Abstract
The production of exopolysaccharide (EPS) was shown to be required for the infection process by rhizobia that induce the formation of indeterminate nodules on the roots of leguminous host plants. In Sinorhizobium meliloti (also known as Rhizobium meliloti) Rm41, a capsular polysaccharide (KPS) analogous to the group II K antigens of Escherichia coli can replace EPS during symbiotic nodule development and serve as an attachment site for the strain-specific bacteriophage φ16-3. The rkpA to -J genes in the chromosomal rkp-1 region code for proteins that are involved in the synthesis, modification, and transfer of an as-yet-unknown lipophilic molecule which might function as a specific lipid carrier during KPS biosynthesis. Here we report that with a phage φ16-3-resistant population obtained after random Tn5 mutagenesis, we have identified novel mutants impaired in KPS production by genetic complementation and biochemical studies. The mutations represent two novel loci, designated the rkp-2 and rkp-3 regions, which are required for the synthesis of rhizobial KPS. The rkp-2 region harbors two open reading frames (ORFs) organized in monocistronic transcription units. Although both genes are required for normal lipopolysaccharide production, only the second one, designated rkpK, is involved in the synthesis of KPS. We have demonstrated that RkpK possesses UDP-glucose dehydrogenase activity, while the protein product of ORF1 might function as a UDP-glucuronic acid epimerase.
Extracellular carbohydrates produced by rhizobia play an important role in the development of nitrogen-fixing symbioses with leguminous plants. Lipochitooligosaccharide signal molecules (Nod factors) are required for the formation of a new plant organ, the nodule, where bacteroids, the endosymbiotic forms of rhizobia, reduce atmospheric nitrogen (7, 34). Additional surface and extracellular components, including lipopolysaccharides (LPS), exopolysaccharides (EPS), capsular polysaccharides (KPS), and cyclic β-glucans, are required for the infection of the nodule tissue by the microsymbiont. The symbiotic phenotype of the LPS- and EPS-defective mutants depends on the type of nodule determined by the host genome (13). For Sinorhizobium meliloti (also known as Rhizobium meliloti), which induces indeterminate nodules on the roots of its symbiotic partner, alfalfa (Medicago sativa), EPS has been reported to be involved in the infection process while the importance of the LPS has not yet been demonstrated. The production of EPS I containing octasaccharide repeating units of seven glucoses and a galactose, which are substituted with pyruvyl, acetyl, and succinyl groups, was shown to be necessary for the development of the infection thread and the invasion process. It has also been shown that S. meliloti has a latent capability to produce a second type of exopolysaccharide, EPS II, containing a glucose-galactose disaccharide subunit carrying pyruvyl and acetyl modifications. A second-site mutation allows several EPS I-nonproducing bacteria to synthesize EPS II and helps to overcome their invasion defect in some host plants, suggesting that EPS II can replace EPS I in the infection process (17).
Recently, it has been demonstrated that some rhizobia possess surface polysaccharides that are analogous to the group II K antigens (KPS) of Escherichia coli (28, 29), and in S. meliloti this type of surface polysaccharide can also replace EPS in the infection process (22, 30). Although the complete chemical structure of the S. meliloti Rm41 KPS has not yet been determined, available data indicate that it is composed of disaccharide repeating units of α-keto-3,5,7,9-tetradeoxy-5,7-diaminonulonosic acid, which is a Kdo (3-deoxy-d-manno-2-octulosonic acid)-like molecule, and of 4-deoxy-4-amino-hexuronic acid (27). The isolated KPS from S. meliloti shows a banding pattern on polyacrylamide gels that corresponds to the different degrees of polymerization that are controlled by the rkpZ gene product (30).
We have isolated a gene cluster of S. meliloti Rm41 (rkp-1, formerly fix-23) that is required for KPS production. Mutants affected in this gene cluster are not able to adsorb the strain-specific bacteriophage φ16-3 and, in an Exo− background, cannot invade the nodule. The rkp-1 region harbors 10 open reading frames (ORFs) that are designated the rkpA to -J genes and whose predicted protein products exhibit a high degree of similarity to the different components and domains of fatty acid synthases as well as to enzymes taking part in the modification and transfer of lipid molecules to capsular polysaccharides (15, 22). These gene products might be involved in the synthesis of a putative lipid anchor of the polysaccharide or, more likely, in the production of a specific lipid carrier required for the biosynthesis of the KPS.
