Abstract
Methods for determining protein–protein interactions in mammalian cells typically rely on single reporter functions and are susceptible to variations between samples particularly in regard to levels of transcription, processing and translation. A method has been developed for determining protein–protein interactions in mammalian cells, which bypasses these variables confounding single reporter assays. The approach utilizes two units of gene expression linked to reporter functions that are interposed by a deactivation–activation unit in such a way that the downstream expression unit is switched off. Hence upstream expression occurs regardless of protein–protein interaction, leading to the production of the upstream reporter. In the event of protein–protein interactions, the downstream expression unit is switched on leading to dual reporter read outs. Thus, the ratio of the two reporter activities provides a measure to determine the efficiency of protein–protein interactions. To access the system we screened a mutant of BMPR2 where the interaction between BMPR-II and LIMK is abrogated. BMPR-II is a type II receptor of the TGFβ superfamily and plays a key role in the pathogenesis of familial pulmonary arterial hypertension. This system has potential for high-throughput screening of libraries (peptide, chemical, cDNA, etc.) to isolate agents that are capable of interfering with highly selective protein–protein interaction.
INTRODUCTION
Protein–protein interactions are common in most biological processes in cells. Perturbed interactions contribute to the development of many pathological states (1–4). Elucidation of protein–protein interactions contributes to the characterization of the function of novel proteins and hence the genes that encode them. Methods for investigation of protein–protein interactions include biophysical, computational, biochemical and genetic approaches. For the identification of multi-protein complexes and to determine their association and dissociation rates together with sites of interactions, methods based on surface plasmon resonance (SPR) (5) and different types of mass spectrometry (MS) have been employed (6–8). These methods are proven to be useful, although purification, sequencing and identification of novel proteins can be limiting especially when present only in small quantities. Computational methods based on various principles, including correlated changes of amino acid sequence between interacting protein domains (9), properties related to interface topology, solvent accessible area (ASA) (10,11) that estimates sites of interaction and primary structure and associated physiochemical properties (12), each can predict interactions. While these predictive methodologies are informative in so much as they provide an indication of the affinity of a given protein for another protein, they all require further experimental validation.
One of the most commonly used methods to determine novel protein–protein interactions is the yeast two-hybrid system (13). This system exploits the fact that transcription factors are comprised of two functional domains, a DNA binding domain and a transcription activation domain. The DNA binding domain recognizes a specific DNA sequence whilst the activation domain facilitates the recruitment of Polymerase II associated transcription complex and initiates transcription of the downstream gene. In this system the protein domains are separated from each other until brought together by two interacting proteins. This system has been widely used to screen pray-expression libraries for proteins that interact with a bait protein. The system is prone to contamination by false positives (interactions that are difficult to validate) and false negatives (interactions that are not detected). Bait proteins that alone activate or repress the expression of the reporter gene can also prove problematic.
To counter a number of inefficiencies associated with the system, alternative approaches have been developed. One such example is the split-ubiquitin system (14). Ubiquitin, a small protein, is necessary for proteosome degradation. Proteins of interest are fused to C- and N-terminal domains of ubiquitin. In the event of protein–protein interactions an active ubiquitin (‘split-ubiquitin’) is reconstituted. Ubiquitin is recognized by ubiquitin specific protease, which leads to the release of reporter protein. Additional technologies, based on the yeast two-hybrid screen are being developed all the time. These include systems based on signalling (15) and dual-bait (16). Each of these modifications has a major limitation as many mammalian proteins are exposed to significant post-translational modifications, important in protein–protein complex formation and function.
In an attempt to overcome these difficulties mammalian versions of the two-hybrid screen have been developed (17–20) but these have been single reporter functions and are hence susceptible to variation between samples of levels of transfection and transcription. We have developed a dual reporter assay system based on yeast two-hybrid screen, which comprised two autonomous units of gene expression. The upstream unit is expressed regardless of protein–protein interactions. In the absence of interacting proteins the downstream unit is switched off. In the event of an interaction, the downstream expression unit is also activated and hence both reporter proteins are produced. Thus, the ratio of the two reporter activities can be used as a reference value to detect mutations or trans-acting factors that might affect protein–protein interactions.
