Abstract
The stabilization of a test plasmid by the proteic, poison-antidote plasmid addiction system (pas) of plasmid pTF-FC2 was host strain dependent, with a 100-fold increase in stability in Escherichia coli CSH50, a 2.5-fold increase in E. coli JM105, and no detectable stabilization in E. coli strains JM107 and JM109. The lethality of the PasB toxin was far higher in the E. coli strains in which the pas was most effective. Models for the way in which poison-antidote systems stabilize plasmids require that the antidote have a much higher rate of turnover than that of the toxin. A decrease in host cell death following plasmid loss from an E. coli lon mutant and a decrease in plasmid stability suggested that the Lon protease plays a role in the rate of turnover of PasA antidote.
Proteic plasmid stabilization systems have been discovered on a number of plasmids and include the ccd system of plasmid F (7), the identical parD/pem and kis/kid systems of plasmids R1 and R100 (1, 16, 19), the parDE system of plasmids RP4/ RK2 (14) and the phd/doc system of phage P1 (10). These systems consist of a long-lived toxin which is expressed at low levels and a short-lived, highly expressed antidote (8). On cell division, if a progeny cell fails to inherit the plasmid, it loses the ability to make the shorter-lived and more abundantly produced antidote and is unable to counter the toxic effects of the poison. As a result, plasmid-free cells are killed or their cell division is inhibited, depending on the type of poison-antidote system.
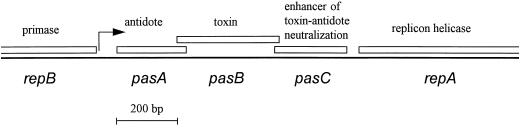
The 12.2-kb mobilizable, broad-host-range plasmid pTF-FC2 (GenBank accession nos. M64981 and M35249 [13]) was originally isolated from Thiobacillus ferrooxidans. This natural hybrid plasmid has a replicon clearly related to those of the IncQ plasmids (e.g., R1162 and RSF1010) (4) and a mobilization region with low but clear similarity to those of the IncP plasmids (e.g., R751 and RK2/R68/RP4) (15). Situated within the IncQ-like replicon and between the repB and repA genes (Fig. 1) is a proteic poison-antidote system named pas (for plasmid addiction system). This system is unusual in that it consists of three genes; pasA encodes an antidote, pasB encodes a toxin (which is bacteriocidal rather than bacteriostatic), and pasC encodes a protein that appears to enhance the neutralizing effect of the antidote (17).
FIG. 1.
Layout of the pTF-FC2 pas showing its location within the plasmid replicon. The positions of the genes are relative to the ClaI site of pTF-FC2 (5).
Efficiency of the pas stability system in Escherichia coli is strain dependent.
The ability of the pTF-FC2 pasABC system to stabilize a heterologous plasmid in E. coli JM105 had previously been shown (17) by cloning the pasABC genes into the unstable, low-copy-number, test plasmid pOU82 (6). We repeated the stability assays in E. coli CSH50 to compare the efficiency of the pTF-FC2 pas to those of other poison-antidote systems in a host strain background identical to that used by other workers (9). It was observed that pas varied in its ability to act as a plasmid stabilization system depending upon which strain (Table 1) was used as host. Plasmid stability was determined by growing plasmid-containing E. coli cells in batch culture for 100 generations without selection in TB (24 g of yeast extract, 12 g of tryptone, and 4 ml of glycerol per 900 ml with 100 ml of sterile 0.17 M KH2PO4–0.72 M K2HPO4 added immediately before use). Aliquots were taken at 20-generation intervals and grown at 37°C overnight in the absence of selection. One hundred colonies were transferred to Luria agar (LA) plates with plasmid selection (ampicillin [100 μg ml−1], chloramphenicol [30 μg ml−1], or kanamycin [50 μg ml−1], as required), and the percentage survival was used to calculate plasmid loss. The stability assay for pOU82 and derivatives was performed as described above, except that aliquots were plated on LA containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. Plasmid-containing cells form blue colonies, whereas plasmid-free cells are white. At least three stability tests were performed for each strain, and the loss frequency was calculated by the method of Gerdes et al. (6). After 100 generations, a test plasmid containing the pasABC genes (pOU-pasABC [Table 1]) was stabilized approximately 2.5-fold in an E. coli JM105 host (Table 2). However, in an E. coli CSH50 host, the pas enhanced plasmid stability about 100-fold. In contrast, the pas was ineffective in enhancing the stability of the test plasmid in an E. coli JM107 host strain or its recA derivative, E. coli JM109. Surprisingly, the base level of pOU82 stability in E. coli JM109 was 10-fold higher than in strain JM107 (Table 2). The fact that E. coli strains JM107 and JM109 are isogenic except for the recA gene implies that the recA system has an effect on the stability of the pOU82 test plasmid. The reason for this increased stability is unknown, but the finding is similar to the finding that mini-RK2 plasmids were threefold more stable in E. coli JM109 than in JM107 (14).