The rkp-1 region does not code for proteins participating in the biosynthesis and polymerization of the KPS precursors. Therefore, to identify novel rkp genes required for these latter functions, a screening for new mutants was carried out. Since most of the rkp mutants isolated so far were resistant to phage φ16-3, new mutations affecting KPS synthesis could be collected by isolating additional phage-resistant derivatives. Here we report the identification of two novel gene clusters, designated the rkp-2 and rkp-3 regions, that are involved in the production of the capsular polysaccharide. Moreover, genetical and biochemical analysis of the rkp-2 region is presented.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, and growth conditions.
Strain AK631 carrying the exoB-631 mutation is a Fix+ Exo− derivative of S. meliloti Rm41 (24). Strains AT211, AT212, AT231, AT232, AT233, AT234, AT578, PP2936, and PP2937 are KPS− and/or LPS− derivatives of AK631 that carry Tn5 and plasmid insertions in the rkp-2 or rkp-3 region, as indicated below (see Fig. 2 and 3). E. coli JM109 was used in the cloning procedures (38) which involved the following plasmids: pUC19 (38), pK18/19 (23), and pOK12 (37).
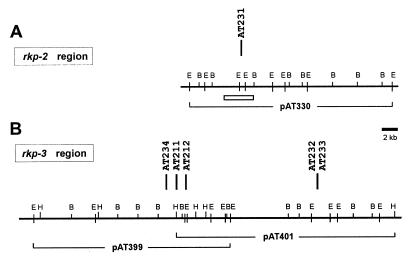
FIG. 2.
Physical-genetic map of the rkp-2 (A) and rkp-3 (B) regions of S. meliloti. Vertical bars represent the Tn5 insertions (with strain numbers) identified in the phage φ16-3-resistant mutants. All mutants were defective in KPS production (see Fig. 1). The physical map was constructed by EcoRI (E), BamHI (B), and HindIII (H) restriction enzymes. Solid lines under the physical map show the genomic DNA insert and the number of the complementing cosmid clones. In panel A, a rectangle indicates the region where the DNA sequence was established. In panel B, mutations Tn5-AT211 and Tn5-AT212 were complemented by both cosmid clones.
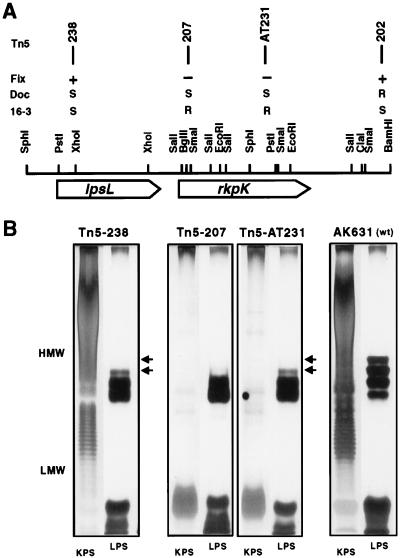
FIG. 3.
Organization of the rkp-2 region. (A) The positions, phenotypes, and insertion numbers of the Tn5 mutations are shown above the physical map and in panel B. Fix, symbiotic phenotype; DOC, DOC sensitivity; 16-3, phage φ16-3 sensitivity. Mutants carrying Tn5-202, Tn5-207, or Tn5-238 were obtained by homologous recombination (see Materials and Methods). (B) KPS and LPS production of the insertion mutants. Phenol-water extracts were separated in two parallel PAGE gels, and the gels were stained either for KPS or for LPS as described in Materials and Methods. Arrows indicate the LPS bands that were altered in the mutants. Wild-type control gels [AK631 (wt)] are also shown. HMW, high molecular weight; LMW, low molecular weight.
Genomic clones pAT330 and pAT399 were isolated from a genomic library prepared by cloning S. meliloti Rm41 total DNA after EcoRI partial digestion into the pLAFR1 cosmid vector (25), while pAT401 is a representative of a similar library prepared by partial HindIII digestion in the pVK102 cosmid vector (26).
Media, antibiotic concentrations, culture conditions for E. coli and S. meliloti strains, and phage sensitivity tests have been described previously (25, 26). The deoxycholic acid (DOC) test was performed as described by Lagares et al. (16).
Isolation of the S. meliloti mutants defective in KPS production and identification of the rkp-2 and rkp-3 regions.