MATERIALS AND METHODS
Plasmid construction
Construction of the reporter plasmids (pTN114 and pTN110) was based on pBLUGA (21), which contains reading frames for the β-galactosidase and luciferase genes. To construct pTN114, a deactivation–activation (D/A) unit was cloned into XhoI/SalI and BglII/BamHI sites of pBPLUGA. The unit comprises a translation termination signal for the upstream reporter followed by a 3′ end trimming and polyA tail addition site (AATAAA), an upstream activation sequence of Saccharomyces cerevisiae (22) linked to a TATA box sequence from adenovirus E1b minimal promoter (23), six copies of binding sites for GAL4 transcription factor (24) and a synthetic mRNA splicing signal for effective expression of the downstream reporter, a 5′-UTR and an in-frame start codon for the downstream reporter. A DNA fragment containing the activation sequences, TATA box, GAL4 binding site and the splicing signal that comprised nucleotides 1–535 of pGAL/lacZ (Invitrogen) was amplified using UASF(Xho) 5′-CCGCTCGAGGGTGAAATAAAGTCGACCCGAGCTCTTACGCGGG-3′ and UASR(BglII) 5′-GAAAGATCTTGCCATGTCTTCGATCTGCAGAATTCC-3′. For cloning purposes the internal XhoI site was replaced by a BamHI site using site directed mutagenesis (25). The plasmid pTN110 was constructed by cloning the XhoI–BamHI fragment that comprised nucleotides 1–202 into SalI and BamHI sites of pBPLUGA. The resulting plasmid did not contain the polyadenylation site and the splicing signal.
The construct pTN111 was made by cloning the Gal4 DNA binding domain (residues 1–147) (26) with a N-terminal T7 epitope tag into AflII and KpnI sites of pcDNA3.1 (Invitrogen). The coding region of LIMK1 gene (NM_002314) was sub-cloned into KpnI and XbaI sites of pTN111. Construct pTN112 was constructed by cloning the VP16 transcriptional activation domain (27) into XbaI and ApaI sites of pcDNA3.1 (Invitrogen). A translation termination site was incorporated at the end of VP16 activation domain. The coding sequences of BMPR-II [NM_001204 and NM_033346 for long (LF) and short forms (SF), respectively] and D485G mutant were cloned into BamHI and EcoRI sites of pTN112.
The dual fluorescence reporter was constructed by cloning the deactivation–activation unit into XhoI and BamHI sites of pDsRed-TN24-GFP (Siskoglou and Nasim, unpublished data). This vector was similar to pTN24 (28) except that the genes encoding β-galactosidase and luciferase were replaced by genes encode for fluorescence proteins and the SV40 promoter was replaced by the CMV promoter. The D/A unit is similar to that used in pTN114 except that an additional SV40 polyA site comprising nucleotides 1498–1648 of the DsRed-Express-C1 (Clonetech) was introduced at the 5′ end of the upstream activation sequence.
Gene transfer, cell culture, enzymatic assay, fluorescence microscopy and flow cytometry
HEK-293 (human embryonic kidney) cells were transiently transfected with relevant plasmids using Gene Jammer (Stratagene). Cells were harvested 48 h after transfection and β-galactosidase and luciferase activities were measured using Dual Light System (Applied Biosystems) as described elsewhere (28,29). For live cell imaging, cells were grown as before and the images were taken under a fluorescence microscope (TE 300, Nikon). The images were analysed using Openlab software (Improvision). For single-cell expression, cells expressing both green and red fluorescences were selected using a FACScan flow cytometer (Becton Dickinson) and the fluorescence intensities were measured using Cellquest software.
RESULTS
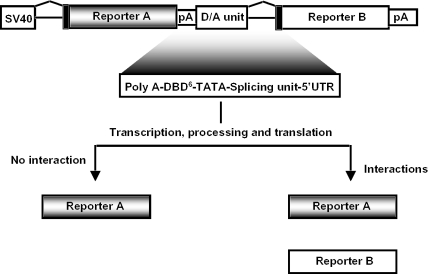
The general principle of the method is outlined in Figure 1. We have constructed a set of reporter constructs in which genes encoding β-galactosidase and luciferase are fused with a recombinant fragment containing the translation termination signal for the upstream reporter, a polyadenylation site, an upstream activation sequence linked to a TATA box sequence from adenovirus E1b minimal promoter, six copies of binding sites for GAL4 transcription factor, a pre-mRNA splicing signal, a 5′-untranslated region (5′-UTR) and in-frame start codon for the downstream reporter. Upon transfection into the mammalian cells, transcription from the SV40 promoter leads to the production of a pre-mRNA which contains a translation termination signal and a polyadenylation signal. Efficient processing and termination (transcription and translation) would result in the production of β-galactosidase protein. In the event of protein–protein interactions, transcription of the luciferase gene would be activated, the mRNA would be exported to the cytoplasm resulting in the production of luciferase protein. Since luciferase activity would be produced only after protein–protein interactions, whereas β-galactosidase is expressed constantly, the ratio of luciferase and β-galactosidase activities would indicate the proportion of cytoplasmic RNA derived from protein–protein interactions.
