Abstract
Background
The stalk traits stalk diameter, stalk length, rind penetrometer resistance and dry matter content are important indicators for measuring lodging resistance.
Results
In this study, 377 inbred lines were used as the basic materials, and four stalk-related traits including stalk diameter, stalk length, rind penetrometer resistance and dry matter content of the third segment of maize, were investigated at the tasseling, grain filling, and maturity stages. 461,053 high-quality SNPs which were obtained by whole genome resequencing were used for genome-wide association study. As a result of mixed linear model analysis (P < 9.77 × 10–6), 29 significant SNPs related to traits were detected, accounting for 7.19% -15.03% of phenotypic variation, among which 4, 1, 4 and 20 SNPs were found related to rind penetrometer resistance, stalk diameter, stalk length, and dry matter content respectively. Most candidate genes are related to plant element structure, signal transduction mechanisms, inorganic ion transport and metabolism, nucleotide transport and metabolism, and transporter enzyme families. Comparing mixed linear model with generalized linear model, a total of 12 candidate genes were detected repeatedly, during which the candidate gene Zm00001d014449 were detected 5 times, with a phenotypic variation interpretation rate of 9.95% -10.84%. This gene is mainly expressed in cells with active cell division and tissue differentiation, and is involved in the formation of stalk vascular bundles and the synthesis of cell walls. Another candidate gene, Zm00001d005300, encodes the transcription factor MYB44, which regulates the dependence of salt stress signal phosphorylation, can effectively inhibit the accumulation of destructive reactive oxygen species, and has a certain resistance to non-biotic stress. In addition, this study also found that 10 unknown functional genes can be further Functional verification.
Conclusions
This study helps to deepen the understanding of the genetic basis of traits related to maize stalk lodging resistance, and provides theoretical guidance for future maize lodging resistance breeding.
Keywords: Maize, Stalk traits, Lodging resistance, Dry matter content, Genome-wide association study
Background
Maize (Zea mays L.) is one of the main food crops in the world and plays a crucial role in Chinese agricultural production [1]. In recent years, lodging has become a key factor restricting maize production, seriously affects maize yield and quality [2]. The maize lodging resistance is influenced by multiple factors, such as plant height (PH), ear height (EH), stalk diameter (SD), stalk length (SL), rind penetrometer resistance (RPR) and dry matter content (DMC), which are all significantly correlated with lodging resistance [3]. Domestic and international research on the stalk lodging resistance mostly focuses on the third internode. The stalk diameter, stalk length, epidermal puncture strength, and dry matter content can well reflect the lodging resistance of stalk and have strong correlations with lodging resistance [4].
The genetic effects of maize stalk traits are controlled by complex polygenetic quantitative traits, with both additive and dominant effects playing important roles. In the study of rind penetrometer resistance, by using genome-wide association analysis method in a natural population, scholars have detected 16 SNPs significantly related to stalk thrust resistance, and obtained 5 association candidate genes [2]; using the same method in segregating populations, two scholars found the presence of QTLs in the marker intervals SNP1950-SNP1953 and phi031-bnlg1702 on chromosome 6, respectively, and expressed them stably in multiple environments [5, 6]; some scholars have found that QTLs for rind penetrometer resistance are clustered in the chromosome segments 1, 3, and 5 of maize [7], and are associated with lignin synthesis and phenylpropanoid pathway genes [8]. However, some scholars have found that in addition to chromosome 5 or 6, QTLs for rind penetrometer resistance are distributed throughout the entire genome [9, 10], and these QTLs explain phenotypic variations ranging from 4.4% to 18.9% [11]. In terms of stalk diameter research, using isolated populations, some scholars have found that there is a main QTL in the 114.3–131.5cM region of chromosome 9 in multiple sites [7]; furthermore, through the study of the infiltration line population constructed with Ruminant Grass, 5 significant stalk-diameter associated loci were detected on chromosomes 1,2,5,6 and 7, 13 associated candidate genes were screened around the loci with the highest phenotypic contribution rate of 8.32% [12]; some scholars have also used natural populations to detect 12 SNP loci significantly associated with stalk diameter and identified 2 candidate genes [13]; 20 stalk diameter thickness consistent QTL regions were also obtained through map integration and meta-analysis methods, resulting in two candidate genes GRMZM2G307588 and GRMZM2G089836 for stalk diameter [14]. In terms of stalk length traits, Zhang Jianhua et al. [15] used 79 DH populations as materials to locate QTLs for stalk length on chromosome 3, bnlg1144-phi036. Li Fang et al. [12] detected significant SNP loci for stalk length on chromosomes 1 and 4.
Most of the previous studies focused on analyzing traits such as stalk puncture strength in isolated populations, but there are limited researches on stalk dry matter content in populations with wide diversity. In the present study, we used a GWAS population with wide diversity composed of 377 inbred lines for genetic analysis on stalk lodging resistance related traits. This study is aimed to explore candidate SNPs/genes controlling stalk lodging resistance at the molecular level, provide theoretical basis for genetic improvement of stalk lodging resistance traits, and further provide references for cultivating new maize varieties with strong lodging resistance.
Results
Phenotypic analysis of lodging resistance related traits in maize stalk
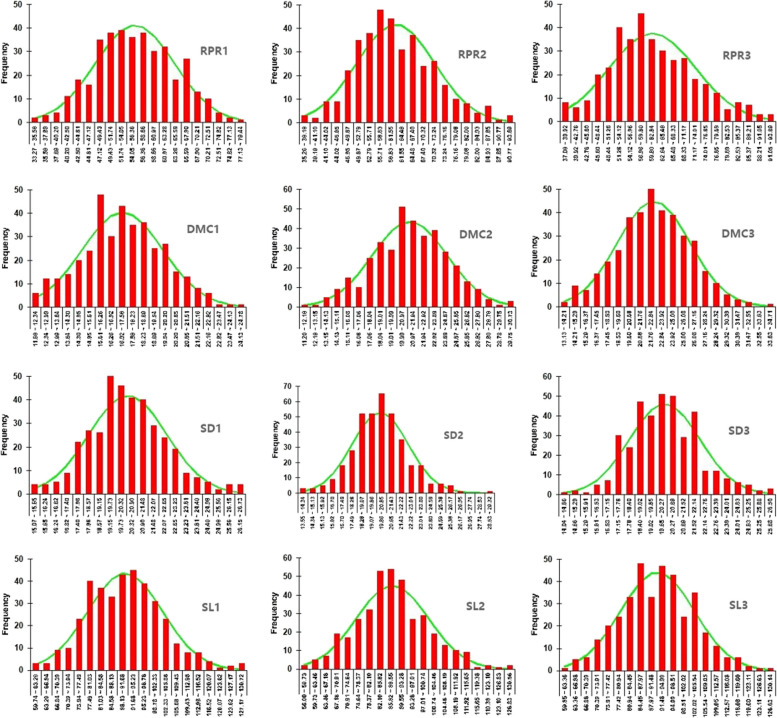
The basic description statistics and frequency distribution of the correlation between stalk lodging resistance in different growth stages of maize are shown in Table 1 and Fig. 1. The variation range of each character is large and the phenotypic diversity is relatively rich. The absolute value of skewness and kurtosis of each character is less than 1. Combining with the histogram of character distribution frequency, it basically presents a normal distribution trend and conforms to the typical quantitative character characteristics. The generalized heritability of stalk lodging resistance related traits at different growth stages in different years is more than 60%, indicating that these traits are mainly affected by genetic factors.
