % (324/1364) were farmers. Most of the participants, 84.7 % (1155/1364), were selected from the adult outpatient department (OPD), and 40.9 % (558/1364) had known underlying medical conditions (Table 1).
Table 1.
Background characteristics of study participants.
| Characteristics | Count (N = 1364) | % |
|---|---|---|
| Sex (Female) | 708 | 52.0 |
| Age, Median (IQR) | 32 (24–50) | |
| Residence, Urban | 827 | 60.1 |
| Educational status | ||
| No formal education | 426 | 31.2 |
| Primary school | 380 | 27.9 |
| High school | 315 | 23.1 |
| College (above high school) | 243 | 17.8 |
| Occupation | ||
| Farmer | 324 | 23.8 |
| Housewife | 250 | 18.3 |
| Student | 222 | 16.3 |
| Unemployed | 189 | 13.9 |
| Other, specify | 172 | 12.6 |
| Daily laborer | 86 | 6.3 |
| Others * | 61 | 4.5 |
| School teacher | 60 | 4.4 |
| Type of service unit | ||
| Adult OPD | 1155 | 84.7 |
| Emergency Unit | 146 | 10.7 |
| IPD | 59 | 4.3 |
| Under -Five OPD | 4 | 0.3 |
| Contact with confirmed or suspected COVID-19 | 99 | 7.3 |
| Having recent COVID-19-related symptoms (cough, fever, pain, etc.) | 422 | 31 |
| Ever been tested for COVID-19 | 129 | 9.5 |
| Underlying medical condition | 558 | 40.9 |
| Ever received COVID-19 vaccination | 64 | 4.7 |
Others* (Security/police/military, Healthcare worker, Bus/taxi/bajaj driver, Bank accountant, and Hairdresser)
3.2. qRT-PCR and Ag-RDT positivity rates and associated factors
Overall, 6.5 % (89/1364) and 17.74 % (242/1364) of the patients tested positive for SARS-CoV-2 with Ag-RDTs and qRT-PCR, respectively. qRT-PCR and Ag-RDTs positivity rates were higher among patients who had contacts with confirmed or suspected COVID-19 cases (64.1 % Vs. 13.4 %; P < 0.001 for PCR and 21.1 % Vs. 4.9 %; P < 0.001 for Ag-RDTs). Those who had recent COVID-19-related symptoms (29.1 % Vs. 12.7 %; P < 0.001 for qRT-PCR and 13.9 % Vs. 2.2 %; P < 0.001 for the rapid test (Table 2) had a higher positivity rate.
Table 2.
qRT-PCR and rapid antigen test positivity rate in various clinical conditions (N = 1364).
| Clinical category | Status | qRT-PCR |
Antigen test |
||
|---|---|---|---|---|---|
| Positivity rate (%) | P-value | Positivity rate | P-value | ||
| Contact with confirmed or suspected COVID-19 case | Yes | 64.1 | <0.001* | 21.1 | <0.001* |
| No | 13.4 | 4.9 | |||
| Ever received the COVID-19 vaccine | Yes | 23.4 | 0.428 | 11.9 | 0.022* |
| No | 17.5 | 5.8 | |||
| Known underlying medical condition | Yes | 11.2 | <0.001* | 4.6 | 0.033* |
| No | 22.3 | 7.1 | |||
| Recent COVID-19 related symptoms (<1 month) | Yes | 29.1 | <0.001* | 13.9 | <0.001* |
| No | 12.7 | 2.2 | |||
| Fever | Yes | 36.7 | 0.011* | 18.9 | 0.014* |
| No | 25 | 11.4 | |||
| Cough | Yes | 30.7 | 0.229 | 13.6 | 0.725 |
| No | 24.8 | 14.8 | |||
| Shortness of breath | Yes | 15.3 | 0.001* | 10.1 | 0.123 |
| No | 33.2 | 15.2 | |||
| Fatigue | Yes | 19.6 | 0.016* | 9.2 | 0.059 |
| No | 32.1 | 15.5 | |||
| Myalgia/arthralgia | Yes | 36.9 | 0.034* | 21.6 | 0.001* |
| No | 26.3 | 11.1 | |||
| Nasal congestion/sneezing | Yes | 23.9 | 0.414 | 4.6 | 0.021* |
| No | 29.7 | 15.1 | |||
| New onset loss of smell or taste | Yes | 42.1 | 0.064 | 20.4 | 0.149 |
| No | 27.8 | 13.2 | |||
| Sore throat | Yes | 22.4 | 0.277 | 8.9 | 0.162 |
| No | 29.9 | 14.7 | |||
| Headache | Yes | 33.3 | 0.067 | 15.9 | 0.171 |
| No | 25.2 | 11.9 | |||
* Statistically significant for P < 0.05.
