Abstract
The 3′ cleavage generating non-polyadenylated animal histone mRNAs depends on the base pairing between U7 snRNA and a conserved histone pre-mRNA downstream element. This interaction is enhanced by a 100 kDa zinc finger protein (ZFP100) that forms a bridge between an RNA hairpin element upstream of the processing site and the U7 small nuclear ribonucleoprotein (snRNP). The N-terminus of Lsm11, a U7-specific Sm-like protein, was shown to be crucial for histone RNA processing and to bind ZFP100. By further analysing these two functions of Lsm11, we find that Lsm11 and ZFP100 can undergo two interactions, i.e. between the Lsm11 N-terminus and the zinc finger repeats of ZFP100, and between the N-terminus of ZFP100 and the Sm domain of Lsm11, respectively. Both interactions are not specific for the two proteins in vitro, but the second interaction is sufficient for a specific recognition of the U7 snRNP by ZFP100 in cell extracts. Furthermore, clustered point mutations in three phylogenetically conserved regions of the Lsm11 N-terminus impair or abolish histone RNA processing. As these mutations have no effect on the two interactions with ZFP100, these protein regions must play other roles in histone RNA processing, e.g. by contacting the pre-mRNA or additional processing factors.
INTRODUCTION
Histone RNA processing, an endonucleolytic cleavage reaction generating non-polyadenylated mature mRNA 3′ ends, is one of the most important steps regulating the expression of animal replication-dependent histone mRNAs [reviewed in (1–3)]. As their name implies, the abundance of these mRNAs is controlled in parallel with S phase and DNA synthesis in metazoans. In mammalian cells, histone gene transcription is regulated ∼5-fold. However, the most important contribution to cell cycle regulation is made by post-transcriptional events. Both RNA 3′ end processing and histone mRNA stability are cell cycle-regulated (4), and both of these regulatory events are mainly controlled by the fluctuating abundance of an RNA-binding protein (5) known as hairpin-binding protein (HBP) (6) or stem–loop binding protein (7). These post-transcriptional controls allow for the concerted regulation of all transcripts from ∼50 replication-dependent histone genes that are encoded by mammalian genomes.
HBP binds to an RNA hairpin embedded in a 26 nt evolutionarily conserved sequence that ends at the RNA cleavage site. Thus, HBP can bind both to histone pre-mRNAs and to mature mRNAs and thereby control both the nuclear processing reaction and later cytoplasmic events [reviewed in (2,3)]. The role of HBP in the processing reaction is to enhance the binding of a second cleavage factor, the U7 small nuclear ribonucleoprotein (snRNP), to another conserved pre-mRNA sequence located a few nucleotides downstream of the processing site (8–10). The U7 snRNP binds to this histone downstream element (HDE) by RNA base pairing between the 5′ end of U7 snRNA and the histone pre-mRNA (11,12). The extent of this base pairing is different for the various non-allelic histone pre-mRNAs, and thus a stabilization of this interaction by HBP may be very important for some pre-mRNAs and less so for others (8,9,13).
How this additional tethering of the U7 snRNP to the histone pre-mRNA by HBP may be achieved has become clearer in recent years, owing to the identification and molecular characterization of additional processing factors. On the one hand, Dominski, Marzluff and co-workers (14) identified a 100 kDa protein containing 18 C2H2 zinc finger repeats (ZFP100, also known as ZFN473 or KIAA1141 protein) which can bind to an RNP complex consisting of HBP and a histone RNA hairpin, but not to either component of this complex alone. ZFP100 was shown to participate in the tethering of the U7 snRNP to the histone pre-mRNA processing substrate, presumably by interacting with an unidentified component of the U7 snRNP.
On the other hand, our laboratory has recently characterized the polypeptide composition of the U7 snRNP. These studies revealed that the U7 snRNP has a unique structure (15–17). The spliceosomal U1, U2, U4 and U5 snRNPs each contain seven small proteins, the Sm proteins B/B′, D1, D2, D3, E, F and G (18,19). These Sm proteins, as well as the related Sm-like (Lsm) proteins, contain two conserved amino acid motifs (the Sm motifs 1 and 2) that form a specific tertiary structure, the Sm fold or Sm domain [reviewed in (18)]. Moreover, the seven spliceosomal Sm proteins form a heptameric ring that associates with a conserved U-rich single-stranded sequence in the snRNAs, the so-called Sm-binding site. This assembled ring is called the Sm core structure. In comparison with the spliceosomal snRNAs, U7 snRNA has a non-canonical Sm-binding site (20,21). Moreover, two of the standard Sm proteins, D1 and D2, are replaced in its Sm core structure by U7-specific Lsm proteins, Lsm10 and Lsm11 (15,16). Whereas the 14 kDa Lsm10 protein is a rather common member of the Sm/Lsm protein family, Lsm11 has an unusual architecture. With ∼40 kDa (apparent molecular mass on gels ∼45–50 kDa), it is very large for an Sm/Lsm protein. This extra mass is found in a long N-terminal extension (170 amino acids) before and in a long intervening sequence (138 amino acids) between its two conserved Sm motifs. Interestingly, the N-terminal region is dispensable for the specific incorporation of the protein into the U7 Sm core but essential for histone RNA processing (16). Additionally, we found that this N-terminal region of Lsm11 can bind ZFP100 (16), indicating that the additional tethering of the U7 snRNP is achieved by the formation of a molecular bridge between HBP bound to the histone hairpin, ZFP100, and Lsm11 which is a component of the U7 snRNP [reviewed in (17)].
In the present study, we have more precisely characterized the interaction between ZFP100 and Lsm11. In addition, we wanted to know whether the N-terminus of Lsm11 contributes to histone RNA processing only by its binding to ZFP100, or whether a separate processing function can be identified. Surprisingly, we find that Lsm11 and ZFP100 can engage in not only one but in two distinct mutual interactions. The N-terminus of Lsm11 binds to the zinc finger repeats of ZFP100 but does not interact with an N-terminal part of ZFP100 that is lacking zinc finger repeats. In contrast, this N-terminal region of ZFP100 interacts with full-length (FL) Lsm11, but also with an Lsm11 truncation mutant lacking the N-terminal region or with the unrelated Sm D2 protein. This indicates that this second interaction may be formed with Sm domains in general. In cell extracts, this second interaction is sufficient for a specific recognition between ZFP100 and the U7 snRNP. Moreover, we have introduced clustered point mutations in four evolutionarily conserved amino acid stretches within the N-terminus of Lsm11. One of these mutations strongly abrogates the ability of the protein to participate in histone RNA processing, although the mutated protein can still form an Sm core with U7 snRNA. Mutations in two other motifs lead to a partial loss of processing activity. Interestingly, these mutations do not affect the binding to ZFP100, indicating that the corresponding motifs contribute to histone RNA processing by a different mechanism.
MATERIALS AND METHODS
Plasmids
Murine Lsm11 clones
The following subclones of Lsm11 cDNA in the pcDNA3-HA vector (15) have been described previously (16): HA-mLsm11FL (FL protein), HA-mLsm11ΔN140 (first 140 amino acids deleted) and HA-mLsm11Δsp77 (amino acids 246–322 deleted, i.e. a major part of the spacer between the Sm motifs). The region encoding the first 136 amino acids (up to a SmaI restriction site) was cloned similarly into pcDNA3-HA to yield HA-mLsm11N136.
