Abstract
Maintenance of telomere integrity requires the dynamic interplay between telomerase, telomere-associated proteins and DNA repair proteins. These interactions are vital to suppress DNA damage responses and changes in chromosome dynamics that can result in aneuploidy or other transforming aberrations. The interaction between the DNA repair protein Ku and the RNA component of telomerase (TLC1) in Saccharomyces cerevisiae has been shown to be important for maintaining telomere length. Here, we sought to determine whether this interaction was conserved in higher eukaryotes. Although there is no sequence similarity between TLC1 and the RNA component (hTR) of human telomerase, we show that human Ku70/80 interacts with hTR both in vitro and in a cellular context. Specifically, Ku70/80 interacts with a 47 nt region of the 3′ end of hTR, which resembles the stem–loop region of the yeast Ku70/80 binding domain on TLC1. Furthermore, utilizing immunoprecipitation/RT–PCR experiments, we show that Ku interacts with hTR in cell lines deficient in the human telomerase reverse transcriptase protein (hTERT), suggesting that this interaction does not require hTERT. These data suggest that Ku interacts directly with hTR, independent of hTERT, providing evidence for the conservation of the interaction between Ku and telomerase RNA among various species and provide significant insight into how Ku is involved in telomere maintenance in higher eukaryotes.
INTRODUCTION
Telomeres are nucleoprotein structures found at the ends of linear chromosomes that protect chromosomal DNA from degradation, recombination and detrimental fusion events (1). In humans, telomeres are composed of telomeric DNA, consisting of tandem repeats of short sequences (TTAGGG), that are synthesized by the enzyme telomerase (1). The human telomerase enzyme is minimally composed of the reverse transcriptase, hTERT and the RNA component, hTR. hTERT utilizes hTR as a template to add the TTAGGG repeats onto the 3′ ends of the chromosome (1,2). In addition to this role as a polymerase, the hTERT/hTR complex also cooperates with a conglomerate of other proteins to form the nucleoprotein ‘cap’ at chromosome termini. These proteins include telomeric DNA-binding proteins, such as TRF1 and TRF2 (3), and DNA repair proteins, such as the Mre11/Rad50/Nbs1 complex (4). Maintenance of this telomere cap involves an intricate, highly regulated network of protein–protein, protein–DNA and protein–RNA interactions, which are crucial for the protection of the genome. Misregulation of this process has been associated with cellular senescence and transformation to a malignant state (5).
The DNA-dependent protein kinase, DNA-PK, has been linked to telomere maintenance (4,6,7). DNA-PK, which is composed of the catalytic subunit DNA-PKcs, and the dimeric DNA-binding regulatory subunits Ku70/80, is required for repair of DNA double-strand (ds) breaks via the non-homologous end-joining (NHEJ) pathway (8). Mouse cells deficient in DNA-PKcs show high levels of chromosome end-to-end fusion (6); moreover, cells deficient in both DNA-PKcs and Terc (mouse hTR) exhibit accelerated rates of telomere shortening compared with cells solely deficient in Terc, suggesting a functional interaction between DNA-PKcs and telomerase in sustaining telomere length (9). Mouse cells lacking Ku70 or Ku80 also display a higher rate of chromosomal end-to-end fusion events (10,11). Inactivation of a single allele of Ku80 in human somatic cells resulted in a similar phenotype (12). Ku70/80 associates with telomeric DNA (13) and binds to TRF1 and TRF2 (14,15). Moreover, Ku has been reported to associate with hTERT and telomerase activity (16). However, the precise biological consequence, the biochemical nature of these interactions and the role of DNA-PK in telomere maintenance remain unknown.
It has been shown in budding yeast that Ku70/80 (yKu70/80) interacts with a stem–loop region of TLC1, and yeast harboring a yKu80 allele that is defective for TLC1 binding possesses shortened telomeres (17). A more recent study demonstrates that the interaction of yKu70/80 with TLC1 is required to recruit two subunits of yeast telomerase (Est1p and Est2p) to telomeres during S phase, when optimal telomere elongation is occurring (18). These observations suggest that the interaction of yKu70/80 with TLC1 is important for maintaining the telomere length. In this study, we have investigated the evolutionary conservation of this interaction and we demonstrate that human Ku interacts with hTR, the RNA component of human telomerase both in vitro and in vivo.
MATERIALS AND METHODS
Electrophoretic mobility shift assays (EMSAs)
Native DNA-PKcs and Ku70/80 were purified to homogeneity from HeLa cells as described previously (19). The EMSA binding reactions were carried out in 10 μl volume containing 100 mM NaCl, 10 mM Tris, pH 7.5, 5% (w/v) glycerol, 1 mM MgCl2, 0.8 U of RNAse OUT (Invitrogen) and 0.33 pmol of 32P-labeled hTR probe. The amount of purified proteins used in each EMSA reaction is indicated in the figure legends. Following a 10 min incubation at 25°C, the reactions were fractionated on 5% non-denaturing PAGE, in 0.5× TBE for 90 V for 3 h. For the smaller truncated hTRs, the gels were run for 1 h.
For the EMSA experiments with DNA, a double-stranded 36 bp blunt-ended oligonucleotide corresponding to the sequence of a non relevant gene (PIG3) was used as the radiolabeled probe. The DNA probe was end-labeled with 32P using T4 kinase as described previously (20), and approximately 0.2 pmol of labeled probe was used for the binding reactions. For the DNA gel shift, the gels were run for 1 h.
The EMSA gels were dried on Whatman 3MM paper and exposed to X-ray films. Addition of competitor RNA, DNA or antibodies to the EMSA reactions, either before or 5 min after the radiolabeled probe, did not alter the results. The monoclonal antibodies used included anti-Ku70 and anti-Ku80 (Ab5 and Ab2, NeoMarkers) and anti-Myc (Santa Cruz Biotechnology).