Figure 1.
The dual-light reporter system for determining protein–protein interactions is based on two reporter genes, which are fused via a recombinant fragment containing a synthetic deactivaion/activation (D/A) unit. The unit comprises polyadenylation signal(s), six copies of Gal4 DNA binding site, a TATA box, a synthetic splicing unit. Upon transfection into mammalian cells the reporter A is transcribed under the control of SV40 promoter, while transcription of the reporter B is switched off. In the event of protein–protein interactions, transcription of the reporter B is activated and both reporters are expressed. 5′-UTRs are indicated by filled bars while the pA denotes polyadenylation signals.
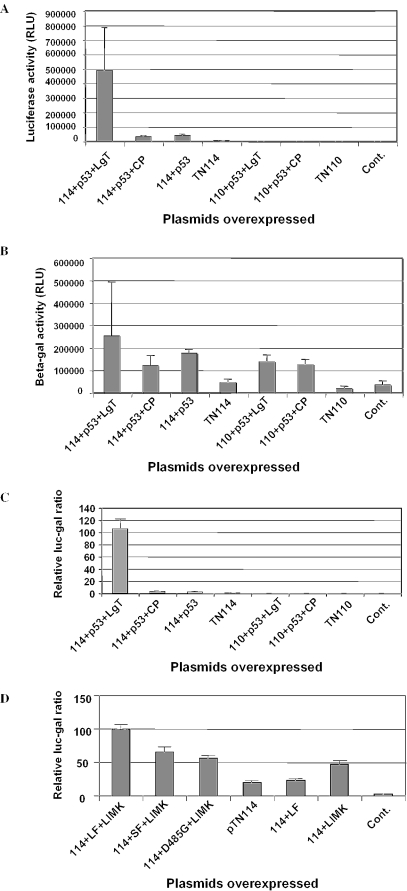
To test the ability of the assay system, HEK 293 cells were transfected with a reporter plasmid pTN114. A second plasmid (pTN110) was generated as identical to pTN114 except for the absence of signals for polyadenylation and splicing. After 48 h cells were harvested and β-galactosidase and luciferase activities were measured. Cells transfected with either plasmid produced only β-galactosidase activity (Figure 2B). Upon co-transfection with plasmids encode for the murine p53 (pCR2.1/p53 produced a Gal4-p53 fusion protein) and SV 40 large T antigen (pCR2.1/LgT produced a VP16-LgT fusion protein), the plasmid pTN114 was able to produce both β-galactosidase and luciferase activities, whereas pTN110 failed to show any luciferase activity (Figure 2A and B). The plasmids encoding p53 and LgT were purchased from Invitrogen for their known interactions. Co-transfection with plasmids containing the p53 and the polyoma viral coat protein (CP) (pCR2.1/VP16-CP generated by Invitrogen produced a VP16-CP fusion protein), failed to produce a significant amount of luciferase activity (Figure 2A). The luciferase and β-galactosidase activities were highly variable between experiments whereas when the data was expressed as the ratio of luc–gal activities, the variability was significantly reduced (Figure 2C). The presence of luciferase protein was confirmed by western blotting using Luciferase HRP conjugate antibody (Ab Cam, UK) (data not shown).
Figure 2.
Analyses of the gal–luc reporter system to determine protein–protein interactions in mammalian cells. The reporter construct along with relevant plasmids was co-transfected into HEK 293 cells and activities of luciferase (A) and β-galactosidase (B) were measured. The ratio of both reporter activities was normalized to a value of 100 (C) with p53 and LgT and (D) with LIMK1 and BMPR2. The standard deviations are indicated by error bars.
