Abstract
Background
Rapid diagnostic tests (RDTs) that detect Plasmodium falciparum histidine-rich protein-2 (PfHRP2) are exclusively deployed in Uganda, but deletion of the pfhrp2/3 target gene threatens their usefulness as malaria diagnosis and surveillance tools.
Methods
A cross-sectional survey was conducted at 40 sites across four regions of Uganda in Acholi, Lango, W. Nile and Karamoja from March 2021 to June 2023. Symptomatic malaria suspected patients were recruited and screened with both HRP2 and pan lactate dehydrogenase (pLDH) detecting RDTs. Dried blood spots (DBS) were collected from all patients and a random subset were used for genomic analysis to confirm parasite species and pfhrp2 and pfhrp3 gene status. Plasmodium species was determined using a conventional multiplex PCR while pfhrp2 and pfhrp3 gene deletions were determined using a real-time multiplex qPCR. Expression of the HRP2 protein antigen in a subset of samples was further assessed using a ELISA.
Results
Out of 2435 symptomatic patients tested for malaria, 1504 (61.8%) were positive on pLDH RDT. Overall, qPCR confirmed single pfhrp2 gene deletion in 1 out of 416 (0.2%) randomly selected samples that were confirmed of P. falciparum mono-infections.
Conclusion
These findings show limited threat of pfhrp2/3 gene deletions in the survey areas suggesting that HRP2 RDTs are still useful diagnostic tools for surveillance and diagnosis of P. falciparum malaria infections in symptomatic patients in this setting. Periodic genomic surveillance is warranted to monitor the frequency and trend of gene deletions and its effect on RDTs.
Keywords: Malaria, Rapid diagnostic tests, Plasmodium falciparum, Histidine rich protein, pfhrp2 and pfhrp3 Gene deletion
Background
Malaria remains a public health problem in Uganda [1]. Transmission occurs throughout the year with peak transmission occurring between in May–June and November–December. Although the entire population remains at risk of malaria infections, transmission is heterogenous across regions [2]. Recent evidence has shown epidemiological transition such as a shift in parasitaemia from children under 5 to those aged 2–15 years as well as variations in transmission and parasite prevalence at sub-national levels [2, 3]. Although Plasmodium falciparum is still the predominant species and accounts for over 90% of malaria infections, Plasmodium malariae, Plasmodium ovale and Plasmodium vivax are also present [2]. Malaria case management that involves test and treat is a key intervention for identification and clearance of parasites in infected cases [4]. Although microscopy examination of blood smears is the gold standard diagnostic method for malaria, the use of HRP2 RDTs accounts for up to 90% of total malaria testing in Uganda [3].
Despite the current malaria control measures, the country continues to experience rise in malaria cases with frequent and protracted epidemics [3]. New biological threats have also emerged posing new challenges to country malaria control programmes. In Uganda, there are recent reports of emerging malaria parasites that evade detection by the routinely used malaria HRP2-based RDTs due to pfhrp2 gene deletions [5–7] and those that evade treatment due to reduced sensitivity to artemisinin-based combination therapy (ACT) mediated by genetic mutations in the parasite’s kelch13 propeller gene [8–11]. The occurrence of parasites that fail to express the HRP2 protein antigen due to deletion of pfhrp2/3 genes has been known to cause false negative RDT results affecting malaria case management in many settings in Africa, the Amazon and India [6, 12–19]. Once the pfhrp2/3 genes are deleted, the parasites do not express the HRP2 protein that is the principal target for RDTs resulting in false negative results [5, 20, 21]. The implication is that infected individuals remain untreated and continue transmitting parasites as infectious reservoirs but also remain at risk of progressing to severe malaria and death. HRP2-detecting RDTs are the main method for malaria diagnosis at all health facilities in the study areas of Acholi, Lango, W. Nile and Karamoja. Previous genomic surveillance has confirmed the presence of pfhrp2/3 gene deletions in parasite populations in multiple locations in Uganda. The World Health Organization (WHO) recommends continuous surveillance to monitor the levels of deletions in areas where they have been confirmed as well as assess if the mutant parasites have emerged in other regions [5, 7].
The northern regions of Acholi, Karamoja, W. Nile and Lango in Uganda covered by this survey have traditionally remained high transmission areas at holoendemic levels compared to other malaria endemic parts of the country [22]. The HRP2 RDTs account for > 80% of total routine malaria testing in this setting. The emergence and spread of parasites with pfhrp2 gene deletions will pose a serious threat to the test and treat strategy and likely increase malaria transmission and burden in these areas. Therefore, there is an urgent need to assess the occurrence of parasite populations with pfhrp2 gene deletions and extent of spread in Northern regions to inform malaria test policy. The current genomic surveillance focused on investigation of pfhrp2/3 gene deletions in symptomatic patients as recommended by the WHO [5].
