We read with interest the work by Park and colleagues, which attempted to elucidate the composition of neuronal intranuclear inclusions (NIIs), central to the pathology of neuronal intranuclear inclusion disease (NIID) [1]. NIID is a clinically heterogeneous neurodegenerative disorder characterised by these intranuclear eosinophilic ubiquitinated inclusions in both neuronal and non-neuronal cells [2]. Using different proteomic approaches to study compositionally biased regions, which have traditionally been elusive to analysis due to their inherent insolubility, the authors identified hornerin, a serine-rich protein, to be a major component of the inclusions [1].
The molecular aetiology of NIID had remained unresolved for decades since its first pathological characterisation until recently, when a GGC repeat expansion in the 5’UTR of the human-specific NOTCH2NLC gene mainly associated with disease in the East Asian population was discovered [3, 4]. This abnormal expansion of GGC repeats has since heralded a new disease entity of polyglycine disorders [5], with evidence for canonical translation of the repeat into a pathogenic polyglycine-containing protein that co-localises with p62-positive NIIs in NIID [6]. However, NIID is genetically heterogeneous, with the GGC repeat expansion in NOTCH2NLC being rare in Europeans [7].
Thus, Park and colleagues rightfully assessed NII composition in the post-mortem brain of an individual of European (Finnish) ancestry with juvenile-onset NIID, not associated with the NOTCH2NLC repeat expansion [7, 8], to gain further insight into the currently unknown molecular mechanism of disease within European individuals. While hornerin deposits were detected within the inclusions, a heterozygous missense variant in the hornerin (HRNR) gene exon 3: NM_001009931.3: c.3023 G > C, p.(Ser1008Thr) was the only variant found on whole exome sequencing, although in silico analysis and a Finnish allele frequency of 0.001748 (within gnomAD v.3.1.2 [9]) deemed it to be unlikely pathogenic.
In order to investigate the genetic basis, extrapolating from the formation of hornerin within the inclusions of the one European case by Park et al. [1], we screened for HRNR variants in a large series of ten additional historical cases of pathologically confirmed NIID in patients of European ancestry (confirmed on genotypying), in whom the causative GGC repeat expansion in NOTCH2NLC was not found (Table 1) [7]. Furthermore, we also reviewed HRNR variants in an European patient with antemortem diagnosis of NIID associated with GGC repeat expansion in NOTCH2NLC [7] as well as confirmation in the index case reported by Park and colleagues [7]. We used polymerase chain reaction (PCR) to amplify the 446 base pair region of HRNR containing the index variant using conditions by Park et al. [1] followed by Sanger sequencing to review the targeted sequence (Additional file 1: Methods).
Table 1.
Demographics, clinical presentation and pathological findings of cases of neuronal intranuclear inclusion disease (NIID) examined, with resulting analysis for HRNR variants
| ID | Age of onset | Age at death | Sex | Family history | Country of origin | Clinical Diagnosis/Presentation pre-biopsy | Main pathological findings and site of pathology | HRNR variant | Estimated number of GGC repeats in NOTCH2NLC | |
|---|---|---|---|---|---|---|---|---|---|---|
| Allele 1 | Allele 2 | |||||||||
| 1 | 17 | 24 | M | Yes | UK | Young-onset parkinsonism and dysautonomia | Widespread neuronal hyaline intranuclear inclusions immunoreactive for ubiquitin and p62 (brain) | No variants detected | 21 | - |
| 2 | 33 | 46 | M | Yes | Australia | Slowly progressive motor and sensory neuronopathy with ataxia | Eosinophilic neuronal intranuclear inclusions (brain) | No variants detected | 22 | 28 |