Clustered point mutations in conserved sequence motifs within the N-terminus of Lsm11 (22) were created by a fusion PCR strategy, resulting in the replacement of the tripeptides PLL, YES, PER or MPL, respectively, by three consecutive alanines. Each of these mutants was recloned into pcDNA3-HA. These mutants were named according to their original sequence (e.g. PLL, YES…).
For bacterial expression, the regions encoding the first 136 amino acids of Lsm11 wild-type (wt) as well as the PLL, YES and PER mutants were cloned into the pGex4T3 vector (Amersham Bioscience). For the MPL mutant and an appropriate wt Lsm11 control, a region corresponding to the first 157 amino acids was amplified by PCR and similarly cloned into pGex4T3.
Human ZFP100 clones
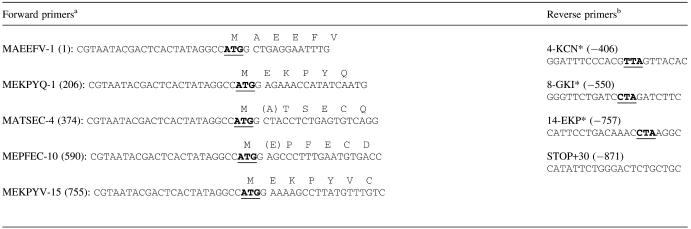
A pSP64 plasmid encoding human ZFP100 (14) (a gift from Z. Dominski, UNC Chapel Hill) was digested with NcoI or NcoI/XhoI, and appropriate restriction fragments were cloned in the pIVEX2.4d vector (Roche) for in vitro translation. Additional ZFP100 fragments to be used for in vitro transcription/translation were generated by PCR with Pfu DNA polymerase and cloned into the pCRII-TOPO vector (Invitrogen). The primers used are shown in Table 1. Each of the upstream primers contained a T7 RNA polymerase promoter and an in-frame start codon (underlined). Downstream primers contained a translation stop codon. The clones were cleaved with Asp718 I or EcoRV, depending on the orientation of the insert, prior to in vitro transcription/translation. For bacterial expression, an NcoI fragment encoding the first 169 amino acids was cloned into pGex4T3.
Table 1.
Primers for PCR-generated deletions of ZFP100
aThe forward primers are named according to the first five amino acids, the first intact zinc finger repeat (after the hyphen) and the first amino acid number (in brackets) of ZFP100 which are encoded by the respective translation product. These primers each contain a T7 RNA polymerase promoter followed by an in-frame start codon (bold underlined). Amino acids shown in brackets were altered with respect to the natural ZFP100 sequence so that the start codon could be followed by a G residue (27).
bThe internal reverse primers are designated with respect to the last intact zinc finger repeat (preceding the hyphen), the last three amino acids preceding the stop codon (asterisk), and the last amino acid of ZFP100 encoded by the respective translation product (in brackets). In-frame stop codons were introduced with the primers (bold underlined). STOP+30 (−871) designates a primer complementary to a sequence downstream of the natural stop codon.
A clone encoding the Kid-1 zinc finger protein (ZFN354a) from the rat in a pET-21a backbone (a gift from E. O'Leary and J. V. Bonventre, Harvard Medical School) was used directly for in vitro transcription/translation.
All clones were verified by DNA sequencing. Details of the constructs are available on request.
Glutathione S-transferase (GST) pull-down assays
To study protein–protein interactions in vitro, recombinant GST (negative control) or GST fusion proteins were isolated from Escherichia coli BL21 Gold transfected with pGex4T3 (negative control) or with pGex4T3-derived plasmids containing either the wt sequence or different clustered point mutations in the context of the Lsm11 N-terminus, or the N-terminus of ZFP100. The purified proteins were coupled to glutathione beads and incubated with [35S]methionine-labelled proteins obtained by coupled in vitro transcription/translation in rabbit reticulocyte lysate (TnT kit; Promega). The in vitro translated proteins were encoded by pcDNA3-HA-derived plasmids for Lsm11 and by pIVEX2.4d- or pCRII-TOPO-derived plasmids for ZFP100.
Each binding reaction contained ∼1.2 nmol of GST (∼36 μg) or GST fusion protein (∼55 μg) and ∼65 fmol of radiolabelled in vitro translated binding substrate (∼3.5–4.1 ng). The beads and proteins were incubated in phosphate-buffered saline (PBS) supplemented with 0.1% NP40 at 4°C for 2 h with gentle agitation on a wheel. Subsequently, the beads were washed with PBS/NP40 and the bound and input materials were analysed by SDS–PAGE and detected on a Storm 820 PhosphorImager (Amersham).
Interaction studies of HA-tagged proteins in 293-T cell extracts
Human 293-T cells were cultured as described previously (15). For transfection, they were grown to 50–60% confluency in 10 cm dishes and transfected with 10 μg of the pcDNA3-HA-derived plasmids complexed with Lipofectamine (Life Technologies). Cells were harvested 48 h post transfection. The preparation of small-scale whole cell extracts, immunoprecipitations and precipitations with biotinylated oligonucleotides complementary to the 5′ ends of U7 or U1 snRNA were performed as described previously (15). In some experiments, the extract from one 10 cm dish (100 μl) was incubated with 100 μg of RNase A (Sigma) for 20 min at 30°C prior to immunoprecipitation. Proteins were resolved on 12% high-TEMED SDS–polyacrylamide gels (23), analysed by western blots with appropriate antibodies (see below) and developed by the enhanced chemiluminescence method (Amersham).
Antibodies
For immunoprecipitation, we used monoclonal antibodies against Sm proteins B/B′, D1 and D3 (Y12) (24) or against the HA tag (Roche). Antibodies to GST-ZFP1–169 were raised in rabbits and affinity-purified by binding to the recombinant protein immobilized on nitrocellulose filters (15). These purified antibodies detect a single ∼100 kDa protein in western blots of whole cell extract from human 293-T cells (data not shown). Sm proteins or ZFP100 were detected in western blots by indirect immunostaining with species-specific antibodies (anti-mouse or anti-rabbit, respectively) coupled to horseradish peroxidase (HRP) (Promega); HA-tagged proteins were detected directly with anti-HA antibody (Roche) coupled to HRP.
GST pull-down assays with HeLa nuclear extract
HeLa cells were harvested by trypsinization and nuclear extracts were prepared as described previously (15). The nuclear extracts were incubated with immobilized recombinant GST (negative control) or GST-ZFP1001–169 (∼1.2 nmol each) for 2 h as described above. The beads were then washed and split into two equal portions. One half of the beads was subjected to 12% SDS–PAGE and western blotting with Y12 anti-Sm and anti-Lsm11 antibodies (16). The other half of the beads was used for RNA extraction at 65°C with a buffer containing 10 mM Tris–HCl, pH 8, 5 mM EDTA, 0.5% SDS, 1 mg proteinase K and 4 μg glycogen. The obtained RNAs were extracted twice with phenol:chloroform:isoamyl alcohol, followed by an ethanol precipitation. Reverse transcriptions were then performed with the StrataScript RT system (Stratagene) and specific 3′ primers for U1, U2 and U7 snRNAs (Table 2). A subsequent PCR was performed with Taq polymerase (Roche) and the specific primers listed in Table 2. RT and PCR conditions were selected for each primer pair and are available on request.