Mutations in the type II receptors for bone morphogenetic protein (BMPR-II), a member of the TGF-β receptor family underlie the majority of inherited forms of primary pulmonary hypertension (PPH) (30) and familial pulmonary arterial hypertension (31). BMPR-II is a multi-domain protein containing extracellular, transmembrane, kinase and cytoplasmic tail domains. The mutations are dispersed all over the functional domains and are likely to impinge upon receptor mediated function. A number of proteins have been shown to interact with BMPR-II (8). The interactions between LIM kinase 1 (LIMK1) and BMPR-II were initially discovered by yeast two-hybrid screening and confirmed by immunoprecipitation in mammalian cells (32). We wished to examine whether we could employ the reporter system to examine the interactions of LIMK1 and BMPR-II. This would provide the basis of a rapid assay system to investigate the effects of mutations in the BMPR2 on receptor-mediated function.
We employed the system to screen mutants of BMPR-II where protein–protein interactions might be abrogated. We constructed a series of constructs where the coding sequence of LIM kinase 1 gene was fused with a N-terminal DBD and the BMPR2 gene was fused with an activation domain at the C-terminal end. Both proteins were either T7 or myc tagged so that they could be easily purified. Co-transfection of plasmids encoding both proteins along with the reporter plasmid (pTN114) into the mammalian cells enabled both proteins to interact with each other giving rise to a luc–gal ratio (Figure 2D). Co-transfection of plasmids encode for the LIMK1 and SF of the BMPR2 or missense mutation produced significantly reduced luc–gal ratio (Figure 2D). The expression of LIMK1 and BMPR-II proteins was confirmed using antiT7 and anti-myc antibodies, respectively, by means of western blotting (data not shown).
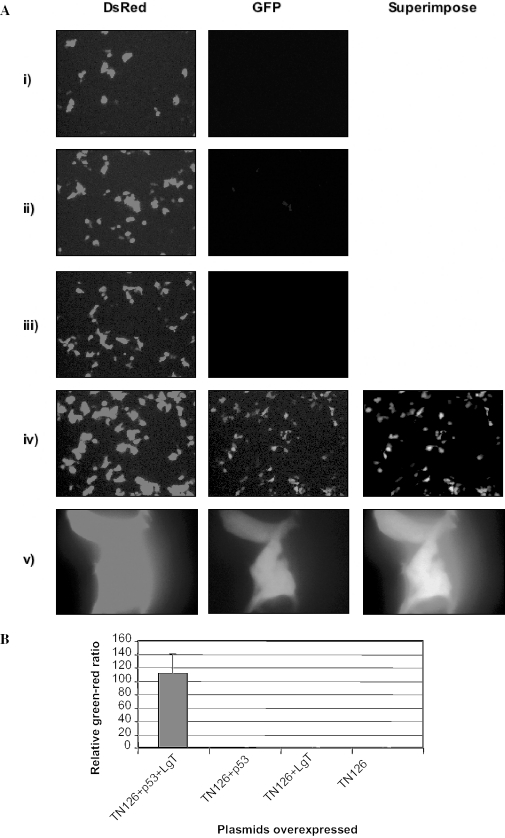
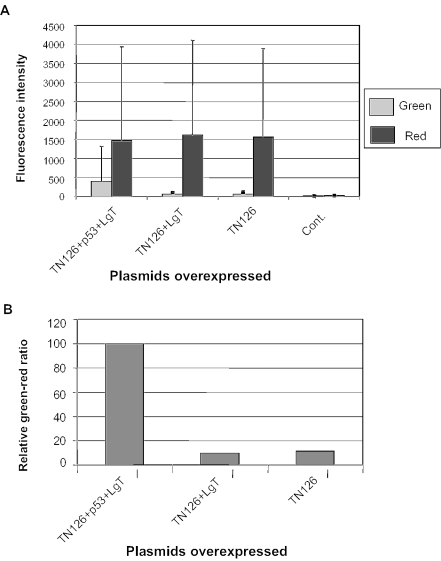
In order to establish a system for high-throughput screening of libraries (chemical, peptide, cDNA, etc.) a dual fluorescence reporter was constructed using genes encoding red (DsRed Express) and green fluorescence proteins (GFP). DsRed Express and GFP were chosen as their excitation peaks are 488 and 557 nm, respectively and therefore they can be easily distinguished under a fluorescence microscope. In addition, each is a highly stable protein, exhibiting no detectable photoinstability with high signal-to-noise ratio (33) and a shorter maturation time than that of other fluorescence proteins including DsRed. Upon transfection into the mammalian cells the reporter plasmid produced only the upstream reporter (DsRed Express) (Figure 3A). Co-transfections of plamids containing either p53 or LgT gene were unable to initiate transcription of the green fluorescence protein. Overexpression of both genes (in this case p53 and LgT) initiated transcription of the GFP. Expanded view revealed that both proteins were co-localized. We investigated the degree of interactions in a living cell. Cells expressing both proteins were randomly selected and fluorescence intensities were measured. Quantification of the fluorescence ratio indicated that the extent of interactions was uniform in a population of cells (Figure 3B). We also determined the extent of interaction in single cell. Significant amount of green fluorescence intensity was observed when cells were transfected with both p53 and LgT genes but not with the LgT alone (Figure 4A and B), a finding consistent with the data obtained from fluorescence microscopy.