Synthesis of hTR RNA truncations
Various truncations of hTR were generated using methods described previously (21). Briefly, 5′ (containing the T7 polymerase promoter sequence) and 3′ primers corresponding to the hTR cDNA sequence were used to amplify the desired cDNA sequence encoding the hTR truncation; the PCR product was then cloned into pUC19 for in vitro transcription reactions (MEGAscript T7; Ambion) (21). For example, for the 404–451 hTR truncation, the 5′ primer used was 5′-GGG AAG CTT TAA TAC GAC TCA CTA TAG GAT TCC CTG AGC TGTG-3′ and the 3′ primer utilized was 5′-GCA TGT GTG AGC CGA GTC-3′. To generate the radiolabeled hTR probes, the appropriate cDNA template was linearized with EcoRI and used for in vitro transcription reactions in the presence of α32P-UTP (3000 Ci/mmol; Amersham Biosciences). The subsequent hTR RNA products were purified using MEGAclear kit (Ambion) as per the manufacturer's guide, and quantitated using a BioPhotometer (Eppendorf, VWR).
Immunoprecipitation and RT–PCR
The three cell lines used for these experiments included two human embryonic kidney cell lines, 293T and HA5, and an SV40 transformed human fibroblast cell line, GM847. Asynchronously grown human cells were harvested, pelleted and resuspended with ∼10 times the pellet volume of CHAPS Lysis buffer [100 mM NaCl, 10 mM Tris, pH 7.5, 10% (w/v) glycerol, 1% CHAPS, 1 mM MgCl2, 5 mM β-mercaptoethanol, 0.5 U/ml RNAse OUT plus protease inhibitors, CompleteMini EDTA free (Boehringer Mannheim)] for 30 min on ice; the lysates were then centrifuged at 16 000 g for 30 min at 4°C, to obtain whole cell extracts for immunoprecipitation experiments (22). For each immunoprecipitation reaction, ∼4 × 106 cells were used. Approximately 5 μg of each antibody was pre-coupled to 25 μl of a 50% slurry of protein G agarose beads by incubating for 1 h at 4°C on a rotator. Whole cell extracts were pre-cleared with protein G agarose, and then added to the antibody coated beads; the mixtures were then subjected to constant rotation for 4 h at 4°C. The subsequent immune complexes were washed three times with CHAPS Lysis buffer. Approximately 10% of the immune complex beads were removed and analyzed by western blots. RNA was then extracted from the remaining amount of beads with phenol, and then with chloroform–isoamyl alcohol (24:1), ethanol precipitated, and redissolved in water. RNA was also isolated from the same number of cells using Trizol reagent (Invitrogen) according to the manufacturer's guide.
The resulting RNA preparations were analyzed for the presence of hTR by RT–PCR using primers and the methodology as described in (23). The reverse transcription reactions were carried out using SuperScriptII One-Step RT–PCR with Taq polymerase (Invitrogen). Purified FL-hTR was used as positive control for the RT–PCR. A control PCR for the β-2-Microglobulin (β2M) gene was also performed based on a previously described protocol (24). The expected PCR products for hTR and β2M are 102 and 300 bp, respectively. Western Blots of the immune complexes were performed using monoclonal antibodies against Ku80 (Ab2, Neomarkers).
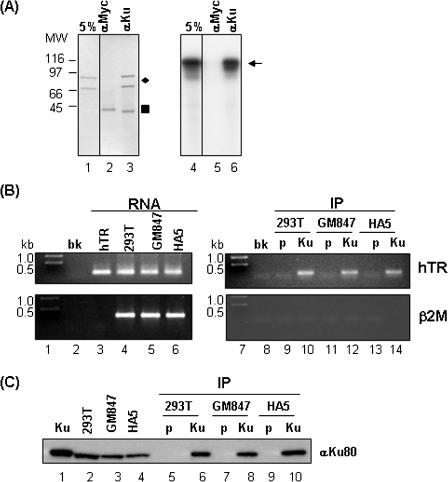
For immunoprecipitation experiments shown in Figure 6A, ∼5 μg of purified Ku70/80 was incubated with 2 pmol of radiolabeled FL-hTR under EMSA conditions for 15 min at 25°C. The reactions were diluted to a volume of 100 μl with CHAPS Lysis buffer, and added to antibody-coated protein G beads, and subjected to constant rotation for 2 h at 4°C. The beads were then washed twice with CHAPS Lysis buffer, and loaded onto SDS–PAGE, Coomassie blue stained and analyzed by autoradiography.
Figure 6.