Random transposon Tn5 mutagenesis of S. meliloti AK631 was carried out by using the mobilizable suicide plasmid pSUP1011 (35). The kanamycin- and streptomycin-resistant transconjugants were selected on minimal medium, and aliquots (1 × 108 to 5 × 108 cells) from 10 mutagenized populations (2,000 to 3,000 transconjugants/population) were infected by bacteriophage φ16-3 (at a multiplicity of infection 10) and plated. Phage-resistant colonies were tested in phage adsorption experiments as described previously (26). For the isolation of the complementing cosmid clones, the genomic libraries were introduced into the rkp mutants and plant assays were used to identify the clones carrying the wild-type genes as described earlier (24).
Directed Tn5 and plasmid integration mutagenesis.
Tn5 mutagenesis of the cosmid clone pAT330 was carried out as described earlier (26). The isolated Tn5 insertions were introduced into the AK631 genome by homologous recombination by the method of Ruvkun and Ausubel (31). Positions of the insertions (Tn5-202, Tn5-207, and Tn5-238) are shown below (see Fig. 3A).
For plasmid integration mutagenesis, DNA fragments were cloned into vector pK18 and derivatives obtained after single homologous recombination with pAT330 were isolated in a Rec+ E. coli strain. The resultant constructions were introduced into the S. meliloti genome in the same way as the Tn5 insertions were, by selecting for the antibiotic (kanamycin) resistance of pK18.
DNA manipulation.
Standard procedures, including DNA isolation, restriction enzyme digestion, radioactive labelling of DNA, agarose gel electrophoresis, DNA ligation, and transformation of E. coli, were performed by using conventional methods (32).
For sequencing, appropriate restriction fragments were subcloned and their nucleotide sequences were determined by the dideoxy nucleotide chain termination method (33) by using [35S]dATP and the T7 sequencing kit from Pharmacia. Additional sequence information was obtained from the Tn5-flanking regions of different insertions by using a Tn5-specific oligonucleotide primer (14).
Computer analysis of the sequence data was carried out by using the PC/Gene microcomputer software (designed by Amos Bairoch, IntelliGenetics) and the Genetics Computer Group (GCG) software package of the University of Wisconsin (8). Amino acid similarity searches were performed by using the Blast programs (1) at the National Center for Biotechnology Information, National Library of Medicine, Bethesda, Md., and the FASTA program (21).
Plant tests.
Assays for the symbiotic properties of the mutant S. meliloti strains were carried out with alfalfa (Medicago sativa L. Nagyszénási) plantlets as described earlier (24).
Preparation and analysis of polysaccharides.
Extraction of the surface polysaccharides by a modified hot phenol-water method and DOC-polyacrylamide gel electrophoresis (PAGE) analysis of the samples were carried out as described elsewhere (15, 28).
Cell fractionation.
S. meliloti cells were harvested from a 500-ml overnight culture and washed with 100 mM Tris-HCl buffer (pH 8.5) containing 1 mM MgCl2. The pellet was then suspended in 10 ml of buffer containing 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 20 μg of DNase I and RNase A per ml. Bacteria were disrupted by passing them three times through a French press (12,000 lb/in2). The suspension was centrifuged at 15,000 × g for 30 min to remove unbroken cells, and the supernatant was centrifuged at 140,000 × g for 2 h. Protein concentrations of the supernatants (soluble fraction) were estimated by using the Bio-Rad (Richmond, Calif.) assay with bovine serum albumin as a standard.
UDP-glucose dehydrogenase assay.
Enzyme activity was assayed with a Shimadzu spectrophotometer in the double-beam mode at 25°C to measure the rate of change of optical density at 340 nm caused by the NAD reduction accompanying the oxidation of UDP-glucose to UDP-glucuronic acid. The assay mixture contained 100 mM Tris-HCl buffer (pH 8.5), 1 mM MgCl2, 5 mM dithiothreitol, 2 mM NAD, 1 mM UDP-glucose, and 200 to 400 mg of the soluble protein fractions in a 1-ml volume. One unit of UDP-glucose dehydrogenase is defined as the amount of enzyme that catalyzes the reduction of 1 micromole of NAD per minute (18).
Nucleotide sequence accession number.
A DNA sequence of 3,673 bp has been registered in the EMBL, GenBank, and DDBJ Nucleotide Sequence Databases under accession no. AJ222661. Coordinates in this publication are identical with those of the database record.
RESULTS
Identification of novel regions required for KPS production.