Table 2.
Primers for RT–PCR of U1, U2 and U7 snRNAs
| snRNA | Primer | Sequence |
|---|---|---|
| U1 | hU1 5′ | 5′-ATACTTACCTGGCAGGGGAGATACCATGATCACG |
| hU1 3′ | 5′-CAGGGGAAAGCGCGAACGCAGTCCCCCACTACC | |
| U2 | hU2 5′ | 5′-ATCGCTTCTCGGCCTTTTGGCTAAGATCAAG |
| hU2 3′ | 5′-GTGCACCGTTCCTGGAGGTACTGCAATACC | |
| U7 | hU7 5′ | 5′-GCATAAGCTTAGTGTTACAGCTCTTTTAGAATTTGTC |
| hU7 3′ | 5′-CGTAGAATTCAGGGGCTTTCCGGTAAAAAGCCAG |
Processing experiments
The functionality of Lsm11 mutants in histone RNA 3′ end processing was tested by an assay involving a chimeric histone pre-mRNA/U7 snRNA molecule (called 12/12-U7 RNA) that was injected into Xenopus laevis oocytes (16,25). Capped, unlabelled mRNAs encoding various HA-tagged mLsm11 proteins were transcribed in vitro from linearized plasmids using T7 RNA polymerase and subsequently polyadenylated using yeast poly(A) polymerase (USB Corp./Amersham) according to the manufacturer's instructions. The mRNAs were injected into the oocyte cytoplasm, followed by incubation at 18°C for 20 h to allow for protein expression.
After a second cytoplasmic injection with capped and radiolabelled chimeric 12/12-U7 RNA, the oocytes were further incubated for 3 h. Oocyte extracts were prepared and subjected to immunoprecipitations with anti-Sm (Y12) or anti-HA antibodies. RNA from the pellet fraction was recovered by phenol extraction and analysed on a 12% denaturing polyacrylamide gel. As control, the HA-tagged proteins were analysed for expression and for incorporation into snRNPs by western blotting with anti-HA antibody with or without prior immunoprecipitation with Y12 antibody.
RESULTS
The N-terminus of Lsm11 binds to the zinc finger repeats of ZFP100
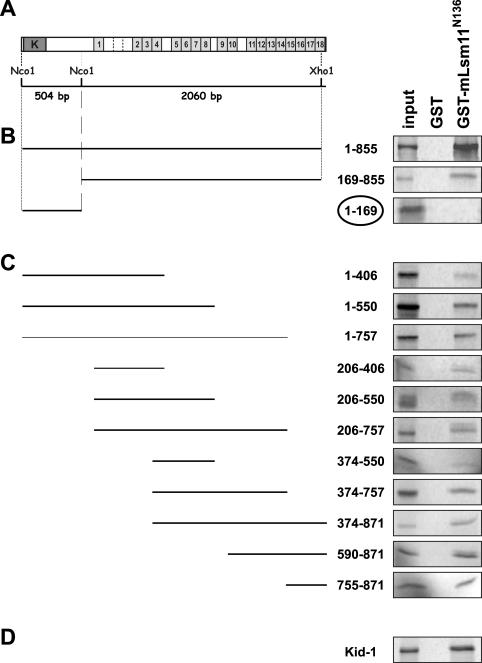
Dominski, Marzluff and co-workers (14) have shown that U7 snRNA can be immunoprecipitated from nuclear extracts by anti-ZFP100 antibodies, suggesting that an interaction between these two components of the histone RNA processing machinery exists in vivo. More specifically, we have demonstrated the ability of the N-terminus of Lsm11 to interact with ZFP100 in vitro (16). Here, we attempted to map the region within ZFP100 that is responsible for this interaction by using the same methodology. In other words, we analysed the precipitation of in vitro translation products corresponding to various parts of ZFP100 (Figure 1A) with either GST-mLsm11N136 (the N-terminal extension of Lsm11 fused to GST) or GST (negative control) which were immobilized on glutathione sepharose beads.
Figure 1.
The N-terminus of Lsm11 (GST-mLsm11N136) binds to the C2H2 zinc finger repeats of ZFP100. (A) Structure of ZFP100. Dark grey box labelled ‘K’, KRAB domain; light grey boxes, C2H2 zinc finger repeats; imperfect repeats are delineated by stippled lines. (B) GST pull-down assays with ZFP1–855, ZFP169–855 and ZFP1–169 encoded by subcloned NcoI and NcoI/XhoI restriction fragments as indicated in (A). (C) GST pull-down assays performed with ZFP100 truncations obtained by PCR (see Materials and Methods). The numbers indicate the ranges of amino acids of FL ZFP100 that are present in the various truncations. (D) GST pull-down assay performed with the C2H2 zinc finger protein Kid-1, a renal transcription factor from rat that is not related to histone RNA processing. All templates were linearized and subjected to coupled in vitro transcription/translation in the presence of [35S]methionine. The translation products were incubated with GST-mLsm11N136 or GST (negative control) immobilized on glutathione sepharose beads. The beads were washed, and the bound material was analysed by SDS–PAGE and autoradiography. Input, 1/10 the amount used in the binding assays was analysed directly. Note that only ZFP1–169 encoding the N-terminus of ZFP100 lacking zinc finger repeats but containing the KRAB domain does not bind to GST-mLsm11N136, whereas all fragments encoding zinc finger repeats bind efficiently.
We initially tested the binding of ZFP100 fragments that had been obtained by restriction fragment subcloning. One of these, the near FL protein ZFP1–855, had already been used in our previous study (16). In agreement with the results obtained there, ZFP1–855 was precipitated by GST-mLsm11N136 but not by GST (Figure 1B). When ZFP1–855 was subdivided into two parts, the N-terminus devoid of zinc fingers but containing a Krüppel-associated box (KRAB) domain (ZFP1–169) and the C-terminal part encompassing all but the last of the zinc fingers (ZFP169–855) behaved differently. The zinc finger-containing fragment ZFP169–855 bound efficiently to GST-mLsm11N136, whereas the N-terminal ZFP1–169 showed no binding. Moreover, none of the proteins showed any binding to GST alone. These results indicated that the binding site of ZFP100 for the Lsm11 N-terminus resides in the zinc finger repeat region.