Interaction of Ku70/80 with hTR in vitro and in vivo. (A) Purified Ku70/80 was incubated with radiolabeled FL-hTR, and immunoprecipitated with monoclonal antibodies against Ku70/80 or Myc; the immune complexes were washed and fractionated on SDS–PAGE, stained with Coomassie blue (left panel) or processed by autoradiography (right panel). Lanes 1 and 4 each represent 5% of the input. The presence of Ku70/80 is indicated by the diamond (left panel), and the presence of the radiolabeled FL-hTR probe is indicated by the arrow (right panel). The filled-in square indicates IgG heavy chain proteins from the immunoprecipitation (left panel). Molecular weight markers in kDa are shown in the Coomassie blue stained gel (left panel). (B) Whole cell extracts were prepared from human 293T, GM847 or HA5 cells and immunoprecipitated with mouse pre-immune or antibodies to Ku70/80; RNA was extracted from 90% of the subsequent immune complexes and subjected to RT–PCR as described in Materials and Methods. The presence of hTR (top panel) and β2M (bottom panel) is indicated by 102 and 300 bp PCR products, respectively. Lane 1 contains the DNA size marker in kb, lane 2 contains no template PCR control (bk), and lane 3 contains 0.1 μg of FL-hTR subjected to the same RT–PCR protocol. Lanes 4–6 contain the RT–PCR of ∼1% of the total RNA isolated from an equal number of cells used for each of the immunoprecipitation experiments to serve as positive controls for the presence of hTR and β2M. Lane 7 contains DNA size marker in kb, and lane 8 contains the no template PCR control (bk). Lanes 9–14 represent the results of the RT–PCR of the pre-immune (p) or anti-Ku70/80 (Ku) immune complexes isolated from the different cell lines. (C) The presence of Ku in the immune complexes used for the RT–PCR in (B) was confirmed by western blot analysis using mouse monoclonal anti-Ku80. Lane 1 contains 0.1 μg of purified Ku70/80. Lanes 2–4 represents 1% of the input cell extract used for the immunoprecipitation experiments. Lanes 5–10 contains ∼10% of the total immune complexes (‘p’ for pre-immune, ‘Ku’ for anti-Ku70/80) isolated from the various cell lines used for extracting RNA for the RT–PCR experiment.
RESULTS
Interaction of Ku70/80 with hTR
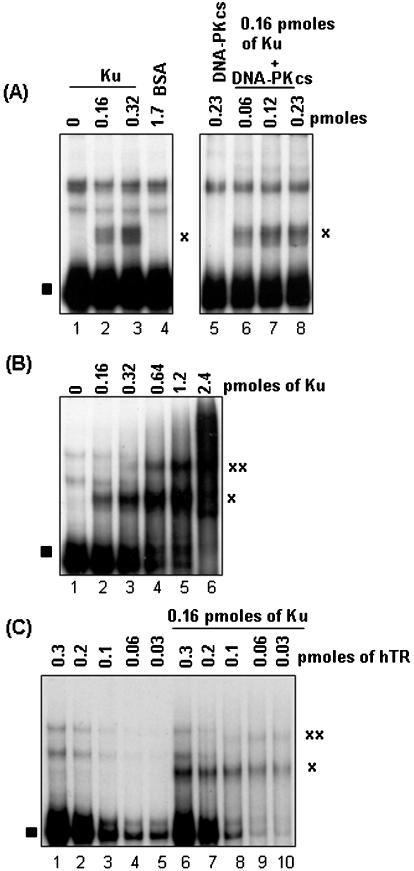
In budding yeast, genetic studies show that yKu70/80 interacts with a 48 nt (nucleotides 288–335) stem–loop region of the RNA template (TLC1) of yeast telomerase (17). In vitro studies show that purified yKu70/80 interacts with three tandem copies of this 48 nt stem–loop region of TLC1. To determine whether or not the telomerase RNA/Ku interaction seen in yeast is conserved in higher eukaryotes, full-length (FL)-hTR was transcribed in vitro in the presence of α32P-UTP, purified, and tested for interaction with Ku70/80 and DNA-PKcs (purified from HeLa cells) in EMSA studies (25). As shown in Figure 1A, a slower migrating form of hTR (indicated by the ‘x’) appeared in the presence of purified Ku70/80 but not with purified DNA-PKcs (compare lanes 2 and 3 with lane 5). The addition of increasing amounts of purified DNA-PKcs did not significantly alter the migration of the putative Ku70/80–hTR ribonucleoprotein complex (Figure 1A, lanes 6–8), implying that under these conditions DNA-PKcs does not interact with Ku bound hTR. Increasing the NaCl concentration up to 300 mM and addition of up to 20 μM ATP and 50 μM MgCl2 in the EMSA binding reactions did not affect the putative Ku-hTR complex (data not shown). The preparation of Ku70/80 used in these experiments is shown in Figure 6A (lane 1).
Figure 1.
Interaction of Ku with hTR. (A) For EMSA binding reactions, 0.16 or 0.32 pmol (lanes 2 and 3) of purified Ku70/80 was incubated with 0.33 pmol of radiolabeled FL-hTR for 10 min at RT and the reaction products were resolved on non-denaturing PAGE. The resulting gel was dried and exposed to film. Purified DNA-PKcs (0.06–0.23 pmol) was also tested to determine its effect on the Ku70/80–hTR complex (lanes 6–8). Lane 1, FL-hTR probe alone; lane 4, 1.7 pmol of BSA; and lane 5, 0.23 pmol of DNA-PKcs with FL-hTR probe. The ‘x’ indicates the putative Ku–hTR complex, and the filled square indicates free FL-hTR probe. (B) A constant amount of radiolabeled FL-hTR (0.33 pmol) was incubated with increasing amounts (as indicated in lanes 2–6) of purified Ku70/80 under previously described conditions and the reaction products were processed as in (A). The two predominant ribonucleoprotein complexes are indicated with ‘x’ and ‘xx’. (C) A constant amount of purified Ku70/80 (0.16 pmol) was incubated with decreasing amounts of radiolabeled FL-hTR (lanes 6–10) under the same binding conditions, with the subsequent reactions processed as in (A). Lanes 1–5, decreasing amount of FL-hTR (0.3–0.03 pmol) probe alone. The complexes formed are indicated by ‘x’ and ‘xx’.
