Abstract
Coenzyme Q (CoQ, ubiquinone) is an essential cellular cofactor composed of a redox-active quinone head group and a long hydrophobic polyisoprene tail. How mitochondria access cytosolic isoprenoids for CoQ biosynthesis is a longstanding mystery. Here, via a combination of genetic screening, metabolic tracing and targeted uptake assays, we reveal that Hem25p—a mitochondrial glycine transporter required for haem biosynthesis—doubles as an isopentenyl pyrophosphate (IPP) transporter in Saccharomyces cerevisiae. Mitochondria lacking Hem25p failed to efficiently incorporate IPP into early CoQ precursors, leading to loss of CoQ and turnover of CoQ biosynthetic proteins. Expression of Hem25p in Escherichia coli enabled robust IPP uptake and incorporation into the CoQ biosynthetic pathway. HEM25 orthologues from diverse fungi, but not from metazoans, were able to rescue hem25Δ CoQ deficiency. Collectively, our work reveals that Hem25p drives the bulk of mitochondrial isoprenoid transport for CoQ biosynthesis in fungi.
Coenzyme Q (CoQ, ubiquinone) is a redox-active lipid that functions as an essential cofactor for multiple cellular processes, including oxidative phosphorylation, pyrimidine biosynthesis, fattyacid oxidation and ferroptotic defence1,2,3,4. Deficiencies in CoQ underlie multiple human pathologies, all of which have limited therapeutic options5,6. Despite being discovered more than 65 years ago, substantial knowledge gaps persist regarding how CoQ is synthesized and distributed throughout the cell, thereby limiting the development of interventional strategies7,8,9.
CoQ is composed of a fully substituted quinone ring and a long hydrophobic tail consisting of repeating isoprene units (Fig. 1a). CoQ is synthesized on the matrix side of the inner mitochondrial membrane, beginning with the precursors 4-hydroxybenzoate (4-HB) and isoprenoid pyrophosphates10,11. These precursors are each formed in the cytosol: 4-HB is derived from tyrosine or the shikimate pathway, and the isoprenoid pyrophosphates are produced by the mevalonate pathway8. Previous work has shown that mitochondria are able to import 4-HB and the isoprenoid isopentenyl pyrophosphate (IPP) for CoQ biosynthesis in vitro12,13. However, the protein(s) that enable this transport have remained elusive for decades. Previous efforts to identify the proteins necessary for CoQ production—largely by leveraging the inability of yeast lacking CoQ to grow in respiratory conditions—have failed to identify candidate transporters8,14,15, suggesting that there is either redundancy in the system or that the required transporter is essential for growth under common conditions16.
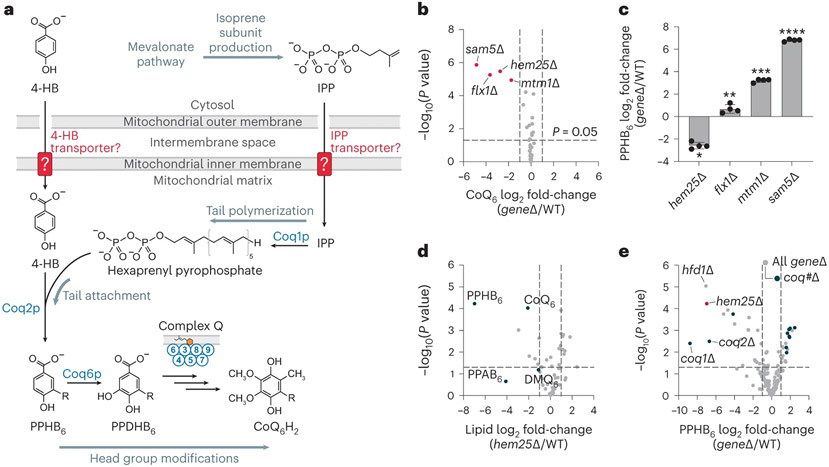
Fig. 1: A targeted genetic screen identifies Hem25p as a potential transporter of CoQ precursors.
a, Schematic of CoQ biosynthesis in S. cerevisiae. 4-HB, 4-hydroxybenzoate; IPP, isopentenyl pyrophosphate; PPHB6, polyprenyl-hydroxybenzoate; PPDHB6, polyprenyl-dihydroxybenzoate. b, Relative CoQ abundance in all geneΔ strains compared to wild type (WT) versus statistical significance. Hits with significantly decreased levels of CoQ are highlighted (mean, n = 3 biologically independent samples, two-sided Student’s t-test). c, Relative PPHB levels in each of the hits from b (*P = 3.1 × 10−6 WT versus hem25Δ, **P = 0.0139 WT versus flx1Δ, ***P = 4.1 × 10−8 WT versus mtm1Δ, ****P = 1.3 × 10−10 WT versus sam5Δ; mean ± s.d., n = 4 biologically independent samples, two-sided Student’s t-test). d, Relative lipid abundances in hem25Δ yeast compared to WT versus statistical significance with CoQ and CoQ biosynthetic intermediates highlighted. PPAB6, polyprenyl-aminobenzoate; DMQ6, demethoxy-coenzyme Q. e, Relative PPHB abundances versus statistical significance across all Y3K geneΔ strains, with hem25Δ, hfd1Δ and coq#Δ strains highlighted. For d and e, raw data from the Y3K dataset33 (respiration-RDR (respiration deficiency response) condition) are displayed as the mean from three biologically independent samples, with two-sided Student’s t-test used for both panels. Numerical data are available as Source data.
In this Article we identify Hem25p (Ydl119cp), a mitochondrial glycine carrier, as the primary IPP transporter for CoQ biosynthesis. Loss of Hem25p reduces the production of early CoQ intermediates in mitochondria, resulting in decreased CoQ abundance. We show that the role of Hem25p in CoQ biosynthesis is distinct from its role in glycine transport and haem biosynthesis. Ectopic expression of Hem25p in Escherichia coli is sufficient to drive IPP transport across the bacterial membrane. Collectively, our data establish Hem25p as an IPP transporter connecting the mevalonate pathway to mitochondrial CoQ biosynthesis.