To locate the binding site for the Lsm11 N-terminus more precisely, we amplified various parts of the ZFP coding sequence using forward primers equipped with T7 promoters and translation start codons and reverse primers introducing premature stop codons or a reverse primer annealing to a sequence downstream of the natural ZFP100 stop codon (Table 1, Figure 1 and Materials and Methods). These PCR products were cloned, the resulting plasmids were subjected to coupled in vitro transcription/translation, and the 35S-labelled translation products were tested for their binding activity. Surprisingly, all these truncated proteins bound to immobilized GST-mLsm11N136 but not to GST, although the binding seemed to be less efficient for some shorter polypeptides, e.g. ZFP374–550 (Figure 1C). This suggested that the binding was not associated with a specific subset of zinc fingers, but that any combination of zinc finger repeats (fragments containing as few as four repeats were tested) could bind to the N-terminus of Lsm11. As a control for specificity, we tested Kid-1 (ZNF354A), a renal transcription factor from the rat which, like ZFP100 (ZNF473), is a member of the large family of KRAB domain-containing C2H2 zinc finger proteins (26). The two proteins share ∼40% amino acid identity and an additional ∼15% similarity over their entire length with the zinc finger regions being somewhat more related. We found that Kid-1 also bound to immobilized GST-mLsm11N136 but not to GST (Figure 1D). In contrast, Sm D2 did not bind to GST-mLsm11N136 (data not shown). Thus, it appears that the N-terminus of Lsm11 has a general affinity for C2H2 zinc finger domains.
A second interaction can be formed between the N-terminus of ZFP100 and proteins containing an Sm domain
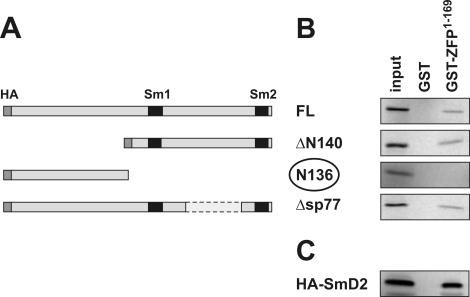
As the N-terminus of ZFP100 showed no binding to the N-terminus of Lsm11, we wanted to know whether it could interact with other parts of Lsm11. For this purpose, GST-ZFP1–169 or GST (negative control) was immobilized on glutathione beads and incubated with in vitro translation products corresponding to various parts of Lsm11 (Figure 2A).
Figure 2.
The N-terminus of ZFP100 binds to the C-terminal Sm domain of Lsm11. (A) Structure of various constructs containing murine Lsm11. An HA-tag introduced at the N-terminus and the Sm motifs are shown in dark grey and black, respectively. (B) GST pull-down assays. The 35S-labelled Lsm11 (FL) and its mutants were generated in reticulocyte lysate and incubated with equal amounts of GST or GST-ZFP100 N-terminal fragment (GST-ZFP1–169). (C) GST pull-down assay with HA-tagged Sm D2. The bound 35S-labelled proteins were analysed by SDS–PAGE and detected by PhosphorImager. Input, 1/10 the amount used in the binding assays. Note that only N136 does not bind to GST-ZFP1–169, whereas all fragments encoding the two Sm motifs bind efficiently.
These experiments indeed revealed that GST-ZFP1–169, but not GST alone, could bind to mLsm11FL (Figure 2B, top panel). In contrast, GST-ZFP1–169 did not bind to the Lsm11 N-terminus (mLsm11N136; third panel), confirming the result of the reciprocal experiment shown in Figure 1B. As expected from this result, GST-ZFP1–169 also bound efficiently to a deletion of Lsm11 lacking the N-terminus (mLsm11Δ140; second panel). Furthermore, deleting 77 amino acids from the spacer separating the two Sm motifs of Lsm11 (mLsm11Δsp77) still allowed for binding to GST-ZFP1–169 (bottom panel).
These results indicated that the C-terminal half of Lsm11 containing the two Sm motifs was the main interaction site for GST-ZFP1–169. We, therefore, wanted to see whether this interaction was specific for Lsm11 or whether it could also occur with another Sm/Lsm protein. Thus, we analysed Sm D2, which is a component of spliceosomal snRNPs but not of U7 snRNPs, for its binding to GST-ZFP1–169. Surprisingly, Sm D2 also efficiently bound to GST-ZFP1–169, but not to GST alone (Figure 2C). This result suggested that the ZFP100 N-terminus primarily binds to Sm domains without discriminating between different Sm/Lsm proteins, at least not between Lsm11 and Sm D2. As we have shown above that the N-terminus of Lsm11 binds to ZFP100's zinc fingers, but not to its N-terminus (Figure 1), this means that the two proteins can make two separate binding contacts.
Four phylogenetically conserved sequences in the N-terminus of Lsm11 are not important for ZFP100 binding
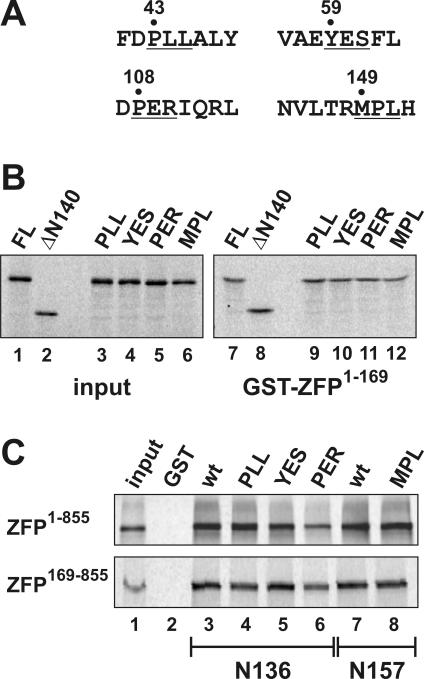
Since the N-terminus of Lsm11 that interacts with the zinc finger repeats of ZFP100 (16) (Figure 1) contains four patches of phylogenetically conserved amino acids (22) (see our Lsm11 database at http://www.izb.unibe.ch/res/schuehome/schuemperli/Lsm11.html), we decided to investigate whether any of these sequences are required for binding to ZFP100. To this end, three consecutive amino acids in each conserved sequence were converted to alanines (Figure 3A) in the context of a vector containing the Lsm11 open reading frame fused in frame to an N-terminal HA tag. We then analysed whether the HA-tagged mutant proteins, when translated in vitro in reticulocyte lysate, could still be precipitated by GST-ZFP1–169 bound to glutathione sepharose beads. Indeed, all four mutant proteins were precipitated as well as the wild-type FL protein (Figure 3B, compare lanes 9–12 with 7). This was an expected finding for three of the mutants, because the mutated sequences were within the region deleted in HA-mLsm11ΔN140, and the latter showed full binding to GST-ZFP1–169 (lane 8). However, the MPL mutant, which is located downstream of amino acid 140, also showed normal binding to the N-terminus of ZFP100 (Figure 3B, lane 12). Thus, we conclude that one of the two interactions between Lsm11 and ZFP100, which links the N-terminus of ZFP100 to the Sm domain of Lsm11, is not affected by any of the clustered point mutations.
Figure 3.
Conserved amino acid sequences in the N-terminus of Lsm11 are not important for binding to ZFP100. (A) Mutagenesis of four phylogenetically conserved amino acid sequences in the N-terminus of Lsm11 (22) (see our Lsm11 database at http://www.izb.unibe.ch/res/schuehome/schuemperli/Lsm11.html). Amino acids that were mutated to alanines in the murine Lsm11 cDNA are underlined and the amino acid position of the first one is indicated. The mutants were named according to the original sequence (i.e. PLL, YES…). (B) The 35S-labelled Lsm11 (FL) and its mutants were generated in reticulocyte lysate and incubated with equal amounts of GST (data not shown, but no signals were obtained) or GST-ZFP100 N-terminal fragment (GST-ZFP1–169). For a description of the Lsm11 FL and ΔN140, see Figure 2A. (C) The 35S-labelled ZFP fragments ZFP1–855 (upper panel) and ZFP169–855 (lower panel) were generated in reticulocyte lysate and incubated with beads loaded with GST (lane 2) or GST-tagged Lsm11 amino acids 1–136 (lanes 3–6) or 1–157 (lanes 7,8) that contained either the wt sequence or the indicated mutations.