3.3. Sensitivity and specificity of Ag-RDTs
The Ag-RDTs test has a very low sensitivity (33.3 %) but a very high specificity (99.3 %) for detecting active SARS-CoV-2 infection (Table 3).
Table 3.
Overall Sensitivity, specificity, and predictive value of rapid antigen test compared with qRT-PCR (N = 1364).
| Measurements | % | [95 % Conf. Inter.] |
|
|---|---|---|---|
| Lower | Upper | ||
| Sensitivity | 33.3 % | 31.0 % | 36.0 % |
| Specificity | 99.3 % | 98.8 % | 99.7 % |
| Positive predictive value | 91.0 % | 89.5 % | 92.5 % |
| Negative predictive value | 87.4 % | 85.6 % | 89.1 % |
| Prevalence | 17.7 % | 15.7 % | 19.8 % |
The sensitivity remains low in those with minimal COVID-19 symptoms. The sensitivity of the Ag-RDTs (Panbio) increased in patients who had typical clinical manifestations of COVID-19, such as shortness of breath (73.3 %), all three major symptoms together (fever + cough + dyspnea) (71.4 %), and myalgia and arthralgia (65.9 %). Specificity remains greater than 98 % in all cases (Table 4).
Table 4.
Sensitivity, specificity, Positive Predictive values (PPV), and Negative Predictive values (NPV) of rapid antigen test in different clinical scenarios (N = 1364).
| Category | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| Total Participants | 33.3 | 99.3 | 91 | 87.3 |
| Contact with confirmed or suspected COVID-19 cases | 34.7 | 100 | 100 | 46.2 |
| Recent COVID-19-related symptoms | 52 | 98.7 | 94.1 | 83.4 |
| Fever | 57.4 | 97.8 | 93.9 | 79.8 |
| Shortness of breath | 73.3 | 98.8 | 91.7 | 95.3 |
| Cough | 51.1 | 98.6 | 94.1 | 82 |
| Loss of taste or smell | 50 | 95.5 | 88.9 | 72.4 |
| Myalgia and arthralgia (Body aches) | 65.9 | 98.6 | 96.4 | 83.1 |
| Runny nose | 27.3 | 100 | 100 | 81.4 |
| Sore throat | 45.5 | 97.4 | 83.3 | 86 |
| Fatigue | 55 | 100 | 100 | 90.1 |
| Headache | 50.7 | 97.8 | 91.9 | 79.9 |
| Cough + fever + SOB | 71.4 | 100 | 100 | 89.5 |
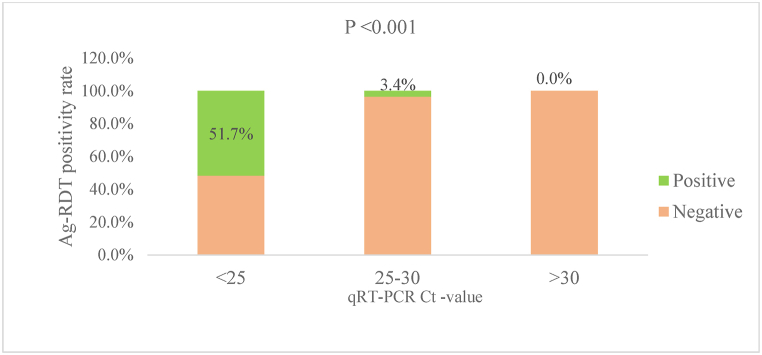
The participants with qRT-PCR positive samples (N = 242) had an overall mean cycle threshold (Ct) value of 24.6 (SD = 2.2). The mean Ct-value in participants who were positive with both Ag-RDT and qRT-PCR (n = 81) was 22.9 (95 % CI: 22.6, 23.2). The Ag-RDT positivity rate was 51.7 % among patients with Ct of <25, 3.4 % among those with Ct of 25–30, and 0 among those with Ct > 30 (P-value <0.001), indicating a very high correlation with viral load (Fig. 1). In fact, 96.3 % of Ag-RDT-positive cases had Ct < 25.