Results
A screen for carriers in CoQ biosynthesis identifies Hem25p
To identify mitochondrial transporters important for CoQ production, we created a custom panel of Saccharomyces cerevisiae gene knockout (KO) (geneΔ) strains, each lacking an established or potential mitochondrial transporter. This panel comprised geneΔ strains for all 35 members of the SLC25 family of mitochondrial transporters, which are responsible for nearly all known metabolite transport into mitochondria17, and seven poorly characterized mitochondrial inner-membrane proteins that might possess transporter function18,19. We grew strains in triplicate under fermentative conditions and monitored their ability to produce CoQ via targeted electrochemical detection coupled to HPLC (HPLC-ECD). Using this strategy, we identified four strains with significantly decreased CoQ levels (Fig. 1b). Of these strains, two lacked transporters (Flx1p and Sam5p) known to carry substrates required for the later head-group-modifying steps of CoQ biosynthesis (cofactors FAD and S-adenosylmethionine, respectively)20,21,22,23. The third lacked Mtm1p, the yeast orthologue of the glutathione transporter SLC25A3924,25,26. Cells lacking Mtm1p demonstrate elevated mitochondrial manganese levels and disrupted iron–sulfur cluster biogenesis, both of which can compromise late CoQ biosynthesis7,27,28,29. The final strain lacked Hem25p, a glycine carrier with no established connection to CoQ30,31.
To further prioritize these candidates, we then measured the levels of polyprenyl-hydroxybenzoate (PPHB). PPHB and, to a lesser extent, polyprenyl-aminobenzoate (PPAB) are the first intermediates of CoQ produced within S. cerevisiae mitochondria and are known to accumulate when the downstream pathway is disrupted. We reasoned that loss of cofactor transporters important for enzymes in the later stages of CoQ biosynthesis would likewise cause an accumulation of PPHB, whereas loss of a 4-HB or IPP transporter would prevent PPHB formation32. Of the four candidates, only the hem25Δ strain had decreased levels of PPHB (Fig. 1c). This finding is consistent with data from our recent systematic analysis of mitochondrial protein functions, in which the hem25Δ strain exhibited decreased levels of CoQ and the prenylated CoQ precursors PPHB and PPAB (Fig. 1d)33. In reanalysing data from all 176 strains in that study (which included few transporter KO strains), hem25Δ exhibited PPHB depletion comparable to strains lacking the established early-stage CoQ proteins Hfd1p (which produces 4-HB) and Coq1p and Coq2p (which collectively produce PPHB and PPAB) (Fig. 1e). Intriguingly, we find that data from large-scale chemical genomic screens also support a link between HEM25, the mevalonate pathway, and statin sensitivity (Fig. 2a,b), suggesting unexplored connections to isoprenoid biology34. Indeed, our treatment of hem25Δ yeast with atorvastatin further reduced CoQ levels, resulting in a growth defect on non-fermentable carbon sources (Fig. 2c,d). Given these results, we proceeded to investigate a CoQ-related role for Hem25p in biochemical depth.
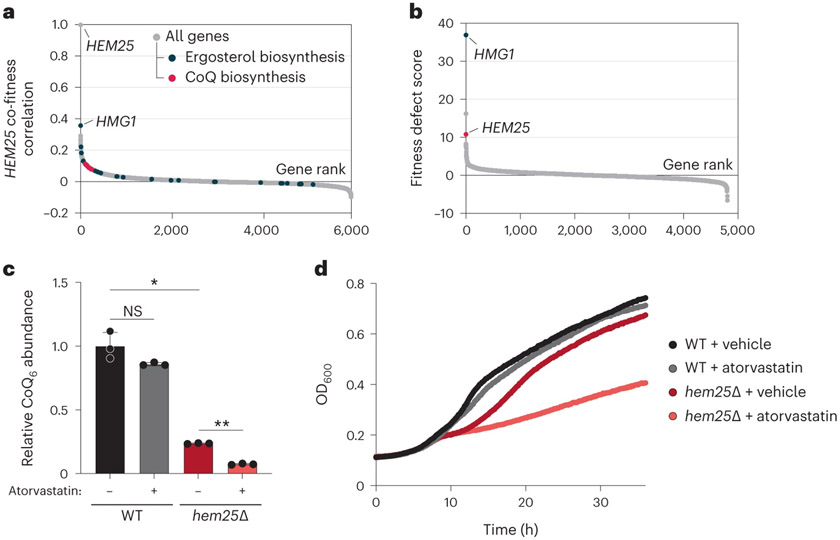
Fig. 2: Chemical genomic screens link HEM25 to the mevalonate pathway.
a, Gene fitness correlations with HEM25. Genes involved with ergosterol and CoQ biosynthesis are highlighted. b, Fitness ranking of geneΔ strains in the presence of 78.68 μM atorvastatin. c, Relative CoQ abundances of WT and hem25Δ cells treated with either vehicle (DMSO) or 10 μM atorvastatin (*P = 0.0002 DMSO-treated WT versus DMSO-treated hem25Δ, **P = 1.1 × 10−6 DMSO-treated hem25Δ versus atorvastatin-treated hem25Δ, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test; NS, not significant). ALA was present in the growth medium to support haem biosynthesis. d, Growth assay of WT and hem25Δ yeast in synthetic complete medium containing 0.1% (wt/vol) glucose, 3% (wt/vol) glycerol and 300 μM ALA. The media were further supplemented with either vehicle (DMSO) or atorvastatin, as indicated (mean, n = 3 biologically independent samples). Under these conditions, yeast enter respiratory growth after approximately 8 h. Raw data for a and b are from the HIPHOP chemogenomics database34. Fitness defect scores reflect the homozygous deletion profiles for atorvastatin (compound SGTC_2648). Numerical data are available as Source data.
Hem25p has distinct roles in haem and CoQ production
Hem25p was previously established as a mitochondrial glycine transporter necessary for haem biosynthesis (Fig. 3a)30,31. Imported glycine condenses with succinyl-CoA to form the haem precursor aminolevulinic acid (ALA). ALA is then exported out of the mitochondrial matrix to the cytosol, where it proceeds along the haem biosynthetic pathway. Yeast lacking Hem25p can still produce low levels of haem31, suggesting that mitochondria contain a secondary glycine carrier. However, its identity remains unknown.