TABLE 1.
Bacteria, plasmid vectors, and pas constructs used in this study
| Strain, plasmid, or construct | Genotype or descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| JM105 | thi rpsL endA sbcB15 hspR4 Δ(lac-proAB) [F′ traD36 proAB lacIqΔM15] | 20 |
| JM107 | thi endA gyrA96 hsdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqΔM15] | 20 |
| JM109 | recA1 thi endA gyrA96 hsdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqΔM15] | 20 |
| 71/18 | thi supE Δ(lac-proAB) [F′ traD36 proAB lacIqΔM15] | 20 |
| CSH50 | rpsL Δ(lac-pro) | 12 |
| CSH50-Iq | rpsL Δ(lac-pro) [F′ traD36 proAB lacIqΔM15] | This work |
| SG22025 | Δlac rcsA166::mini-kan parent of SG22093 and SG22095 | S. Gottesman (11) |
| SG22093 | Δlac rcsA166::mini-kan clpP1::cat | S. Gottesman (11) |
| SG22095 | Δlac rcsA166::mini-kan lon-146::mini-Tn10 | S. Gottesman (11) |
| Plasmids | ||
| pACYC184 | p15a replicon, Cmr Tcr | 3 |
| pKK223-3 | ColE1 replicon, Aprtac | 2 |
| pOU82 | R1 replicon, AprlacZYA | 6 |
| pKG399 | pSC101 replicon, Tcr (see Fig. 3) | 9 |
| pKGCm | pACYC184 replicon, Cmr (see Fig. 3) | This work |
| pas-containing constructs | ||
| pTac-pasB | pKK223-3 replicon, Apr, pas region 1518–1816b | 17 |
| pTac-pasAB | pKK223-3 replicon, Aprpas region 1316–1816 | 17 |
| pTac-pasABC | pKK223-3 replicon, Aprpas region 1316–2028 | 17 |
| pOU-pasABC | pOU82 replicon (R1), Apr, pas region 1158–2027 | 17 |
| pOU-pasABC*c | pOU82 replicon (R1), Apr, pas region 1158–2027 | 17 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; tac, trp-lac hybrid promoter.
The pas region numbers are the nucleotide positions relative to those of the ClaI-PstI fragment of pTV100 (5).
C* indicates a T insertion at 1833 bp to inactivate the pasC product (17).
TABLE 2.
Loss frequency of plasmids from E. coli host strains after 100 generations of growth
| Strain | Loss frequency
|
Approximate fold increase in stability | |
|---|---|---|---|
| No stability genes present (pOU82) | Stability genes present (pOU-pasABC) | ||
| JM105 | 2 × 10−2 | 9 × 10−3 | 2.5 |
| JM107 | 5 × 10−2 | 5 × 10−2 | 1 |
| JM109 | 5 × 10−3 | 4 × 10−3 | 1 |
| CSH50 | 2 × 10−2 | 3 × 10−4 | 100 |
In previous work (17), we showed that inactivation of PasC (through the introduction of a frameshift mutation in the pasC gene) resulted in an increase in the toxicity of the PasA-PasB poison-antidote complex (see also Fig. 2). We therefore examined how inactivation of PasC affects the ability of the pas to stabilize the test plasmid in different E. coli hosts. In E. coli JM105 (pOU-pasABC*), plasmid stability was about the same (loss frequency of 3 × 10−2) as that of the pOU82 test plasmid and considerably less than that of pOU-pasABC (loss frequency of 9 × 10−3), whereas in E. coli CSH50, inactivation of pasC reduced the ability of pas to stabilize the test plasmid from about 100- to 2.5-fold. Inactivation of pasC had little effect in E. coli JM107 or JM109.
