Abstract
Pseudomonas aeruginosa nalB mutants which hyperexpress the MexAB-OprM multidrug efflux system produce reduced levels of several extracellular virulence factors known to be regulated by quorum sensing. Such mutants also produce less acylated homoserine lactone autoinducer PAI-1, consistent with an observed reduction in lasI expression. These data suggest that PAI-1 is a substrate for MexAB-OprM, and its resulting exclusion from cells hyperexpressing MexAB-OprM limits PAI-1-dependent activation of lasI and the virulence genes.
Pseudomonas aeruginosa is an opportunistic human pathogen characterized by an innate resistance to a wide array of antimicrobial agents. Once attributed to a highly impermeable outer membrane (24), this property is now recognized to result from the operation of broadly specific drug efflux pumps which act synergistically with low outer membrane permeability to elicit multidrug resistance (20). One such efflux system, encoded by the mexAB-oprM operon (11, 34, 35), expels a range of antibiotics, including tetracycline, chloramphenicol, quinolones, β-lactams, novobiocin, macrolides, and trimethoprim (11, 14, 17, 18). Although expressed in wild-type cells (7), the operon is hyperexpressed in nalB mutants (36), which display markedly elevated levels of resistance to substrate antibiotics.
Homologues of this system have been reported for P. aeruginosa (mexCD-oprJ [33]; mexEF-oprN [15]), Escherichia coli (acrAB-tolC) (9, 20), Neisseria gonorrhoeae (mtrCDE) (12), and Burkholderia cepacia (ceoA-ceoB-opcM) (4, 5). An oprM gene probe was used to demonstrate the presence of oprM homologues in Burkholderia pseudomallei and Pseudomonas putida (2), suggesting the presence of such systems in these organisms as well. Although the aforementioned systems all play a role in resistance to clinically relevant antibiotics, the likely natural function has been addressed only with respect to the E. coli AcrAB and N. gonorrhoeae MtrCDE systems, which appear to play a role in the export of toxic environmental lipids or hydrophobic agents (e.g., bile salts) (12, 19, 44).
During studies intended to elucidate the natural function of the MexAB-OprM efflux system, including the identification of natural, cell-associated substrates, we noted an inverse correlation between the presence of mexAB-oprM in P. aeruginosa and the production of the blue-green pigment pyocyanin, a virulence factor in this organism (6). (Intriguingly, a similar observation was made regarding the mexEF-oprN operon: strains expressing this system were demonstrably pyocyanin deficient compared to strains lacking this system [15]). A more detailed study subsequently revealed that this effect on pyocyanin was due to the apparent influence of MexAB-OprM on autoinducer (AI) levels, pyocyanin production being AI dependent (16).
AIs are a family of acylated homoserine lactones found in a number of gram-negative bacteria whose accumulation in the growth medium mirrors cell density, triggering the expression of certain target genes upon reaching a critical AI (i.e., cell) concentration (10). Quorum sensing, as this process is now known, involves an AI synthase, which produces AI destined for release into the growth medium, and a transcriptional activator, which acts in concert with the AI upon its reentry into cells to activate target genes in response to increases in bacterial cell density (10). Two homoserine lactone AIs have been characterized in detail for P. aeruginosa, N-(3-oxo)-dodecanoyl-l-homoserine lactone (29) (also called PAI-1 [31]) and N-butanoyl-l-homoserine lactone (30) (also called PAI-2 [10]), synthesized by the products of the lasI (28) and rhlI (vsmI) (16, 25) genes, respectively. Together with their cognate quorum-sensing regulators, LasR (31) and RhlR (26) (also called VsmR [16]), these act to stimulate production of a number of extracellular virulence factors in P. aeruginosa (16, 31). We report here that hyperexpression of MexAB-OprM compromises production of PAI-1 and, thus, expression of LasR-LasI-dependent virulence factors. Apparently, reentry of PAI-1 is prevented by the efflux activity of MexAB-OprM, leading to a reduction in intracellular PAI-1 and, thus, reduced expression of PAI-1-dependent genes.