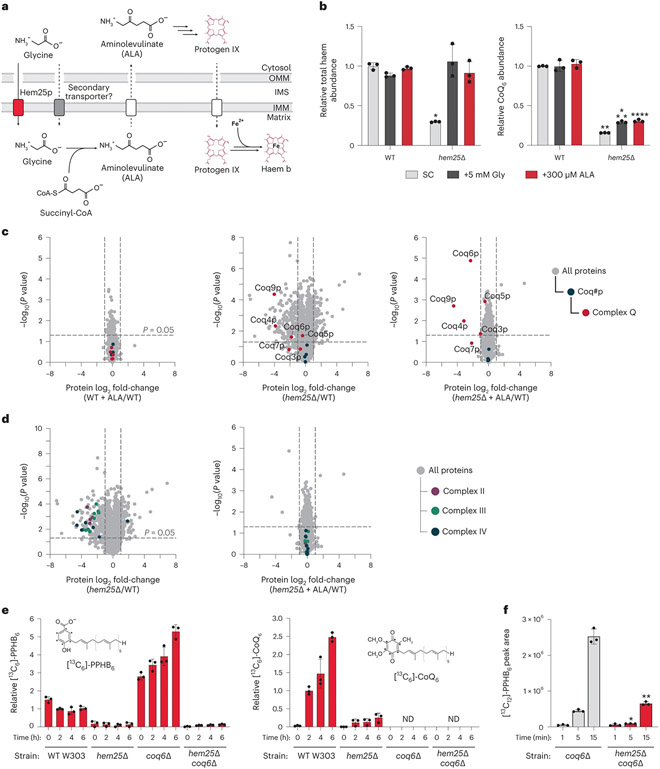
Fig. 3: Hem25p drives CoQ production independently of its role in haem biosynthesis.
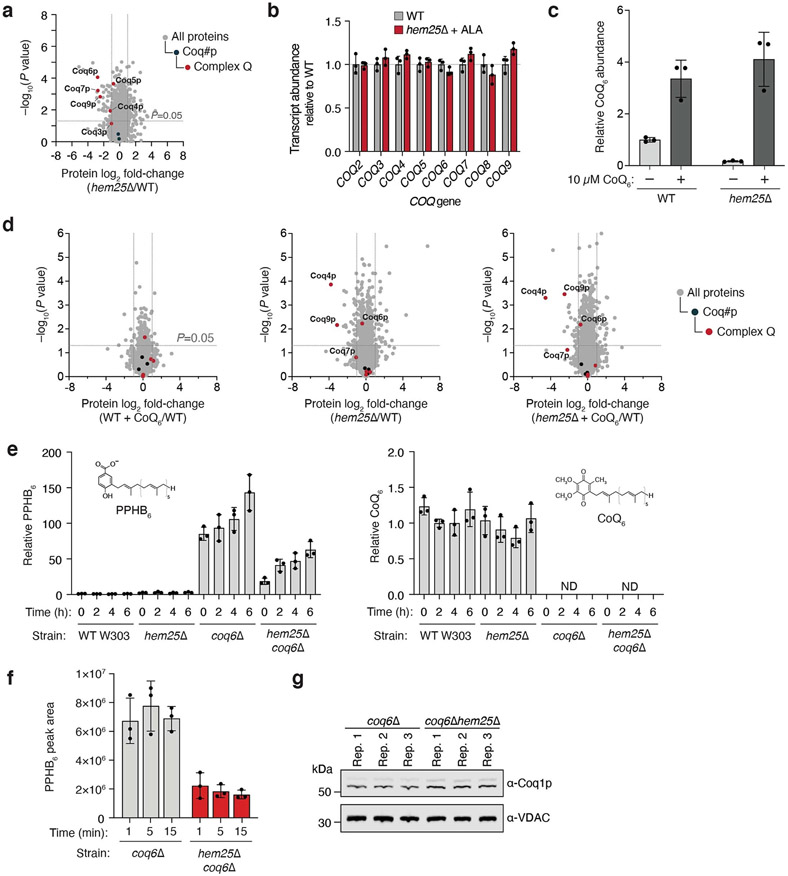
a, Overview of haem biosynthesis and the role of Hem25p. OMM, outer mitochondrial membrane; IMS, inter membrane space; IMM, inner mitochondrial membrane. b, Relative total haem and CoQ abundances in WT and hem25Δ yeast grown in synthetic complete (SC) medium with and without glycine and ALA supplementation (*P = 1.01 × 10−5 WT SC haem versus hem25Δ SC haem, **P = 7.56 × 10−10 WT SC CoQ versus hem25Δ SC CoQ, ***P = 8.47 × 10−8 WT SC CoQ versus hem25Δ+Gly CoQ, ****P = 2.22 × 10−7 WT SC CoQ versus hem25Δ+ALA CoQ; mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). c, Relative protein abundances of WT cells with ALA, hem25Δ cells without ALA, and hem25Δ cells with ALA supplementation compared to WT cells without ALA supplementation versus statistical significance. CoQ biosynthetic proteins and complex Q proteins are highlighted (mean, n = 3 biologically independent samples, two-sided Student’s t-test). d, Relative protein abundances of hem25Δ cells ± ALA supplementation compared to WT cells without ALA supplementation versus statistical significance. Components of the electron-transport chain are highlighted (mean, n = 3 biologically independent samples, two-sided Student’s t-test). e, Relative abundance of de novo synthesized [13C6]-PPHB and [13C6]-CoQ in WT, hem25Δ, coq6Δ and coq6Δhem25Δ cells grown in –pABA SD, 3% (wt/vol) glycerol and 50 μM [phenyl-13C6]-4-HB (mean ± s.d., n = 3 biologically independent samples). ND, not detected. f, Normalized abundance of de novo synthesized [13C12]-PPHB in isolated coq6Δ and coq6Δhem25Δ mitochondria (*P = 5.73 × 10−5 WT versus hem25Δ at 5 min, **P = 0.0001 WT versus hem25Δ at 15 min; mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). Numerical data are available as Source data.
To determine whether the effect of Hem25p on CoQ levels is related to its role in haem production, we supplemented the hem25Δ strain with either ALA or high levels of glycine, both of which have been shown to bypass the requirement of Hem25p for haem production30,31. Without supplementation, hem25Δ cells had markedly reduced levels of haem and CoQ (Fig. 3b). Consistent with previous work, both glycine and ALA supplementation fully rescued haem in the hem25Δ strain to wild-type (WT) levels. However, supplementation had a minimal effect on CoQ levels, demonstrating distinct roles for this transporter. Notably, the residual levels of CoQ in hem25Δ suggest that other transporters may enable CoQ precursor uptake. This may explain why Hem25p has been difficult to identify by screens for respiratory incompetence, as very little CoQ is required for yeast to survive on non-fermentable carbon sources35.