Table 1.
Statistical analysis of lodging resistance related traits in maize stalk at different growth stages
| Trait | Range | Mean | CV (%) | Skewness | Kurtosis | Broad-sense heritability |
|---|---|---|---|---|---|---|
| RPR1(N/mm2) | 33.27-79.44 | 55.77 | 15.1 | 0.07 | -0.4 | 0.766 |
| RPR2(N/mm2) | 35.27-93.69 | 61.34 | 17.16 | 0.46 | 0.15 | |
| RPR3(N/mm2) | 37.08-93.89 | 61.37 | 18.48 | 0.4 | -0.1 | |
| DMC1(%) | 11.68-24.78 | 17.27 | 14.11 | 0.06 | -0.31 | 0.643 |
| DMC2(%) | 11.20-30.73 | 21.16 | 15.89 | -0.05 | -0.06 | |
| DMC3(%) | 13.13-34.71 | 22.57 | 15.99 | 0.02 | -0.04 | |
| SD1(mm) | 15.07-26.73 | 20.38 | 10.26 | 0.36 | 0.37 | 0.648 |
| SD2(mm) | 13.55-29.32 | 20.13 | 11.1 | 0.26 | 0.98 | |
| SD3(mm) | 14.04-26.50 | 20.12 | 10.05 | 0.35 | 0.43 | |
| SL1(mm) | 59.74-130.72 | 90.78 | 13.46 | 0.34 | 0.21 | 0.714 |
| SL2(mm) | 56.00-130.56 | 88.21 | 14.05 | 0.32 | 0.36 | |
| SL3(mm) | 59.85-130.14 | 91.71 | 13.03 | 0.14 | -0.04 |
Fig. 1.
Frequency distribution map of lodging resistance related traits in maize stalk
Analysis of variance and correlation among various traits of the tested inbred lines
The analysis of variance for lodging resistance related traits in 377 inbred line materials (Table 2) showed that the genotype effects of traits such as RPR, DMC, SD, SL reached a highly significant level (P < 0.01), indicating significant genetic variation in lodging resistance related traits in the natural population. In addition, the correlation analysis results between various traits showed (Table 3) that there was a close correlation and mutual promotion between traits such as RPR, DMC, SD, and SL in maize stalk lodging resistance. In 2016 and 2017, the correlation between each trait was basically the same. Over the course of two years, RPR reached a highly significant positive correlation with DMC, SL traits (P < 0.01), while RPR was positively correlated with SD, but did not reach a significant level. DMC showed a highly significant negative correlation with SD, while DMC showed a highly significant positive correlation with SL.
Table 2.
Variance analysis of various traits in different maize inbred lines
| Trait | Variation | Sum of squares | DF | SD | F |
|---|---|---|---|---|---|
| RPR | Environment | 7840.1967 | 2 | 3920.0983 | 161.596** |
| Genotype | 98467.414 | 376 | 261.8814 | 10.795** | |
| Error | 18242.541 | 752 | 24.2587 | ||
| DMC | Environment | 5675.8017 | 2 | 2837.9008 | 788.143** |
| Genotype | 8672.4047 | 376 | 23.0649 | 6.406** | |
| Error | 2707.7603 | 752 | 3.6007 | ||
| SD | Environment | 16.3361 | 2 | 8.1681 | 5.173** |
| Genotype | 3872.2911 | 376 | 10.2986 | 6.522** | |
| Error | 1187.448 | 752 | 1.5791 | ||
| SL | Environment | 2482.4769 | 2 | 1241.2385 | 29.275** |
| Genotype | 135669.28 | 376 | 360.8225 | 8.51** | |
| Error | 31884.682 | 752 | 42.3998 |
Note: * or * * indicate a significant correlation at the 0.05 or 0.01 level.
Table 3.
Correlation analysis of various traits in different maize inbred lines
| Trait | RPR | DMC | SD | SL |
|---|---|---|---|---|
| RPR | 1 | 0.55** | 0.07 | 0.34** |
| DMC | 0.56** | 1 | -0.22** | 0.17** |
| SD | 0.05 | -0.36** | 1 | 0.05 |
| SL | 0.15** | 0.13** | -0.03 | 1 |
Note: The lower left represents the correlation between various traits in 2016, the upper right represents the correlation between various traits in 2017, and * or * * indicates a significant correlation at the 0.05 or 0.01 level.
Genome-wide association study of stalk puncture intensity and dry matter content at different growth stages of maize
The genome-wide association study was conducted using GLM and MLM models by Tassel 5.0, and the Manhattan map and QQ map were drawn (Fig. 2 and Fig. 3). The inbred line population used in this study has rich genetic basis, which will cause a certain deviation from the confidence interval on the QQ map. SNPs exceeding the threshold are sites significantly related to the target traits of this experiment. The GLM model was used to analyze the correlation between stalks at different growth stages over two years. A total of 41 significantly associated SNPs loci were detected on 10 chromosomes of maize (Table 4). 3, 9, 7, 2, 14, 2, 0, 1, 1, and 2 SNPs were detected on chromosomes 1–10 respectively. A total of 18 SNPs (RPR1: 4, RPR2: 7, RPR3: 7) were detected related to RPR, as well as 19 SNPs (DMC1: 3, DMC2: 3, DMC3: 13) related to DMC, 4 SNPs (SL2: 3, SL3: 1) related to SL, while explaining the phenotypic variation ranged from 8.67% to 14.96%, with the highest phenotypic interpretation rate being SL2 on chromosome 10 at 70,989,765 and the lowest being RPR2 on chromosome 1 at 51,729,873. Using the MLM model to perform correlation analysis on stalk traits at different growth stages over two years, a total of 29 significantly correlated SNPs loci were detected on 10 chromosomes of maize, with 1, 4, 3, 2, 16, 0, 1, 1, 0, and 1 SNPs being detected on chromosomes 1–10 respectively. A total of 4 SNPs (RPR1: 1, RPR2: 1, RPR3: 2) were detected related to RPR, as well as 20 SNPs (DMC2: 3, DMC3: 17) related to DMC, 4 SNPs (SL2: 3, SL3: 1) related to SL, and 1 SNP (SD3: 1) related to SD, explaining a phenotypic variation range of 7.19% -15.03%, with SL2 in chromosome 10_70989765 accounting for the highest phenotypic interpretation rate, and RPR2 on chromosome 7 accounting for the lowest.