Figure 3.
Analyses of the dual fluorescence reporter into HEK 293 cells. (A) The expression of GFP was suppressed in the absence of interacting proteins as shown, where cells were transfected with (i) reporter alone, or co-transfected with (ii) p53 and (iii) LgT. The expression of the GFP is activated when cells were co-transfected with plasmids encoding the p53 and CP (iv and v). (B) Fluorescence intensities of both proteins were measured and normalized as mentioned in Figure 2. Mean and standard deviations were derived from a pool of 30 to 40 randomly selected cells.
Figure 4.
Single-cell assay for the dual fluorescence reporter. Cells transfected with the reporter along with the respective plasmids as mentioned above. Cells with both fluorescence activities were selected using a fluorescence activated cell sorter and their intensities were determined (A) and normalized (B). The mean and standard deviations were derived from 300 to 400 cells.
DISCUSSION
Since the conventional methods based on the yeast two-hybrid system for assaying protein–protein interactions in mammalian cells are susceptible to numerous variables, we developed a system that bypassed the variables that affect single reporter assays. One of the important features of this system is that a synthetic deactivation–activation unit is created which comprises signals for transcription, polyadenylation, processing and translation. Signals for polyadenylation, transcription and translation termination are introduced such that the upstream reporter is expressed. Upon protein–protein interactions the Pol II transcription machinery is recruited upstream of the second reporter, hence enabling the production of the second reporter. We have previously showed that there is a good correlation between the level of transcript and luciferase–galactosidase ratio (28).
We have analysed the inter-experiment variability for expression of the reporter proteins and found substantial scatter of the data points. However, by taking advantage of constitutive expression of reporter A, we can correct the expression of reporter B to this reference standard. Hence, the ratio of two reporter activities show marked reduction in the signal variability (Figure 2C). We have tested the dual-light reporter in several ways to show that it reflects the efficiency of protein–protein interactions. The highest ratio of luciferase and galatosidase activities was observed when co-transfection was carried out using plasmids encoding the p53 with LgT and the BMPR2 with LIMK1 (Figure 2C and 2D), as these proteins interact with each other (32,34,35). The lowest ratio was observed when either the reporter (pTN114) was transfected alone or co-transfected with plasmids encoding the p53 with CP and the LIMK1 with short or mutant form of BMPR2. The interactions among these proteins are very weak (32,35). We observed that the interaction between LIMK1 and BMPR-II was weaker than that of LgT and p53, possibly due to mislocalization of BMPR-II. Luciferase activity was completely abolished when the polyadenylation and splicing signals were ablated (pTN110). Co-transfections of the p53 and LgT had no discernible effects possibly due to transcriptional interference, a process that is known to affect the expression of downstream gene (36). In this case, upon transcription from the SV40 promoter a gal–luc fusion RNA was produced. The presence of translation termination signal at the end of β-galactosidase gene prompted the production of β-galactosidase protein. We would assume that in the case of pTN114, transcription termination occurred at poly A sites located at the end of both reporter genes (37). Both RNAs were polyadenylated, bound by Poly A binding protein (PABP), circularized by interacting with translation initiation complex (38) and thereby acted as autonomous units of expression.
A potential complication of the approach was that of transcriptional interference, which might affect recruitment of transcription complex for the second reporter. This could be easily addressed by inserting multiple termination sites (36) at the end of first reporter as transcription termination occurs through a non-processive mechanism (39). We have not determined the level of promoter suppression by which the expression of the downstream reporter could be affected but again this could be tackled by interposing MAR/SAR elements (40) between the adjacent units. Although we have not encountered the problems of autoactivation of library screening (41), it is unlikely to affect the usefulness of the method, as long as a second independent method (i.e. pull-down, immunoprecipitation) is used to validate the most interesting candidates that emerge from a screen.