To further probe Hem25p’s distinct roles in haem and CoQ biosynthesis, we performed whole-cell proteomic analyses on the hem25Δ strain. Unsurprisingly, disruption of Hem25p resulted in widespread protein changes, with 185 proteins showing a greater than twofold decrease in abundance versus WT cells. Of these significantly decreased proteins, 121 were mitochondrial, including select CoQ biosynthetic proteins (Fig. 3c). Reanalysis of our previous large-scale proteomics data revealed a similar proteomic response (Extended Data Fig. 1a)33. Remarkably, hem25Δ cells supplemented with ALA exhibited a decrease in only four proteins, all of which are enzymes in the later stages of the CoQ biosynthetic pathway (Fig. 3c). Furthermore, this reduction in CoQ biosynthetic proteins was independent of transcript levels, which remained unchanged (Extended Data Fig. 1b). Previous studies have demonstrated that CoQ or a late CoQ intermediate is essential for stabilizing the CoQ biosynthetic complex (complex Q, CoQ synthome) responsible for the later, head-group-modifying reactions and that multiple subunits of this complex are degraded in its absence36,37. To investigate the effect of CoQ specifically on complex Q stability, we repeated our proteomic analyses using cells supplemented with CoQ6. Despite increasing the total cellular CoQ abundance of WT and hem25Δ cells to similar levels (approximately fourfold higher than unsupplemented WT cells), the levels of complex Q proteins remained decreased (Extended Data Fig. 1c,d). These results suggest that Hem25p supports the production of a stabilizing intermediate, consistent with a role in transporting IPP or 4-HB. Importantly, our proteomic analyses showed an overall decrease in respiratory chain complexes in hem25Δ cells that was completely restored to WT levels upon ALA supplementation (Fig. 3d). This is consistent with previous reports of Hem25p’s effect on respiratory chain stability, and further demonstrates Hem25p’s distinct roles in haem and CoQ biosynthesis30,38.
To assess Hem25p’s contribution to CoQ production, we monitored the incorporation of heavy-isotope-labelled 4-HB ([phenyl-13C6]-4-HB) into the CoQ pathway in hem25Δ cells supplemented with ALA using a custom mass spectrometry (MS) analysis. Incorporation of labelled 4-HB into PPHB and CoQ was markedly decreased in the hem25Δ strain compared to the WT strain, again suggesting that Hem25p enables PPHB production (Fig. 3e and Extended Data Fig. 1e). To minimize any potentially confounding results from decreased complex Q levels in hem25Δ, we also compared these results to those from a coq6Δ background strain. Coq6p, a hydroxylase, is the first enzyme to act upon PPHB once formed20. Yeast lacking Coq6p are unable to hydroxylate PPHB, leading to decreased levels of complex Q proteins, disrupted CoQ biosynthesis, and an accumulation of labelled PPHB20,37 (Fig. 3e). This strain thus enables us to isolate a potential role for Hem25p upstream of Coq6p. Indeed, the coq6Δ PPHB accumulation was greatly decreased in the coq6Δhem25Δ strain, providing additional evidence that Hem25p enables PPHB production, probably by providing one or both precursors required for its formation (Fig. 3e).
To directly measure Hem25p’s contribution to CoQ biosynthesis in vitro, we cultured the coq6Δ and coq6Δhem25Δ strains with ALA, isolated their mitochondria, and monitored their ability to form PPHB in the presence of 4-HB, MgCl2 and [1,2-13C2]-IPP. Consistent with our whole-cell results, disruption of Hem25p greatly reduced the ability of mitochondria to generate labelled PPHB (Fig. 3fand Extended Data Fig. 1f,g). Thus, even with ALA supplementation, cells and mitochondria lacking Hem25p exhibit a major deficiency in generating CoQ and CoQ intermediates. Collectively, these results show that Hem25p directly contributes to CoQ biosynthesis and that this role is independent of its established function in haem biosynthesis.
Hem25p enables bacterial isoprene transport
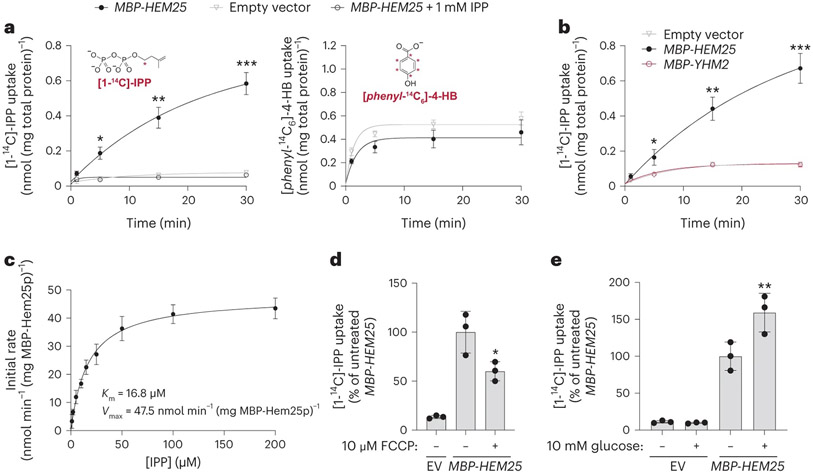
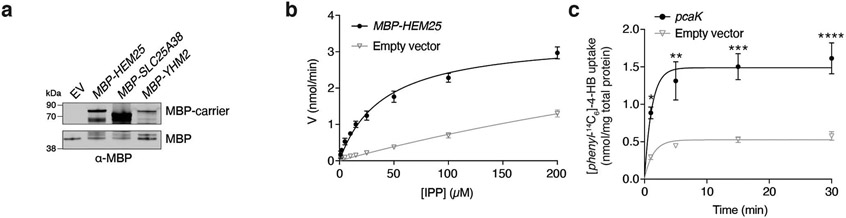
Our above results suggest a model whereby Hem25p transports a precursor—either 4-HB or IPP—into the mitochondrial matrix for CoQ biosynthesis. To test this directly, we generated a construct encoding an in-frame fusion of the periplasmic maltose-binding protein (MBP) with Hem25p, enabling expression at the E. coli plasma membrane39,40. Control (empty vector) E. coli cells, or those expressing MBP-Yhm2p, the mitochondrial citrate and oxoglutarate carrier41, exhibited little to no ability to take up [1-14C]-IPP (Fig. 4a,b and Extended Data Fig. 2a). However, cells expressing MBP-Hem25p demonstrated a clear, time-dependent [1-14C]-IPP uptake that was inhibited by excess unlabelled IPP (Fig. 4a). This IPP import was saturatable with a Michaelis constant (KM) of 16.8 μM and a maximum velocity (Vmax) of 47.5 nmol min−1 mg−1 MBP-Hem25p (Fig. 4c and Extended Data Fig. 2b). In contrast, no 4-HB import was detected in cells expressing MBP-Hem25p (Fig. 4a). Uptake of 4-HB was only seen when the known bacterial 4-HB transporter PcaK was expressed (Extended Data Fig. 2c)42. The addition of FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone) and glucose resulted in a decrease and increase of IPP uptake, respectively, suggesting that the proton motive force contributes to IPP import (Fig. 4d,e).