In gel shift assays with dsDNA, Ku70/80 has been shown to form multiple protein–DNA complexes, depending on the amount of Ku70/80 and the length of dsDNA (20). This is believed to represent the ability of Ku70/80 to bind to the dsDNA end, and translocate along the DNA in an ATP-independent manner (20). To see whether a similar phenomenon exists with Ku–RNA interactions, we incubated radiolabeled FL-hTR with increasing amounts of Ku. As shown in Figure 1B, two predominant slower migrating forms of ribonucleoprotein complexes begin to appear with increasing amounts of Ku70/80 (Figure 1B, indicated by ‘x’ and ‘xx’), with concomitant lost of free labeled FL-hTR. Consistently, the same two ribonucleoprotein complexes appeared when a constant amount of Ku was incubated with decreasing amounts of labeled FL-hTR (Figure 1C, lanes 6–10, indicated by ‘x’ and ‘xx’). It would premature at this time to speculate on the molecular nature of these multiple forms of the Ku-hTR complexes, but it likely represents multiple Ku molecules loaded onto a single hTR molecule, or dimerization of hTR molecules bound with Ku.
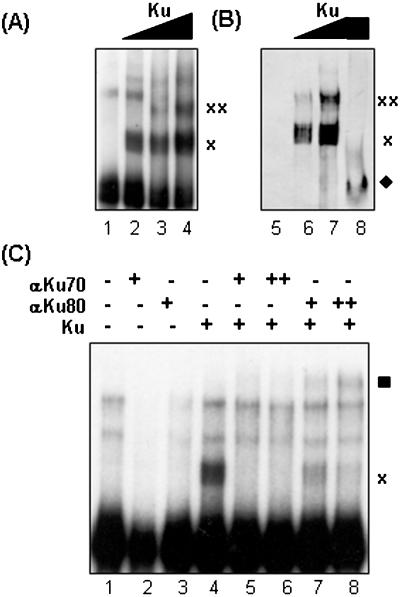
To confirm the presence of Ku70/80 within these higher order ribonucleoprotein complexes, an EMSA gel (Figure 2A) was transferred to PVDF membrane and analyzed by western blot analysis with monoclonal antibodies against Ku70/80 (Figure 2B). As indicated in Figure 2B, Ku70/80 is present in ribonucleoprotein complexes indicated by ‘x’ and ‘xx’. Furthermore, monoclonal antibodies to Ku70 inhibited Ku70/80–hTR complex formation (Figure 2C, lanes 5 and 6), while antibodies to Ku80 retarded the Ku70/80–hTR complex (Figure 2C, lanes 7 and 8). Monoclonal antibodies to Myc had no effect on the hTR–Ku70/80 complex (data not shown). These observations indicate the presence of Ku70/80 in these protein–RNA complexes and suggest that the epitope for the Ku70 antibody may reside in the Ku70 region that interacts with hTR, and therefore ablated the formation of Ku–hTR complex. Conversely, the Ku80 antibody may recognize an epitope outside of the Ku–hTR interaction interface, and thus did not disrupt the Ku–hTR complex. Together, these results suggest that Ku70/80 binds directly to the RNA template of human telomerase.
Figure 2.
Ku and hTR form higher order protein–RNA complexes. (A) The radiolabeled FL-hTR probe was incubated by itself (lane 1) or with 0.32, 0.64 or 1.2 pmol of Ku70/80 (lanes 2, 3 and 4, respectively). As indicated by ‘x’ and ‘xx’, multiple slower forms of radiolabeled hTR appeared with increasing amounts of Ku. (B) Another EMSA gel with FL-hTR probe alone (lane 5), 0.64 and 1.2 pmol of Ku and FL-hTR probe (lanes 6 and 7) or 1.2 pmol Ku alone (lane 8) was transferred to PVDF membrane to be analyze for the presence of Ku in the protein–RNA complexes by western blot with monoclonal antibodies to Ku70/80 (lanes 5–8). The putative Ku70/80–hTR complexes are indicated by ‘x’ and ‘xx’, and the migration position of Ku70/80 in the absence of radiolabeled FL-hTR is indicated by the filled diamond (lane 8). (C) 0.1 (+) and 0.2 μg (++) of monoclonal antibodies to Ku70 (lanes 5 and 6) or Ku80 (lanes 7 and 8) were incubated with 0.16 pmol of purified Ku70/80, prior to the addition of radiolabeled FL-hTR probe. After 10 min incubation at RT, the reactions were then fractionated on non-denaturing PAGE and processed by autoradiography. Monoclonal antibodies to Ku70 ablated the Ku–hTR complex (lanes 5 and 6), while monoclonal antibodies to Ku 80 ‘super-shifted’ the Ku–hTR complex (indicated by filled square, lanes 7 and 8). The ‘x’ indicates the migration position of FL-hTR–Ku complex. The contents of the remaining lanes are as follows: lane 1, FL-hTR probe alone; lane 2, anti-Ku70 with FL-hTR probe; lane 3, anti-Ku80 with FL-hTR probe; and lane 4, Ku with FL-hTR probe.