Strains used in this study are listed in Table 1. Luria (L) broth (Difco), pyocyanin production broth (6), and peptone tryptic soy broth (27) have been described previously. Assays for the exoproducts pyocyanin (6), elastase (27), and casein protease (13) have been previously described. A nalB derivative of streptomycin-resistant PAO1 strain K1171 (designated K1168) was selected on L agar containing 0.4 μg of ciprofloxacin and 100 μg of carbenicillin per ml as described previously and was screened for nalB-type multidrug resistance (43) and OprM hyperexpression by a Western immunoblotting procedure with an OprM-specific antiserum (42). The use of a streptomycin-resistant strain was necessitated by the need to subsequently introduce a mexAB-oprM deletion (via conjugation; see below) into the nalB strain via a procedure involving streptomycin counterselection of the donor strain. For the construction of mexAB-oprM deletion strain K1169, vector pELCT04 was constructed. First, the mercury resistance Ω Hg interposon from pHP45::Ω Hg (8) was cloned into HindIII-restricted pK18mobsacbB (38) on a 4.6-kb HindIII fragment, to yield pELCT02. The previously constructed mexAB-oprM deletion fragment was then cloned from vector pRSP14 (43) into pELCT02 on a 1.4-kb BamHI fragment to yield pELCT04. All manipulations were carried out with E. coli DH5α. Following transformation (37) of pELCT04 into E. coli S17-1, the vector was mobilized into P. aeruginosa nalB strain K1168 via conjugation as described previously (34) and pELCT04-containing P. aeruginosa was selected on L agar supplemented with 15 μg of HgCl2 per ml (to select the vector) and 10 μg of tetracycline per ml (to counterselect E. coli S17-1). HgCl2-resistant colonies were recovered and streaked for single colonies on L agar containing 10% (wt/vol) sucrose. Sucrose-resistant colonies were screened for loss of HgCl2 resistance (and kanamycin resistance), and those carrying the mexAB-oprM deletion were identified following PCR amplification of chromosomal DNA with Taq DNA polymerase as described previously (43). Where indicated, AIs PAI-1 and PAI-2, synthesized as described previously (29, 30), were included in the culture medium at a final concentration of 0.5 to 5 μM. In cross-streaking experiments, bacteria were streaked onto the surfaces of L agar plates at right angles so that areas of bacterial growth approached but did not contact. Pyocyanin production was then assessed visually on plates, although control experiments confirmed that the pigment observed was pyocyanin. This involved the recovery of pigmented agar, following the removal of bacterial cells, and extraction and assay of pyocyanin as described above. AI levels were quantitated by previously described bioassays (29, 30) following the extraction of AIs from cell-free culture supernatant with ethyl acetate (29). Synthetic PAI-1 and PAI-2 were used to construct a standard dose-response curve, which permitted the quantification of the extracted AIs based on the results of the bioassay (29). Expression of lasI was assessed with a plasmid-borne lasI-lacZ fusion vector (40). β-Galactosidase assays were carried out as described previously (22) with cells cultured in pyocyanin production broth.
TABLE 1.
Bacterial strains
| Strain | Descriptiona | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 (K767)b | Prototroph | |
| OCR1 | PAO1 (K767) nalB | 21 |
| K784 | Spontaneous Smr derivative of OCR1 | 36 |
| K1170 | K784 ΔmexAB-oprM | This study |
| PAO1 (K867)bc | Prototroph; Cmr | B. H. Iglewski |
| K1171 | Spontaneous Smr derivative of K867 | This study |
| K1168 | K1171 nalB | This study |
| K1169 | K1168::ΔmexAB-oprM | This study |
| PDO100 | PAO1 (K867) ΔrhlI::Tn501 | 3 |
| PAO-JP1 | PAO1 (K867) ΔlasI::tet | B. H. Iglewski |
| E. coli | ||
| DH5α | endA hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 1 |
| S17-1 | thi pro hsdR recA Tra+ | 41 |
Cmr, chloramphenicol resistant; Tra+, mobilizes nonconjugative mob-carrying vectors.