Fig. 2.
Genome-wide association study of stalk lodging resistance under GLM model Manhattan map and QQ map. From the inside to the outside, the SNP sites identified in the RPR, DMC, SD and SL of maize in different years are represented respectively; The red highlights represent the significant SNP loci of each trait at 0.01 levels, respectively; Figure b shows the QQ chart of stalk related traits of maize in different years
Fig. 3.
Genome-wide association study of stalk lodging resistance under MLM model Manhattan map and QQ map. Note in the figure is shown in Fig. 2
Table 4.
Significant locus information on the correlation of maize stalk lodging resistance
| Trait | Marker | Chr | Model | Pos | p | marker_Rsq% |
|---|---|---|---|---|---|---|
| RPR1 | 1_5925472 | 1 | GLM | 5925472 | 9.62E-07 | 10.248 |
| RPR2 | 1_51729873 | 1 | GLM | 51729873 | 6.17E-07 | 7.19 |
| SL2 | 1_105656256 | 1 | GLM | 105656256 | 8.67E-08 | 12.758 |
| RPR2 | 2_33979628 | 2 | GLM | 33979628 | 5.66E-07 | 8.478 |
| RPR3 | 2_73833386 | 2 | GLM | 73833386 | 8.37E-07 | 10.155 |
| DMC1 | 2_146172998 | 2 | GLM | 146172998 | 8.96E-07 | 7.729 |
| RPR3 | 2_156417807 | 2 | GLM | 156417807 | 5.07E-07 | 9.308 |
| RPR2 | 2_162859353 | 2 | GLM | 162859353 | 5.34E-07 | 8.939 |
| RPR2 | 2_162876327 | 2 | GLM | 162876327 | 2.48E-07 | 8.909 |
| RPR3 | 2_162876327 | 2 | GLM | 162876327 | 3.64E-07 | 9.596 |
| DMC2 | 2_168529088 | 2 | GLM | 168529088 | 6.57E-08 | 12.234 |
| DMC2 | 2_170637158 | 2 | GLM | 170637158 | 1.85E-07 | 9.83 |
| RPR3 | 3_12279836 | 3 | GLM | 12279836 | 7.85E-07 | 8.754 |
| RPR2 | 3_50985869 | 3 | GLM | 50985869 | 1.30E-08 | 8.933 |
| RPR3 | 3_50985869 | 3 | GLM | 50985869 | 2.01E-08 | 9.606 |
| RPR2 | 3_119840946 | 3 | GLM | 119840946 | 4.30E-07 | 8.151 |
| RPR2 | 3_119865757 | 3 | GLM | 119865757 | 3.58E-07 | 7.427 |
| RPR3 | 3_175959350 | 3 | GLM | 175959350 | 9.79E-07 | 8.615 |
| DMC3 | 3_204520247 | 3 | GLM | 204520247 | 5.54E-08 | 11.482 |
| RPR1 | 4_178358908 | 4 | GLM | 178358908 | 4.40E-08 | 10.212 |
| DMC1 | 4_193185052 | 4 | GLM | 193185052 | 4.15E-07 | 10.146 |
| DMC3 | 5_47645572 | 5 | GLM | 47645572 | 2.85E-07 | 9.473 |
| DMC3 | 5_47711262 | 5 | GLM | 47711262 | 2.20E-07 | 10.724 |
| DMC3 | 5_47726081 | 5 | GLM | 47726081 | 3.93E-07 | 10.314 |
| DMC3 | 5_47727153 | 5 | GLM | 47727153 | 1.69E-07 | 10.393 |
| DMC3 | 5_47747099 | 5 | GLM | 47747099 | 9.02E-07 | 9.871 |
| DMC3 | 5_47750439 | 5 | GLM | 47750439 | 6.37E-07 | 10.293 |
| DMC3 | 5_47752006 | 5 | GLM | 47752006 | 1.17E-07 | 9.95 |
| DMC3 | 5_47800595 | 5 | GLM | 47800595 | 9.66E-09 | 13.012 |
| DMC2 | 5_47800595 | 5 | GLM | 47800595 | 9.70E-07 | 9.958 |
| DMC3 | 5_47876150 | 5 | GLM | 47876150 | 2.95E-07 | 10.036 |
| DMC3 | 5_47880130 | 5 | GLM | 47880130 | 4.12E-07 | 10.631 |
| DMC3 | 5_47925170 | 5 | GLM | 47925170 | 4.67E-07 | 10.485 |
| SL3 | 5_160469150 | 5 | GLM | 160469150 | 9.40E-07 | 9.172 |
| RPR1 | 5_217360437 | 5 | GLM | 217360437 | 4.47E-07 | 8.571 |
| RPR1 | 6_66775760 | 6 | GLM | 66775760 | 3.72E-08 | 11.587 |
| RPR3 | 6_169956647 | 6 | GLM | 169956647 | 3.69E-07 | 9.647 |
| DMC3 | 8_159862794 | 8 | GLM | 159862794 | 1.33E-07 | 11.605 |
| DMC1 | 9_106888912 | 9 | GLM | 106888912 | 1.44E-07 | 8.732 |
| SL2 | 10_70989765 | 10 | GLM | 70989765 | 8.78E-08 | 15.028 |
| SL2 | 10_117358884 | 10 | GLM | 117358884 | 9.72E-07 | 10.855 |
| SL2 | 1_105656256 | 1 | MLM | 105656256 | 1.54E-06 | 12.397 |
| RPR3 | 2_73833386 | 2 | MLM | 73833386 | 7.57E-06 | 10.913 |
| DMC2 | 2_168529088 | 2 | MLM | 168529088 | 7.07E-06 | 10.684 |
| DMC2 | 2_170637158 | 2 | MLM | 170637158 | 1.18E-06 | 10.648 |
| SL2 | 2_176741537 | 2 | MLM | 176741537 | 7.49E-06 | 9.657 |
| RPR2 | 3_50985869 | 3 | MLM | 50985869 | 3.65E-06 | 9.437 |
| DMC3 | 3_200027631 | 3 | MLM | 200027631 | 7.06E-06 | 10.199 |
| DMC3 | 3_204520247 | 3 | MLM | 204520247 | 4.34E-07 | 12.685 |
| RPR3 | 4_185698633 | 4 | MLM | 185698633 | 2.88E-06 | 11.224 |
| SD3 | 4_201423345 | 4 | MLM | 201423345 | 9.69E-06 | 8.848 |
| DMC3 | 5_47645572 | 5 | MLM | 47645572 | 1.76E-06 | 10.081 |
| DMC3 | 5_47711262 | 5 | MLM | 47711262 | 1.81E-06 | 11.216 |
| DMC3 | 5_47716318 | 5 | MLM | 47716318 | 6.60E-06 | 10.094 |
| DMC3 | 5_47726081 | 5 | MLM | 47726081 | 2.61E-06 | 10.954 |
| DMC3 | 5_47727153 | 5 | MLM | 47727153 | 9.38E-07 | 10.829 |
| DMC3 | 5_47735500 | 5 | MLM | 47735500 | 8.41E-06 | 10.894 |
| DMC3 | 5_47744387 | 5 | MLM | 47744387 | 7.54E-06 | 10.115 |
| DMC3 | 5_47747099 | 5 | MLM | 47747099 | 6.11E-06 | 10.369 |
| DMC3 | 5_47750439 | 5 | MLM | 47750439 | 2.95E-06 | 10.845 |
| DMC3 | 5_47752006 | 5 | MLM | 47752006 | 7.82E-07 | 10.413 |
| DMC3 | 5_47800595 | 5 | MLM | 47800595 | 7.17E-08 | 13.681 |
| DMC2 | 5_47800595 | 5 | MLM | 47800595 | 4.85E-06 | 10.702 |
| DMC3 | 5_47876150 | 5 | MLM | 47876150 | 1.31E-06 | 10.497 |
| DMC3 | 5_47880130 | 5 | MLM | 47880130 | 3.47E-06 | 10.949 |
| DMC3 | 5_47925170 | 5 | MLM | 47925170 | 2.56E-06 | 11.001 |
| SL3 | 5_160469150 | 5 | MLM | 160469150 | 6.24E-06 | 9.498 |
| RPR1 | 7_167499865 | 7 | MLM | 167499865 | 9.77E-06 | 8.674 |
| DMC3 | 8_159862794 | 8 | MLM | 159862794 | 1.97E-06 | 11.998 |
| SL2 | 10_70989765 | 10 | MLM | 70989765 | 3.791E-06 | 14.962 |