Specificity of the interaction of Ku with hTR
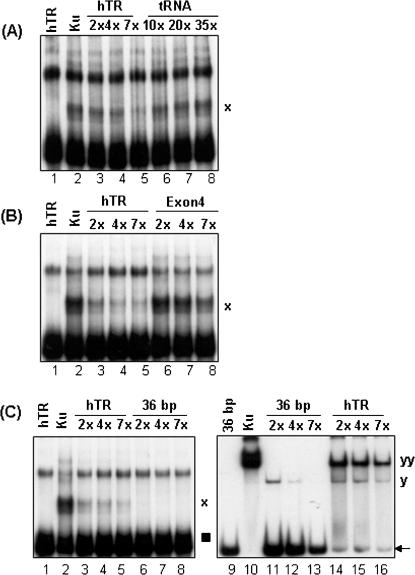
The interaction of Ku70/80 with dsDNA and its role in activating DNA-PKcs has been well characterized. Current models for NHEJ suggest that Ku70/80 initially associates with ends of dsDNA breaks and recruits DNA-PKcs (8). However, recent reports demonstrate that in human cells, the phosphorylation of RNA helicase A and hnRNP proteins by DNA-PK is RNA-dependent and that a number of unidentified RNA molecules can bind to Ku (26). As well, Ku70/80 has been shown, in vitro, to preferentially associate with RNA molecules with the sequence motif of GCUUUCCCANNNAC (N being any nucleotide) derived from the HIV TAR RNA (20,27). However, the physiological relevance of these interactions remains unknown. To test the specificity of the interaction of Ku70/80 with hTR, we performed competition assays with unlabeled FL-hTR, tRNA and an unrelated RNA sequence, Exon 4 of the PIG3 gene (24) and dsDNA. The Ku70/80–hTR complex signal diminished with the addition of molar excess amounts of unlabeled FL-hTR (Figure 3A, lanes 3–5), but not with tRNA (Figure 3A, lanes 6–8). Similarly, Exon 4 RNA at equimolar amounts with unlabeled FL-hTR did not compete for Ku binding to labeled FL-hTR (Figure 3B, compare lanes 3–5 with lanes 6–8), suggesting that Ku has a higher affinity for FL-hTR compared with Exon 4. On the other hand, at equal molar amounts, a 36 bp blunt-ended dsDNA oligonucleotide competed better than the unlabeled FL-hTR for the binding of Ku (Figure 3C, compare lanes 3–5 with lanes 6–8). This apparent higher affinity of Ku for dsDNA compared with FL-hTR was recapitulated in reciprocal experiments using labeled 36 bp DNA (Figure 3C, lanes 9–16). Similar results were obtained regardless of whether the unlabeled competitors were added together or 5 min after the addition of the radiolabeled probes (data not shown). These data suggest that under these EMSA conditions, Ku prefers to associate with FL-hTR compared with other types of RNA, but has a higher affinity for dsDNA (to be discussed later). Inspection of the hTR nucleotide sequence reveals that it does not contain the aforementioned previously characterized Ku binding RNA motif (27), suggesting that the molecular determinants of the Ku binding region in hTR are unique, likely involving RNA secondary structures (see below).
Figure 3.
Specificity of the Ku–hTR interaction. (A) Purified Ku70/80 (0.16 pmol) was incubated with radiolabeled FL-hTR probe (lane 2) in the presence of unlabeled FL-hTR (lanes 3–5) or tRNA (lanes 6–8). The molar excess amount of unlabeled RNA competitor compared with labeled FL-hTR used in each instance was as indicated. Lane 1, free FL-hTR probe; ‘x’, the migration position of the FL-hTR–Ku70/80 complex. (B) Radiolabeled FL-hTR was incubated with purified Ku70/80 (0.16 pmol, lane 2) in the presence of unlabeled FL-hTR (lanes 3–5) or the mRNA product of exon 4 of the PIG3 gene (exon 4, 196 nt in length) (lanes 6–8). The molar excess amount of the unlabeled RNA competitor compared with FL-hTR used in each instant is indicated. Lane 1, free FL-hTR probe; and ‘x’, the FL-hTR–Ku70/80 complex. (C) Left panel: Ku70/80 (0.16 pmol) was incubated with FL-hTR probe (lane 2) in the presence of unlabeled FL-hTR (lanes 3–5) or a 36 bp dsDNA oligonucleotide (lanes 6–8). The amount of unlabeled competitor in molar excess amounts is indicated. The hTR–Ku70/80 complex is indicated by the ‘x’ and the free FL-hTR probe is indicated by the filled square. Right panel: Ku70/80 (0.1 pmol) was incubated with radiolabeled 36 bp dsDNA (0.20 pmol) alone (lane 10) or together with molar excess amounts of unlabeled 36 bp dsDNA (lanes 11–13) or FL-hTR (lanes 14–16), and fractionated on non-denaturing gels and processed as before. The Ku–DNA complexes are indicated by ‘y’ and ‘yy’, and the free 36 bp dsDNA probe is indicated by the arrow. Lane 1, free FL-hTR probe; and lane 9, free 36 bp dsDNA probe.