Fig. 4: Hem25p enables IPP import in bacteria.
a, Time course of 50 μM [1-14C]-IPP (left) or 50 μM [phenyl-14C6]-4-HB (right) uptake by E. coli cells expressing MBP-Hem25p. Cells carrying the empty expression vector and competition by excess unlabelled IPP were used as controls (*P = 0.0023, **P = 0.0006, ***P = 0.0001 empty vector versus MBP-HEM25, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). b, Time course of 50 μM [1-14C]-IPP uptake by E. coli cells expressing MBP-Hem25p, MBP-Yhm2p, or the empty vector (*P = 0.0165, **P = 0.0012, ***P = 0.0003 empty vector versus MBP-HEM25, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). c, Steady-state kinetics of [1-14C]-IPP uptake. For each time point, the corresponding empty vector control was subtracted. Data reflect the mean ± s.d. from three biologically independent samples. d,e, Relative E. coli [1-14C]-IPP uptake in the presence of 10 μM FCCP (d) or 10 mM glucose (e). IPP uptake levels reflect 30 min of incubation time and are relative to the respective vehicle-treated MBP-HEM25 (*P = 0.0419 MBP-HEM25 ± FCCP, **P = 0.0346 MBP-HEM25 ± glucose, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). Numerical data are available as Source data.
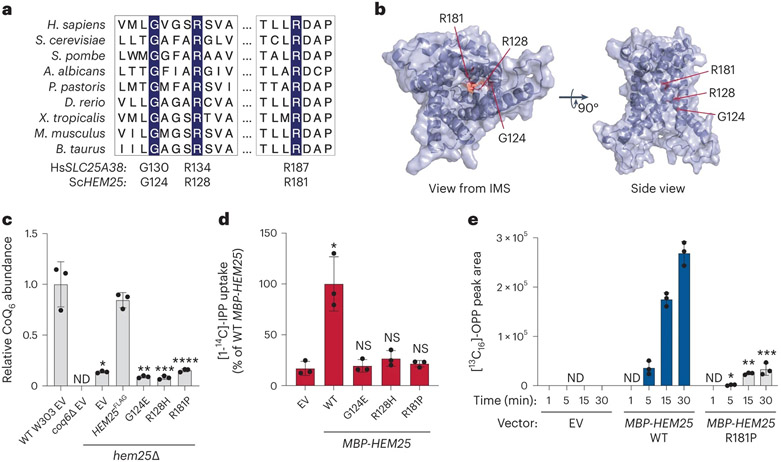
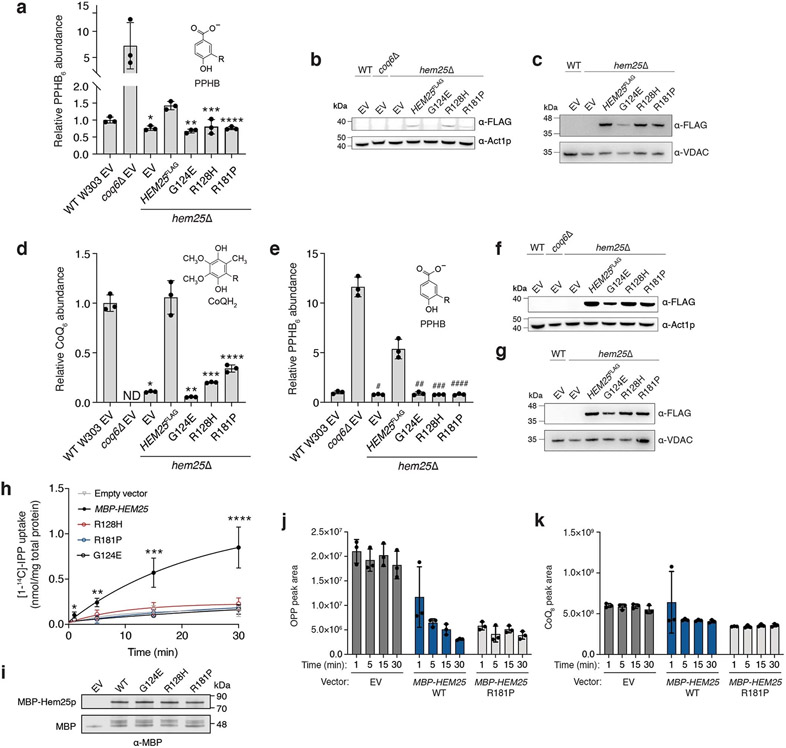
To ensure that IPP uptake was the result of Hem25p function, we generated three Hem25p point mutants: G124E, R128P and R181P. All three residues are highly conserved in HEM25 orthologues, with R128 and R181 located in the common substrate-binding site of mitochondrial carriers (Fig. 5a,b)43. These specific mutations were identified in the human orthologue SLC25A38 from cases of congenital sideroblastic anaemia (due to haem deficiency)41. Thus, given their pathogenicity and detrimental effect on carrier function, we introduced the corresponding Hem25p mutants into hem25Δ yeast and tested their ability to rescue CoQ abundance. When expressed from the endogenous HEM25 promoter, none of the three mutants was able to rescue CoQ levels (Fig. 5c and Extended Data Fig. 3a-c). Constitutive overexpression of the mutants resulted in only a partial rescue in the R128H and R181P mutants (Extended Data Fig. 3d-g), suggesting that these residues are important for Hem25p’s IPP transport function. Consistent with our CoQ rescue observations, none of the mutants demonstrated IPP transport when expressed in bacteria (Fig. 5d and Extended Data Fig. 3h,i).