Further analysis revealed that 21 SNPs related to stalk lodging resistance traits were detected more than twice by independent association analystis using GLM and MLM models (Table 4), suggesting that these loci carry important genes that can stably regulate stalk related traits in different environments. Among them, four SNPs were detected more than twice by independent association analyses at different growth stages with the same stalk related traits. It is speculated that these loci may carry important genes in the growth process of corn stalk related traits, which are continuously expressed and play a role in multiple growth stages. Using GLM and MLM models, 20 SNPs related to the same stalk traits were detected more than twice by independent association analyses, indicating that candidate genes carried by these loci are more likely to be stably expressed in maize stalk related traits. These SNPs that can be significantly correlated with stalk related traits multiple times have high confidence (HC) Among them, HC-SNP site 5_47800595 was found to be significantly correlated with DMC four times during multiple growth stages in different models, and during 5_47645572-5_47925170 segments, there are 27 significant SNPs found by different models that are associated with DMC. This segment is a region where DMC related genes are enriched. Loci adjacent to 3_50985869 segment were found to be significantly correlated with RPR traits three times. The segment 2_73833386 and 2_162876327 were found to be significantly correlated with RPR twice, respectively. Loci adjacent to 1_105656256、5_160469150 and 10_70989765 were found significantly correlateed with SL. Loci adjacent to 2_168529088、2_170637158、3_204520247 and 8_159862794 were found to be significantly correlated with DMC twice. This study has not yet found the same significant SNP for different stalk traits under model analysis, that is, no one cause multiple effect significant loci were found.
Analysis of genes associated with stalk related traits
Based on the correlation analysis between two models, candidate genes were screened and predicted within a 2.5kb range upstream and downstream of 70 significant SNPs which were associated with stalk related traits. Combined with gene functional annotation, 34 candidate genes were identified (Table 5). Among them, 22 and 12 candidate genes were obtained by the GLM model test and the MLM model test respectively, 12 candidate genes were detected more than twice. These genes may be high-confidence association genes for stalk related traits. Most of the candidate genes discovered are related to plant growth and development, carbohydrate transport and metabolism, signal transduction mechanism proteins, synthase family and cell growth regulation mechanism functions, signal transduction mechanism, inorganic ion transport and metabolism, nucleotide transport and metabolism, and transporter enzyme family. At the same time, this study also found 10 unknown functional genes, namely Zm00001d028925, Zm00001d005183, Zm00001d040577, Zm00001d041441, Zm00001d014448, LOC103641209, Zm00001d025403, LOC109945001, Zm00001d014448 and LOC103641209.
Table 5.
Prediction information of candidate gene
| Trait | Marker | marker_Rsq | Chr | Pos | p | Phenotypic variation explanation rate% | Gene accession number | Analysis model | Gene annotation |
|---|---|---|---|---|---|---|---|---|---|
| RPR2 | 1_51729873 | 0.0719 | 1 | 51729873 | 6.17E-07 | 7.19 | Zm00001d028924,Zm00001d028925 | GLM | NifU-like protein 1 chloroplastic;Uncharacterized protein |
| RPR2 | 2_33979628 | 0.08478 | 2 | 33979628 | 5.66E-07 | 8.48 | Zm00001d003157 | GLM | S-adenosylmethionine decarboxylase proenzyme S-adenosylmethionine decarboxylase alpha chain S-adenosylmethionine decarboxylase beta chain |
| RPR2 | 2_162859353 | 0.08939 | 2 | 162859353 | 5.34E-07 | 8.94 | Zm00001d005182 | GLM | Charged multivesicular body protein 2a |
| RPR2 | 2_162876327 | 0.08909 | 2 | 162876327 | 2.48E-07 | 8.91 | Zm00001d005183 | GLM | Uncharacterized protein |
| RPR3 | 2_162876327 | 0.09596 | 2 | 162876327 | 3.64E-07 | 9.60 | Zm00001d005183 | GLM | Uncharacterized protein |
| DMC2 | 2_168529088 | 0.10684 | 2 | 168529088 | 6.57E-08 | 12.23 | Zm00001d005300,Zm00001d000967,Zm00001d005301 | GLM | MYB transcription factor, Transcription factor MYB44;NA;DNA binding |
| DMC2 | 2_168529088 | 0.12234 | 2 | 168529088 | 7.07E-06 | 10.68 | Zm00001d005300, Zm00001d000967, Zm00001d005301 | MLM | MYB transcription factor, Transcription factor MYB44;NA;DNA binding |
| DMC2 | 2_170637158 | 0.0983 | 2 | 170637158 | 1.85E-07 | 9.83 | Zm00001d005351 | GLM | Arogenate dehydratase, 4.2.1.91 |
| DMC2 | 2_170637158 | 0.10648 | 2 | 170637158 | 1.18E-06 | 10.65 | Zm00001d005351 | MLM | Arogenate dehydratase, 4.2.1.91 |
| RPR3 | 3_12279836 | 0.08754 | 3 | 12279836 | 7.85E-07 | 8.75 | Zm00001d039695 | GLM | S-adenosyl-L-methionine-dependent methyltransferase superfamily protein |
| RPR2 | 3_50985869 | 0.08933 | 3 | 50985869 | 3.65E-06 | 9.44 | LOC109945001 | MLM | Unknow |
| RPR2 | 3_50985869 | 0.09606 | 3 | 50985869 | 1.30E-08 | 8.93 | Zm00001d040577 | GLM | Uncharacterized protein |
| RPR3 | 3_50985869 | 0.09437 | 3 | 50985869 | 2.01E-08 | 9.61 | Zm00001d040577 | GLM | Uncharacterized protein |
| RPR2 | 3_119865757 | 0.07427 | 3 | 119865757 | 3.58E-07 | 7.43 | Zm00001d041441 | GLM | Unknow |
| RPR3 | 3_175959350 | 0.08615 | 3 | 175959350 | 9.79E-07 | 8.62 | Zm00001d042665 | GLM | Transcription repressor MYB6 |
| DMC3 | 3_204520247 | 0.11482 | 3 | 204520247 | 5.54E-08 | 11.48 | Zm00001d043592,Zm00001d043593 | GLM | SPA1-related 3;CESA4 (CELLULOSE SYNTHASE A4) |