Then, we analysed whether the other interaction, taking place between the N-terminus of Lsm11 and the zinc finger region of ZFP100, was affected in any of these mutants. To this end, we cloned N-terminal fragments from each mutant into pGex4T3 to produce and purify GST-tagged proteins in E.coli. For the PLL, YES and PER mutations, the resulting proteins corresponded in size to GST-mLsm11N136. However, as the MPL site is located downstream from amino acid 136, we cloned the MPL mutation and an equivalent wt control as a fragment corresponding to amino acids 1–157. All these GST-tagged proteins were used in GST pull-down assays to analyse the binding of in vitro translated ZFP1–855 or ZFP169–855.
The results of these binding assays indicated no significant reduction with three of the mutants (Figure 3C, lanes 4, 5 and 8). In contrast, the binding of the PER > AAA mutant to both ZFP1–855 and ZFP169–855 was slightly reduced (lane 6). However, additional experiments in which the amounts of GST fusion proteins recovered after the precipitation and washing steps were monitored by Ponceau red staining indicated that a lower amount of PER mutant protein had been used, although identical amounts based on Lowry assay results had been pipetted (data not shown). Thus, we conclude that none of the four mutants significantly affects either type of binding to ZFP100.
The failure to detect an effect of these mutations on the interaction with ZFP100 could have been due to the binding conditions used to perform the binding assays (see Materials and Methods), in particular the large excess of GST-tagged N-termini of Lsm11 compared with the in vitro translated proteins. However, when the binding assays presented in Figure 3C were repeated with a smaller (40-fold) molar excess of GST-tagged proteins, identical results were obtained (data not shown). Moreover, experiments shown in Figure 4B below demonstrate that the four mutant proteins, after in vivo expression, still interact with ZFP100.
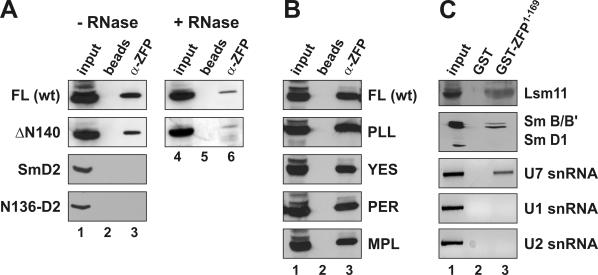
Figure 4.
Determinants of ZFP100–U7 snRNP interactions in mammalian cell extracts. (A) The various HA-tagged proteins indicated on the left were expressed in human 293-T cells by transient transfection. Whole cell extracts were subjected to immunoprecipitation with affinity-purified antibodies directed against the N-terminal 169 amino acids of human ZFP100 (α-ZFP). The samples were subjected to SDS–PAGE, blotted, and the HA-tagged proteins were revealed by anti-HA antibody. For the two top panels, the extracts were incubated in the absence (lanes 1–3) or presence of RNase A (lanes 4–6) prior to immunoprecipitation (see Materials and Methods). The samples for each protein were processed in the same experiment and analysed on the same gel, but an intermediate lane was excised from the picture. FL, full-length murine Lsm11 (HA-mLsm11FL); ΔN140, mLsm11 lacking the first 140 amino acids (HA-mLsm11Δ140); N136-D2, first 136 amino acids of mLsm11 fused to Sm D2 (16). Beads, precipitation by protein G sepharose beads without antibody; input, 1/20 of original extract. (B) Wild-type HA-tagged Lsm11 and the clustered point mutants indicated on the right (for their sequence see Figure 3) were expressed in human 293-T cells and their ability to interact with ZFP100 was assessed by immunoprecipitation with α-ZFP as in (A). (C) Nuclear extract from HeLa cells was subjected to precipitation with glutathione beads coupled to GST-ZFP1–169 (lane 3) or GST alone (lane 2) and analysed for several snRNP components as indicated on the right. Lsm11 was detected by affinity-purified anti-Lsm11 antibodies (16); Sm B/B′ and D1 by the monoclonal anti-Sm antibody Y12; and snRNAs U1, U2 and U7 by RT–PCR as described in Materials and Methods. Input, 1/10 of original extract.
Determinants of ZFP100–U7 snRNP interactions in cellular extracts
Several of the above observations motivated us to further analyse these interactions in cellular extracts. In particular, we wanted to determine whether both interactions play a role for the recognition between ZFP100 and the U7 snRNP. For example, it was unclear whether the N-terminus of ZFP100 could only recognize monomeric Sm domains or whether it could bind to fully assembled Sm/Lsm rings. Moreover, it was important to investigate whether the apparent promiscuity for other proteins containing similar domains (Sm or C2H2 zinc finger domains, respectively) that had been observed under these in vitro conditions could also be seen in cellular extracts.
Thus, we expressed different HA-tagged versions of Lsm11 or other Sm proteins in 293-T cells by transient transfection and analysed the ability of anti-ZFP100 antibodies to co-immunoprecipitate these HA-tagged proteins from whole cell extracts. The full-length HA-mLsm11FL was readily precipitated by the anti-ZFP100 antibodies (Figure 4A, top panel, lane 3) (data not shown), whereas protein G sepharose beads either alone (lane 2) or coupled to unrelated antibodies (data not shown) did not precipitate HA-mLsm11FL. This confirmed previous findings of Dominski, Marzluff and co-workers (14) who showed that a different anti-ZFP100 antibody could co-precipitate U7 snRNA from HeLa cell nuclear extract. We similarly expressed and analysed HA-tagged mLsm11Δ140, which should be able to form only one of the two interactions characterized above. Despite the fact that this protein lacks the N-terminus and is non-functional in histone RNA processing [see below and (16)], it still gets incorporated into U7 snRNPs in vivo (16). Interestingly, HA-mLsm11Δ140 was also precipitated by the anti-ZFP100 antibodies (Figure 4A, second panel, lane 3).
Since the interactions in these extracts could be mediated by the base pairing between histone pre-mRNA and U7 snRNA, we also analysed the precipitation of the same two HA-tagged proteins from extracts that had been treated with RNase A. For both proteins, the precipitation by the anti-ZFP100 antibodies was less efficient than in extract incubated in the absence of RNase A (Figure 4A, compare lanes 6–3). This suggested that the interaction was strongly enhanced by RNA, presumably by the histone pre-mRNA and U7 snRNA. Nevertheless, both proteins still showed a residual co-precipitation with ZFP100. In particular, the residual co-precipitation of HA-mLsm11Δ140 suggested that the interaction between the N-terminus of ZFP100 and the Sm domain of Lsm11 (and possibly other Sm/Lsm proteins) can occur in the context of a fully assembled snRNP and may contribute to the binding between ZFP100 and the U7 snRNP in cell extracts.