Methods
Study design
This was a health facility-based surveillance targeting symptomatic malaria patients seeking health care as recommended by the WHO protocol for surveillance of pfhrp2/3 deletions.
Study area and setting
The survey was conducted in four regions of Acholi, Karamoja, W. Nile and Lango in northern Uganda (Fig. 1). The four regions covered by the survey are well designed demographic health survey (DHS) clusters or enumeration areas that are periodically used for the national malaria indicator surveys in the country. Malaria transmission across all the four regions is intense and stable at holoendemic levels, with parasite prevalence ranging from 13–34% as determined by blood smear microscopy (Fig. 1) [22].
Fig. 1.
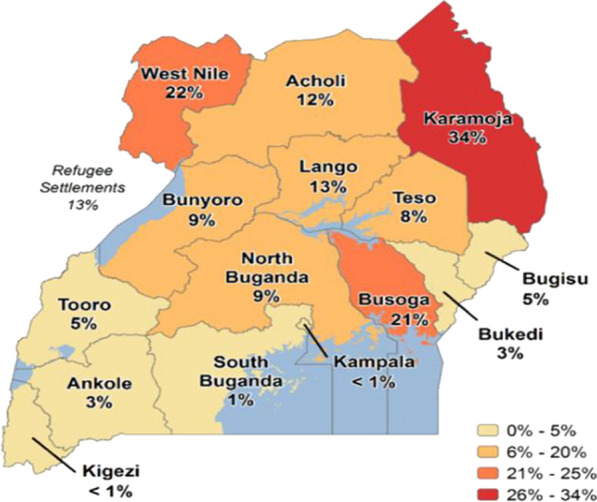
Malaria parasite prevalence by blood smear microscopy per region from population-based surveys in the study areas [22]
Population
The survey targeted symptomatic individuals of all age groups, suspected to have malaria who seek treatment and care at health facilities within the survey regions. Symptomatic status was based on fever defined as axillary temperature of ≥ 37.50 C.
Sampling
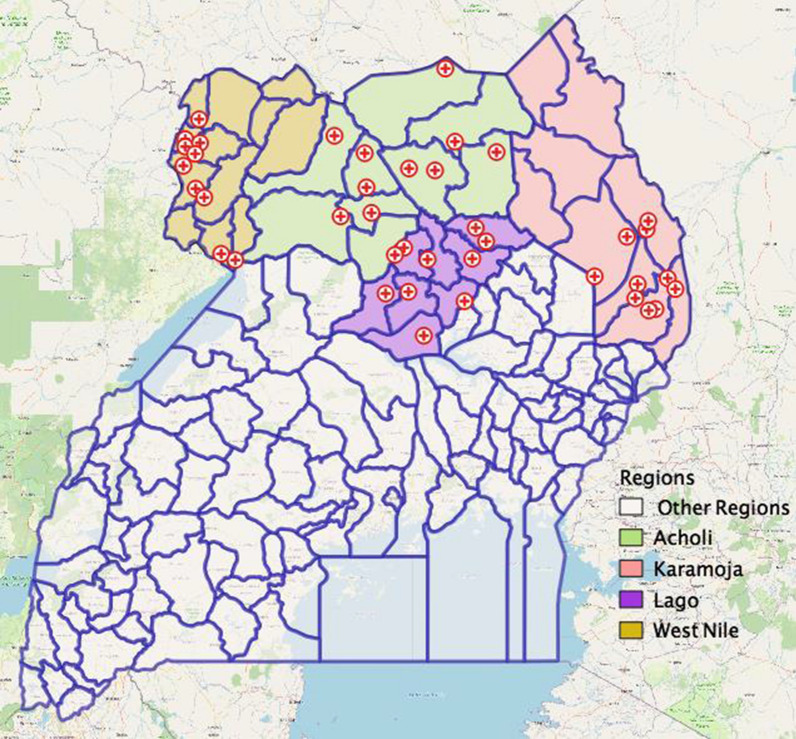
Health facilities that served as survey sites were selected in accordance with the WHO protocol for pfhrp2/3 gene deletions surveillance [23]. Briefly, a total of ten facilities were randomly selected from each region (domain) making a total of forty facilities across the four survey regions (Fig. 2).
Fig. 2.
Location of survey sites
From each selected facility, a total of thirty-seven malaria confirmed positive blood samples from symptomatic individuals were collected on filter papers and transported to the central laboratory in Kampala for processing.
Survey procedures and recruitment
At each facility, a designated staff explained the survey procedures and administered a consent form to eligible patients who expressed willingness to enroll. A questionnaire was used to collect patient demographics. Consenting patients were screened for malaria using two malaria rapid diagnostic tests, an HRP2 detecting and a pLDH detecting RDT. Dried blood spots (DBS) were collected from all patients with pLDH positive results.