Fig. 1.
SARS-CoV-2 Ag-RDT positivity rate based on qRT-PCR cycle threshold value (N = 1364).
The overall agreement of the Ag-RDTs with qRT-PCR was 87.6 %. The Cohen's Kappa (The Scale reliability coefficient) of the Panbio Ag-RDTs was 0.66 compared with qRT-PCR. In contrast, qRT-PCR detected 91 % (81/89) of Ag-RDT positive cases, and Ag-RDT detected only 33.3 % (81/243) of qRT-PCR cases. Table 4 presents the yield of Ag-RDTs in different clinical scenarios.
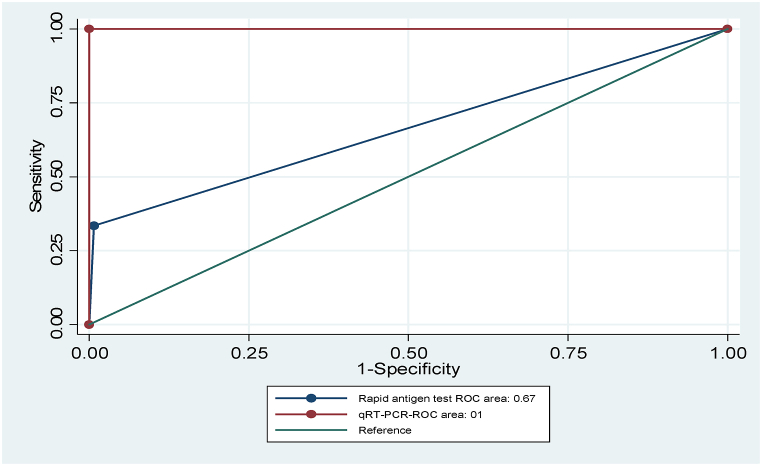
The area under the receiver operating characteristic (ROC) curve (AUC) of Ag-RDT compared with qRT-PCR was 0.67 (95 %, CI:0.63–0.69) (Fig. 2).
Fig. 2.
Three hypothetical ROC curves representing the diagnostic accuracy of the RT-PCR (red lines; AUC = 1) on the upper and left axes in the unit square, a typical rapid antigen ROC curve (grey line curve; AUC = 0.67), and a reference line corresponding to random chance (green line curve; AUC = 0.5). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
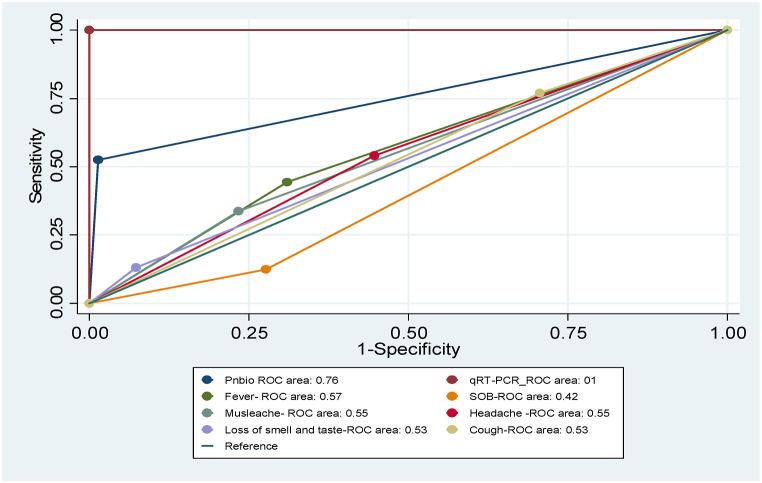
The AUC-ROC of Ag-RDT improves to 0.76 (95 % CI, 0.71–0.80) as the tests focused on suspected cases with COVID-19 typical clinical presentations like fever, shortness of breath, loss of smell and taste, and muscle weakness (Fig. 3).