Fig. 5: Hem25p function is required for bacterial IPP import.
a, Multiple sequence alignment of HEM25 orthologues with residues mutated in congenital sideroblastic anaemia highlighted. b, Predicted structure of Hem25p showing the location of the disease-related residues. IMS, intermembrane space. c, Relative CoQ abundances in hem25Δ yeast carrying WT and mutant HEM25FLAGconstructs. Levels are relative to WT yeast carrying the empty expression vector (*P = 8.58 × 10−5, **P = 6.67 × 10−5, ***P = 6.78 × 10−5, ****P = 9.73 × 10−5 HEM25FLAG versus mutants or empty vector, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). ND, not detected. d, Relative [1-14C]-IPP uptake by E. coli cells expressing WT or mutant MBP-Hem25p. Uptake levels reflect 30 min of incubation time and are relative to that of WT MBP-Hem25p (*P = 0.0063 empty vector versus WT HEM25, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). NS, not significant. e, Normalized abundance of de novo synthesized [13C16]-OPP in E. coli cells expressing the empty vector (EV), WT MBP-Hem25p or the R181P MBP-Hem25p mutant (*P = 0.0116 WT versus R181P MBP-Hem25p at 5 min, **P = 4.1 × 10−5 WT versus R181P MBP-Hem25p at 15 min, ***P = 0.0001 WT versus R181P MBP-Hem25p at 30 min, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). Numerical data are available as Source data.
To differentiate between bona fide IPP transport and mere binding of IPP to the extracellular portion of Hem25p, we exploited the fact that E. coli harbour a complete intracellular CoQ biosynthetic pathway. Using a modified version of our E. coli uptake assay, we monitored the import and incorporation of [1,2-13C2]-IPP into this pathway. Escherichia coli expressing MBP-Hem25p demonstrated a time-dependent accumulation of labelled octaprenylphenol (OPP), a bacterial CoQ biosynthetic intermediate (Fig. 5e and Extended Data Fig. 3j,k). No labelled OPP was detected in cells carrying the empty vector. Similarly, labelled OPP production was greatly reduced in cells expressing the R181P mutant. Taken together, our results show that Hem25p is sufficient to transport IPP into the mitochondrial matrix for CoQ biosynthesis.
Hem25p’s role in CoQ biosynthesis is restricted to fungi
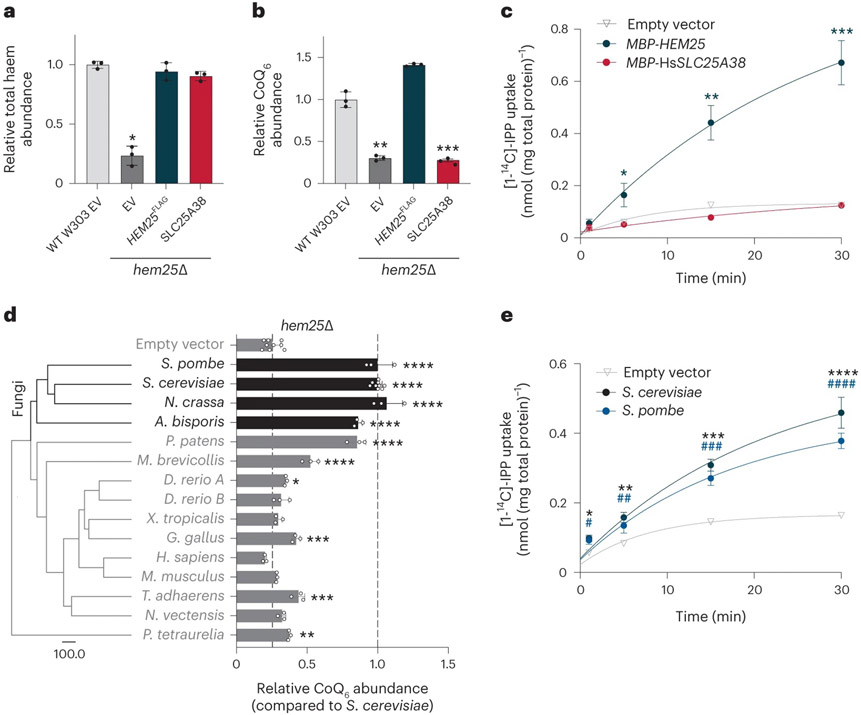
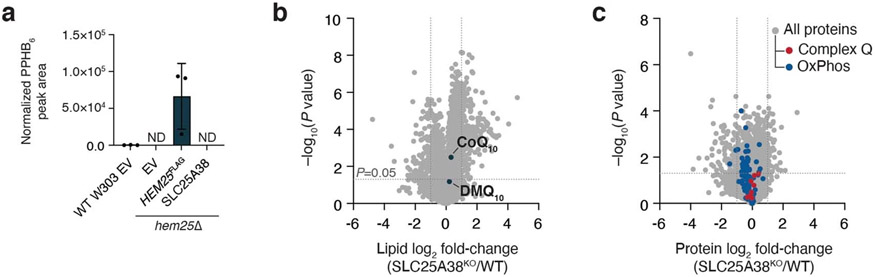
Hem25p exhibits high overall sequence conservation with human SLC25A38 (33% identity, 45% similarity), an established glycine transporter whose disruption results in congenital sideroblastic anaemia44,45. Given Hem25p’s function in yeast CoQ biosynthesis, we next tested whether the human SLC25A38 also doubles as an IPP transporter. Expression of the human SLC25A38 in the hem25Δ strain restored total haem levels, but had no effect on CoQ (Fig. 6a,b and Extended Data Fig. 4a). Consistently, when expressed in bacteria, MBP-SLC25A38 failed to transport IPP (Fig. 6c). Thus, despite sharing many of the same substrate-binding residues as Hem25p, only the glycine transport function is conserved in human SLC25A38. This is further supported by our recent MITOMICS dataset, where the human HAP1 cells lacking SLC25A38 had normal levels of CoQ and complex Q members (Extended Data Fig. 4b,c)46.