| DMC3 | 3_204520247 | 0.12685 | 3 | 204520247 | 4.34E-07 | 12.69 | Zm00001d043592,Zm00001d043593 | MLM | SPA1-related 3;CESA4 (CELLULOSE SYNTHASE A4) |
| RPR1 | 4_178358908 | 0.10212 | 4 | 178358908 | 4.40E-08 | 10.21 | Zm00001d052068 | GLM | Pathogenesis-related protein-1-like |
| SD3 | 4_201423345 | 0.08848 | 4 | 201423345 | 9.69E-06 | 8.85 | Zm00001d052804 | MLM | MYB transcription factor, Putative MYB DNA-binding domain superfamily protein |
| DMC3 | 5_47711262 | 0.10724 | 5 | 47711262 | 2.20E-07 | 10.72 | Zm00001d014448 | GLM | Unknow |
| DMC3 | 5_47711262 | 0.11216 | 5 | 47711262 | 1.81E-06 | 11.22 | Zm00001d014448 | MLM | Unknow |
| DMC3 | 5_47716318 | 0.10094 | 5 | 47716318 | 6.60E-06 | 10.09 | Zm00001d014448 | MLM | Unknow |
| DMC3 | 5_47744387 | 0.10115 | 5 | 47744387 | 7.54E-06 | 10.12 | Zm00001d014449 | MLM | Chromo domain-containing protein LHP1,Chromdomain-containing protein CRD101, Chromo domain-containing protein LHP1 |
| DMC3 | 5_47750439 | 0.10293 | 5 | 47750439 | 6.37E-07 | 10.29 | Zm00001d014449 | GLM | Chromo domain-containing protein LHP1,Chromdomain-containing protein CRD101, Chromo domain-containing protein LHP1 |
| DMC3 | 5_47750439 | 0.10845 | 5 | 47750439 | 2.95E-06 | 10.85 | Zm00001d014449 | MLM | Chromo domain-containing protein LHP1,Chromdomain-containing protein CRD101, Chromo domain-containing protein LHP1 |
| DMC3 | 5_47752006 | 0.0995 | 5 | 47752006 | 1.17E-07 | 9.95 | Zm00001d014449 | GLM | Chromo domain-containing protein LHP1,Chromdomain-containing protein CRD101, Chromo domain-containing protein LHP1 |
| DMC3 | 5_47752006 | 0.10413 | 5 | 47752006 | 7.82E-07 | 10.41 | Zm00001d014449 | MLM | Chromo domain-containing protein LHP1,Chromdomain-containing protein CRD101, Chromo domain-containing protein LHP1 |
| SL3 | 5_160469150 | 0.09172 | 5 | 160469150 | 9.40E-07 | 9.17 | Zm00001d014451 | GLM | Glutamate receptor 3.4 |
| SL3 | 5_160469150 | 0.09498 | 5 | 160469150 | 6.24E-06 | 9.50 | Zm00001d014451 | MLM | Glutamate receptor 3.4 |
| RPR1 | 5_217360437 | 0.08571 | 5 | 217360437 | 4.47E-07 | 8.57 | Zm00001d018247 | GLM | Zinc finger protein |
| RPR3 | 6_169956647 | 0.09647 | 6 | 169956647 | 3.69E-07 | 9.65 | Zm00001d039094 ;Zm00001d039095 | GLM | 3-ketoacyl-CoA synthase, 2.3.1.-;Uncharacterized protein |
| SL2 | 10_70989765 | 0.14962 | 10 | 70989765 | 3.79E-06 | 14.96 | LOC103641209 | MLM | Unknow |
| SL2 | 10_70989765 | 0.15028 | 10 | 70989765 | 8.78E-08 | 15.03 | LOC103641209 | GLM | Unknow |
| SL2 | 10_117358884 | 0.10855 | 10 | 117358884 | 9.72E-07 | 10.86 | Zm00001d025403 | GLM | Unknow |
Identification and functional analysis of high-confidence associated genes
Using two models, a total of 12 candidate genes were detected more than twice. Among them, Zm00001d014449 was found to be associated with DMC five times, with a phenotypic variation interpretation rate of 9.95% -10.84%. This gene originates from the assembly of CRD101 and is associated with the LHP1 gene, which is mainly expressed in cells undergoing cell division and tissue differentiation, such as in vascular bundle synthesis, which is of great significance for maintaining cell and tissue structural specificity. Zm00001d014449 also participates in regulating the expression of a large number of flowering genes. Zm00001d014448 was found to be associated with DMC three times, with a phenotypic variation explanation rate of 10.09% -11.22%. The function of this gene is unknown. LOC103641209 was found to be associated with SL twice, with phenotypic variation explanatory rates of 15.03% and 14.96%. The function of this gene is unknown. Zm00001d005183 was found to be associated with the RPR twice, with phenotypic variation explanatory rates of 8.91% and 9.60%. The function of this gene is unknown. Zm00001d005300, Zm00001d000967, and Zm00001d005301 were found twice to be associated with DMC, with phenotypic variation explanatory rates of 10.68% and 12.23%. Zm00001d005300 encodes the transcription factor MYB44, Persak Helene et al. [16] found that MYB44 regulates the dependence of salt stress signal phosphorylation, can effectively inhibit the accumulation of destructive reactive oxygen species, and has a certain resistance to abiodic stress. No functional reports have been found on the Zm00001d000967 gene. Zm00001d005301 is related to the DNA binding domain. Zm00001d005351 was found twice to be associated with DMC traits, with phenotypic variation explanatory rates of 9.83% and 10.65%. This gene is involved in amino acid synthesis, encodes pre phenylalanine dehydrogenase, and matches the biosynthetic pathways of phenylalanine, tyrosine, and tryptophan. Zm00001d014451 was found twice to be associated with SL, with phenotypic variation explanatory rates of 9.17% and 9.50%. This gene belongs to the glutamic acid receptor (GLR) family gene in maize, and may be related to the opening or closing of ion channels. Under environmental stress, gene expression significantly decreases [17]. Zm00001d040577 was found twice to be associated with the RPR, with phenotypic variation explanatory rates of 9.43% and 9.61%, and no characteristic protein was found to express by this gene. Zm00001d043592 and Zm00001d043593 were found to be related to DMC traits twice, with phenotypic variation interpretation rates of 11.48% and 12.69%. This gene expresses cellulose synthase A4, which is a specific signal intermediate of Phytochrome a, and is used as a light dependent inhibitor of photomorphogenesis in Arabidopsis seedlings [18].