Eligibility criteria
All malaria suspected symptomatic individuals who presented to the selected facilities and provided consent to participate were included in the survey. Samples with negative pLDH results were excluded.
Ethical consideration
The survey protocol was approved by the National council of science and technology (UNCST) and the Makerere University School of Public Health research and ethics committee. A consent form translated into the four indigenous languages spoken in the four different survey regions was administered for all patients before recruitment into the survey.
Rapid diagnostic tests (RDTs)
At the health facility, two different RDTs were used and a dried blood spot collected from each patient recruited in the survey. The RDTs used were SD Bioline malaria Ag P.f Cat. 05FK50 (Abbott Diagnostics Korea Inc.) that has the Pf (HRP2) line only and the SD Bioline Malaria Ag Pf (HRP2/pLDH) Cat. 05FK90 – (Abbott Diagnostics Korea Inc.) that has a separate HRP2 and a pf-LDH test line. These tests were done simultaneously to test for malaria in symptomatic febrile patients (based on axillary temperature of > 37.5 ºC). Both RDT were used following the manufacturer’s instructions. Dried blood spot was collected from each patient recruited in the survey.
Parasite DNA extraction
The dried blood spots were shipped to the collaborating institution, the Australia Defense Forces Malaria and Infectious Diseases Institute (ADFMIDI), a WHO Collaborating Centre for Malaria and a member of the WHO laboratory network for molecular testing to detect pfhrp2/3 deletions, for parasite genomic analysis. From each DBS sample, 3 discs of dried blood spot were punched into sterile tubes. DNA was extracted using QIAamp DNA Mini Kits and a QIAcube Connect (QIAGEN, Crawley, UK) according to the manufacturer’s instructions. Samples were eluted into a volume of 100 µL with nuclease free water.
Characterization of pfhrp2 and pfhrp3 deletions in samples
A published multiplex qPCR method that amplifies a fragment each of pfhrp2, pfhrp3, pfldh and human tubulin (htb) genes simultaneously [24] was used to determine pfhrp2 and pfhrp3 status in collected patient samples. Published primers, probes and cycling conditions were used with slight modifications in the probe fluorescence dye and quencher combinations including changing quencher on the pfhrp2 probe to BHQ1 and pfldh probe to BHQ2. The multiplex qPCR assay uses a Quantinova Multiplex PCR kit master mix (QIAGEN) and was carried out on a Mic qPCR cycler (Bio Molecular Systems). The assay includes two internal controls: pfldh for the quality of parasite DNA and human tubulin gene for the efficiency of DNA extraction process. Serial diluted DNA (1, 0.1, 0.01 and 0.001 ng/µL) from a laboratory strain 3D7 (no gene deletions) was used in each run to establish standard curve for quantitation. Laboratory lines with gene deletions such as Dd2 (pfhrp2-deleted), HB3 (pfhrp3-deleted) and 3BD5 (double pfhrp2/3 deleted) were also included in each PCR run as controls.
Cqhtb values were used as DNA extraction control from DBS samples with a Cqhtb value > 30 considered invalid due to inefficient extraction. Samples with no detectable pfldh, or Cqpfldh > 35 were also classified as invalid due to insufficient quality of parasite DNA (either due to low parasite counts or no parasites). Samples with ΔCq (Cqpfhrp2—Cqpfldh and Cqpfhrp3—Cqpfldh) values ≥ 3, or not detected at all are classified as pfhrp2 and pfhrp3 deleted [24].
Serological analysis to confirm double pfhrp2/3 deletions
HRPs and pLDH antigen levels were measured for samples classified as single and double pfhrp2/3 deletions using antigen specific ELISAs (Quantimal Celisa Pf HRP2 Assay kit, KM 810 and Quantimal Celisa Pf pLDH Assay kit, KM7, Cellabs, Australia) to confirm non-expressions of HRPs while expressing pLDH. A subset of samples without gene deletions were also measured by ELISA as controls.
Molecular species diagnosis
A conventional multiplex PCR targeting species specific 18S rRNA gene [25] was performed to confirm Plasmodium spp (P. falciparum, P. vivax, P. malariae, P. ovale) for all samples.
Data analysis
Using a data tool, patient demographics and variables were collected from the symptomatic patients at survey sites. The data tool was a hardcopy questionnaire administered and completed by the study staff with a carbon copy to ensure its backup. All the completed hardcopy data tools were stored in lockable cupboards and later transported to Kampala for entry. All data were entered and managed in one central Excel database. Data quality checks were done to check for and correct any inconsistencies. Data analysis was done with STATA Ver 14, College Station, TX, USA: StataCorp LP. Descriptive analysis was done to describe the participant baseline characteristics and determine proportions of parasitemia and gene deletions in the samples. ArcGIS software version 10.8, Environmental Systems Research Institute (ESRI), CA, USA was used to map the locations where all blood samples were collected across the survey regions.