Fig. 3.
Hypothetical ROC curves representing the diagnostic accuracy of the qRT-PCR and Ag-RAT in different clinical scenarios.
4. Discussion
Our finding showed that compared to qRT-PCR, Panbio COVID-19 Ag-RDT exhibited a much lower sensitivity but a greater specificity for detecting active SARS-CoV-2 infection. qRT-PCR and rapid antigen tests had better yield among patients with recent contact with confirmed or suspected COVID-19 cases and those with one or more COVID-19-related symptoms. The Ag-RDT test also showed a better yield among patients with high viral load (Ct < 25).
The manufacturer reported the specificity of Panbio COVID-19 Ag as 99.8 % (95%CI: 98.65%–100 %) and the overall sensitivity as 98.1 % (95%CI: 93.2%–99.8 %) (23). Our study showed very low sensitivity (33.3 %) but comparable specificity (99.3 %) compared with the manufacturer's report. Further analysis among patients with three major COVID-19 symptoms (cough, fever, and shortness of breath) showed an improved sensitivity (71.4 %) and specificity (100 %). These findings are similar to what has been reported by Scohy et al. [28] and Niclot et al. [29] and are explained by the tests' capacity to detect only those with high viral load. As the symptoms depend on the viral load, the yield of these laboratory tests is expected to be better among symptomatic individuals. It is also important to note that febrile patients and those reporting myalgia/arthralgia had higher positivity rates for both tests in our study. No strong association was observed between specific COVID-19-related symptoms and test positivity rates.
The sensitivity of the Ag-RDTs in our study is more than that was reported by Scohy et al. (30.2 %) [28] and Niclot et al. (50.0 %) [29]. Scohy et al. performed tests on randomly selected samples from patients admitted to different referral hospitals, while Niclot et al. collected samples randomly from the general population. They conducted the tests during the pandemic's early stages (April 2020), which might have led to low sensitivity due to the low incidence of the infection by then.
In general, Ag-RDTs have better performance during pre-symptomatic and early symptomatic phases of SARS-CoV-2 infection when the viral load is high (Ct values ≤ 25 or >106 genomic virus copies/mL [ [30,31]]. A previous study has shown SARS-CoV-2 Ag-RDT sensitivity of 100 % in patients with high viral load (Ct < 25) [32]. Although the sensitivity of Ag-RDT improved to 51.7 % among those with CT < 25 (specificity of 96.7 %) in our study, the finding falls short of the expectations. Our study's Panbio Ag also had a lower sensitivity and specificity than the viral load-based and duration of illness-based LUMIPULSE antigen test study [ [34,35]]. Compared with the study from Thailand, it again had a lower sensitivity but better specificity [33].
In line with our study, SARS-CoV-2 diagnostics accuracy tests (comparison of Ag-RDT with qRT-PCR) conducted in Germany [15] and Italy [36] showed similar low sensitivity and higher specificity. Positive Predictive Value (PPV) and Negative Predictive Value (NPV) were also almost identical to our study, implying the need to use both tests for confirmatory diagnosis. Similar sensitivity, specificity, PPV, and NPV, as in our investigation, were also observed in the Kuwait City population-level screening for SARS-CoV-2 infection [37].
Differently from our study, SARS-CoV-2 diagnostics accuracy tests conducted with Panbio (Ag-RDT) and qRT-PCR in Margalla Hospital, Pakistan showed higher sensitivity (94.3 %) and lower specificity (37.9 %) [18]. This may be explained by the fact that they only included participants with symptoms in the infection's early stages (3–4 days of onset of symptoms). In our study, we have included both symptomatic and asymptomatic cases.