Two different PAO1 strains were used in this study and are distinguished by the laboratory designations K767 and K867.
This PAO1 strain displays substantial resistance to chloramphenicol, in contrast to PAO1 strain K767.
Cultures of P. aeruginosa PAO1 (strain K767) elicited a blue-green pigment, reminiscent of pyocyanin, during growth on L agar (Table 2). Indeed, extraction of this pigment from agar plates and subsequent spectrophotometric examination confirmed it as pyocyanin (data not shown). Interestingly, the nalB derivative of this strain, OCR1, lacked this pigmentation (Table 2). When both strains were cultured on the same L-agar plate, however, OCR1 growth in the vicinity of PAO1 strain K767 growth was pigmented (Table 2). This suggested that the nalB strain was deficient in pyocyanin production and that PAO1 strain K767 produced something that restored pyocyanin production in OCR1. Similarly, a second PAO1 strain, K867, and its streptomycin-resistant derivative, K1171, were also pyocyanin proficient in L broth, while a nalB derivative of K1171, strain K1168, was pyocyanin deficient (Table 2). Moreover, elimination of mexAB-oprM in either OCR1 or K1168 (yielding K1170 and K1169, respectively) restored pyocyanin production (Table 2). Thus, there appeared to be an inverse correlation between levels of mexAB-oprM expression and pyocyanin production by P. aeruginosa.
TABLE 2.
Pyocyanin production by P. aeruginosa
| Strain | Efflux genotype | Pyocyanin productiona | Cross-feeds OCR1b |
|---|---|---|---|
| K767 | Wild type | + | + |
| OCR1 | nalB | − | − |
| K784 | nalB | − | − |
| K1170 | nalB ΔmexAB-oprM | + | ND |
| K1171 | Wild type | + | ND |
| K1168 | nalB | − | ND |
| K1169 | nalB ΔmexAB-oprM | + | ND |
| PAO-JP1 | Wild type | − | − |
| PDO100 | Wild type | − | + |
Assessed visually following growth on L agar. +, produces pyocyanin; −, no pyocyanin produced.
The indicated strains were streaked at right angles to OCR1 on L agar, and production of pyocyanin by OCR1 was assessed as described above in the vicinity of the cross-streaked strain. +, cross-feeds OCR1; −, does not cross-feed OCR1; ND, not determined.
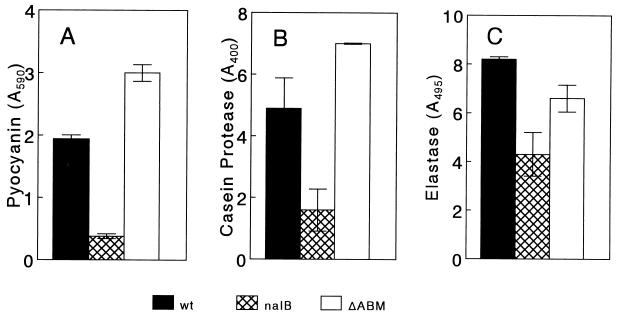
Although a possible explanation for the above-mentioned influence of mexAB-oprM on pyocyanin production was that nalB strains expel a precursor necessary for pyocyanin production, the latter being a substrate for MexAB-OprM, the observation that PAO1 could, in effect, cross-feed OCR1 (Table 2), restoring pyocyanin production, argued against this. Intriguingly, cells overexpressing the mexAB-oprM operon (e.g., OCR1) failed to elicit this cross-feeding phenomenon, while those deficient in or with reduced (such as the wild type) mexAB-oprM expression were proficient at cross-feeding (Table 