To isolate genetic determinants involved in the biosynthesis of the precursors as well as in the polymerization of the rhizobial KPS, random Tn5 mutagenesis was carried out in the S. meliloti Rm41 derivative strain AK631. For selection of potential mutants, we used phage φ16-3, which had previously been shown to be unable to adhere to the surfaces of the KPS-defective strains harboring a mutation in the rkp-1 region (26). In phage inactivation experiments, 20 isolates resistant to phage φ16-3 were assayed for their capacity to adsorb to the phage particles. Ten mutants were not able to bind the phages. These strains were further screened in genetic complementation experiments for mutations affecting as-yet-unidentified rkp regions or genes. The phage adsorption capability of six mutants (AT211, AT212, AT231, AT232, AT233, and AT234) was not restored by the cosmid clone pPP428 carrying the rkp-1 region; therefore, these mutants were retained for further biochemical and genetic studies.
The LPS and KPS were extracted with hot phenol-water from the mutants, and the samples were analyzed on polyacrylamide gels containing DOC as the detergent (DOC-PAGE). All six mutants exhibited characteristic changes in the KPS banding (Fig. 1), and one of them (AT231) produced altered LPS as well (see Fig. 3B). In plant nodulation assays, all of the mutants were ineffective in nitrogen fixation (Fix−). Moreover, light microscopic analysis of the ineffective symbiotic nodules revealed that they were incapable of infecting the nodules (Inf−) (data not shown).
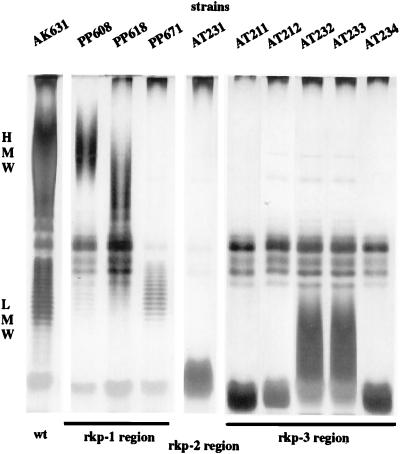
FIG. 1.
KPS production by the rkp mutants. The phenol-water-extracted material from bacteria was analyzed by PAGE. For KPS-specific analysis, the gel was silver stained without oxidation. KPS production of the mutants carrying insertions Tn5-618 and Tn5-608 was reported earlier (15). The six new mutants produce no high-molecular-weight (HMW) KPS, and the lower-molecular-weight (LMW) polysaccharides isolated from the mutants show altered banding. wt, wild type.
To isolate the DNA regions carrying the wild-type allele of the genes affected in the rkp mutants, two different genomic libraries were introduced into the mutants, and in plant assays the transconjugants carrying the complementing cosmid clones were reisolated from the Fix+ nodules as described earlier (24). From the analysis of several independent transconjugants, three different DNA regions (cosmid clones) were identified (pAT330, pAT399, and pAT401). The latter two proved to be overlapping as determined by restriction enzyme analysis and hybridization experiments. These novel loci were designated the rkp-2 and rkp-3 regions (Fig. 2). The link between the transposon insertions and the mutant phenotype was verified by isolating the Tn5 insertions from the genomes of the mutants by double homologous recombination with the complementing cosmid clones and then by reintroducing the mutations into the AK631 genome by homogenization. All of these homogenotes were resistant to phage φ16-3 and were Fix−, indicating that the mutant phenotypes are due to Tn5 insertions (Fig. 2).
Genetic analysis of the rkp-2 region.
To delimit the genes in the rkp-2 region, directed lambda::Tn5 mutagenesis was carried out by using the complementing cosmid clone pAT330. Transposon insertions were introduced into the genome of AK631 by homologous recombination, and the recombinants were tested for phage φ16-3 sensitivity and KPS and LPS production, as well as for symbiotic properties. Only one additional transposon insertion (Tn5-207), located in the vicinity of the original mutation (Tn5-AT231), resulted in a phage-resistant phenotype (Fig. 3A). However, during the homogenization, we realized that in some cases (including Tn5-AT231) it was difficult to obtain bona fide homogenotes and, moreover, that these strains formed small colonies. One possible explanation is that the mutations are in genes that affect the viability (fitness) of the cells.