In general, in this study, we found that the sensitivity of the rapid antigen test (Panbio COVID-19 Ag) for diagnosis of COVID-19 in different clinical scenarios is lower than the standard set by WHO, which states that the SARS-CoV-2 antigen test should have an acceptable sensitivity of 80 % and desirable sensitivity of 90 % compared to qRT-PCR [38]. The observed lower sensitivity for asymptomatic individuals and moderate sensitivity in symptomatic cases may be due to a high proportion of asymptomatic subjects in our study. Another possible explanation could be linked to different testing times performed in the early or late phase of infection.
This indicates that using Ag-RDTs alone may give misleading results and misdiagnoses by concealing actual COVID-19 cases. However, the specificity of the COVID-19 Ag-RDT meets the minimum performance requirements of ≥97 % compared with qRT-PCR, as set by the WHO. The introduction of high-specificity tests (>99 %) is preferred in all settings but is of particular importance in low-prevalence settings and general screening [38].
In our study, the sensitivity and PPV of Panbio COVID-19 Ag improved for symptomatic patients (shortness of breath, cough, and fever), which was similar to the study conducted for the cohort of individuals tested to evaluate the quality of the operators performing the test [39].
The area under the Receiver Operator Curve (ROC) of the worldwide diagnostic accuracy metric of the Panbio COVID-19 Ag test in our study was found to be 0.67. This result suggests that Panbio COVID-19 Ag is sufficient in identifying non-diseased illnesses compared to qRT-PCR. A perfect diagnostic test has an AUC of 1.0. at the same time, a non-discriminating test has an area of 0.5 [[40], [41], [42]].
In this study, the scale reliability coefficient (Cohen's kappa) of the rapid antigen in comparison with the gold standard (qRT-PCR) was 0.66. This indicates that the level of agreement between the tests was substantial (greater than moderate but not nearly perfect). The higher sensitivity of SARS-CoV-2 antigen detection in early infection might be a crucial finding for designing new RDT-based algorithms, particularly important in weaker health systems and low-resource settings, where other high-burden diseases, as in Ethiopia, need to be considered. Based on our findings, Panbio Ag-RDT tests should only be performed in areas with high incidences of infection and for symptomatic people at low risk. A parallel test or SARS-CoV-2 qRT-PCR must be the primary diagnostic test in high-risk cases.
4.1. Strengths and limitations
As a strength, we have enrolled a large sample size and participants with different clinical scenarios. We also performed all the recommended quality control tests. Furthermore, the data collection occurred during one of Ethiopia's major waves of the COVID-19 outbreak, when the prevalence of infection in the population was greater than 5 %. Therefore, we presume the predictive value of our test result was unaffected by disease prevalence. We also included centres with high COVID-19 caseloads in Ethiopia. According to our review and search, this was the first study in Ethiopia that compared the quantitative performance of the current Rapid antigen test with qRT-PCR during major COVID-19 outbreaks in the country. The study's preliminary findings guided national policymakers on how to apply the rapid test at points of care. This study's limitation was that the onset of symptoms was not considered when collecting samples to classify the condition as pre-symptomatic, symptomatic, or late symptomatic of infection.
5. Conclusion
Although Ag-RDT for diagnosing SARS-CoV-2 is a good option as a point-of-care screening tool, it has an overall low sensitivity. Using this test for diagnostic and treatment decisions may lead to a false negative, resulting in misdiagnosis and ultimately bearing to disease spread and poor patient outcomes. Hence, Panbio Ag-RDT, as an initial screening test for SARS-CoV-2 infection, should be limited to settings with a high incidence of infection and for symptomatic patients with a low risk of severe disease. A parallel test or SARS-CoV-2 qRT-PCR must be the primary diagnostic test in high-risk cases.