To investigate the specificity of these interactions, we analysed HA-tagged versions of either Sm D2 or of a fusion between the N-terminus of Lsm11 and D2 (N136-D2) by the same assay. Both of these proteins associate with U1, but not with U7, snRNA in vivo (16). Neither of these proteins could be precipitated by anti-ZFP100 antibodies (Figure 4A, two lower panels). This indicated that ZFP100, in extracts, specifically interacts with U7 snRNPs containing either full-length or N-terminally truncated Lsm11, but not with spliceosomal snRNPs, even if these contain Sm D2 fused to the Lsm11 N-terminus. This latter finding indicated that the interaction between the zinc finger repeats of ZFP100 and the N-terminus of Lsm11 may be relatively inefficient in such extracts or may require additional determinants on the U7 snRNP.
We also wanted to confirm by an independent approach that the clustered point mutations in the conserved N-terminal regions of Lsm11 introduced in Figure 3 above do not affect the binding to ZFP100. We, therefore, expressed wild-type HA-Lsm11 and all four mutant proteins by transient transfection of 293-T cells and analysed the ability of anti-ZFP100 antibodies to co-immunoprecipitate these HA-tagged proteins. As shown in Figure 4B, none of the mutants had a significant effect on the interaction of Lsm11 with ZFP100 as detected by this binding assay.
As a further test for the interactions between Lsm11 and ZFP100, we subjected HeLa cell nuclear extracts to GST pull-down assays. With GST-mLsm11N136-coated glutathione beads, only trace amounts of ZFP100 could be precipitated from the extract, barely more than with beads coupled to GST alone (data not shown). In contrast, GST-ZFP1–169-coated glutathione beads efficiently precipitated Lsm11 (Figure 4B, top panel, lane 3) and Sm B/B′, but not Sm D1 from the extracts (second panel, lane 3), indicating that U7 snRNPs, but not spliceosomal snRNPs, were being precipitated. No precipitation was seen with beads coupled to GST (top two panels, lane 2). RT–PCR assays performed on RNA isolated from the precipitates indicated that U7, but not U1 or U2, snRNA was being precipitated by the GST-ZFP1–169-coated beads (Figure 4B, lower three panels). These results are consistent with the above co-immunoprecipitation experiments in demonstrating that the N-terminus of ZFP100 can specifically bind to the U7 snRNP in cell extracts.
Three of the phylogenetically conserved N-terminal motifs of Lsm11 are important for histone RNA processing
Since the N-terminus of Lsm11, in addition to its interaction with ZFP100, fulfils an essential function in histone RNA 3′ end processing (16), and it contains the four patches of phylogenetically conserved amino acids described above, we decided to investigate whether any of these sequences are required for processing. To this end, capped and polyadenylated mRNAs encoding full-length, HA-tagged Lsm11 with either the wt sequence or with a triple alanine mutation in one of the conserved motifs (see Figure 3A) were synthesized in vitro and injected into Xenopus oocytes. After overnight incubation to allow for translation, a capped, radiolabelled chimeric RNA consisting of a histone pre-mRNA sequence fused to a U7 snRNA moiety (25) (Figure 5A) was injected into the same oocytes. Three hours later, oocyte extracts were prepared, the HA-tagged proteins were precipitated and the associated chimeric RNA was analysed by denaturing gel electrophoresis.
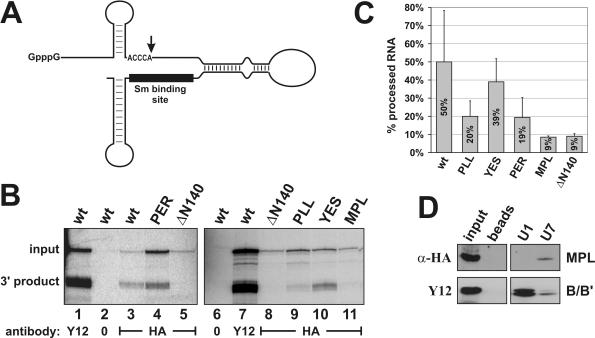
Figure 5.
Three conserved amino acid sequences in the N-terminus of Lsm11 are important for histone RNA 3′ end processing. (A) The chimeric histone–U7 RNA [12/12-U7 RNA (25)] used in (B) contains 49 nt of histone pre-mRNA upstream and 36 nt downstream of the cleavage site (vertical arrow), a connector segment of 28 and 65 nt of U7 RNA sequence. The Sm-binding site is indicated by a black bar. (B) Processing of chimeric histone–U7 RNA in Xenopus oocytes. Synthetic mRNAs encoding HA-tagged versions of mLsm11FL (wt) or of the four mutants (see Figure 3A) were injected into the cytoplasm of X.laevis oocytes. RNA encoding HA-tagged Lsm11 lacking the first 140 amino acids (ΔN140) was injected as a processing-deficient control (16). After overnight incubation to allow for translation of the recombinant proteins, the oocytes were challenged with radiolabelled, chimeric histone–U7 RNA [see (A)]. In the oocytes, this RNA gets cleaved at the histone RNA processing site, dependent upon assembly of a functional Sm/Lsm core at its Sm-binding site. Oocyte extracts were subjected to immunoprecipitation with either Y12 anti-Sm or anti-HA antibodies to enrich for total snRNPs or for particles containing HA-tagged Lsm11, respectively. The radiolabelled RNA was analysed by denaturing polyacrylamide gel electrophoresis and autoradiography. Lanes 2 and 6, HA-mLsm11FL extract (wt) precipitated with beads lacking antibody. (C) Quantitation of the ratios of 3′ processing product:total chimeric RNA determined by PhosphorImager scanning. The results represent averages of 2–3 independent determinations for each construct. Note that the MPL mutant and the N-terminal deletion show a strong defect in processing, but still associate with the chimeric RNA. The PLL and PER mutants are partly deficient in processing. (D) The MPL mutant protein is associated with U7 snRNA in mammalian cell extracts. Whole cell extract from human 293-T cells transiently transfected with the plasmid encoding HA-mLsm11MPL was incubated with biotinylated oligonucleotides complementary to the 5′ ends of either U7 or U1 snRNA and precipitated with magnetic streptavidin beads. The samples were subjected to SDS–PAGE and immunoblotted with anti-HA antibody to detect the HA-mLsm11MPL protein and with Y12 antibody to detect the Sm B/B′ protein as a precipitation control. Beads, precipitation by beads without oligonucleotide; input, 1/10 of original extract. The two panels are taken from the same gel, but an intermediate lane was excised from the picture.
The rationale of this assay is that processing of the chimeric RNA depends on the assembly of a U7-specific Sm/Lsm core at the Sm-binding site of the U7 snRNA moiety (16,25). If the RNA associated with a given HA-tagged Lsm11 protein has been cleaved in the oocyte, this means that the protein is functional in histone RNA processing, because all the other proteins of the processing complex will be wt Xenopus proteins.