Protein–protein interactions have a great potential as a target for therapeutic inoculation. Identification of a small molecule that dimerizes two proteins can be used for induction of cell proliferation (42) and apoptosis (43). On the contrary, small molecule antagonists of proteins can be useful in developing antibiotic, antiviral and antiparasitic drugs (44). The advantages of the dual fluorescence reporter are that the assay is measured directly on the intact cell and therefore does not require cell wall disruption or addition of a substrate and the sensitivity of this assay is comparable with that reported for luc–gal assay. In addition the dual fluorescence assay could be performed in 96- or 386-well format and would be useful to screen chemical or peptide libraries to identify dimerizers or antagonists that affect specific protein–protein interactions. In this case it would be advantageous to set up a stable mammalian cell line by incorporating the fluorescence reporter. The maturation time of the DsRed Express is shorter than that of its DsRed counterpart, which makes the reporter system useful in investigating protein interactions in the early stages of developing embryos.
Although we have only shown that this system would be useful in screening mutations of BMPR-II that are involved in LIMK interactions, there are a number of other human diseases for which it would be beneficial to isolate drugs capable of targeting selected protein–protein interactions. This system in its present form would be useful, for example, in identifying proteins that interact with selenoprotein N. Mutations in the selenoprotein N gene have been implicated in muscular dystrophy (45). The classical two-hybrid screen to identify the interacting proteins is not suitable because yeast is known to contain no selenocysteine incorporation machinery. Other methods, such as pull-down or immunoprecipitation, would be difficult due to the fact that selenocysteine incorporation in mammalian cell line is an inefficient process (46).
We earlier developed a dual-reporter based assay system (28) to determine RNA processing efficiency in mammalian cells. In this system, two reporter activities fused with splicing signals were linked to single gene expression unit. In the event of splicing, the expression unit produced a fusion protein with two reporter functions whereas inefficient splicing produced a protein with single reporter function. The system presented here is based on two autonomous gene expression units where one expression unit is expressed regardless of the expression of the other unit. The other unit is switched on upon efficient protein–protein interactions. We have shown here the usefulness of this principle in upgrading a classical two-hybrid screen. We suggest that this principle may be beneficial for other systems where the activities are limited to single reporter functions.
Acknowledgments
The authors wish to thank Mr Edward C. Schwalbe for critically reading the manuscript and Ms S. L. Wattam for valuable technical assistance. The authors express their sincere gratitude to Dr R. T. Snowden, MRC Toxicology Unit, Leicester for providing access to facilities for flow cytometry analysis. The project was supported by a programme grant from the British Heart Foundation (RG/2000012 to RCT). Funding to pay the Open Access publication charges for this article was provided by the University of Leicester.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berezovska O., Bacskai B.J., Hyman B.T. Monitoring proteins in intact cells. Sci. Aging Knowledge Environ. 2003;2003:PE14. doi: 10.1126/sageke.2003.23.pe14. [DOI] [PubMed] [Google Scholar]
- 2.Cohen F.E., Prusiner S.B. Pathologic conformations of prion proteins. Annu. Rev. Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D.J. The cell biology of beta-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 4.Burke J.R., Enghild J.J., Martin M.E., Jou Y.S., Myers R.M., Roses A.D., Vance J.M., Strittmatter W.J. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nature Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 5.von der Haar T., Hughes J.M., Manjarul Karim M., Ptushkina M., McCarthy J.E. Translation initiation and surface plasmon resonance: new technology applied to old questions. Biochem. Soc. Trans. 2002;30:155–162. doi: 10.1042/0300-5127:0300155. [DOI] [PubMed] [Google Scholar]