Discuss
Comparative analysis of significant SNPs loci related to stalk traits
Twenty-one HC-SNPs were found by GLM and MLM tests, and then a comparative analysis was conducted according to the genetic mapping results of stalk related traits at home and abroad. Xie [4] used 301 inbred lines as experimental materials to conduct GWAS analysis on the puncture strength of corn stalks, and found a significant locus 172,176,633 on chromosome 4, which is close to the significant SNPs locus 4_178358908 related to RPR identified in this study. Liu [19] found that there were multiple QTLs at different growth stages in maize stalk puncture intensity on chromosomes 1 of 49.39Mb-58.5Mb, 2 of 156.86Mb-174.98Mb, 4 of 177.15Mb-189.04Mb, and 5 of 46.08Mb-69.55Mb and 208.1Mb-210.4Mb, respectively. The association analysis results of this study showed that RPR significant SNPs were independently detected at the 51729873 bp locus on chromosome 1, and two differentially expressed genes Zm00001d028924 and Zm00001d028925 were found in the candidate region of this locus. Significant SNPs for RPR and DMC traits were independently detected at 9 loci in the 156417807bp-170637158bp region of chromosome 2. Six associated gene information, including Zm00001d005182, Zm00001d005183, Zm00001d005300, Zm00001d000967, Zm00001d005301, and Zm00001d0053516, were also detected multiple times in this region. Significant RPR loci were independently detected at 178358908bp and 185698633bp on chromosome 4, and the associated gene Zm00001d052068 was found in the candidate region of this locus. Significant DMC trait loci were independently detected at 27 loci in the 47645572bp-47925170bp region of chromosome 5, and two high reliabilities associated genes Zm00001d014448 and Zm00001d014449 were found in the candidate region of this locus, A significant RPR locus was independently detected at the 217360437bp locus, and an associated gene Zm00001d018247 was found at this locus, which coincides with the interval or segment of Liu's research results. Li [20] located lignin significant SNPs near 164498311bp on chromosome 6, 172376137bp on chromosome 3, 200651833bp on chromosome 5, 152112653bp on chromosome 8, and 11457267bp on chromosome 9, respectively, and located hemicellulose significant SNPs near 11457267bp on chromosome 9_ 169624611、3_ 175959350、5_217360437 and DMC trait significance loci 8_ 159862794、9_ 106888912 is overlapping or similar. Shao [7] found that the significant SNPs loci for maize RPR and SD were clustered on chromosome 2 at 54.00Mb-75.00Mb in his study. However, in this study, significant associations with RPR were detected at both 73833386bp loci on chromosome 2 in both analysis models. Ma Qingmei et al. [2]. found significant loci for maize stalk thrust resistance on chromosome 3 at 171756718bp and chromosome 4 at 191414929bp, which were consistent with the significant loci for RPR at 3_175959350 trait and for DMC at 4_193185052 found in this study. The significant SNPs interval of stalk related traits identified in this study overlaps significantly with the correlation traits previously studied, indicating that the results of stalk related traits identified are highly reliable and suitable for further exploration of key genes related to stalk related traits. In addition, the new significant SNPs loci detected in this experiment, which are different from previous studies, especially some high-confidence significant SNPs loci and efficient significant SNPs with a phenotype interpretation rate of over 15%, are of great significance for discovering new significant loci related to stalk traits and further exploring some new associated genes.
Analysis of genes associated with corn stalk related traits
Research has shown that there is a negative correlation between internode length and epidermal puncture strength, while there is a positive correlation between stalk diameter and stalk strength [21]. Some studies have also shown that the length of the third internode at the base of corn is significantly positively correlated with puncture intensity, while the stalk diameter is significantly negatively correlated with puncture intensity [4]. The results of this study indicate that the length of the third internode at the base of corn is significantly positively correlated with the strength of epidermal puncture over two years; the stalk diameter and epidermal puncture intensity had a positive correlation in the two-year experiment, but not significant; there is a highly significant positive correlation between dry matter content and epidermal puncture strength. Some studies have pointed out that not all corn stalk structures have a significant impact on biomass and economic coefficients, and corn basal stalk diameter has the smallest correlation coefficient with biomass, mainly due to the negative effect of stalk tip diameter reflected through basal diameter [22]. In addition, there is a significant positive correlation between stalk diameter and individual plant weight, and a negative correlation with biomass, which has not reached a significant level [23]. In this study, stalk diameter and dry matter content showed a highly significant negative correlation in the two-year data analysis. This may be due to the fact that cellulose and vascular bundles are concentrated around the epidermis in the thick material of the third section of the stalk, and most of the middle of the stalk is pithy and soft, with relatively low dry matter per unit area. The 377 inbred lines used in this study include conventional maize inbred lines, sweet glutinous maize inbred lines, and popcorn inbred lines, with a variety of types and different breeding objectives, which may have an impact on stalk correlation analysis.
Different plant tissues are composed of different types of cells. Thickened thick parietal cell form the strength of plant tissues. Genes involved in cell wall synthesis and metabolism play an important role in the formation of stalk strength. As a complex matrix, cell wall is mainly composed of polysaccharides, proteins and lignin [24, 25]. A variety of enzyme families will directly affect the metabolism and synthesis of cell wall substances. In addition, fatty acid metabolism and lipid metabolism may also affect cell wall synthesis. The structure of cell wall is also affected by the regulation and tissue of multiple transport pathways of dynamic cytoskeleton and cell wall polymer [26]. The corn stalk is mainly composed of cellulose, a small amount of sugar, inorganic salt and water. The lateral of the stalk starts to split, the cambium in the bundle grows and differentiates, and the stalk starts to thicken and extend.