Funding
There is no specific funding for this study. In collaboration with different universities in the region, the Oromia Health Bureau initiated this work to strengthen COVID-19 surveillance and innovative approaches to COVID-19. The research team supervises the testing procedures at the Adama and Nekemte reference laboratories (these two laboratories are under the management of the Oromia Health Bureau). Overall, this study is an effort to generate evidence to support decision-making in fighting COVID-19 in the Oromia region, Ethiopia. Hence, no specific funding was allocated for this study by the Oromia Health Bureau or other organisation.
Data sharing
All the analyzed data was included in the manuscript, and we can share the raw data by removing individual-level identifiers and other sensitive information as requested.
CRediT authorship contribution statement
Dabesa Gobena: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Esayas Kebede Gudina: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Getu Gebre: Writing – review & editing, Validation, Supervision, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Tizta Tilahun Degfie: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Zeleke Mekonnen: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Gudina E.K., Gobena D., Debela T., Yilma D., Girma T., Mekonnen Z., et al. COVID-19 in Oromia Region of Ethiopia: a review of the first 6 months' surveillance data. BMJ Open. 2021;11:1–9. doi: 10.1136/bmjopen-2020-046764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaye B., Khoury S., Cene C.W., Kingue S., N'Guetta R., Lassale C., et al. Socio-demographic and epidemiological consideration of Africa's COVID-19 response: what is the possible pandemic course? Nat. Med. 2020;26:996–999. doi: 10.1038/s41591-020-0960-y. [DOI] [PubMed] [Google Scholar]
- 3.Colombo S., Scuccato R., Fadda A., Cumbi A.J. COVID-19 in Africa: il poco che sappiamo e il molto che ignoriamo. Epidemiol. Prev. 2020;44:408–422. doi: 10.19191/EP20.5-6.S2.146. [DOI] [PubMed] [Google Scholar]
- 4.Lone S.A., Ahmad A. COVID-19 pandemic–an African perspective. Emerg. Microb. Infect. 2020;9:1300–1308. doi: 10.1080/22221751.2020.1775132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulu A., Bekele A., Abdissa A., Balcha T.T., Habtamu M., Mihret A., et al. The challenges of COVID-19 testing in Africa: the Ethiopian experience. Pan Afr Med J. 2021;38:1–4. doi: 10.11604/pamj.2021.38.6.26902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramanathan K., Antognini D., Combes A., Paden M., Zakhary B., Ogino M., et al. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- research that is available on the COVID-19 resource centre - including this for unrestricted research re-use a. 2020:19–21. [Google Scholar]
- 7.Gudina E.K., Tesfaye M., Siraj D., Haileamilak A., Yilma D. 203466-Article Text-508823-1-10-20210202. 2020:180. [Google Scholar]
- 8.Mukwege D., Byabene A.K., Akonkwa E.M., Dahma H., Dauby N., Buhendwa J.P.C., et al. High SARS-CoV-2 seroprevalence in healthcare workers in bukavu, eastern democratic republic of Congo. Am. J. Trop. Med. Hyg. 2021;104:1526–1530. doi: 10.4269/ajtmh.20-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olayanju O., Bamidele O., Edem F., Eseile B., Amoo A., Nwaokenye J., et al. SARS-CoV-2 seropositivity in asymptomatic frontline health workers in Ibadan, Nigeria. Am. J. Trop. Med. Hyg. 2021;104:91–94. doi: 10.4269/AJTMH.20-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sykes W., Mhlanga L., Swanevelder R., Glatt T.N., Grebe E., Coleman C., et al. Prevalence of anti-SARS-CoV-2 antibodies among blood donors in northern cape, KwaZulu-natal, eastern cape, and free state provinces of South Africa in january 2021. Res Sq. 2021 doi: 10.21203/rs.3.rs-233375/v1. [DOI] [Google Scholar]