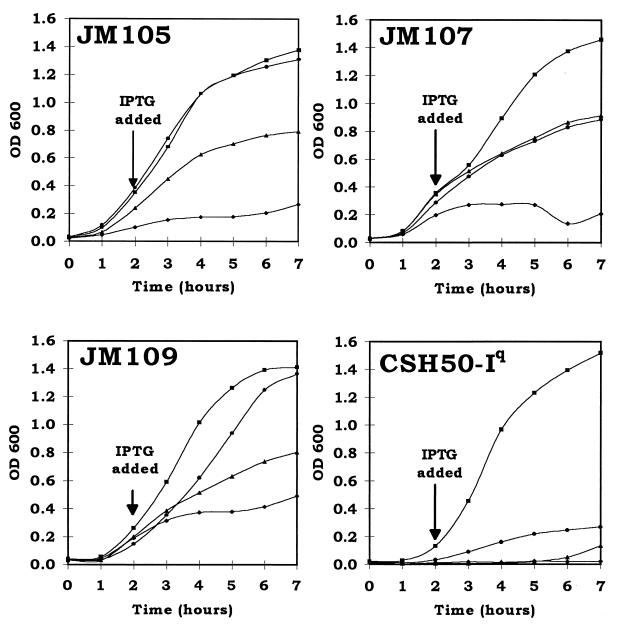
FIG. 2.
Growth curves of E. coli strains overexpressing the pas genes. Each graph shows pKK223-3 (control) (■), pTac-pasB (⧫), pTac-pasAB (▴), and pTac-pasABC (•). Datum points are the means of three separate experiments. OD 600, optical density at 600 nm.
Pas toxicity varies between E. coli host strains.
To investigate the reason for host-strain dependent variation in stability, combinations of the pas genes were cloned behind the tac promoter of a pKK223-3 vector (Table 1). In lacIq strains containing these constructs, tac-controlled expression of the pas genes is induced by isopropyl-β-d-thiogalactopyranoside (IPTG). Since E. coli CSH50 does not contain lacIq, the F′ lacIq-containing episome was transferred from E. coli 71/18 to CSH50 by conjugation. Conjugation was carried out overnight on the surface of a LA plate followed by plating on minimal medium plus streptomycin (100 μg/ml) to select for streptomycin resistance and proline independence. The stability of the test plasmid in this E. coli CSH50-Iq strain was indistinguishable from that of the test plasmid in CSH50. The effect of IPTG-induced pas gene expression on the growth of E. coli host strains JM105, JM107, JM109, and CSH50-Iq is shown in Fig. 2. Not all of the strains were equally sensitive to the PasB toxin. Growth of strain CSH50-Iq was the most severely inhibited by induction of pTac-pasB, with JM105 less inhibited and strains JM107 and JM109 the least inhibited. Expression of the pasA gene (encoding the antidote) relieved the toxic effect of pasB, although in E. coli strain CSH50-Iq, growth inhibition was relieved only slightly. When all three pasABC genes were expressed, the growth rates of all strains increased further, although in no strain did the growth rate reach that of the vector control. IPTG-induced expression of the pasABC system was toxic to all strains, but toxicity was most severe in E. coli CSH50-Iq, which was also the strain in which the pas plasmid stabilization system was most effective.
The effects of combinations of pas genes expressed under the control of a tac promoter on the host strains provided some insight into why pas-mediated plasmid stability varied in strains. The lower growth rate and cell density of E. coli CSH50-Iq containing different combinations of pas genes indicated that in this strain the PasA antidote did not effectively neutralize the toxic effect of PasB toxin even in the presence of PasC. It may be that the greater toxicity of the pas in E. coli CSH50-Iq was why the test plasmid was best stabilized by pas in this strain. Possible reasons for increased PasB toxicity in this strain are that the as yet unidentified cytoplasmic target may be more susceptible to the PasB toxin and that the protease which degrades the antidote may be particularly effective in E. coli CSH50-Iq. Variations in the levels of pas gene expression between strains may also play a role in the efficiency of plasmid stability, but this is less likely. Since the pas is autoregulated (18), differences in pas promoter strength between strains would not be expected to be as important as in a non-self-regulated system. Variations in the rate of transcription would alter the time required to reach self-regulating levels of antidote and toxin but probably not the actual levels reached.
Role of Lon protease in plasmid stabilization.