- 6.Issaq H.J., Veenstra T.D., Conrads T.P., Felschow D. The SELDI-TOF MS approach to proteomics: protein profiling and biomarker identification. Biochem. Biophys. Res. Commun. 2002;292:587–592. doi: 10.1006/bbrc.2002.6678. [DOI] [PubMed] [Google Scholar]
- 7.Causier B. Studying the interactome with the yeast two-hybrid system and mass spectrometry. Mass Spectrom. Rev. 2004;23:350–367. doi: 10.1002/mas.10080. [DOI] [PubMed] [Google Scholar]
- 8.Hassel S., Eichner A., Yakymovych M., Hellman U., Knaus P., Souchelnytskyi S. Proteins associated with type II bone morphogenetic protein receptor (BMPR-II) and identified by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2004;4:1346–1358. doi: 10.1002/pmic.200300770. [DOI] [PubMed] [Google Scholar]
- 9.Pazos F., Helmer-Citterich M., Ausiello G., Valencia A. Correlated mutations contain information about protein–protein interaction. J. Mol. Biol. 1997;271:511–523. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- 10.Jones S., Thornton J.M. Analysis of protein–protein interaction sites using surface patches. J. Mol. Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 11.Jones S., Thornton J.M. Prediction of protein–protein interaction sites using patch analysis. J. Mol. Biol. 1997;272:133–143. doi: 10.1006/jmbi.1997.1233. [DOI] [PubMed] [Google Scholar]
- 12.Bock J.R., Gough D.A. Predicting protein–protein interactions from primary structure. Bioinformatics. 2001;17:455–460. doi: 10.1093/bioinformatics/17.5.455. [DOI] [PubMed] [Google Scholar]
- 13.Fields S., Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 14.Auerbach D., Thaminy S., Hottiger M.O., Stagljar I. The post-genomic era of interactive proteomics: facts and perspectives. Proteomics. 2002;2:611–623. doi: 10.1002/1615-9861(200206)2:6<611::AID-PROT611>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 15.Aronheim A., Zandi E., Hennemann H., Elledge S.J., Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein–protein interactions. Mol. Cell. Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serebriiskii I., Khazak V., Golemis E.A. A two-hybrid dual bait system to discriminate specificity of protein interactions. J. Biol. Chem. 1999;274:17080–17087. doi: 10.1074/jbc.274.24.17080. [DOI] [PubMed] [Google Scholar]
- 17.Eyckerman S., Verhee A., der Heyden J.V., Lemmens I., Ostade X.V., Vandekerckhove J., Tavernier J. Design and application of a cytokine-receptor-based interaction trap. Nature Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 18.Shioda T., Andriole S., Yahata T., Isselbacher K.J. A green fluorescent protein-reporter mammalian two-hybrid system with extrachromosomal maintenance of a prey expression plasmid: application to interaction screening. Proc. Natl Acad. Sci. USA. 2000;97:5220–5224. doi: 10.1073/pnas.97.10.5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H.F., Kiyota T., Chowdhury S., Purisima E., Banville D., Konishi Y., Shen S.H. A mammalian genetic system to screen for small molecules capable of disrupting protein–protein interactions. Anal. Chem. 2004;76:2922–2927. doi: 10.1021/ac035396m. [DOI] [PubMed] [Google Scholar]
- 20.Tavernier J., Eyckerman S., Lemmens I., Van der Heyden J., Vandekerckhove J., Van Ostade X. MAPPIT: a cytokine receptor-based two-hybrid method in mammalian cells. Clin. Exp. Allergy. 2002;32:1397–1404. doi: 10.1046/j.1365-2745.2002.01520.x. [DOI] [PubMed] [Google Scholar]
- 21.Kollmus H., Flohe L., McCarthy J.E. Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res. 1996;24:1195–1201. doi: 10.1093/nar/24.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giniger E., Varnum S.M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 23.Lillie J.W., Green M.R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 24.Marmorstein R., Carey M., Ptashne M., Harrison S.C. DNA recognition by GAL4: structure of a protein–DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 25.Nasim M.T., Eperon I.C., Wilkins B.M., Brammar W.J. The activity of a single-stranded promoter of plasmid ColIb-P9 depends on its secondary structure. Mol. Microbiol. 2004;53:405–417. doi: 10.1111/j.1365-2958.2004.04114.x. [DOI] [PubMed] [Google Scholar]