- 11.Birhanu Z., Ambelu A., Fufa D., Mecha M., Zeynudin A., Abafita J., et al. Risk perceptions and attitudinal responses to COVID-19 pandemic: an online survey in Ethiopia. BMC Publ. Health. 2021;21:1–17. doi: 10.1186/s12889-021-10939-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hailemariam B.W., Zealiyas K., Gutema G., Gebremicael G., Adane S., Tadele S., et al. Performances of four nucleic acid amplification tests for the identification of SARS-CoV-2 in Ethiopia. Sci. Rep. 2022;12:1–7. doi: 10.1038/s41598-022-24411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh G., Nahirniak S., Lamarche Y., Fan E. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- company s public news and information website. Elsevier hereby grants permission to make all its COVID-19-r. 2020:19–21. doi: 10.1093/nsr/nwaa036.6. [DOI] [Google Scholar]
- 14.Hamar Á., Filipánits K., Váradi A., Váradi-Rácz R., Gellén H.O., Futács K., et al. Diagnostic accuracy of SARS-CoV-2 PanbioTM rapid antigen diagnostic tests in a 4,440-case clinical follow-up. Front. Med. 2022;9 doi: 10.3389/fmed.2022.908127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagenhäuser I., Knies K., Rauschenberger V., Eisenmann M., McDonogh M., Petri N., et al. Clinical performance evaluation of SARS-CoV-2 rapid antigen testing in point of care usage in comparison to RT-qPCR. EBioMedicine. 2021;69:1–7. doi: 10.1016/j.ebiom.2021.103455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagenhäuser I., Knies K., Rauschenberger V., Eisenmann M., McDonogh M., Petri N., et al. Clinical performance evaluation of SARS-CoV-2 rapid antigen testing in point of care usage in comparison to RT-qPCR. EBioMedicine. 2021;69:1–7. doi: 10.1016/j.ebiom.2021.103455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favresse J., Gillot C., Oliveira M., Cadrobbi J., Elsen M., Eucher C., et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. J. Clin. Med. 2021;10:1–10. doi: 10.3390/jcm10020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asif S., Ahmed A., Gul A., Abbasi T. Efficacy of PanbioTM covid-19 Ag rapid test in sars-cov-2 detection: comparison with RT-PCR test. J. Ayub Med. Coll. Abbottabad. 2022;34(1):S928–S931. doi: 10.55519/JAMC-04-S4-10113. [DOI] [PubMed] [Google Scholar]
- 19.Anum M.D., Minhas Paul Scheel S., Md Bgmdglmdmsmhmdmjmdmhssrjmdedmmdmhsaghmd Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- Ann. Oncol. 2020:19–20. [Google Scholar]
- 20.Nsoga M.T.N., Kronig I., Rodriguez F.J.P., Sattonnet-Roche P., Silva D Da, Helbling J., et al. Diagnostic accuracy of Panbio rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. PLoS One. 2021;16:1–7. doi: 10.1371/journal.pone.0253321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahar F, Dirawan GD, Samad S, Qomariyah N, Purlinda DE. The epidemiology of COVID-19, attitudes and behaviors of the community during the Covid pandemic in Indonesia. Structure. 2020;10(8) [Google Scholar]
- 22.Klein J.A.F., Krüger L.J., Tobian F., Gaeddert M., Lainati F., Schnitzler P., et al. Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. Med. Microbiol. Immunol. 2021;210:181–186. doi: 10.1007/s00430-021-00710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Table S., Lowering P., Comparing T., Intervention A., Cad M.I. Supplementary Appendix Supplementary Appendix. 2015;6736:1–24. [Google Scholar]
- 24.Treggiari D., Piubelli C., Caldrer S., Mistretta M., Ragusa A., Orza P., et al. SARS-CoV-2 rapid antigen test in comparison to RT-PCR targeting different genes: a real-life evaluation among unselected patients in a regional hospital of Italy. J. Med. Virol. 2022;94:1190–1195. doi: 10.1002/jmv.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strömer A., Rose R., Schäfer M., Schön F., Vollersen A., Lorentz T., et al. Performance of a point-of-care test for the rapid detection of sars-cov-2 antigen. Microorganisms. 2021;9:1–11. doi: 10.3390/microorganisms9010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.COVID-19 Ag rapid test device. Biomed. Saf. Stand. 2022;52:171–172. doi: 10.1097/01.bmsas.0000904520.63753.0b. [DOI] [Google Scholar]