Results
Participants and RDT results
A total of 2435 symptomatic patients were enrolled and screened across 40 surveillance sites in the four survey regions using the HRP2 and pLDH RDT. Out of the 2435 malaria symptomatic patients tested, 1504 (61.8%) were positive for malaria based on the pLDH RDTs, while 1657 (68.0%) were positive on HRP2 RDT (Table 1).
Table 1.
Demography of participants and RDT results (n = 2435)
| Variables | Participants (N) | Proportion Positive (pLDH) N (%) | Proportion positive (HRP2) N (%) | HRP2-/pLDH + N (%) | HRP2 + /pLDH-N (%) | HRP2 + /pLDH + N (%) |
|---|---|---|---|---|---|---|
| Age | ||||||
| < 5 | 664 | 437 (65.8%) | 464 (69.9%) | 16 (2.4%) | 49 (7.4%) | 420 (63.2%) |
| > = 5 | 1771 | 1068 (60.4%) | 1193 (67.4%) | 34 (1.9%) | 164 (9.3%) | 1034 (58.4%) |
| Sex | ||||||
| Male | 856 | 287 (33.8%) | 234 (27.6%) | 17 (2.0%) | 70 (8.3%) | 544 (64.2%) |
| Female | 1579 | 641 (40.6%) | 544 (34.4%) | 45 (2.8%) | 143 (9.1%) | 900 (57.0%) |
| Region | ||||||
| Acholi | 549 | 378 (68.0%) | 372 (67.8%) | 7 (1.8%) | 3 (0.55%) | 371 (67.6%) |
| Lango | 583 | 370 (63.8%) | 448 (76.8%) | 19 (5.1%) | 99 (17.0%) | 351 (60.2%) |
| W. Nile | 676 | 386 (57.1%) | 471 (69.7%) | 13 (3.4%) | 102 (15.1%) | 369 (54.6%) |
| Karamoja | 627 | 370 (59.0%) | 366 (58.4%) | 11 (3.0%) | 9 (1.4%) | 363 (57.8%) |
| Total | 2435 | 1504 (61.8%) | 1657 (68.0%) | 50 (3.3%) | 213 (8.7%) | 1454 (59.6%) |
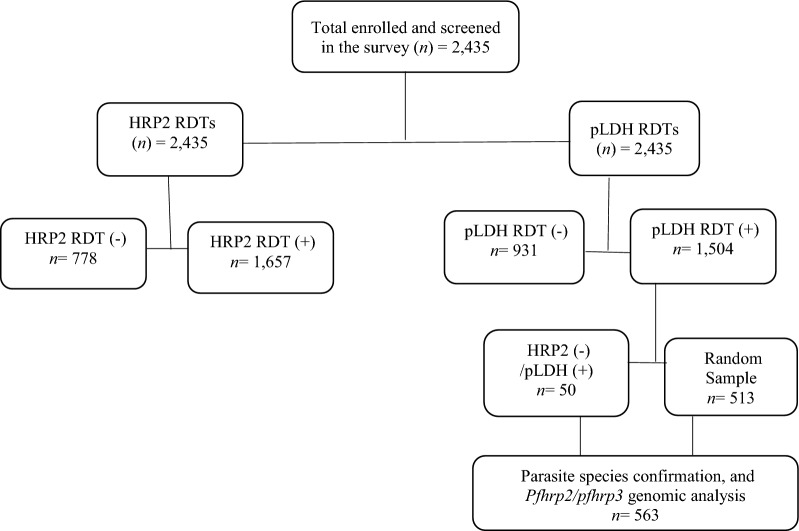
All samples that were pLDH RDT positive were assumed to contain parasites. Of the 1504 positive pLDH RDT samples, 50 were HRP2 (-)/pLDH ( +) discordant samples giving an overall discordance (HRP2 -/pLDH +) rate of 3.3% (50/1504) in the survey (Fig. 3).
Fig. 3.
Survey Flow chat
A random sample of 563 DBS were selected from the 1504 for DNA confirmation of the presence of Plasmodium species and subsequent investigation and characterization of the pfhrp2/3 gene deletions (these included the 50 HRP2-/pLDH + samples). The selected samples represent 23.6% to 27.5% of the total pLDH positive samples from each region. The baseline characteristics of all survey samples (overall and per region) is shown in Tables 1, 2.
Table 2.