2). Given that pyocyanin production is regulated by quorum sensing (3, 16), however, it was likely that strains overexpressing MexAB-OprM (e.g., OCR1 and K1168) were somehow defective in the quorum-sensing process. Moreover, given their inability to cross-feed but their ability to be cross-fed, it was likely that they were defective in a diffusible component of quorum sensing, namely the AI. Consistent with this, pyocyanin produced by K1168 in liquid culture could be increased (by twofold) upon the addition of 1 to 2 μM PAI-1 (data not shown). Initially, the nalB strain K1168 was examined for the production of pyocyanin and additional AI-dependent components, including elastase and casein protease (3), to see if there was, indeed, a general deficiency in quorum sensing in this strain. An examination of pyocyanin (Fig. 1A), casein protease (Fig. 1B), and elastase (Fig. 1C) levels revealed that K1168 produced reduced levels of these compared with levels produced by the parent strain, K1171. Deletion of mexAB-oprM in K1168 restored the production of pyocyanin (Fig. 1A), casein protease (Fig. 1B), and elastase (Fig. 1C) in the resultant strain, K1169, indicating that the quorum-sensing defect identified in nalB strain K1168 resulted from overexpression of the efflux pump and not some other manifestation of the nalB mutation.
FIG. 1.
Production of AI-dependent virulence factors as a function of mexAB-oprM expression. P. aeruginosa K1171 (wild type [wt] for mexAB-oprM), K1168 (nalB), and K1169 (nalB ΔmexAB-oprM; ΔABM) were examined for the production of elastase (values reported have been multiplied by 100), casein protease (values reported have been multiplied by 10), and pyocyanin. Results reported are per milliliter of cells at an A600 of 1.0 and are the means of the results of three separate experiments ± the standard deviations.
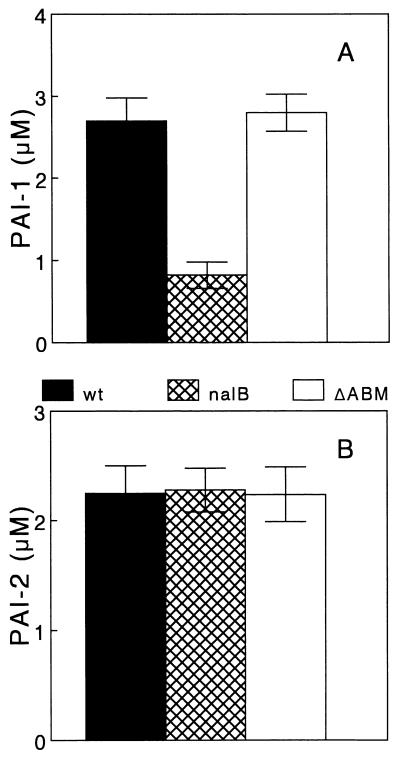
These data strongly argued that nalB strains were AI deficient. To assess this directly, we measured the levels of PAI-1 and PAI-2 in spent culture supernatants of K1171, K1168, and K1169 by previously described bioassays (29, 30). As can be seen in Fig. 2A, nalB strain K1168 consistently produced ca. threefold less PAI-1 than its parent strain, K1171 (wild type with respect to MexAB-OprM). A similar result was observed for a number of independently isolated nalB derivatives of P. aeruginosa (data not shown). Interestingly, this decline in PAI-1 levels was abrogated upon the deletion of the mexAB-oprM efflux genes (see K1169; Fig. 2A), indicating that this reduction was a function solely of MexAB-OprM overproduction. In contrast, PAI-2 levels remained constant in all three strains (Fig. 2B), indicating that PAI-2 production is not influenced by the status of MexAB-OprM.
FIG. 2.