Identification of the region of hTR that interacts with Ku70/80
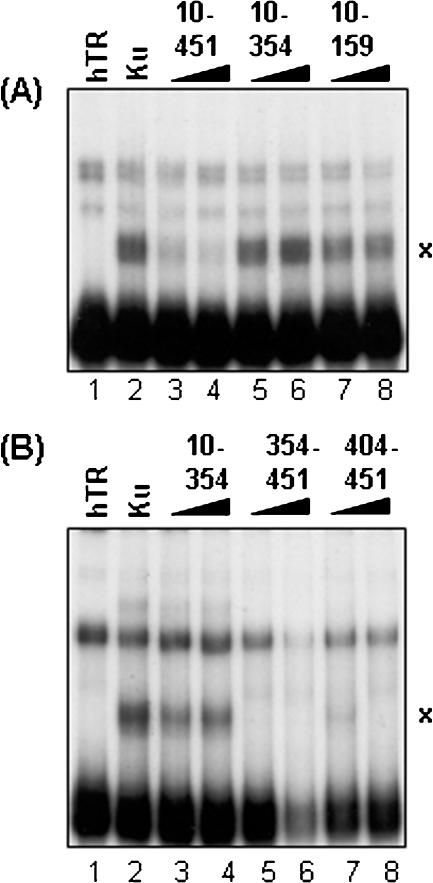
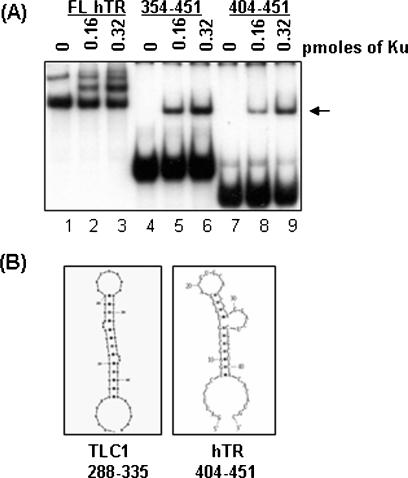
The 48 nt region of the stem–loop of TLC1 that associates with yKu70/80 is located within the 5′ end (nucleotides 288–335) of the 1.3 kb TLC1 RNA molecule (17). Comparison of the nucleotide sequence of hTR with TLC1 shows no sequence similarity, although the topology of several regions, such as the reverse transcriptase interaction domain, appear to be functionally conserved (28,29). To isolate the region of the hTR (451 nt in length) that interacts with Ku70/80, we performed competition assays with truncated forms of hTR. As shown in Figure 4A, nucleotides 10–451 of hTR competed for interaction with Ku70/80, but nucleotides 10–354 or 10–159 were not as effective (compare lanes 5–8 with lanes 3–4). This observation suggested that Ku interacts with the 3′ end of hTR. To further delineate the specific hTR-Ku binding region, two other hTR fragments, comprising nucleotides 354–451 and 404–451 (referred to as hTR-354 and hTR-404, respectively), were generated. As expected, increasing amounts of these hTR truncations in molar excess competed for Ku binding to radiolabeled FL-hTR, but not hTR spanning nucleotides 10–354 (Figure 4B, compare lanes 5–8 with lanes 2 and 3). Ku70/80 was also able to interact with radiolabeled hTR-404 in EMSA (Figure 5A, lanes 8 and 9), suggesting that the minimal region of hTR that is required to interact with Ku resides in the extreme 3′ end of the molecule (Figure 5A, lanes 7–9). In context of the proposed predicted secondary structure for hTR, the region between nucleotides 404 and 451 forms a stem–loop structure similar to that of the Ku binding region seen in yeast TLC1 (29,30). Moreover, according to the M-fold RNA structure prediction program (http://www.bioinfo.rpi.edu/applications/mfold), hTR-404 alone also forms a similar structure (Figure 5B). However, as mentioned earlier, the yKu70/80 binding studies were carried out with three tandem repeats of the TLC1 stem–loop, but under our conditions, only one copy of hTR-404 was necessary for Ku70/80 binding. This suggests that the RNA structural determinants required for Ku70/80 association may be different in higher eukaryotes and require further studies.
Figure 4.
Identification of the region of hTR that interacts with Ku70/80 (A) Various truncations of hTR were generated and tested against FL-hTR for their ability to compete for the interaction with Ku70/80. The molar excess amount used for each unlabeled competitor compared with labeled FL-hTR is as follows: for hTR fragments 10–451, 3.5 and 7 times (lanes 3 and 4); for hTR-10–354, 4 and 8 times (lanes 5 and 6); and for hTR-10–159, 11 and 22 times (lanes 7 and 8). Lane 1, FL-hTR probe alone; and lane 2, FL-hTR probe and Ku70/80. (B) Two other hTR truncations between nucleotides 354–451 were generated and tested in the same competition assays. The amount of each unlabeled hTR fragment used as a competitor (10–354, 354–451 and 404–451) was 3 and 6 times in molar excess compared with labeled FL-hTR (lanes 3–8). Lane 1, FL-hTR probe alone; and lane 2, FL-hTR probe and Ku70/80.
Figure 5.
The minimal region of hTR that interacts with Ku70/80. (A) FL-hTR and two hTR truncations comprising nucleotides 354–451 (lanes 4–6) and 404–451 (lanes 7–9) were radiolabeled with 32P-UTP and tested for their ability to associate with increasing amounts of Ku70/80 (0, 0.16 and 0.32 pmol). The migration position of the FL-hTR probe and Ku–FL-hTR complex is presented in lanes 1–3. The presence of hTR-404–Ku protein complex is indicated by the arrow. (B) Comparison of the structure of the stem–loop region on TLC1 (nucleotides 288–335) from S.cerevisiae [adapted from (17)] and hTR-404 using mfold RNA secondary structure prediction program (http://www.bioinfo.rpi.edu/applications/mfold). hTR+404 forms a similar stem–loop as that seen in TLC1.
Co-immunoprecipitation of Ku70/80 with hTR in a cellular context
As an alternative methodology to explore the nature of the interaction between Ku70/80 and hTR, we utilized an independent assay based on co-immunoprecipitation. Purified Ku70/80 was incubated with radiolabeled FL-hTR under EMSA binding conditions. The reaction products were then subjected to immunoprecipitation with monoclonal antibodies against Ku70/80 or Myc. As shown in Figure 6A, radiolabeled FL-hTR co-immunoprecipitated with the Ku70/80 polypeptides (Figure 6A, lane 6), demonstrating a direct interaction between Ku70/80 and hTR. These data further suggested that this method can be utilized to investigate the interaction of Ku70/80 with hTR in cell extracts.