Number and percentage of samples with valid qPCR results (N = 563)
| Location | Total sample | Samples with valid PCR results | |
|---|---|---|---|
| n | N | % | |
| Acholi | 140 | 93 | 66.4% |
| West Nile | 133 | 113 | 85.0% |
| Karamoja | 135 | 93 | 68.9% |
| Lango | 155 | 117 | 75.5% |
| Total | 563 | 416 | 73.9% |
Real-time multiplex qPCR
Out of the 563 samples run on multiplex qPCR, 73.9% (416/563) gave valid PCR results (Cqhtpb < 30 and/or Cqpfldh < 35) indicating sufficient human DNA and parasite DNA (Table 3). The 147/563 samples (26.1%) giving invalid PCR results (Cqhtb > 30 and/or Cqpfldh > 35) did not have sufficient human and parasite DNA to accurately determine pfhrp2 and pfhrp3 status and were excluded from gene deletion analysis. Invalid qPCRs were related to poor DBS resulting in poor DNA extraction (Cqhtb > 30) for 22 (3.9%) samples while the 125 samples (22.2%) was likely due to low parasite density or no parasites (Cqpfldh > 35).
Table 3.
Number and prevalence of gene deletions in samples with valid PCR results
| Location | Valid PCR | Samples without deletion | ΔCqpfhrp2-pfldh > 3 or not detected (gene deleted parasites > 90% in the sample) | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Acholi | 93 | 93 | 100 | 0 | 0 |
| West Nile | 113 | 113 | 100 | 0 | 0 |
| Karamoja | 93 | 93 | 100 | 0 | 0 |
| Lango | 117 | 116 | 99.1 | 1 | 0.9 |
| Total | 416 | 415 | 99.8 | 1 | 0.2 |
Pfhrp2 and pfhrp3 gene status
Of 416 samples that gave valid PCR results, 1 sample (0.2%, AM44) from Lango was confirmed to have single pfhrp2 deletion (Table 4). This was confirmed also by conventional PCR. Two other samples from Lango (AL42 and AM17) may also contain pfhrp2 deleted parasites but not dominant (< 90%) in the sample (mixed with parasites without deletions) as ΔCqpfhrp2-pfldh values were 1.7 and 1.9, respectively.
Table 4.
Plasmodium spp confirmed by multiplex PCR
| Location | Total samples tested | Samples positive | Plasmodium species composition in the samples (n = 563) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pf | Po | Pf + Pm | Pf + Pm + Po | |||||||
| n | % | n | % | n | % | n | % | |||
| Acholi | 140 | 125 | 125 | 100.0 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| West Nile | 133 | 126 | 124 | 98.40 | 1 | 0.80 | 1 | 0.80 | 0 | 0.00 |
| Karamoja | 135 | 127 | 125 | 98.40 | 0 | 0.00 | 2 | 1.60 | 0 | 0.00 |
| Lango | 155 | 145 | 144 | 99.30 | 0 | 0.00 | 0 | 0.00 | 1 | 0.70 |
| Total | 563 | 523 | 518 | 99.00 | 1 | 0.20 | 3 | 0.60 | 1 | 0.20 |
Pf (Plasmodium falciparum), Po (Plasmodium ovale), Pm (Plasmodium malariae)
Discordant subset: 50 samples were positive on PfLDH and negative on HRP2 band. Only 22/50 samples (44.0%) gave valid qPCR results, i.e. had sufficient human DNA and parasite DNA, of which no pfhrp2 and/or pfhrp3 deletions were detected. 28 samples gave invalid qPCR results due to DBS issues resulting in insufficient human and parasite DNA for analysis while 7 was due to low parasite density or negative for parasites. All 21 samples with DBS issues also gave negative ELISA for both HRP2 and pLDH. Therefore, samples with DBS issues failed to extract DNA and failed to elute proteins. 14/21 samples with DBS issues and 4/7 low parasite samples were also negative for Plasmodium species PCR. Only one sample in this group was determined to have mixed infections (Pf/Po/Pm). It is not clear why samples with DBS issues were enriched in the discordant subset.
ELISA results
HRP2 and pLDH ELISAs were performed for a subset of 125 samples including the single sample determined by both qPCR and conventional PCR as pfhrp2-deleted and two samples may contain pfhrp2-deleted parasites mixed with parasites without gene deletions. 98/125 samples including three pfhrp2-deleted samples gave positive ELISA results for both HRP2 and pLDH. Only 27/125 samples were negative for HRP2 of which 23 were also negative for pLDH and classified by qPCR as having insufficient DNA.
Detection of non-P. falciparum species
Among 563 samples, 523 samples including 416 samples that gave valid multiplex qPCR results gave positive identifications for plasmodium species by PCR. Plasmodium falciparum accounted for 99.0% (518/523). One P. ovale (0.2%), 3 P. falciparum + P. malariae (0.6%) and 1 P. falciparum + P. malariae + P. ovale (0.2%) was detected (Table 4). In this subset of samples that were pfLDH RDT positive, all but one were confirmed to contain P. falciparum infections by PCR.