AI production as a function of mexAB-oprM expression. PAI-1 (A) and PAI-2 (B) levels were measured in cell-free supernatants of 18-h cultures of P. aeruginosa K1171 (wild type for mexAB-oprM; wt), K1168 (nalB), and K1169 (nalB ΔmexAB-oprM; ΔABM) as described in the text. Values reported are the means of results from five separate experiments ± the standard deviations.
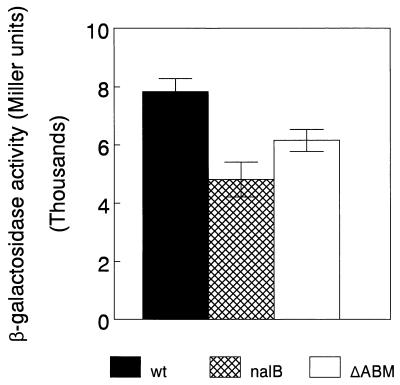
It is apparent from these data that MexAB-OprM hyperexpression yields a specific decline in PAI-1 levels which is correlated with a decline in the levels of several known AI-dependent products. That PAI-1 levels alone were impacted in the nalB strain was, in fact, consistent with the observation that an rhlI mutant, which produces PAI-1 but not PAI-2, was able to cross-feed nalB strain OCR1 (with respect to pyocyanin production) while a lasI mutant (produces no PAI-1) was not (Table 2). One explanation for these observations is that a precursor for PAI-1 synthesis is exported by MexAB-OprM, leading to a reduced synthesis of this AI in MexAB-OprM-overexpressing nalB strains such as K1168. Still, in light of evidence indicating that AIs are synthesized from S-adenosylmethionine and acylated-acyl carrier protein (23, 39), neither of which is a likely candidate for export via MexAB-OprM, this is improbable. Alternatively, PAI-1, but not PAI-2, may be a substrate for the MexAB-OprM efflux system. According to currently accepted models of quorum sensing, whereby AI released by cells in a population accumulates in the extracellular milieu and then diffuses back into the cell to stimulate the expression of cell density-dependent genes, the increased expression of MexAB-OprM in a nalB strain would serve to compromise this reentry of PAI-1. The resulting reduction in PAI-1 accumulation within the cell would limit LasR–PAI-1 formation and subsequent activation of target genes (e.g., elastase and casein protease). Moreover, since lasI expression is also LasR–PAI-1 dependent (31), this would also lead to a reduction in PAI-1 synthesis in a nalB strain. In fact, we did observe a ca. twofold decrease in lasI expression in nalB strain K1168 relative to expression in its parent strain (Fig. 3), consistent with this decline in PAI-1 levels in K1168. Although LasR–PAI-1 does not directly regulate pyocyanin biosynthesis, which appears to be controlled by the RhlRI system (16), rhlRI gene expression is positively regulated by LasR–PAI-1 (32) and lasI mutants are compromised as regards pyocyanin production (7). Thus, any reduction in PAI-1 formation would be expected to yield a decrease in pyocyanin levels.
FIG. 3.
β-Galactosidase activities of 18-h cultures of P. aeruginosa K1171 (wild type for mexAB-oprM; wt), K1168 (nalB), and K1169 (nalB ΔmexAB-oprM; ΔABM) harboring lasI-lacZ fusions. Values reported are the means of results from three separate experiments ± the standard deviations.
What is less clear, given the involvement of LasR–PAI-1 in rhlRI gene expression, is why PAI-2 levels were not altered in a nalB strain. Certainly, in light of the effect on pyocyanin production, a nalB strain is compromised as regards the operation of some component of the rhl quorum-sensing system. Perhaps rhlI expression is less sensitive to changes in LasR–PAI-1 than is that of rhlR. Similarly, since RhlR is required for the expression of rhlI and the production of pyocyanin, the latter may be more affected by any decline in RhlR levels than is rhlI. Moreover, given suggestions that PAI-1 antagonizes PAI-2 association with RhlR (32), a reduction in PAI-1 might lessen this effect, enhancing PAI-2 interaction with available RhlR molecules. Thus, while there might be less RhlR in a nalB strain, what is present might be more active as a result of increased association with PAI-2.