In agreement with our previous experiments (16), wild-type HA-mLsm11FL formed RNPs with the chimeric RNA that were functional in histone RNA processing (Figure 5B, lane 3), whereas the HA-mLsm11ΔN140 mutant with most of the N-terminal extension deleted was non-functional (lanes 5 and 8). Of the four clustered point mutations, the MPL mutant (lane 11) was similarly deficient in histone RNA processing as the N-terminal deletion. Moreover, the PLL (lane 9) and PER (lane 4) mutants showed a reduced processing efficiency. In contrast, the YES mutant (lane 10) supported processing of the chimeric RNA with a similar efficiency as the wt protein. A quantitation of the results averaged over 2–3 independent assays for each of these proteins is shown in Figure 5C.
Notably, the precipitated HA-mLsm11MPL mutant protein was associated with uncleaved chimeric RNA (Figure 5B, lane 11), indicating that although it was deficient in histone RNA processing, it could still form an RNP complex along with the other six members of the U7-specific Sm core. To analyse this more directly, HA-mLsm11MPL was transiently transfected into human 293-T cells. A whole cell extract from these cells was then precipitated with magnetic streptavidin beads after the addition of biotinylated oligonucleotides complementary to either U7 or U1 snRNAs or without oligonucleotide (as a negative control). Indeed, HA-mLsm11MPL was found to be associated with U7, but not with U1 snRNA or beads alone, in a complex with SmB/B′ (Figure 5D). Thus, it behaved similarly as the wild-type HA-mLsm11FL protein and different truncation mutants that contain an intact C-terminal Sm domain (16). Taken together, this means that the MPL > AAA mutation causes a complete loss of processing activity, but does not affect the assembly of the protein into the U7 snRNP. In addition, the PLL > AAA and PER > AAA mutations cause a partial loss of processing activity.
DISCUSSION
Characterization and relative importance of two separate interactions between Lsm11 and ZFP100
One of the aims of this study was to characterize the interaction between the N-terminus of Lsm11 and ZFP100 that we had recently discovered (16). Now, our in vitro binding analyses have revealed that the interaction between these two proteins is more complex than anticipated. On the one hand, we could show that this interaction between the N-terminus of Lsm11 and ZFP100 is directed towards the latter's zinc finger repeats, but cannot be pinpointed to a specific set of zinc fingers (Figure 1). Rather, the fact that an interaction can also be observed with the unrelated Kid-1 protein suggests that the binding is directed towards C2H2 zinc finger repeats in general. Thus, within ZFP100, the various zinc finger repeats can be regarded as redundant binding sites for the N-terminus of Lsm11. Interestingly, the HBP/hairpin complex also interacts with this region of the protein. Dominski, Marzluff and co-workers (14) have localized the region of ZFP100 that efficiently interacts with the HBP/hairpin complex to the zinc fingers 2–8 (see Figure 6). Additionally, certain shorter truncations of this region had a reduced binding activity. If their mapping is correct, then the zinc finger repeats 2–8 may not be able to interact with Lsm11 in the context of a HBP–ZFP100–U7 snRNP complex.
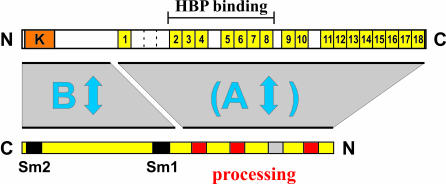
Figure 6.
Model depicting the interactions between Lsm11 and ZFP100 characterized in this paper. To illustrate the interactions, Lsm11 (bottom) has been drawn in the opposite orientation relative to ZFP100 (top). An interaction between the zinc finger repeats 2–8 of ZFP100 and the HBP–histone RNA hairpin complex was previously characterized by Dominski, Marzluff and co-workers (14). The three conserved motifs in the N-terminus of Lsm11, which are important for histone RNA processing (PLL, PER and MPL), are shown in red, the fourth motif (YES) in grey.
On the other hand, we show here that the N-terminus of ZFP100, which lacks zinc finger domains, can also bind Lsm11 (Figure 2). This region of ZFP100 interacts with the Sm domain but not with the N-terminus of Lsm11. Moreover, this binding is not specific for Lsm11, because Sm D2, an Sm protein that does not normally occur in the U7 snRNP (15,16), can also bind to the ZFP100 N-terminus (Figure 2C).
Thus, we are faced with the fact that these two proteins involved in histone RNA processing can undergo not only one but two interactions, and that neither of them is completely specific for the respective binding partner. To simplify the following discussion, we will henceforth call the first interaction A and the second one B (Figure 6).
Unfortunately, it was technically not possible to determine relative affinities for these interactions. Despite intensive efforts, any recombinant proteins containing either the zinc fingers of ZFP100 or the Sm domain of Lsm11 were insoluble, so that we could only work with the small amounts of protein that could be translated in vitro. This meant that all in vitro binding reactions were carried out with an excess of either GST-mLsm11N136 or GST-ZFP1–169, both of which could be obtained as soluble proteins in E.coli. In order to gain more insight into the relevance of these interactions, we therefore investigated whether they could be observed in cell extracts and whether, under these conditions, they would also be relatively non-specific for the respective binding partner. Moreover, for the interaction B, this approach helped us determine whether the ZFP100 N-terminus can recognize Sm domains in the context of fully assembled Sm/Lsm cores or whether it binds to them only as individual subunits.
No strong evidence that the interaction A occurs in cell extracts could be obtained. A precipitation of HeLa nuclear extract with GST-mLsm11N136-coated glutathione beads contained only very low amounts of ZFP, barely more than was precipitated by GST alone (data not shown). In contrast, GST-ZFP1–169-coated glutathione beads efficiently precipitated U7 snRNPs, but not spliceosomal ones, from the nuclear extract (Figure 4B). This experiment proved that the N-terminus of ZFP100 that binds to Sm domains can do so in the context of fully assembled Sm/Lsm cores and, moreover, specifically recognizes the U7 snRNP in nuclear extracts. We also observed a specific recognition of the U7 snRNP by ZFP100 in our co-immunoprecipitation experiments with anti-ZFP100 antibody (Figure 4A) in agreement with previous results by Marzluff, Dominski and co-workers (14). Our further findings with this immunoprecipitation assay indicated that this interaction does not require the presence of the N-terminus of Lsm11 and that spliceosomal snRNPs containing a fusion between the N-terminus of Lsm11 and Sm D2 are not recognized. Although the interaction between ZFP100 and the U7 snRNP detected in these experiments was strongly stimulated by RNA (presumably reflecting the U7 snRNA:histone pre-mRNA interaction), a weaker binding persisted even after an RNAse treatment, indicating that the interaction between ZFP100 and the Sm core of the U7 snRNP does play a role under these conditions.
All these results strongly suggest that it is primarily the interaction B, occurring between the N-terminus of ZFP100 and the Sm domain of Lsm11 and possibly other Sm/Lsm proteins, which is responsible for the recognition of the U7 snRNP and hence for stabilizing the base pairing between the 5′ end of U7 snRNA and the histone HDE (Figure 6). This leaves us with the question whether the previously identified interaction A is at all relevant for the processing reaction. It is apparently not affected by mutations in the most strongly conserved amino acid sequence motifs in the Lsm11 N-terminus (Figure 3; see below for a more detailed discussion of the mutants). However, the interaction A might still play a role, e.g. if the processing complex undergoes rearrangements during the processing reaction.