Defects in the LPS structure may result in similar effects; therefore, we tested whether the mutations affected LPS production. LPS mutants are sensitive to detergents since their outer membrane forms a less effective barrier against hydrophobic compounds than the wild type does (16); therefore, the insertion mutants were tested for sensitivity to DOC on complete medium containing 1 mg of DOC per ml. The wild-type strain, AK631, showed the same viability on both the control and the DOC-containing media, while the mutants carrying the Tn5 transposon on a 5.5-kb BamHI fragment formed 104 to 105 times fewer colonies in the presence of DOC, indicating that these strains are affected in their LPS production (Fig. 3).
Production of KPS and LPS by these mutants was also analyzed by parallel DOC-PAGE experiments (Fig. 3B). One gel was developed by the silver staining method of Tsai and Frisch (36) to detect changes in the LPS. The second gel was prestained with alcian blue, a cationic dye that specifically binds acidic polysaccharides, and then silver stained without oxidation to detect the KPS-specific bands (see Materials and Methods). The LPS pattern of the mutants differed from that of the control strain (AK631), confirming that the mutants were defective in LPS structure. The KPS production of the mutants correlated with their sensitivity to phage φ16-3; in the phage-resistant strains, no capsular polysaccharide was detectable, while the banding patterns of KPS isolated from the phage-sensitive mutants were indistinguishable from that of AK631.
In plant nodulation assay, all of the KPS-defective mutants exhibited the Fix− phenotype on alfalfa, while strains altered only in LPS production were capable of establishing nitrogen-fixing symbiosis (Fig. 3A).
The rkp-2 region encodes proteins similar to enzymes involved in the metabolism of nucleotide diphospho-sugars.
To identify the functions of the genes and gene products involved in the formation of both LPS and KPS, appropriate overlapping restriction fragments of the SphI-BamHI fragment (Fig. 3A), as well as the flanking regions of Tn5 from the insertion mutants, were subcloned and the entire region was sequenced. Computer-aided analysis of the sequence data revealed the presence of two ORFs transcribed in the same direction (Fig. 3A). Codon preference analysis confirmed that the codon usage in these ORFs is in accordance with most of the S. meliloti genes sequenced to date. Weak similarities to the E. coli promoter consensus were found upstream of both ORFs. ORF1 was followed by short inverted repeats preceding several thymidine residues, while an 11-bp-long inverted repeat was found after the translational stop codon of ORF2. These sequences might serve as transcription termination signals.
The first ORF is located between nucleotide positions 307 and 1332 and starts with an ATG initiation codon. A potential ribosomal binding site is situated 5 bp upstream of the initiation codon. The putative translational product of ORF1 contains 341 amino acid residues and has a calculated molecular mass of 38,082 Da. Comparison of the amino acid sequence with protein (SwissProt, PIR) and pretranslated nucleic acid (GenBank) databases revealed significant similarities to different epimerases involved in the epimerization of nucleotide diphospho-sugars. The most definite homology was found with the CapI protein (51% identity, 67% similarity), which is involved in the capsular polysaccharide production of Staphylococcus aureus and which participates in the biosynthetic pathway leading to the synthesis of N-acetyl-aminogalacturonic acid (19). Similar scores resulted from comparisons with a putative epimerase encoded in the chain length determinant (cld) region of E. coli (2) and the UDP-galacturonic acid epimerase protein encoded by the cap1J gene of Streptococcus pneumoniae (20). A limited, 22.9% amino acid identity was also found with the S. meliloti ExoB protein which had previously been shown to posses UDP-glucose epimerase activity (6).
The second ORF is 1,314 bp in length and starts at nucleotide position 1479 with an ATG initiation codon that is preceded by a putative ribosomal binding site sequence. The predicted protein consists of 437 amino acid residues and has a calculated molecular size of 47,229 Da. This protein exhibits a high degree of similarity to different nucleotide diphospho-sugar dehydrogenases. The highest amino acid sequence identities (more than 50%) were detected with UDP-glucose dehydrogenases identified in other organisms, like a Synechocystis sp. (12).
The sequence analysis of the DNA regions flanking the Tn5 insertions (Fig. 3) revealed that only mutations in ORF2 resulted in KPS−, φ16-3 phage-resistant, and Fix− phenotypes; therefore, the corresponding gene was designated rkpK; mutations in ORF1 affected only the production of LPS, and this ORF is referred to hereafter as a rhizobial LPS gene, lpsL.
Operon analysis by plasmid integration mutagenesis.