The high-confidence candidate genes Zm00001d040805, Zm00001d010201, Zm00001d021805, Zm00001d029050, Zm00001d033080, and Zm00001d041438 associated repetitively with SNPs markers in this study have been validated in previous studies, and are related to stalk puncture intensity and stalk correlation. They are involved in regulating metabolic substances, cell wall formation, and material transport, and can further affect stalk differentiation and cortical development. In addition, this study also found some new gene information related to stalk correlation, which is important to maintain the structure of histiocyte and participate in the regulation of root growth regulators and coenzyme families. In particular, Zm00001d014449 is associated with LHP1 gene, which is involved in vascular bundle synthesis, and is important to maintain the specificity of cell and tissue structure; Zm00001d010201 expresses MYB6 transcription inhibitors related to the secondary wall. Previous studies [4, 27] have confirmed that the MYB transcription factor family can be used as stalk strength related genes for in-depth research; Zm00001d029860 participates in the metabolism of 3-hydroxyisobutyl CoA hydrolase, which affects cell wall synthesis through biological processes such as lipid transport and metabolism. Further in-depth research is needed in the next step.
Conclusion
In this study, 461053 high-quality SNPs were used to conduct genome-wide association study on the third internode related traits of 236 maize inbred lines. With GLM and MLM, 70 significant SNPs were found to be related to stalk lodging resistance in a two-year environment, with a phenotypic variation explanation rate of 8.76% -15.03%. Among them, 21 loci were found in independent association analyses at each growth stage more than twice by different model tests, and these loci were found belonging to HC-SNP. Between 5_47645572-5_ 47925170 segments, 27 SNPs were found by different model tests to be significantly associated with DMC. The result suggests that this segment is the region where DMC related genes are enriched. Combined with gene functional annotation, a total of 34 candidate associated genes were identified among which 12 candidate genes were detected more than twice. Most of these genes are related to plant growth and development, carbohydrate transport and metabolism, signal transduction mechanisms, and synthesis transporter enzyme families. This study helps to deepen the understanding of the genetic basis of maize stalk lodging resistance related traits, and provides theoretical guidance for future maize lodging resistance breeding.
Materials and methods
Test materials and design
The natural test population materials composed of 377 inbred lines were selected from the domestic backbone population and the major international dominant group. A total of 400 materials of different germplasm types were collected from the International Maize and Wheat Improvement Center (CIMMYT) in Mexico, and 377 stable germplasm materials were retained through the first year of breeding. 377 materials include 231 common maize inbred lines, 58 popcorn inbred lines and 88 waxy maize inbred lines. The test materials were planted in the experimental park of the Hebi Academy of Agricultural Sciences (Hebi, Henan Province) in the summer of 2016 and 2017. The random block design is adopted in three replicates. Each plot consisted of two rows which were 5m in length, 0.6 m from the next row, with seed spacing of 0.2m. The field growth is managed according to normal agricultural production.
Genotype and genome-wide association analysis
Two hundred thirty-six materials were randomly selected from the population and genomic DNA was extracted using the magnetic bead method, and then the DNA concentration, purity, and integrity were tested. Qualified DNA uses the re sequencing method to carry out genome simplified sequencing. After comparing the sequence to the B73 genome, identify the SNP, removing markers with deletion rate greater than 60% and minimum allele frequency lower than 0.05. After quality control, 461053 high-quality SNP markers were obtained for genome-wide association study (Fig. 4). At the P < 0.01 level, the significance of the association between SNP markers and stalk thrust resistance was determined.
Fig. 4.
Chromosome density map containing 461,053 SNP markers
Taking into account population structure and kinship, the generalized linear model (GLM) and mixed linear model (MLM) of TASSEL V5.0 software [28] were used to conduct correlation analysis on the stalk correlation at different growth stages, while the population structure-principal component analysis (PCA) was considered as a fixed effect, and kinship was considered as a random effect. According to the published Bonferroni corrected threshold GLM model with the same association group, P < 0.05/n was selected, while MLM model was selected with P < 1/n, n = number of markers [29]. Therefore, the threshold GLM model threshold for this study is P < 9.79 × 10–7, MLM model threshold is P < 9.77 × 10–6.
Phenotypic data measurement method and statistical method
The related characters of the third above-ground stalk (stalk diameter (16SD1, 16SD2 and 16SD3 at the stage of tasseling, grain filling and maturing) were measured at the stage of tasseling, grain filling and maturing, and the number of other stalk characters in 2016 was similar to that in 2017). Select three representative plants with the same growth condition in each growth period, cut the third above-ground internode completely, and take a stalk strength appliance (Zhejiang Topp Instrument Co., Ltd., model YYD-1B) to measure the puncture resistance of corn stalk in the middle quickly, and the unit is Newton (N). Each sample shall be measured for 3 times and the mean value shall be taken. Use the digital vernier caliper to measure the stalk diameter and internode length. After measurement, the sample is blanched for 30 min at 105 °C in an air drying oven, and then dried to constant weight at 65 °C, and the dry matter content is calculated. The mean value of the three replicates represents the phenotypic value of the traits at the growth stage.
The software EXCEL2007 and DPS7.05 were used for statistical correlation analysis and heritability calculation. Calculate group PCA and Kinship using TASSEL V5.0 software. Use R "CMplot" software package to draw correlation graph (https://github.com/YinLiLin/CMplot).
Prediction of candidate genes
According to the public database MaizeGDB (https://www.maizegdb.org/genome/), the sequence information of the B73 reference genome (B73 RefGenv4) is used as a reference to obtain the genes in the range of 2.5Kb in the upstream and downstream that are significantly associated with SNPs as the candidate genes for stalk correlation in the third internode of maize. Annotate and predict the function of candidate genes in Uniprot protein database (https://www.uniprot.org/uniprotkb/).
Acknowledgements
We thank professor Chunhua Mu of Shandong Academy of Agricultural Sciences for providing the maize population.
Authors’ contributions
BTW, MLY, GWQ and JFC designed this study. HG, JW developed the populations. ZHW recorded the data. BTW analyzed the data. BTW and MLY drafted the manuscript. HWL, HG, and JFC revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Henan Provincial Key Science and Technology Project "Creation of Germplasm Resources and New Variety Breeding and Application of High-Yield and Multi-Resistance Maize" (2022010202); Henan Innovative Talent Project (22HASTIT040); Henan Provincial Science and Technology Project "Analysis of Genetic Basis and Application of Stalk Quality Traits by GWAs" (202102110248).
Availability of data and materials
The datasets generated during the current study are available in the [GVM000531] repository, [https://bigd.big.ac.cn/gvm/getProjectDetail?Project=GVM000531].
Declarations
Ethics approval and consent to participate
No ethics approval was required. The authors declare that the experimental methods conducted in this study complied with current Chinese laws and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bangtai Wang and Meili Yang contributed equally to this work.
Contributor Information
Guiwen Qin, Email: qgw6666@126.com.