Discussion
This is the first survey in Uganda conducted following the WHO protocol: 37 symptomatic malaria patients enrolled per facility and 10 facilities in each region; a double RDT screening method was used, and DBS collected for parasite genomic analysis [23]. Based on the WHO protocol, the survey design and sample size are good for determining if prevalence of gene deletions causing false negative RDT results is a major threat to the utility of RDTs in Uganda. The molecular analyses focused on the discordant set (n = 50) and a randomly selected set of concordant samples (n = 513). This strategy is aligned with the WHO recommendations. Following this protocol, a single sample with pfhrp2 deletion only was identified in one of 4 regions surveyed. Two other pfhrp2-deleted parasites were detected in mixture with wild type parasites suggesting gene deleted parasites are circulating in Lango region. These single pfhrp2-deleted parasites are unlikely to have a major impact on the utility of HRP2 based RDTs because of: (1) low prevalence, (2) single pfhrp2-deleted parasites still expressed measurable HRP proteins likely due to cross reactivity with HRP3, (3) no gene deletions were detected from the discordant set of samples. This was also supported by the markedly higher HRP2 RDTs positive rate than that of pLDH in symptomatic patients of this study. Therefore, there is no immediate need to switch away from HRP2-detecting RDTs in Uganda as HRP2-detecting RDTs are generally more sensitive and heat durable than pLDH-detecting RDTs. However, as mathematical modelling have shown that once gene deleted parasites exist, the prevalence will rise rapidly under the continued use of HRP2-based RDTs [26]. Therefore, while no need to change RDTs at the moment, continued genomic surveillance across the country is required.
The prevalence of 0.2% pfhrp2 deletion observed in this study among a subset of symptomatic individuals is relatively low implying limited threat to the utility of HRP2 RDTs in this setting. The assumption was that this prevalence also apply to the entire sample set of the study, as the subset of samples undergone genomic analysis were randomly selected representation 24–29% of samples collected from each region. This prevalence is relatively lower than what was previously detected in Eastern and Western regions Uganda [5]. This difference in prevalence of pfhrp2/3 deletions between regions may be explained by the differences in the volumes of RDTs and the duration within which the RDTs have been in use since introduction. Historically, initial pilot and feasibility studies of malaria RDT use were conducted in the mid-western and eastern Uganda followed by the actual RDTs introduction, deployment and scale up to other regions in a phased manner. The emergence of pfhrp2/3 gene deletions may occur first in areas with long term use of RDTs as this mutation allows the parasite to evade detection and survive and contribute to transmission. Regionally, the prevalence of pfhrp2-deleted parasites seen in the survey is also lower when compared to those reported in other endemic countries in Africa such as Eritrea [13], Ethiopia [16] and Ghana [12].
The WHO pfhrp2/3 surveillance protocol recommends a switch of malaria RDTs from HRP2 to those targeting alternatives antigens, such as LDH when prevalence of gene deletions causing false negative RDT results exceeds the 5% cut off [23, 27]. The low prevalence of single gene deletions observed in the survey implies that the HRP2 RDTs tests are likely to detect the majority of P. falciparum malaria infections in symptomatic patients in this setting. However, continuous pfhrp2/3 surveillance is recommended to monitor the trends and extent of pfhrp2/3 deletions.
Historically, genomic characterization of pfhrp2/3 deletions was done by conventional PCR that amplifies the exon 1 and exon 2 of the two genes and detected by gel electrophoresis. In recent years, several new molecular based approaches have been developed and adopted for the detection and characterization of pfhrp2/3 gene deletion providing more streamlined and robust analysis and yielding more accurate results. In this survey, a published multiplex qPCR method was used that amplifies a fragment each of pfhrp2, pfhrp3, pfldh and human tubulin (htb) genes simultaneously [24]. The sample identified to have pfhrp2 deletion was also confirmed by the conventional PCR. The WHO pfhrp2/3 protocol also recommends proof of failure to express HRP2 protein antigen in isolates classified as pfhrp2/3 gene deleted. However, it is well established that single pfhrp2- deleted parasites often test positive for HRP2 protein due to cross reactivity with HRP3. Indeed, the single sample determined by qPCR as pfhrp2-deleted was positive on HRP2 ELISA suggesting the presence of HRP3 protein. As there was only one sample confirmed of having single pfhrp2-deleted parasite in the entire sample set, there was no value to perform further ELISA on this set of samples.