Sequence analysis revealed that the two ORFs that are transcribed in the same direction are separated by 150 bp between the translation stop codon of lpsL and the ATG initiation codon of rkpK. To determine whether the transcription of the two ORFs is coupled or whether rkpK represents a monocistronic transcription unit, plasmid integration mutagenesis was carried out. To perform this mutagenesis, the 665-bp HincII fragment, carrying the 3′ part of lpsL, the intergenic region with the putative promoter, as well as the 5′ part of rkpK, was cloned into the pK18 vector in both orientations (AT593 and AT594). These constructs were then introduced into the AK631 genome as described in Materials and Methods. The resultant mutants (PP2936 and PP2937) were sensitive to phage φ16-3 and had a Fix+ phenotype, suggesting that KPS production was not affected by these mutations. Since it is unlikely that in both strains a vector-specific promoter would drive the transcription of the rkpK gene, we concluded that there is a promoter located in the intergenic region. These results are in agreement with previous findings where mutations in lpsL induced by either Tn5 insertion or plasmid integration had no polar effect on rkpK, i.e., the KPS production in the lpsL mutants was not affected.
RkpK is a UDP-glucose dehydrogenase.
Soluble protein preparations from the wild-type and different mutant bacteria were assayed for UDP-glucose dehydrogenase activity as described in Materials and Methods. The specific enzyme activities measured were 1.82 and 0.97 mU mg−1 min−1 in the control strain AK631 and in a plasmid integration mutant of ORF1 (AT578), respectively. In strains carrying the Tn5-AT231 or Tn5-207 insertion in the rkpK gene, no UDP-glucose dehydrogenase activity could be detected. When the complemeting cosmid clone, pAT330, was introduced into the rkpK mutants, the enzyme activity was restored and it was about ninefold higher than that of the wild-type strain.
DISCUSSION
In this paper, we report on the isolation of novel genomic regions required for the capsular polysaccharide production of S. meliloti Rm41. This surface component had previously been shown to be involved in the attachment of the strain-specific bacteriophage φ16-3 to the bacterial surface and to be able to replace EPS during symbiotic nodule development (22, 26). In contrast to EPS II, KPS production of S. meliloti is independent of the presence of a second mutation, like mucR or expR, as in the case of EPS II (10, 39), and KPS can replace EPS in the invasion process not only on alfalfa and Melilotus albus but also on all hosts tested to date (M. sativa subsp. valria, Medicago media, Medicago truncatula, Melilotus officinalis, and Trigonella coerulea) (26).
The genetic determinants required for the synthesis of extracellular polysaccharides in S. meliloti have already been identified (4, 11), but much less data are available on those required for KPS production. The rkpA to -J genes in the chromosomal rkp-1 (formerly fix-23) region code for proteins that may participate in the synthesis, modification, and transfer of an as-yet-unknown lipophilic molecule which might serve as a specific lipid carrier during KPS biosynthesis (15, 22), while the rkpZ gene harbored by a megaplasmid affects the degree of KPS polymerization (30). No genes that are involved in the synthesis and polymerization of the precursors, as well as in the export of the mature polymer, have been described so far.
Our earlier observation (26) that mutations in the rkp-1 region abolished phage φ16-3 adsorption offered a simple way to identify other genetic determinants of KPS synthesis. We screened for mutants defective in KPS production in a phage-resistant population obtained after random transposon Tn5 mutagenesis of S. meliloti AK631. Several novel mutants isolated in this way were shown to be defective in both KPS production and nodule invasion. The mutations affected two novel rkp gene clusters that were designated the rkp-2 and rkp-3 regions.