We next prepared whole cell extracts from asynchronously growing human embryonic kidney 293T cells, which contain wild-type hTR and hTERT (31), and performed immunoprecipitations with mouse pre-immune anti-sera and monoclonal antibodies to Ku70/80. RNA was isolated from the subsequent immune complexes by phenol/chloroform extraction and subjected to RT–PCR for the presence of hTR, or β2M mRNA as a control. As shown in Figure 6B, antibodies to Ku70/80, but not to mouse pre-immune anti-sera, immunoprecipitated hTR from whole cell extracts prepared from 293T cells (Figure 6B, lanes 9 and 10). As a positive control for the presence of hTR and β2M, ∼1% of the total amount of RNA isolated from an equal number of cells used for the immunoprecipitations was subjected to the same RT–PCR (Figure 6B, left panel, lanes 1–6).
To eliminate the possibility that the co-immunoprecipitation of Ku with hTR is mediated through hTERT (16), we performed similar experiments using two other cell lines, HA5 (human embryonic kidney cells) and GM847 (human fibroblast), which are proficient in hTR but are deficient in hTERT (32,33). As shown in Figure 6B, Ku70/80 co-immunoprecipitated with hTR from HA5 and GM847 cell extracts (Figure 6B, top panel, lanes 11–14). As expected, none of these immune complexes contained β2M mRNA (Figure 6B, bottom panel). The presence of Ku70/80 in the immune complexes was confirmed by western blots (Figure 6C, lanes 6, 8 and 10). This strategy of immunoprecipitation followed by RT–PCR was utilized to show the in vivo interaction of yeast TLC1 and Ku (18). In combination with the previous in vitro binding data, these data suggest that Ku and hTR reside in the same complex in a cellular context, and that the interaction between Ku and hTR is direct and occurs independent of hTERT.
DISCUSSION
In this study, we show that human Ku70/80 interacts with hTR, the RNA template of human telomerase both in vitro and in vivo. Minimally, Ku70/80 can interact with a region of 47 nt in the 3′ end of hTR and the predicted secondary structure of this region appears to be similar to the Ku70/80 binding region of yeast TLC1. However, the precise structural requirements of these interactions remain to be explored. Moreover, in a cellular context, Ku interacts with hTR in cell lines deficient for the reverse transcriptase, hTERT, suggesting that the interaction between Ku and hTR occurs independent of hTERT.
Based on our EMSA data, it appears that Ku preferentially binds hTR compared with other types of RNA, but has a higher affinity for dsDNA. This is not surprising given the role of Ku in DNA repair. Stellwagen and co-workers (17) identified a yKu80 allele (referred to as yKu80-135i) that harbors a 15 nt in-frame insertion in the N-terminus of yKu80 that does not bind TLC1. This mutation maps to conserved residues in human Ku80 and is located adjacent to the DNA-binding region of the Ku70/80 heterodimer (17,34), implying that the interaction between Ku with RNA and Ku with DNA may be mutually exclusive. Consistent with this notion, yKu80-135i yeast strains show no DNA repair defect, but show shortened telomeres, revealing that the DNA-binding function of Ku for NHEJ and TLC1 association for telomere maintenance can be separated.
Given that the TLC1 binding deficient mutation for yKu70/80 resides in the yKu80 allele, it suggests that the yKu80 subunit is responsible for mediating the interaction with telomerase RNA (17). However, efforts to determine which human Ku subunit interacts with hTR utilizing Far Northern and UV cross-linking experiments were inconclusive (data not shown). Based on our immunoprecipitation data (Figure 6A), it is very likely that Ku interacts with hTR as a heterodimer. Nevertheless, it remains to be determined which subunit of human Ku mediates the interaction with hTR.
Budding yeast strains harboring the yKu80-135i mutation or tlc1Δ48 (TLC1 lacking 48 nt Ku binding stem–loop) show accelerated loss of telomere length, which is not attributed to loss of telomerase activity but to reduced kinetics of telomere addition (17). This can be attributed to the reduced association of two yeast telomerase subunits (Est1p and Est2p) to the telomere during telomere synthesis, revealing that the interaction of yKu70/80 with TLC1 is crucial for recruiting the active reverse transcriptase complex (18). In higher eukaryotes, mouse cells lacking Ku70 and human cells carrying an inactivated Ku80 allele also show accelerated telomere shortening (12,35). Human Ku70/80 has also been shown to immunoprecipitate with hTERT and telomerase activity (16), consistent with a role in telomere length regulation. Combined with the data presented, it seems likely that Ku may promote telomere elongation either by recruiting hTR to chromosome ends or by stabilizing hTR/hTERT complexes once they form at the ends. Interestingly, as mentioned earlier, the yKu80-135i or tlc1Δ48 yeast strain did not show DNA repair defects, revealing that the function of Ku70/80 in NHEJ and in telomere maintenance can be separated. A more intriguing observation was the fact that Ku immunoprecipitated with hTR from HA5 and GM847 cell lines, which do not express the hTERT transcript or protein (32,33). Although neither of these cell lines contains the hTERT protein, there are very significant differences between the two; HA5 cells are an SV40 transformed cell line with a finite lifespan with no detectable telomerase activity (33), while GM847 cells are immortalized cells that rely on the ALT (alternative lengthening of telomeres) pathway to maintain its telomere length (32,36). This suggests the exciting possibility that interaction of Ku and hTR may have roles in addition to telomere length maintenance in higher eukaryotes.
To our knowledge, this is the first biochemical characterization of human Ku showing that it interacts directly with hTR. This is significant in that it implies a strong evolutionary conservation of the interaction of Ku with the telomerase RNA template and suggests that the function of Ku in DNA repair and in telomere maintenance may also be separable in higher eukaryotes. Thus, our data have provided a novel avenue of studies to specifically define the physiological role of Ku70/80 at mammalian telomeres.