Limitations
Out of the entire survey population, only a random sample were analysed by molecular and serological methods to detect pfhrp2/3 gene deletions and HRP2 protein expression respectively. However, significant impact on prevalence is not expected as a good proportion of samples was analysed per region (130 ~ 150 samples per region). A proportion of the HRP2-/pLDH + discordant samples gave invalid qPCR results mainly due to DBS issues resulting in insufficient human and parasite DNA for analysis. While most of the other samples eluted well and gave good quality DNA, nearly half of the DBS in the discordant set failed to lyse despite extending incubation in lysis buffer at 85° C from 15 to 30 min and 4 °C overnight. The issues encountered with the dried blood spots (DBS) particularly in the discordant set prevented molecular analysis for ~ 45% discordant samples to be analysed and this could have led to potential risk of underestimation of pfhrp2 gene deletion in this discordant sample set.
Conclusion and implications for the national malaria control programme
This study provides the first evidence of pfhrp2 deletion in P. falciparum parasite populations circulating in Northern Uganda and the first survey to be conducted in accordance with the WHO surveillance protocol for pfhrp2/3 deletions in Uganda. Overall, these findings show limited presence of pfhrp2/3 gene deletions causing false negative RDT results in this setting to be below the 5% WHO recommended cut-off required for switch of RDTs. The low prevalence observed implies limited threat of pfhrp2/3 gene deletions suggesting that the HRP2 RDTs are still useful diagnostic tools to support malaria surveillance and case management in symptomatic patients in this setting. However, confirmed presence of single pfhrp2 gene deletion in this parasite population underscores the need to conduct periodic genomic surveillance to monitor the frequency and trend of pfhrp2/3 gene deletions and its effect in this setting.
Acknowledgements
We thank the field study teams for leading the field activities and sample collections at the facilities. We are grateful to the study participants who participated in the survey. We thank Mr. David Matsiga for the support in processing the samples.
Abbreviations
- HRP2:
Histidine rich protein 2
- pfhrp2:
Plasmodium falciparum Histidine rich protein 2 gene
- pfhrp3:
Plasmodium falciparum Histidine rich protein 3 gene
- RDT:
Rapid diagnostic test
- ACT:
Artemisinin-based combination therapy
- PCR:
Polymerase chain reaction
- WHO:
World Health Organization
- DNA:
Deoxyribonucleic acid
- DBS:
Dried blood spots
- WBC:
While blood cells
- NAAT:
Nucleic acid amplification tests
- pLDH:
Parasite lactate dehydrogenase
- qPCR:
Quantitative Polymerase Chain Reaction
- ELISA:
Enzyme Linked Immunosorbent Assay
- DHS:
Demographic Health Program
Author contributions
BBA designed the study. BBA, DS, JT, CP did the genomic and serological analysis, BBA, MN, EA, IS, QC supported the data analysis. BBA drafted the manuscript. BBA, SN, JC, MK, QC reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported, in whole or in part, by the Bill & Melinda Gates Foundation Grant Number: INV-031515. The grant is awarded to BBA as the Principal investigator. The molecular analysis of parasites was funded by the US DoD Armed Forces Health Surveillance Division, Global Emerging Infections Surveillance Branch (AFHSD/GEIS), PROMIS ID P0111_22_AF. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
Data for this pfhrp2 and pfhrp3 study is available upon request to the corresponding author. All data files related to this work have been uploaded as additional files to the manuscript.
Declarations
Ethics approval and consent to participate
Ethical approval for the study was obtained from the Makerere University School of Public of Health Research and Ethics Committee, the Uganda National Council of Science and Technology.
Consent for publication
All authors read and approved the manuscript for publication.
Competing interests
All authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO . World Malaria Report 2021. Geneva: World Health Organization; 2021. [Google Scholar]
- 2.National Malaria Control Division . National Malaria Control Strategic Plan. Uganda: Kampala; 2021. [Google Scholar]
- 3.Health Management Information System 2023. Uganda.