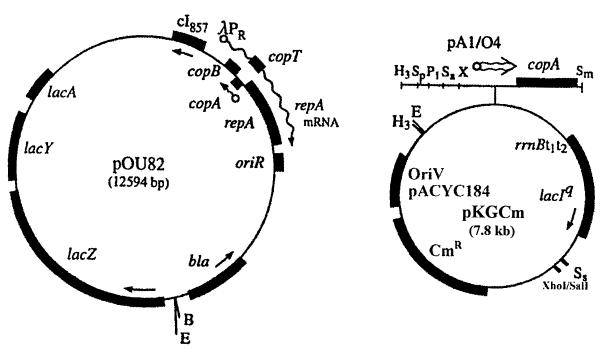
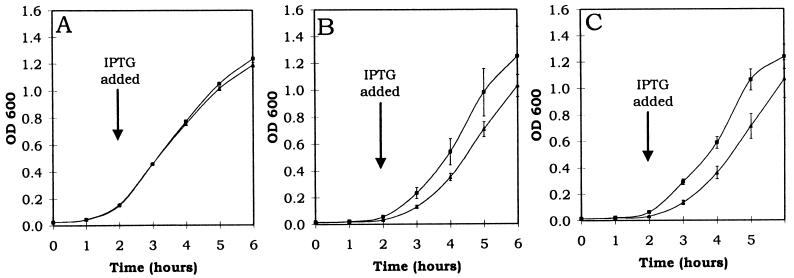
Plasmid proteic stabilization systems operate on the principle that the antidote is subject to a much higher rate of turnover than the toxin (8). A pOU82-pKG339-based conditional replication system (9) was used to determine which E. coli protease is involved in the selective degradation of the PasA antidote. Since one of the E. coli protease mutants was resistant to tetracycline (Table 1), the tetracycline resistance marker and pSC101 replicon of pKG339 were replaced by the chloramphenicol resistance marker and the p15a replicon of pACYC184 to produce plasmid pKGCm. When the copA gene of pKGCm is provided in trans to pOU82, replication of pOU82 can be halted by IPTG induction of the pA1/O4 promoter (Fig. 3). Therefore, addition of IPTG to E. coli (pOU-pasABC) cells will result in arrest of plasmid replication, and plasmid-free cells containing the toxin-antidote complex will result. The protease responsible for selective degradation of the antidote will digest the antidote, which will no longer inhibit the toxin, resulting in cell death. Functional antidote will persist in E. coli mutants deficient in the antidote-degrading protease, and cell death will not occur. Plasmid pOU82 and the pas-containing pOU82-based plasmid (pOU-pasABC) were transformed into E. coli SG22093 clpP1 and SG22095 lon mutants and into E. coli SG22025, a protease-proficient parental strain of these mutants, each of which contained a coresident pKGCm plasmid. The growth of all strains following the addition of 2 mM IPTG is shown in Fig. 4. Growth of the E. coli (pOU-pasABC) lon mutant strain was similar to that of the same strain containing pOU82 (Fig. 4A), suggesting that in this strain the antidote was long-lived. In contrast, growth of the E. coli (pOU-pasABC) clpP mutant and E. coli (pOU-pasABC) protease-proficient strains was reduced relative to that of the same strains containing the pOU82 control plasmid (Fig. 4B and C). The observation that growth was not reduced by forced plasmid loss in an E. coli lon mutant, whereas there was a decrease in growth following the loss of plasmids expressing the pasABC genes in lon-proficient strains, suggests that the antidote protein was stable in the lon mutant and that the Lon protease is involved in the degradation of the PasA antidote.
FIG. 3.
pOU82-pKGCm conditional replication system based on the pOU82-pKG339 system (modified from reference 9 with permission of the publisher). The pACYC replicon and chloramphenicol resistance markers of pKGCm have replaced the pSC101 replicon and tetracycline resistance marker of pKG339. Addition of IPTG results in expression of copA from the pA1/O4 promoter, and CopA inhibits replication of the pOU82 R1 replicon. Restriction site abbreviations: B, BamHI; E, EcoRI; H3, HindIII; Sp, SphI; P1, PstI; Sa, SalI; X, XhaI; Sm, SmaI; Ss, SspI.
FIG. 4.
Growth curves of protease mutants with different plasmids. Growth curves of E. coli lon mutant SG22095 (A), E. coli clpP1 mutant SG22093 (B), and E. coli protease mutant parent strain SG22025 (C) containing pOU82 and pKGCm (control) (■) or pOU-pasABC and pKGCm (▴) are shown. OD 600, optical density at 600 nm.