Acknowledgments
The authors thank Karl Riabowol for critical comments on the manuscript, Dierdre Lobb for excellent technical assistance, Chris Nicholls for assistance with the RT–PCR procedure and the members of Lees-Miller laboratory for helpful discussions. S.P.L.-M. is a Scientist of the Alberta Heritage Foundation for Medical Research (AHFMR), an Investigator of the Canadian Institutes for Health Research (CIHR) and holds the Engineered Air Chair in Cancer Research. T.L.B. is a Scholar of the AHFMR and a New Investigator of CIHR. This work was funded by grant 69022 (to S.P.L.-M. and T.L.B) from the CIHR, and grants (to T.L.B) from National Cancer Institute of Canada and Terry Fox Foundation. Funding to pay the Open Access publication charges for this article was provided by the CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 2.Vega L.R., Mateyak M.K., Zakian V.A. Getting to the end: telomerase access in yeast and humans. Nature Rev. Mol. Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 3.Cech T.R. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 4.d'Adda di Fagagna F., Teo S.H., Jackson S.P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 5.Cong Y.S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilley D., Tanaka H., Hande M., Kurimasa A., Li G., Oshimura M., Chen D. DNA-PKcs is critical for telomere capping. Proc. Natl Acad. Sci. USA. 2001;98:15084–15088. doi: 10.1073/pnas.261574698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goytisolo F.A., Samper E., Edmonson S., Taccioli G.E., Blasco M.A. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 2001;21:3642–3651. doi: 10.1128/MCB.21.11.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lees-Miller S.P., Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Espejel S., Franco S., Sgura A., Gae D., Bailey S.M., Taccioli G.E., Blasco M.A. Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. EMBO J. 2002;21:6275–6287. doi: 10.1093/emboj/cdf593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espejel S., Franco S., Rodriguez-Perales S., Bouffler S.D., Cigudosa J.C., Blasco M.A. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 2002;21:2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samper E., Goytisolo F.A., Slijepcevic P., van Buul P.P., Blasco M.A. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myung K., Ghosh G., Fattah F.J., Li G., Kim H., Dutia A., Pak E., Smith S., Hendrickson E.A. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol. Cell. Biol. 2004;24:5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianchi A., de Lange T. Ku binds telomeric DNA in vitro. J. Biol. Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- 14.Hsu H.L., Gilley D., Galande S.A., Hande M.P., Allen B., Kim S.H., Li G.C., Campisi J., Kohwi-Shigematsu T., Chen D.J. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song K., Jung D., Jung Y., Lee S., Lee I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 16.Chai W., Ford L.P., Lenertz L., Wright W.E., Shay J.W. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 2002;277:47242–47247. doi: 10.1074/jbc.M208542200. [DOI] [PubMed] [Google Scholar]
- 17.Stellwagen A.E., Haimberger Z.W., Veatch J.R., Gottschling D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher T.S., Taggart A.K., Zakian V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nature Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 19.Goodarzi A.A., Lees-Miller S.P. Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair (Amst.) 2004;3:753–767. doi: 10.1016/j.dnarep.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 20.Dynan W.S., Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beattie T.L., Zhou W., Robinson M.O., Harrington L. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell. 2000;11:3329–3340. doi: 10.1091/mbc.11.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beattie T.L., Zhou W., Robinson M.O., Harrington L. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 2001;21:6151–6160. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabuy E., de Ridder L. Telomerase activity and expression of telomerase reverse transcriptase correlated with cell proliferation in meningiomas and malignant brain tumors in vivo. Virchows Arch. 2001;439:176–184. doi: 10.1007/s004280100466. [DOI] [PubMed] [Google Scholar]
- 24.Nicholls C.D., Shields M.A., Lee P.W., Robbins S.M., Beattie T.L. UV-dependent alternative splicing uncouples p53 activity and PIG3 gene function through rapid proteolytic degradation. J. Biol. Chem. 2004;279:24171–24178. doi: 10.1074/jbc.M401049200. [DOI] [PubMed] [Google Scholar]
- 25.Bachand F., Triki I., Autexier C. Human telomerase RNA–protein interactions. Nucleic Acids Res. 2001;29:3385–3393. doi: 10.1093/nar/29.16.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S., Schlott B., Gorlach M., Grosse F. DNA-dependent protein kinase (DNA-PK) phosphorylates nuclear DNA helicase II/RNA helicase A and hnRNP proteins in an RNA-dependent manner. Nucleic Acids Res. 2004;32:1–10. doi: 10.1093/nar/gkg933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo S., Dynan W.S. Characterization of the RNA binding properties of Ku protein. Biochemistry. 1998;37:1336–1343. doi: 10.1021/bi972100w. [DOI] [PubMed] [Google Scholar]
- 28.Dandjinou A.T., Levesque N., Larose S., Lucier J.F., Abou Elela S., Wellinger R.J. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 29.Zappulla D.C., Cech T.R. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J.L., Blasco M.A., Greider C.W. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 31.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 32.Kilian A., Bowtell D.D., Abud H.E., Hime G.R., Venter D.J., Keese P.K., Duncan E.L., Reddel R.R., Jefferson R.A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 33.Counter C.M., Avilion A.A., LeFeuvre C.E., Stewart N.G., Greider C.W., Harley C.B., Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertuch A.A., Lundblad V. Which end: dissecting Ku's function at telomeres and double-strand breaks. Genes Dev. 2003;17:2347–2350. doi: 10.1101/gad.1146603. [DOI] [PubMed] [Google Scholar]
- 35.d'Adda di Fagagna F., Hande M.P., Tong W.M., Roth D., Lansdorp P.M., Wang Z.Q., Jackson S.P. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 36.Bryan T.M., Marusic L., Bacchetti S., Namba M., Reddel R.R. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum. Mol. Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]