Jiafa Chen, Email: chenjiafa2005@163.com.
References
- 1.Ma YJ, Bao JX, Gao YX, Li YN, Qin WX, Wang YB, Long Y, Li JP, Dong ZY, Wan XY. Genome- wide association analysis of plant height and ear height related traits in maize. Acta Agronomica Sinica. 2023;49(03):647–661. [Google Scholar]
- 2.Ma QM, Pei YB, Chen DB, Song XY. Genome-wide association analysis of maize stalk anti-thrust. Scientia Agricultura Sinical. 2017;50(24):4671–4678. [Google Scholar]
- 3.Ren AR, Wu LF, Guan HH, Yang S, Jian LQ, Liu YF, Du YQ, Wang J, Guo JJ, Chen JT. Stalk fiber related traits and lodging resistance correlation analysis under different planting density in maize. J Plant Genet Resour. 2017;18(4):653–664. [Google Scholar]
- 4.Xie LY. Genome-wide association studies and important gene mining of maize stalk strength related traits, Ph. D. Dissertation of Shandong Agricultural University, 2021 (In Chinese)
- 5.Liu XG. Genomic selection for yield-relevant traits and stalk strength in maize, Postdoctoral Thesis of the Chinese Academy of Agricultural Sciences, 2020. (In Chinese)
- 6.Fang HY, Li W, Dan Y, Xia H, Guang H, Jin FX. QTL analysis for Rind Penetrometer Resistance (RPR) in maize under different plant densities. J Maize Sci. 2017;25(1):52–56. [Google Scholar]
- 7.Sao C. QTL mapping for stalk diameter and rind penetrometer resistance in maize, Master's Thesis of Yangzhou University, 2022. (In Chinese)
- 8.Flint-Garcia SA, Darrah L, McMullen M, Hibbard BE. Phenotypic versus marker-assisted selection for stalk strength and second-generation European corn borer resistance in maize. Theor Appl Genet. 2003;107(7):1331–1336. doi: 10.1007/s00122-003-1387-9. [DOI] [PubMed] [Google Scholar]
- 9.Hu HX, Liu WX, Fu ZY, Homann L, Technow F, Wang HW, Song CL, Li ST, Melchinger AE, Chen SJ. QTL mapping of stalk bending strength in a recombinant inbred line maize population. Theor Appl Genet. 2013;126(9):2257–2266. doi: 10.1007/s00122-013-2132-7. [DOI] [PubMed] [Google Scholar]
- 10.Jason AP, Sherry AF, Natalia DL, Michael DM, Shawn MK, Edward SB. The genetic architecture of maize stalk strength. PLos One. 2017;8(6):67066. doi: 10.1371/journal.pone.0067066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li K, Yan JB, Li JS, Yang XH. Genetic architecture of rind penetrometer resistance in two maize recombinant inbred line populations. BMC Plant Biol. 2014;14(1):152. doi: 10.1186/1471-2229-14-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F, Liu ZH, Hu JX, Xiao RJ, Zhu XQ, Xu Y, Deng M, Li RL, Luo HB, Huang C. Dissecting the genetic basis of stem length and stem diameter in maize (Zea mays) using a maize-teosinte introgression line population. J Agric Biotechnol. 2021;29(02):216–223. [Google Scholar]
- 13.Wang K. Genome-wide association analysis and candidate gene prediction of plant architecture related traits in maize, Master's Thesis of Yunnan University, 2022. (In Chinese)
- 14.Liu FP, Li W, Fang HY, Li L, Jin FX, Lin W. Analysis of meta-QTL and candidate genes related to stalk diameter in maize. J Northeast Agric Sci. 2019;44(5):30–33. [Google Scholar]
- 15.Zhang JH, Plant height, internodes, ear traits, general combining ability analysis and their QTLs mapping using double haploid lines of maize, Master's Thesis of Hebei Agricultural University, 2009. (In Chinese)
- 16.Persak H, Pitzschke A. Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and per sensitivity to abiotic stress. Int J Mol Sci. 2014;15(2):2517–2537. doi: 10.3390/ijms15022517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou SH, Zhang L, Lv Q, Huang JG. Identification and analysis of GLR family genes in maize. J Maize Sci. 2021;29(2):35–42. [Google Scholar]
- 18.Laubinger S, Hoecker U. The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J. 2003;35(3):373–385. doi: 10.1046/j.1365-313X.2003.01813.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu XG, Hu XJ, Li K, Liu ZF, Wu YJ, Wang HW, Huang CL. Genetic mapping and genomic selection for maize stalk strength. BMC Plant Biol. 2020;20(1):196. doi: 10.1186/s12870-020-2270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K. Genome-wide association study of cell wall components and forage quality in maize stalk. Postdoctoral Thesis of the Chinese Academy of Agricultural Sciences, 2017 (In Chinese).
- 21.Zhang Y. Multi-locus genome-wide association study reveals the genetic architecture of stalk lodging resistance-related traits in maize. Front Plant Sci. 2018;9:611. doi: 10.3389/fpls.2018.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan HH, Yang GJ, Zhang GS, Feng HK, Wang YJ. Selecting corn agronomic traits in breeding high potential yield: effects of stem structure. 3rd International Conference on Education, Management and Computing Technology (ICEMCT 2016).
- 23.Liu Q, Fu P, Liu XB, He C, Sun QZ. Effects of planting density on plant characters and biomass yield of six silage corn cultivars. China Dairy Cattle. 2015;5:54–55. [Google Scholar]
- 24.Martin MJ, Russell WA. Correlated responses of yield and other agronomic traits to recurrent selection for stalk quality in a maize synthetic 1. Crop Sci. 1984;24:746–750. doi: 10.2135/cropsci1984.0011183X002400040028x. [DOI] [Google Scholar]
- 25.Zhang B. The plant cell wall: Biosynthesis, construction, and functions. Plant Biol. 2021;63:251–272. doi: 10.1111/jipb.13055. [DOI] [PubMed] [Google Scholar]
- 26.Bashline L. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol Plant. 2014;7:586–600. doi: 10.1093/mp/ssu018. [DOI] [PubMed] [Google Scholar]
- 27.Wu LF, Zheng YX, Jiao FC, Wang M, Zhang J, Zhang ZQ, Huang YQ, Jia XY, Zhu LY, Zhao YF, Guo JJ, Chen JT. Identification of quantitative trait loci for related traits of stalk lodging resistance using genome-wide association studies in maize (Zea mays L.) BMC Genomic Data. 2022;23(1):76. doi: 10.1186/s12863-022-01091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–5. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 29.Wang MY. Genome-wide association study (GWAS) of resistance to head smut in maize. Plant Sci. 2012;196:125–131. doi: 10.1016/j.plantsci.2012.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available in the [GVM000531] repository, [https://bigd.big.ac.cn/gvm/getProjectDetail?Project=GVM000531].