Given that efflux gene hyperexpression compromises PAI-1 and AI-dependent virulence gene expression, it will be of interest to determine whether nalB strains are less virulent, despite their increased multidrug resistance. Moreover, the impact of MexAB-OprM on quorum sensing, possibly by compromising the reentry of PAI-1 into cells, represents the first P. aeruginosa-associated process or substrate which is influenced by this efflux system. Still, given the broad substrate specificity of MexAB-OprM, it is highly unlikely that the export of a quorum-sensing-related molecule such as PAI-1 would be a specific function of MexAB-OprM, and therefore any modulation of quorum sensing in response to MexAB-OprM is probably a secondary effect of its primary and hitherto unidentified primary role in the cell.
Acknowledgments
This work was supported by an operating grant to K.P. from the Canadian Cystic Fibrosis Foundation and a Cystic Fibrosis Foundation (United States) grant to L.P. K.E. holds a Medical Research Council of Canada studentship. R.S. is a Natural Sciences and Engineering Research Council of Canada (NSERC) Postdoctoral Fellow. E.T. was supported by a summer studentship from the Canadian Cystic Fibrosis Foundation. K.P. is an NSERC University Research Fellow.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 2.Bianco N, Neshat S, Poole K. Conservation of the multidrug resistance efflux gene oprM in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:853–856. doi: 10.1128/aac.41.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns J L, Ptritzlaff C, Barry J, Charron M, Cieri M. Program and abstracts of the 98th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1998. Cloning and sequence analysis of a multiple antibiotic efflux operon from Burkholderia cepacia, abstr. V-108; p. 531. [Google Scholar]
- 5.Burns J L, Wadsworth C D, Barry J J, Goodall C P. Nucleotide sequence analysis of a gene from Burkholderia (Pseudomonas) cepacia encoding an outer membrane lipoprotein involved in multiple antibiotic resistance. Antimicrob Agents Chemother. 1996;40:307–313. doi: 10.1128/aac.40.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox C D. Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun. 1986;52:263–270. doi: 10.1128/iai.52.1.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, K., and K. Poole. Unpublished data.
- 8.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 11.Gotoh N, Tsujimoto H, Poole K, Yamagishi J-I, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 13.Kessler E, Israel M, Landshman N, Chechick A, Blumberg S. In vitro inhibition of Pseudomonas aeruginosa elastase by metal-chelating derivatives. Infect Immun. 1982;38:716–723. doi: 10.1128/iai.38.2.716-723.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koehler T, Kok M, Michea-Hamzehpour M, Plesiat P, Gotoh N, Nishino T, Curty L K, Pechere J-C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koehler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 16.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 17.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X-Z, Zhang L, Srikumar R, Poole K. β-Lactamase inhibitors are substrates of the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:399–403. doi: 10.1128/aac.42.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 20.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in Gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 21.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 72–74. [Google Scholar]
- 23.More M I, Finger D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum sensing autoinducer using defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsner U, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochsner U A, Koch A K, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohman D E, Cryz S J, Iglewski B H. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 29.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesci E C, Iglewski B H. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 1997;5:132–135. doi: 10.1016/S0966-842X(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 32.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 34.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 35.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schaefer A, Tauch A, Jaeger W, Kalinowski J, Thierbach G, Puehler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 39.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Generation of cell-to-cell signals in quorum-sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seed P C. Ph.D. thesis. Rochester, N.Y: University of Rochester; 1996. [Google Scholar]
- 41.Simon R, Priefer U, Puehler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 42.Srikumar R, Kon T, Gotoh N, Poole K. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob Agents Chemother. 1998;42:65–71. doi: 10.1128/aac.42.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srikumar R, Li X-Z, Gotoh N, Poole K. The inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts in Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]