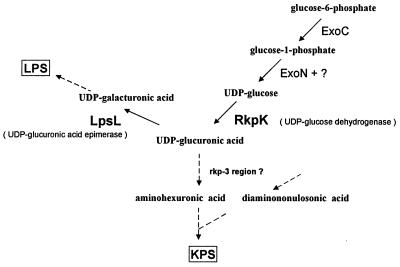
Genetic analysis of the rkp-2 region revealed the presence of two ORFs which are transcribed in the same direction. The phenotypes of the different Tn5 and plasmid integration mutants indicated that transcription of these ORFs is independent, i.e., they form monocistronic transcription units. Interestingly, the analysis of the mutants of this region showed that while both genes are required for normal LPS production, only the second ORF (rkpK) is involved in the synthesis of KPS. The predicted protein products of the two ORFs exhibited a high degree of similarity to enzymes involved in the metabolism of nucleotide diphospho-sugars. The product of ORF1 (lpsL) showed a high degree of homology to epimerases encoded by the capI and cap1J genes of Staphylococcus aureus and Streptococcus pneumoniae, respectively. These epimerases are involved in the formation of UDP-galacturonic acid (derivatives) (19, 20). RkpK had a high degree of identity with different UDP-glucose dehydrogenases. By using a biochemical assay, we demonstrated that RkpK functions as a UDP-glucose dehydrogenase. Our results suggest that these two enzymatic activities are probably required for the formation of UDP-galacturonic acid from UDP-glucose, which is the source of galacturonic acid during LPS synthesis. The disaccharide repeating unit of the KPS in S. meliloti Rm41 is composed of 3,5,7,9-tetradeoxy-5,7-diamino-2-nonulonosic acid and of an as-yet-undetermined 4-deoxy-4-amino-hexuronic acid which might be 4-deoxy-4-amino-glucuronic acid (27). Based on the biochemical data obtained here as well as on previous reports, we concluded that the biosynthesis of the latter component involves enzymatic steps catalyzed by the RkpK protein as well as by the protein products of different exo genes (Fig. 4). As the first step, glucose-6-phosphate is converted to glucose-1-phosphate by the phosphoglucomutase enzyme encoded by the exoC gene. The mutation in this gene affects the synthesis of most of the rhizobial polysaccharides (EPS I, EPS II, LPS, and cyclic β-glucan) including KPS, since, in contrast to other exo mutants of S. meliloti Rm41, the exoC mutant is Fix−, indicating the lack of KPS (5). The reversible conversion of glucose-1-phosphate to UDP-glucose is catalyzed by the UDP-glucose pyrophosphorylase encoded by the exoN gene. Since the exoN mutation does not inhibit EPS production completely, a second enzymatic activity that can replace ExoN must be present in S. meliloti (3). Thus, the amount of KPS produced by an exoN mutant is expected to be somewhat smaller than that of the wild type. UDP-glucose is oxidized by the RkpK protein to form UDP-glucuronic acid, which is in turn converted to the aminohexuronic acid by enzymes that might be encoded in the rkp-3 region.
FIG. 4.
Proposed role of rkpK and lpsL genes in the KPS and LPS biosynthesis of S. meliloti.
In contrast to the results with the rkp-2 region, the random mutagenesis experiment resulted in more mutations in the rkp-3 region and the mutations are situated within a relatively large DNA region covered by two cosmid clones. These results suggest that the rkp-3 region may carry the majority of the genes necessary for the biosynthesis of the disaccharide subunits of KPS. Our preliminary Tn5 mutagenesis and sequencing results support this idea. The K antigens of rhizobia determined so far show strain-specific variation, although they possess a common structural motif (9), suggesting that several rkp genes in the newly identified regions are probably strain specific and uncommon even in the different S. meliloti isolates. The structure of KPS and the presence of different rkp genes in the microsymbiont may be important determinants of the successful invasion of a given host plant.
ACKNOWLEDGMENTS
We are grateful to E. Sárai and Z. Liptai for skillful technical assistance.
This work was supported by grant OTKA T 016674 and by the U.S.-Hungarian Science and Technology Joint Fund no. 513.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bastin D A, Stevenson G, Brown P K, Haase A, Reeves P R. Repeat unit polysaccharides in bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 3.Becker A, Kleickmann A, Keller M, Arnold W, Pühler A. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 4.Becker A, Rüberg S, Küster H, Roxlau A A, Keller M, Ivashina T, Cheng H-P, Walker G C, Pühler A. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J Bacteriol. 1997;179:1375–1384. doi: 10.1128/jb.179.4.1375-1384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzoska P M, Signer E R. lpsZ, a lipopolysaccharide gene involved in symbiosis of Rhizobium meliloti. J Bacteriol. 1991;173:3235–3237. doi: 10.1128/jb.173.10.3235-3237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buendia A M, Enenkel B, Köplin R, Niehaus K, Arnold W, Pühler A. The Rhizobium meliloti exoZ/exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol Microbiol. 1991;5:1519–1530. doi: 10.1111/j.1365-2958.1991.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 7.Denarie J, Debelle F. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsberg L S, Reuhs B L. Structural characterization of the K antigens from Rhizobium fredii USDA257: evidence for a common structural motif, with strain-specific variation, in the capsular polysaccharides of Rhizobium spp. J Bacteriol. 1997;179:5366–5371. doi: 10.1128/jb.179.17.5366-5371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glazebrook J, Walker G C. A novel exopolysaccharide can function in place of the Calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 11.Gonzales J U, York G M, Walker G C. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene. 1996;179:141–146. doi: 10.1016/s0378-1119(96)00322-8. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 13.Kannenberg E L, Brewin N J. Host-plant invasion by Rhizobium: the role of cell-surface components. Trends Microbiol. 1994;2:277–283. doi: 10.1016/0966-842x(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 14.Kereszt A, Slaska-Kiss K, Putnoky P, Banfalvi Z, Kondorosi A. The cycHJKL genes of Rhizobium meliloti involved in cytochrome c biogenesis are required for “respiratory” nitrate reduction ex planta and for nitrogen fixation during symbiosis. Mol Gen Genet. 1995;247:39–47. doi: 10.1007/BF00425819. [DOI] [PubMed] [Google Scholar]
- 15.Kiss E, Reuhs B L, Kim J S, Kereszt A, Petrovics G, Putnoky P, Dusha I, Carlson R W, Kondorosi A. The rkpGHI and -J genes are involved in capsular polysaccharide production by Rhizobium meliloti. J Bacteriol. 1997;179:2132–2140. doi: 10.1128/jb.179.7.2132-2140.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagares A, Caetano-Anollés G, Niehaus K, Lorenzen J, Ljunggren H D, Pühler A, Favelukes G. A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation in alfalfa. J Bacteriol. 1992;174:5941–5952. doi: 10.1128/jb.174.18.5941-5952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leigh J A, Walker G C. Exopolysaccharides of Rhizobium: synthesis, regulation and symbiotic function. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 18.Lin C S, Lin N T, Yang B Y, Weng S F, Tseng Y H. Nucleotide sequence and expression of UDP-glucose dehydrogenase gene required for the synthesis of xanthan gum in Xanthomonas campestris. Biochem Biophys Res Commun. 1995;207:223–230. doi: 10.1006/bbrc.1995.1176. [DOI] [PubMed] [Google Scholar]
- 19.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the synthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz R, Mollerach M, Lopez R, Garcia E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrovics G, Putnoky P, Reuhs B, Kim J, Thorp T A, Noel K D, Carlson R W, Kondorosi A. The presence of a novel type of surface polysaccharide in Rhizobium meliloti requires a new fatty acid synthase-like gene cluster involved in symbiotic nodule development. Mol Microbiol. 1993;8:1083–1094. doi: 10.1111/j.1365-2958.1993.tb01653.x. [DOI] [PubMed] [Google Scholar]
- 23.Pridmore R D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 24.Putnoky P, Grosskopf E, Ha D T C, Kiss G B, Kondorosi A. Rhizobium fix genes mediate at least two communication steps in symbiotic nodule development. J Cell Biol. 1988;106:597–607. doi: 10.1083/jcb.106.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putnoky P, Kondorosi A. Two gene clusters of Rhizobium meliloti code for early essential nodulation functions and a third influences nodulation efficiency. J Bacteriol. 1986;167:881–887. doi: 10.1128/jb.167.3.881-887.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putnoky P, Petrovics G, Kereszt A, Grosskopf E, Ha D T C, Banfalvi Z, Kondorosi A. Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J Bacteriol. 1990;172:5450–5458. doi: 10.1128/jb.172.9.5450-5458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuhs B L. Acidic capsular polysaccharides (K antigens) of Rhizobium. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: International Society for Molecular Plant-Microbe Interactions; 1997. pp. 331–336. [Google Scholar]
- 28.Reuhs B L, Carlson R W, Kim J S. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J Bacteriol. 1993;175:3570–3580. doi: 10.1128/jb.175.11.3570-3580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuhs, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. Kumar Kolli, and S. G. Pueppke.Sinorhizobium fredii and S. meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 30.Reuhs B L, Williams M N V, Kim J S, Carlson R W, Côté F. Suppression of the Fix− phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J Bacteriol. 1995;177:4289–4296. doi: 10.1128/jb.177.15.4289-4296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruvkun G B, Ausubel F M. A general method for site-directed mutagenesis in procaryotes. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultze M, Kondorosi E, Ratet P, Buire M, Kondorosi A. Cell and molecular biology of Rhizobium-plant interactions. Int Rev Cytol. 1994;156:1–75. [Google Scholar]
- 35.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 36.Tsai C, Frisch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 37.Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhan H J, Levery S B, Lee C C, Leigh J A. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc Natl Acad Sci USA. 1989;86:3055–3059. doi: 10.1073/pnas.86.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]