| 3 | 60 s | 67 | F | No | Australia | Unknown presentation | Eosinophilic intranuclear inclusions in pyramidal cells (brain) | c.3236 G > A, p.(Glu1054Lys), synonymous c.3346 C > T | 15 | 20 |
| 4 | 52 | 72 | F | No | Australia | Slowly progressive primary lateral sclerosis | Cortical neuronal and astrocytic intranuclear inclusions (brain) | No variants detected | 15 | 23 |
| 5 | 11 | 21 | F | Yes (monozygotic twin) | Finland | Ataxia, seizures, and extrapyramidal symptoms | Inclusion bodies in most nerve cell types of central and peripheral nervous systems, as well as in occasional astrocytes | c.3023 G > C, p.(Ser1008Thr) – confirmation of variant found in the same individual by Park et al.[1] | 19 | 22 |
| 6 | 49 | 62 | F | Yes | Spain | Ataxia | Intranuclear hyaline inclusions in neurons and glia in widespread areas of the brain immunoreactive for ubiquitin (brain) | No variants detected | 15 | 25 |
| 7 | 82 | 84 | F | Yes | Spain | Dementia | Intranuclear hyaline inclusions in neurons and glia in widespread areas of the brain immunoreactive for ubiquitin (brain) | No variants detected | 16 | 23 |
| 8 | 26 | - | F | No | USA | Unknown presentation | Pathological changes in keeping with NIID (brain) | No variants detected | 17 | 23 |
| 9 | 84 | - | M | No | USA | Alzheimer’s disease, ataxia | Intranuclear hyaline inclusions in neurons and glia in widespread areas of the brain (brain) | c.3236 G > A, p.(Glu1054Lys), synonymous c.3346 C > T | 15 | 19 |
| 10 | 69 | - | M | No | USA | Diagnosed clinically with NIID | Neuronal intranuclear inclusions (brain) | No variants detected | 14 | 27 |
| 11 | 80 | - | M | No | USA | Unknown presentation | Neuronal intranuclear inclusions (brain) | c.3236 G > A, p.(Glu1054Lys), synonymous c.3346 C > T | 19 | - |
| 12 | 51 | N/A | F | No | Ukraine | Recurrent encephalopathy and migraines | NOTCH2NLC repeat expansion positive NIID: Antemortem biopsy contains p62 positive intranuclear inclusions (skin) | No variants detected | 19 | 92–106 |
All cases were previously investigated for the NOTCH2NLC GGC repeat expansion with sizing of the repeat sequence through repeat-primed PCR [7]. NIID cases 1 to 11 were not associated with repeat expansion in NOTCH2NLC. Case 12 was the only case found to have a GGC repeat expansion in NOTCH2NLC to be associated with NIID, with repeat sizing from Oxford Nanopore Technologies long-read sequencing. Case 5 is the case investigated by Park and colleagues [1, 8]
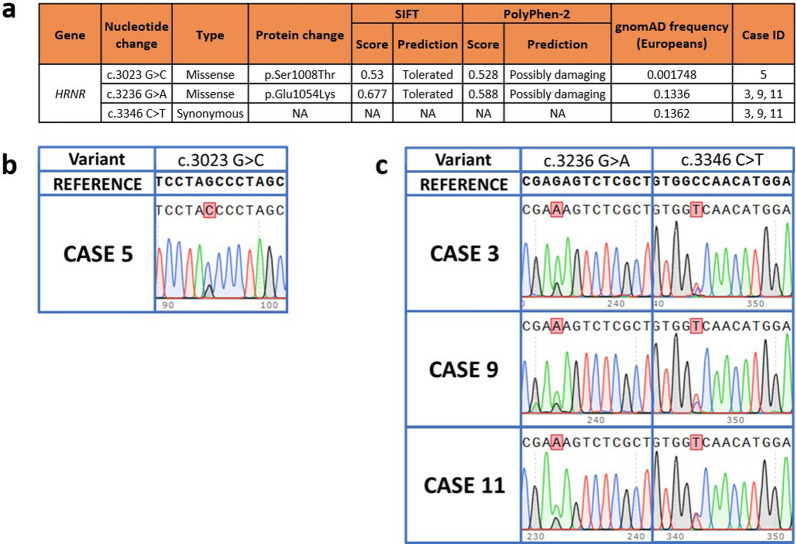
The previously reported p.(Ser1008Thr) variant in HRNR was verified in DNA extracted from heart tissue of the index case using this approach. However, none of the other ten pathologically confirmed NIID cases harboured the same reported HRNR variant (Table 1) despite sharing the common characteristic of an absent pathogenic NOTCH2NLC repeat expansion and pathological presence of NIIs. Out of these cases, three further European NIID cases diagnosed pathologically through post-mortem brain examination (Cases 3, 9 and 11 in Table 1) were found to have two variants in HRNR: a missense variant (c.3236 G > A, p.(Glu1054Lys)) and a synonymous variant (c.3346 C > T) (Fig. 1). However, in silico analysis and prevalent European population frequencies [9] (0.1336 and 0.1362 for the missense and synonymous variants respectively) suggest that these are unlikely to be pathogenic candidates (Fig. 1). As expected, for the patient in which NOTCH2NLC repeat expansion was found to be associated with NIID (Case 12), no HRNR variants were detected on Sanger sequencing. Moreover, the expression of HRNR is not enriched within the central nervous system with low human brain region-specific expression, as exemplified in the Genotype-Tissue Expression (GTEx) project [10].
Fig. 1.