Fig. 6: Hem25p’s role in CoQ biosynthesis is conserved in fungi.
a,b, Haem (a) and CoQ (b) rescue by SLC25A38 in hem25Δ yeast. Levels are relative to the WT strain carrying the empty expression vector (*P = 0.0001, **P = 0.0002, ***P = 0.0001, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). c, Time course of 50 μM [1-14C]-IPP uptake by E. coli cells expressing MBP-SLC25A38 and MBP-Hem25p. Uptake by cells carrying the empty expression vector is included as a control (*P = 0.0165, **P = 0.0012, ***P = 0.0003 empty vector versus MBP-HEM25, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). d, Phylogenetic analysis of Hem25p orthologues and relative CoQ abundance in hem25Δ cells expressing these orthologues. The scale bar represents evolutionary distance, indicating the number of aminoacid substitutions per site. Levels are relative to that of the S. cerevisiae Hem25p (****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05 versus empty vector; mean ± s.d., at least three biologically independent samples, two-sided Student’s t-test). e, Time course of [1-14C]-IPP uptake by E. coli cells expressing S. cerevisiae Hem25p, S. pombe Hem25p, or the empty vector (*P = 0.0016, **P = 0.001, ***P = 9.51 × 10−5, ****P = 0.0003 empty vector versus S. cerevisiae Hem25p, #P = 0.0074, ##P = 0.0153, ###P = 0.0005, ####P = 8.48 × 10−5 empty vector versus S. pombe Hem25p, mean ± s.d., n = 3 biologically independent samples, two-sided Student’s t-test). Numerical data are available as Source data.
We next sought to better understand the evolutionary divergence of Hem25p and SLC25A38 function. HEM25 is highly conserved among opisthokonts. Thus, we used the PANTHER database to compile a list of HEM25 orthologues from a number of model organisms across this group47. We expressed each orthologue in hem25Δ yeast and measured their ability to rescue CoQ levels. Full rescue was largely limited to fungal orthologues, with little to no rescue with metazoan orthologues (Fig. 6d). Phylogenic analysis supports this functional split, showing an early evolutionary event resulting in separate fungal and metazoan branches (Fig. 6d)47. To validate Hem25p’s role in fungi, we expressed the Schizosaccharomyces pombe orthologue in bacteria and measured IPP uptake. Consistent with our rescue experiment, Hem25p from S. pombe and S. cerevisiae—two highly evolutionarily diverse yeast species—demonstrated similar levels of IPP uptake (Fig. 6e). Collectively, our results show that the mitochondrial glycine carrier Hem25p is required for IPP transport in fungi, and will empower subsequent studies to identify IPP transporters in metazoans.
Discussion
In this Article we identify Hem25p as a mitochondrial carrier required for IPP transport. Our results reveal a second function for this established glycine transporter and propose a model whereby a single carrier, Hem25p, enables the bulk of mitochondrial haem and CoQ biosynthesis. However, even in the absence of Hem25p, haem and CoQ are still produced at low levels (Fig. 3b). This suggests that functional redundancy is inherent among mitochondrial transporters, and may explain why previous large-scale screens for CoQ biosynthetic proteins failed to identify Hem25p16,17. In our targeted screen, we did not find any strong candidates for secondary transporters, despite having screened every known yeast mitochondrial transporter. It is possible that multiple SLC25 transporters each have low-level IPP transport capability or that a non-SLC25 protein not included in our screen is the secondary transporter. Compensatory mechanisms may also mask subtle changes when a secondary transporter is disrupted. CoQ biosynthetic proteins have been shown to organize into domains at endoplasmic reticulum (ER)-mitochondria contact sites48,49. Given that the mevalonate pathway, which supplies IPP, is partially associated with the ER, this co-localization might suggest that CoQ precursor transport can be facilitated by these contact sites. Thus, it is possible that these domains provide an additional layer of redundancy. Further efforts are needed to dissect the complete mechanisms of isoprenoid pyrophosphate transport.
Our finding that Hem25p enables both glycine and IPP transport suggests that haem and CoQ biosynthesis are coupled in yeast. Although the two biosynthetic pathways are distinct, their end products are each essential components of the electron-transport chain. It is possible that Hem25p synchronizes the production of haem and CoQ to support optimal mitochondrial oxidative phosphorylation. This is especially important in yeast, as the use of different carbon sources (fermentable versus non-fermentable) can result in large shifts in cellular energy demands33. In evolving to transport glycine and IPP despite their chemical and structural differences, yeast can readily adapt to changing nutrient conditions while simultaneously reducing the need to synthesize a second transporter. However, further investigation into the regulation of Hem25p and its effect on electron-transport chain function is needed to support this model.
Hem25p’s ability to support two separate biosynthetic pathways raises the possibility that other mitochondrial transporters can accommodate multiple substrates. Indeed, the yeast Pic2p and its mammalian homologue SLC25A3 were recently shown to transport copper in addition to phosphate—two highly distinct substrates, much like glycine and IPP50,51. In that case, a single carrier enables oxidative phosphorylation by supporting both ATP synthesis and cytochrome-c oxidase activity. Many metabolites still lack a defined transporter, despite evidence of mitochondrial uptake. The fact that yeast and humans have only 35 and 53 SLC25 carriers, respectively, suggests that functional redundancy is widespread17. However, a potential contribution from non-SLC25 carriers, many of which remain unidentified, must also be considered. Thus, a complete and accurate definition of mitochondrial transporters and their substrate specificities is still needed. The development of new experimental technologies and screening approaches will accelerate these characterization efforts.
A previous study identified six SLC25 members that genetically interact with HEM25, including FLX1, MTM1 and SAM538. Interestingly, disruption of these three genes resulted in decreased CoQ levels in our screen (Fig. 1b). FLX1 and SAM5 encode transporters for FAD and SAM, respectively, both of which are necessary cofactors for CoQ biosynthesis20,21,22,23. Mtm1p is the yeast orthologue of SLC25A39, the mammalian mitochondrial glutathione transporter24,25,26. Mtm1p was originally proposed to assist with mitochondrial manganese trafficking. However, it was later suggested that it participates in iron–sulfur cluster biogenesis27,29, consistent with a role in glutathione transport. Cells lacking Mtm1p exhibit elevated mitochondrial manganese levels, which can result in the mismetallation and degradation of Coq7p, the diiron hydroxylase responsible for the penultimate step of CoQ biosynthesis24,25. Additionally, mtm1Δ cells phenocopy disruptions in the biosynthesis of iron–sulfur clusters—cofactors necessary for electron-transfer reactions that enable Coq6p function7. Thus, it is possible that the observed decrease of CoQ in mtm1Δ is a result of Coq7p mismetallation and degradation, dysfunctional Coq6p, or both. These roles are supported by our CoQ and PPHB measurements and may underlie the observed genetic interactions with HEM25. Additional work is needed to confirm the molecular basis of these interactions.