To obtain additional evidence for the involvement of the Lon protease, the stability of pOU-pasABC was tested in each strain in the absence of pKGCm. Strains in which the protease required for selective degradation of the antidote is not present would cause pOU-pasABC to be less stably inherited. In the E. coli clpP mutant and lon-proficient parental strains, the pOU-pasABC loss frequencies were 1.5 × 10−2 and 1.4 × 10−2, respectively, whereas the loss frequency was increased to 3.5 × 10−2 in the E. coli lon mutant. This 2.5-fold decrease in plasmid stability seen in the E. coli lon mutant strain supports the notion that the Lon protease plays a role in the degradation of PasA. The level of stabilization by the pas in the E. coli lon parental strain was comparable to that in E. coli JM105 but less than that found in E. coli CSH50-Iq. This is additional evidence that the Lon protease plays a role in pas-mediated plasmid stability.
Acknowledgments
We are most grateful to Kenn Gerdes for the gift of plasmids pOU82 and pKG339 and to Susan Gottesman for E. coli SG22025, SG22093, and SG22095.
This work was supported by grants from Gencor (now Billiton) Process Research (Randburg, South Africa), the Foundation for Research Development (Pretoria, South Africa), and the THRIP programme of the Department of Trade and Industry.
REFERENCES
- 1.Bravo A, de Torrontegui G, Dìaz R. Identification of the components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci USA. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorrington R A, Rawlings D E. Characterization of the minimum replicon of the broad-host-range plasmid pTF-FC2 and similarity between pTF-FC2 and the IncQ plasmids. J Bacteriol. 1990;172:5697–5705. doi: 10.1128/jb.172.10.5697-5705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorrington R A, Bardien S, Rawlings D E. The broad-host-range plasmid pTF-FC2 requires a primase-like protein for autonomous replication in Escherichia coli. Gene. 1991;108:7–14. doi: 10.1016/0378-1119(91)90481-p. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes K, Larsen J E L, Molin S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985;161:292–298. doi: 10.1128/jb.161.1.292-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaffé A, Ogura T, Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen R B, Gerdes K. Programmed cell death in bacteria:proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 9.Jensen R B, Grohmann E, Schwab H, Dìaz-Orejas R, Gerdes K. Comparison of ccd of F, parDE of RP4, and parD of R1 using a novel conditional replication control system of plasmid R1. Mol Microbiol. 1995;17:211–220. doi: 10.1111/j.1365-2958.1995.mmi_17020211.x. [DOI] [PubMed] [Google Scholar]
- 10.Lehnherr H, Maguin E, Jafri S, Yarmolinsky M B. Plasmid addiction genes of plasmid P1: doc, which causes cell death on curing of the prophage, and phd, which prevents host death when the prophage is retained. J Mol Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 11.Lehnherr H, Yarmolinsky M B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- 13.Rawlings D E, Kusano T. Molecular genetics of Thiobacillus ferrooxidans. Microbiol Rev. 1994;58:39–55. doi: 10.1128/mr.58.1.39-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts R C, Helinski D R. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J Bacteriol. 1992;174:8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohrer J, Rawlings D E. Sequence analysis and characterization of the mobilization region of a broad-host-range plasmid, pTF-FC2, isolated from Thiobacillus ferrooxidans. J Bacteriol. 1992;174:6230–6237. doi: 10.1128/jb.174.19.6230-6237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz-Echevarrìa M J, Berzal-Herranz A, Gerdes K, Díaz-Orejas R. The kis and kid genes of the parD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the co-ordinated action of the Kis and Kid proteins. Mol Microbiol. 1991;5:2685–2693. doi: 10.1111/j.1365-2958.1991.tb01977.x. [DOI] [PubMed] [Google Scholar]
- 17.Smith A S G, Rawlings D E. The poison antidote stability system of the broad-host-range Thiobacillus ferrooxidans plasmid pTF-FC2. Mol Microbiol. 1997;26:261–270. doi: 10.1046/j.1365-2958.1997.6332000.x. [DOI] [PubMed] [Google Scholar]
- 18.Smith A S G, Rawlings D E. Autoregulation of the pTF-FC2 proteic poison-antidote plasmid addiction system (pas) is essential for plasmid stabilization. J Bacteriol. 1998;180:5463–5465. doi: 10.1128/jb.180.20.5463-5465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuchimoto S, Ohtsubo H, Ohtsubo E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988;170:1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yannisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]