- 27.Tosepu R., Effendy D.S., Ahmad L.O.A.I. The first confirmed cases of COVID-19 in Indonesian citizens. Public Health Indonesia. 2020;6(2):70–71. [Google Scholar]
- 28.Scohy A., Anantharajah A., Bodéus M., Kabamba-mukadi B., Verroken A., Rodriguez-villalobos H. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19 . The COVID-19 resource centre is hosted on Elsevier Connect, the company's public news and information. J. Clin. Virol. 2020;129:1–3. [Google Scholar]
- 29.Lambert-clot S., Cuffel A., Pape L., Vauloup-fellous C., Morand-joubert L. Evaluation of a rapid diagnostic assay for detection of. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K., et al. Prospective study of 1308 nasopharyngeal swabs from 1033 patients using the LUMIPULSE SARS-CoV-2 antigen test: comparison with RT-qPCR. Int. J. Infect. Dis. 2021;105:7–14. doi: 10.1016/j.ijid.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Kleines M. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID-19. The COVID-19 resource centre is hosted on Elsevier Connect, the company's public news and information. 2020:2020–2022. [Google Scholar]
- 33.Chaimayo C., Kaewnaphan B., Tanlieng N., Athipanyasilp N., Sirijatuphat R., Chayakulkeeree M., et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 2020;17:1–7. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinnes J., Jj D., Berhane S., Taylor M., Adriano A., Davenport C., et al. 2021. Diagnosis of SARS-CoV-2 Infection (Review)www.cochranelibrary.com [DOI] [Google Scholar]
- 35.Thirion-Romero I., Guerrero-Zúñiga D.S., Arias-Mendoza D.A., Cornejo-Juárez D.D.P., Meza-Meneses D.P., Torres-Erazo D.D.S., et al. Evaluation of Panbio rapid antigen test for SARS‐CoV‐2 in symptomatic patients and their contacts: a multicenter study. Int. J. Infect. Dis. 2021;113:218–224. doi: 10.1016/j.ijid.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treggiari D., Piubelli C., Caldrer S., Mistretta M., Ragusa A., Orza P., et al. SARS-CoV-2 rapid antigen test in comparison to RT-PCR targeting different genes: a real-life evaluation among unselected patients in a regional hospital of Italy. J. Med. Virol. 2022;94:1190–1195. doi: 10.1002/jmv.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alghounaim M., Bastaki H., Bin Essa F., Motlagh H., Al-Sabah S. The performance of two rapid antigen tests during population-level screening for SARS-CoV-2 infection. Front. Med. 2021;8:1–5. doi: 10.3389/fmed.2021.797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Points K. 2021. Antigen-detection in the Diagnosis of SARS-CoV-2 Infection. [Google Scholar]
- 39.Nguyen L.H., Drew D.A., Graham M.S., Joshi A.D., Guo C.G., Ma W., et al. Risk of COVID-19 among front-line healthcare workers and the general community: a prospective cohort study. Lancet Public Health. 2020;5:e475–e483. doi: 10.1016/S2468-2667(20)30164-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obuchowski N.A., Lieber M.L., Wians F.H. ROC curves in Clinical Chemistry: uses, misuses, and possible solutions. Clin. Chem. 2004;50:1118–1125. doi: 10.1373/clinchem.2004.031823. [DOI] [PubMed] [Google Scholar]
- 41.Henderson A.R.H. Assessing test accuracy and its clinical consequences: a primer for receiver operating characteristic curve analysis. Ann. Clin. Biochem. 1993;30:521–539. doi: 10.1177/000456329303000601. [DOI] [PubMed] [Google Scholar]
- 42.Erland L.A.E., Saxena P.K. Melatonin natural health products and supplements: presence of serotonin and significant variability of melatonin content. J. Clin. Sleep Med. 2017;13:275–281. doi: 10.5664/jcsm.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]