It is not evident from our experiments why the interaction B, which does not discriminate between the Sm domains of Lsm11 and Sm D2 in vitro (Figure 2), is specific for the U7 snRNP in nuclear extract (Figure 4B). Since we could not measure relative in vitro binding affinities, we cannot exclude that the ZFP100 N-terminus may have a higher affinity for Lsm11 and/or Lsm10 than for the standard Sm proteins, even though it can bind to Sm D2 when present in excess. Another possible explanation for this additional specificity in cell extracts is that the ZFP100 N-terminus may recognize additional features of the U7 snRNP, e.g. certain determinants on U7 snRNA.
Strong evidence for a ZFP100-independent function of Lsm11 in histone RNA 3′ end processing
The Lsm11 proteins from various species contain four conserved amino acid motifs in their N-terminal regions (22). To study the function of these phylogenetically conserved sequences, we have analysed clustered point mutations in each of them for their effects on histone RNA processing and on the binding of the mutated proteins to ZFP100.
Three of these mutations (MPL, PLL and PER to AAA) reduced the ability of the corresponding proteins to support histone RNA 3′ processing (Figure 5). Nevertheless, these mutant proteins, and in particular the strongly deficient MPL mutant, still associated with chimeric histone–U7 RNA in Xenopus oocytes or with U7 snRNA in mammalian cells. Thus, the formation of a U7-specific Sm/Lsm core is not affected by these mutations, but the mutant proteins indeed function less well in processing.
Similar clustered point mutations in the other conserved motif (YES to AAA) had no phenotype in any of the assays used in this work. It is possible that more extensive mutagenesis could have revealed phenotypic changes for this motif as well. Alternatively, it may have important functions in other aspects of U7 snRNP biology that were not covered by our assays, e.g. during U7 snRNP biogenesis or subcellular localization.
As a first step to elucidate the roles of these important (FDPLLALY and DPERIQRL) and essential (NVLTRMPLH) amino acid sequence motifs, we tested whether the mutant proteins were impaired in their ability to undergo either of the two interactions with ZFP100 characterized in this study. This was clearly not the case under our in vitro binding conditions (Figure 3), but, for the reasons explained above, affinity changes caused by the mutations can not be excluded.
However, another argument indicates that the processing defect of these mutant proteins is not due to an impaired interaction with ZFP100. The processing assay used in this study is not dependent on a stabilization of the U7 snRNA:HDE interaction by HBP and ZFP100. Since the two RNA moieties are covalently linked, the base pairing between the HDE and the U7 moiety is thermodynamically favoured. Moreover, as the particular histone pre-mRNA sequence used can form 16 bp (including G–U wobble pairs) with U7 snRNA (9), the hybrid should be relatively stable at the temperature of 18°C at which the oocytes are incubated. Consistent with this, we have previously found that the processing is not affected when the histone hairpin of the chimeric RNA is mutated such that it cannot bind to HBP (B. Stefanovic and D. Schümperli, unpublished data). Thus, we can indeed conclude that the clustered point mutations affect a processing function of Lsm11 that is independent of its interaction with ZFP100.
At present, it can only be speculated what this ZFP100-independent function of these motifs might be. Perhaps they interact directly with the processing substrate or with another, yet unidentified, processing factor. The availability of the processing-deficient mutant proteins will hopefully allow us to further elucidate this essential function of Lsm11 and the mechanism of histone pre-mRNA cleavage.
Acknowledgments
The authors thank Ramesh S. Pillai for contributing several of the plasmids used in this work, Michael D. Hebert and Oliver Mühlemann for critical comments, and Karin Schranz for technical help. This work was supported by the State of Bern and by Swiss National Science Foundation grant 31-65225.01 to D.S. Funding to pay the Open Access publication charges for this article was provided by Swiss National Science Foundation grants 31-65225.01 and 3100A0-105547 to D.S.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schümperli D. Multilevel regulation of replication-dependent histone genes. Trends Genet. 1988;4:187–191. doi: 10.1016/0168-9525(88)90074-1. [DOI] [PubMed] [Google Scholar]
- 2.Müller B., Schümperli D. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 1997;8:567–576. doi: 10.1006/scdb.1997.0182. [DOI] [PubMed] [Google Scholar]
- 3.Marzluff W.F., Duronio R.J. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 4.Harris M.E., Böhni R., Schneiderman M.H., Ramamurthy L., Schümperli D., Marzluff W.F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitfield M.L., Zheng L.X., Baldwin A., Ohta T., Hurt M.M., Marzluff W.F. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin F., Schaller A., Eglite S., Schümperli D., Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;15:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z.F., Whitfield M.L., Ingledue T.C., Dominski Z., Marzluff W.F. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 8.Streit A., Wittop Koning T.H., Soldati D., Melin L., Schümperli D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993;21:1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spycher C., Streit A., Stefanovic B., Albrecht D., Wittop Koning T.H., Schümperli D. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski Z., Zheng L.X., Sanchez R., Marzluff W.F. Stem–loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaufele F., Gilmartin G.M., Bannwarth W., Birnstiel M.L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986;323:777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- 12.Bond U.M., Yario T.A., Steitz J.A. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 13.Melin L., Soldati D., Mital R., Streit A., Schümperli D. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominski Z., Erkmann J.A., Yang X., Sanchez R., Marzluff W.F. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem–loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 2002;16:58–71. doi: 10.1101/gad.932302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pillai R.S., Will C.L., Lührmann R., Schümperli D., Müller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillai R.S., Grimmler M., Meister G., Will C.L., Lührmann R., Fischer U., Schümperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schümperli D., Pillai R.S. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol. Life Sci. 2004;60:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kambach C., Walke S., Nagai K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 1999;9:222–230. doi: 10.1016/s0959-440x(99)80032-3. [DOI] [PubMed] [Google Scholar]
- 19.Will C.L., Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 20.Grimm C., Stefanovic B., Schümperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefanovic B., Hackl W., Lührmann R., Schümperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azzouz T.N., Schümperli D. Evolutionary conservation of the U7 small nuclear ribonucleoprotein in Drosophila melanogaster. RNA. 2003;9:1532–1541. doi: 10.1261/rna.5143303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Will C.L., Kastner B., Lührmann R. Analysis of ribonucleoprotein interactions. In: Higgins S.J., Hames B.D., editors. RNA Processing. Vol. 1. Oxford: Oxford University Press; 1994. pp. 141–177. [Google Scholar]
- 24.Lerner E.A., Lerner M.R., Janeway C.A., Steitz J.A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stefanovic B., Wittop Koning T.H., Schümperli D. A synthetic histone pre-mRNA-U7 small nuclear RNA chimera undergoing cis cleavage in the cytoplasm of Xenopus oocytes. Nucleic Acids Res. 1995;23:3152–3160. doi: 10.1093/nar/23.16.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witzgall R., O'Leary E., Gessner R., Ouellette A.J., Bonventre J.V. Kid-1, a putative renal transcription factor: regulation during ontogeny and in response to ischemia and toxic injury. Mol. Cell. Biol. 1993;13:1933–1942. doi: 10.1128/mcb.13.3.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981;9:5233–5262. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]