Characteristics of variants detected in HRNR in neuronal intranuclear inclusion disease (NIID). a Table showing in silico predictions of all variants detected across 12 NIID samples. Sorting Intolerant from Tolerant (SIFT) (https://sift.bii.a-star.edu.sg/) predicts if a substitution at the amino acid level affects protein function with scores ranging from 0 to 1. A variant is predicted damaging to protein function if the score is ≤ 0.05 and tolerated if the score is > 0.05. Polymorphism Phenotyping version 2 (PolyPhen-2) (http://genetics.bwh.harvard.edu/pph2/) is a tool that predicts the possible effect of an amino acid substitution on protein function, with scores ranging from 0 (most probably benign) to 0.999 (most probably damaging). b The c.3023 G > C variant detected in Case 5, but not in any other cases, verifies the findings from Park and colleagues[1]. This variant of interest is highlighted in the chromatogram. c Missense variant c.3236 G > A and synonymous variant c.3346 c > T found in cases 3, 9 and 11. These variants of interest are highlighted in the chromatogram
Taken together, these findings support those of Park and colleagues, albeit in a larger cohort of NOTCH2NLC-negative NIID in patients of European ancestry. The molecular basis of disease in these cases, which are genetically distinct from East Asian NIID cases, is unlikely to be secondary to single nucleotide variation within HRNR. It should be noted that while the identification of hornerin as a major component of NIIs in this Finnish case [8] is of interest in providing further molecular insight into the pathogenesis of NOTCH2NLC repeat-negative NIID, further direct identification of NII composition in other such molecularly undetermined cases [7] is essential in moving towards establishing the underlying aetiology. The identification of a common genetic explanation for European NIID has thus far remained elusive due to the lack of large pedigrees, a likely complex variant that has eluded conventional sequencing techniques, paucity of antemortem diagnostic clues (as seen in East Asian NIID) and the clinical and genetic heterogeneity of disease. As such, the overarching clue to driving a molecular diagnosis may lie in the accurate pathological characterisation of such disorders, as attempted by Park and colleagues [1], in order to decipher convergent mechanisms for pathogenesis.
Supplementary Information
Additional file 1. Supplementary Methods.
Acknowledgements
The authors thank the participants and their families for their generous donations, without which this work would not have been possible.
Abbreviations
- NIL
Neuronal intranuclear inclusions
- NIID
Neuronal intranuclear inclusion disease
- PCR
Polymerase chain reaction
Author contributions
HL, HH and ZC conceived and designed the study. HL, EKG, HM, ND, KZ, KM, CA, WYY and SE performed experimental analyses for the study. ZJ and MJH provided pathological interpretation. CT, JH, TR, TL, MD, DWD, KAJ, EG, GGK, GH, DBR, IB, LMP, EM, PJT, ASW, NCF, NWW, AJL, and MJH all provided pathological samples, or patient data. ZC, HH, MR and AT supervised the project. All authors discussed the results and contributed to the final manuscript.
Funding
This work was funded by Leonard Wolfson Foundation grant 157793 and Medical Research Council grant MR/S01165X/1. ZC was funded by a Leonard Wolfson Clinical Research Fellowship (Grant number 157793). ZJ is supported by the Department of Health’s NIHR UCLH/UCL Biomedical Research Centre’s funding scheme. AT is a Medical Research Council Clinician Scientist (MR/S006753/1). MR is supported through the award of a Tenure Track Medical Research Council Clinician Scientist Fellowship (MR/N008324/1).
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by UCL Queen Square Institute of Neurology Institutional Review Board.
Consent for publication
The participants have provided consent for publication of data.
Competing interests
The authors have no financial and non-financial competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Park H, Yamanaka T, Toyama Y, Fujita A, Nirasawa T, Murayama S, et al. Hornerin deposits in neuronal intranuclear inclusion disease: direct identification of proteins with compositionally biased regions in inclusions. Acta Neuropathol Commun. 2022;10(1):1–17. doi: 10.1186/s40478-022-01333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sone J, Mori K, Inagaki T, Katsumata R, Takagi S, Yokoi S, et al. Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain. 2016;139(12):3170–3186. doi: 10.1093/brain/aww249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sone J, Mitsuhashi S, Fujita A, Mizuguchi T, Hamanaka K, Mori K, et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet. 2019;51(8):1215–1221. doi: 10.1038/s41588-019-0459-y. [DOI] [PubMed] [Google Scholar]
- 4.Ishiura H, Shibata S, Yoshimura J, Suzuki Y, Qu W, Doi K, et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nature Genet. 2019;51(8):1222–1232. doi: 10.1038/s41588-019-0458-z. [DOI] [PubMed] [Google Scholar]
- 5.Liufu T, Zheng Y, Yu J, Yuan Y, Wang Z, Deng J, et al. The polyG diseases: a new disease entity. Acta Neuropathol Commun. 2022;10(1):79. doi: 10.1186/s40478-022-01383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boivin M, Deng J, Pfister V, Grandgirard E, Oulad-Abdelghani M, Morlet B, et al. Translation of GGC repeat expansions into a toxic polyglycine protein in NIID defines a novel class of human genetic disorders: the polyG diseases. Neuron. 2021;109(11):1825–35.e5. doi: 10.1016/j.neuron.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Yan Yau W, Jaunmuktane Z, Tucci A, Sivakumar P, GaglianoTaliun SA, et al. Neuronal intranuclear inclusion disease is genetically heterogeneous. Ann Clin Transl Neurol. 2020;7(9):1716–1725. doi: 10.1002/acn3.51151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haltia M, Somer H, Palo J, Johnson WG. Neuronal intranuclear inclusion disease in identical twins. Ann Neurol. 1984;15(4):316–321. doi: 10.1002/ana.410150403. [DOI] [PubMed] [Google Scholar]
- 9.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Methods.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.