- 4.WHO . Malaria Treatment Guidelines. Geneva: World Health Organization; 2010. [Google Scholar]
- 5.Agaba BB, Anderson K, Gresty K, Prosser C, Smith D, Nankabirwa JI, et al. Molecular surveillance reveals the presence of pfhrp2 and pfhrp3 gene deletions in Plasmodium falciparum parasite populations in Uganda, 2017–2019. Malar J. 2020;19:300. doi: 10.1186/s12936-020-03362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agaba BB, Yeka A, Nsobya S, Arinaitwe E, Nankabirwa J, Opigo J, et al. Systematic review of the status of pfhrp2 and pfhrp3 gene deletion, approaches and methods used for its estimation and reporting in Plasmodium falciparum populations in Africa: review of published studies 2010–2019. Malar J. 2019;18:355. doi: 10.1186/s12936-019-2987-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosco AB, Nankabirwa JI, Yeka A, Nsobya S, Gresty K, Anderson K, et al. Limitations of rapid diagnostic tests in malaria surveys in areas with varied transmission intensity in Uganda 2017–2019: implications for selection and use of HRP2 RDTs. PLoS ONE. 2020;15:e0244457. doi: 10.1371/journal.pone.0244457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asua V, Conrad MD, Aydemir O, Duvalsaint M, Legac J, Duarte E, et al. Changing prevalence of potential mediators of aminoquinoline, antifolate, and artemisinin resistance across Uganda. J Infect Dis. 2021;223:985–994. doi: 10.1093/infdis/jiaa687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balikagala B, Fukuda N, Ikeda M, Katuro OT, Tachibana SI, Yamauchi M, et al. Evidence of artemisinin-resistant malaria in Africa. N Engl J Med. 2021;385:1163–1171. doi: 10.1056/NEJMoa2101746. [DOI] [PubMed] [Google Scholar]
- 10.Conrad MD, Asua V, Garg S, Giesbrecht D, Niaré K, Smith S, et al. Evolution of partial resistance to artemisinins in malaria parasites in Uganda. N Engl J Med. 2023;389:722–732. doi: 10.1056/NEJMoa2211803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tumwebaze PK, Conrad MD, Okitwi M, Orena S, Byaruhanga O, Katairo T, et al. Decreased susceptibility of Plasmodium falciparum to both dihydroartemisinin and lumefantrine in northern Uganda. Nat Commun. 2022;13:6353. doi: 10.1038/s41467-022-33873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amoah LE, Abankwa J, Oppong A. Plasmodium falciparum histidine rich protein-2 diversity and the implications for PfHRP 2-based malaria rapid diagnostic tests in Ghana. Malar J. 2016;15:101. doi: 10.1186/s12936-016-1159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berhane A, Anderson K, Mihreteab S, Gresty K, Rogier E, Mohamed S, et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2. Eritrea Emerg Infect Dis. 2018;24:462–470. doi: 10.3201/eid2403.171723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berhane A, Russom M, Bahta I, Hagos F, Ghirmai M, Uqubay S. Rapid diagnostic tests failing to detect Plasmodium falciparum infections in Eritrea: an investigation of reported false negative RDT results. Malar J. 2017;16:105. doi: 10.1186/s12936-017-1752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berzosa P, González V, Taravillo L, Mayor A, Romay-Barja M, García L, et al. First evidence of the deletion in the pfhrp2 and pfhrp3 genes in Plasmodium falciparum from Equatorial Guinea. Malar J. 2020;19:99. doi: 10.1186/s12936-020-03178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golassa L, Messele A, Amambua-Ngwa A, Swedberg G. High prevalence and extended deletions in Plasmodium falciparum hrp2/3 genomic loci in Ethiopia. PLoS ONE. 2020;15:e0241807. doi: 10.1371/journal.pone.0241807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koita OA, Doumbo OK, Ouattara A, Tall LK, Konare A, Diakite M, et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg. 2012;86:194–198. doi: 10.4269/ajtmh.2012.10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parr JB, Verity R, Doctor SM, Janko M, Carey-Ewend K, Turman BJ, et al. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis. 2017;216:36–44. doi: 10.1093/infdis/jiw538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N. Prevalence of pfhrp2 and/or pfhrp3 gene geletion in Plasmodium falciparum population in eight highly endemic States in India. PLoS ONE. 2016;11:e0157949. doi: 10.1371/journal.pone.0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. 2014;3:283. doi: 10.1186/1475-2875-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO . False-negative RDT results and implications of new reports of P. falciparum histidine-rich protein 2/3 gene deletions. Geneva: World Health Organization; 2017. [Google Scholar]
- 22.National Malaria Control Division . National Malaria Indicator Survey (MIS) Kampala: Uganda; 2019. [Google Scholar]
- 23.WHO . Protocol for surveillance of pfhrp2 and pfhrp3 gene deletions. Geneva: World Health Organization; 2017. [Google Scholar]
- 24.Grignard L, Nolder D, Sepúlveda N, Berhane A, Mihreteab S, Kaaya R, et al. A novel multiplex qPCR assay for detection of Plasmodium falciparum with histidine-rich protein 2 and 3 (pfhrp2 and pfhrp3) deletions in polyclonal infections. EBioMedicine. 2020;55:102757. doi: 10.1016/j.ebiom.2020.102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol. 2003;97:131–137. doi: 10.1179/000349803125002977. [DOI] [PubMed] [Google Scholar]
- 26.Watson OJ, Slater HC, Verity R, Parr JB, Mwandagalirwa MK, Tshefu A, et al. Modelling the drivers of the spread of Plasmodium falciparum hrp2 gene deletions in sub-Saharan Africa. Elife. 2017;6:e25008. doi: 10.7554/eLife.25008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO . Response plan to pfhrp2 gene deletions. Geneva: World Health Organization; 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data for this pfhrp2 and pfhrp3 study is available upon request to the corresponding author. All data files related to this work have been uploaded as additional files to the manuscript.