- 26.Sadowski I., Bell B., Broad P., Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 28.Nasim M.T., Chowdhury H.M., Eperon I.C. A double reporter assay for detecting changes in the ratio of spliced and unspliced mRNA in mammalian cells. Nucleic Acids Res. 2002;30:e109. doi: 10.1093/nar/gnf108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasim M.T., Chernova T.K., Chowdhury H.M., Yue B.G., Eperon I.C. HnRNP G and Tra2beta: opposite effects on splicing matched by antagonism in RNA binding. Hum. Mol. Genet. 2003;12:1337–1348. doi: 10.1093/hmg/ddg136. [DOI] [PubMed] [Google Scholar]
- 30.Trembath R.C., Harrison R. Insights into the genetic and molecular basis of primary pulmonary hypertension. Pediatr. Res. 2003;53:883–888. doi: 10.1203/01.PDR.0000061565.22500.E7. [DOI] [PubMed] [Google Scholar]
- 31.Newman J.H., Trembath R.C., Morse J.A., Grunig E., Loyd J.E., Adnot S., Coccolo F., Ventura C., Phillips J.A., III, Knowles J.A., et al. Genetic basis of pulmonary arterial hypertension: current understanding and future directions. J. Am. Coll. Cardiol. 2004;43:33S–39S. doi: 10.1016/j.jacc.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 32.Foletta V.C., Lim M.A., Soosairajah J., Kelly A.P., Stanley E.G., Shannon M., He W., Das S., Massague J., Bernard O., et al. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J. Cell Biol. 2003;162:1089–1098. doi: 10.1083/jcb.200212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finley K.R., Davidson A.E., Ekker S.C. Three-color imaging using fluorescent proteins in living zebrafish embryos. Biotechniques. 2001;31:66–70. doi: 10.2144/01311st02. ,72. [DOI] [PubMed] [Google Scholar]
- 34.Li B., Fields S. Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J. 1993;7:957–963. doi: 10.1096/fasebj.7.10.8344494. [DOI] [PubMed] [Google Scholar]
- 35.Luker G.D., Sharma V., Pica C.M., Prior J.L., Li W., Piwnica-Worms D. Molecular imaging of protein–protein interactions: controlled expression of p53 and large T-antigen fusion proteins in vivo. Cancer Res. 2003;63:1780–1788. [PubMed] [Google Scholar]
- 36.Proudfoot N.J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986;322:562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- 37.Osheim Y.N., Proudfoot N.J., Beyer A.L. EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell. 1999;3:379–387. doi: 10.1016/s1097-2765(00)80465-7. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy J.E. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeung G., Choi L.M., Chao L.C., Park N.J., Liu D., Jamil A., Martinson H.G. Poly(A)-driven and poly(A)-assisted termination: two different modes of poly(A)-dependent transcription termination. Mol. Cell. Biol. 1998;18:276–289. doi: 10.1128/mcb.18.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villemure J.F., Savard N., Belmaaza A. Promoter suppression in cultured mammalian cells can be blocked by the chicken beta-globin chromatin insulator 5′HS4 and matrix/scaffold attachment regions. J. Mol. Biol. 2001;312:963–974. doi: 10.1006/jmbi.2001.5015. [DOI] [PubMed] [Google Scholar]
- 41.Fashena S.J., Serebriiskii I., Golemis E.A. The continued evolution of two-hybrid screening approaches in yeast: how to outwit different preys with different baits. Gene. 2000;250:1–14. doi: 10.1016/s0378-1119(00)00182-7. [DOI] [PubMed] [Google Scholar]
- 42.MacCorkle R.A., Freeman K.W., Spencer D.M. Synthetic activation of caspases: artificial death switches. Proc. Natl Acad. Sci. USA. 1998;95:3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amara J.F., Clackson T., Rivera V.M., Guo T., Keenan T., Natesan S., Pollock R., Yang W., Courage N.L., Holt D.A., et al. A versatile synthetic dimerizer for the regulation of protein–protein interactions. Proc. Natl Acad. Sci. USA. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gadek T.R., Nicholas J.B. Small molecule antagonists of proteins. Biochem. Pharmacol. 2003;65:1–8. doi: 10.1016/s0006-2952(02)01479-x. [DOI] [PubMed] [Google Scholar]
- 45.Moghadaszadeh B., Petit N., Jaillard C., Brockington M., Roy S.Q., Merlini L., Romero N., Estournet B., Desguerre I., Chaigne D., et al. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nature Genet. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- 46.Nasim M.T., Jaenecke S., Belduz A., Kollmus H., Flohe L., McCarthy J.E. Eukaryotic selenocysteine incorporation follows a nonprocessive mechanism that competes with translational termination. J. Biol. Chem. 2000;275:14846–14852. doi: 10.1074/jbc.275.20.14846. [DOI] [PubMed] [Google Scholar]