Our results corroborate previous in vitro experiments showing that imported IPP is sufficient to generate CoQ’s polyprenyl tail (Fig. 3f)12,13. We did not assess whether other isoprenoid pyrophosphates, such as dimethylallyl pyrophosphate (DMAPP), geranyl pyrophosphate (GPP) or farnesyl pyrophosphate (FPP), can be imported. Coq1p, a mitochondrial prenyltransferase, has shown an ability to use GPP and FPP in isolated membrane preparations52, but whether these isoprenoids can be transported into mitochondria remains unknown. Moreover, it has been proposed that a common cytosolic FPP pool supplies precursors for CoQ and other isoprenoids, but this also remains unresolved53,54,55. Although our investigation shows that yeast mitochondria can bypass this cytosolic FPP pool by importing and incorporating IPP alone, we cannot exclude the contribution of other isoprenoid pyrophosphate species to CoQ biosynthesis.
Despite being the closest orthologue to Hem25p, SLC25A38 did not facilitate IPP transport, nor did it functionally rescue CoQ levels in hem25Δ (Fig. 6b,c). We also did not identify any reports of CoQ deficiency in patients with SLC25A38 mutations. At first glance, this seems surprising given that both carriers share many of the same key binding-site residues (Fig. 5a)43. Moreover, we found these conserved residues to be important for Hem25p stability or substrate binding (Fig. 5c,d and Extended Data Fig. 3b,c). However, our experimental and phylogenetic analyses show a clear divergence between fungal and metazoan orthologues, with IPP import limited to fungal orthologues (Fig. 6c-e). It is perhaps likely that the emergence of multicellular and multiorgan animals necessitated a higher level of regulation of haem and CoQ biosynthesis, such that a common carrier was no longer sufficient. Consistent with this model, human SLC25A38 demonstrates preferential expression in early erythroid cells, and both zebrafish orthologues are strongly associated with haematopoiesis and haematopoietic tissues31,44. Given that CoQ is produced in nearly every mammalian cell type, we would expect ubiquitous expression from a mammalian isoprenoid transporter. Additional differences in both the haem and CoQ biosynthetic pathways have been identified between yeasts and metazoans56,57,58, and more work is needed to dissect the physiological underpinnings of these differences.
The divergence in Hem25p and SLC25A38 functions implies the existence of a separate metazoan IPP transporter. Such an occurrence would not be unique to IPP, with NAD being the most recent example of a substrate having distantly related fungal and metazoan transporters59,60,61,62. Our identification, here, of the primary fungal IPP transporter lays the foundation for future efforts to identify the functional metazoan orthologue.
Methods
Yeast strains and cultures
The S. cerevisiae haploid strain W303 (MATa leu2 trp1 can1 ura3 ade2 his3) was used. Single (geneΔ) and double (geneΔgeneΔ) deletion strains were generated using standard homologous recombination63. Open reading frames were replaced with the kanMX6, His3MX6 or Trp1 cassettes and confirmed by polymerase chain reaction (PCR) assay64,65.
For the targeted screen, cells were grown in yeast extract–peptone–dextrose (YPD) medium consisting of 1% (wt/vol) yeast extract (‘Y’; Research Products International, RPI), 2% (wt/vol) peptone (‘P’; RPI) and 2% (wt/vol) dextrose (‘D’; Fisher). YP medium without dextrose was sterilized by an automatic autoclave. Glucose was sterile-filtered (0.22-μm pore size, VWR) and added to sterile YP before use. For all other cultures, synthetic complete (SC) or synthetic dropout (SD) medium was used, containing yeast nitrogen base (YNB) with ammonium sulfate and without amino acids (US Biological), the corresponding dropout mix (US Biological) and the indicated carbon source. pABA−medium contained YNB without amino acids and without pABA (Formedium), Complete Supplement Mixture (Formedium) and the indicated carbon source. All synthetic yeast media were sterile-filtered (0.22-μm pore size, VWR) before use. Where indicated, ALA or glycine was added to medium before sterile filtration.
For measurements in respiration33, starter cultures (3 ml, SC or SD, 2% dextrose (2%D)) were inoculated with individual colonies and incubated (30 °C, 230 r.p.m., 14–16 h). Cell density was measured as an optical density at 600 nm (OD600) and converted to cells ml−1 (1 OD = 1 × 107 cells ml−1). Respiratory medium (100 ml, SC or SD, 0.1%D, 3% glycerol (3%G)) in a sterile 250-ml Erlenmeyer flask was inoculated with 2.5 × 106 yeast cells. Samples were incubated (30 °C, 230 r.p.m.) for 25 h, a time point that corresponds to early respiratory growth.
Lipid extraction
Lipid extractions were performed essentially as described previously66. Cells were collected by centrifugation (4,000g, 5 min, room temperature (r.t.)). The supernatant was removed and the cells washed with water (600 μl). Cells were pelleted again (15,000g, 30 s, r.t.) and the supernatant removed. Cell pellets were either used immediately for lipid extraction or snap-frozen in liquid nitrogen (LN2) and stored at −80 °C until analysis. Frozen pellets were thawed on ice before extraction. For lipid extraction, 150 mM KCl (50 μl) was added to each sample, followed by ice-cold methanol (600 μl; with 1 μM CoQ8 as an internal standard). Glass beads (100 μl; 0.5 mm; BioSpec) were then added and the samples were vortexed (10 min, 4 °C) to lyse the cells. Ice-cold petroleum ether (400 μl; Sigma) was added to extract the lipids, and the samples were vortexed again (3 min, 4 °C). Samples were centrifuged (1,000g, 3 min, r.t.) and the top petroleum ether layer was collected in a new tube. The petroleum ether extraction was repeated a second time, with the petroleum ether layer from the second extraction combined with that from the first. The extracted lipids were dried under argon before being resuspended in 2-propanol (50 μl) and transferred to an amber glass vial (Sigma; QSertVial, 12 × 32 mm, 0.3 ml).
Targeted yeast genetic screen
Yeast from a −80 °C glycerol stock were struck onto YPD plates and incubated (30 °C, 48 h). Starter cultures of YPD (3 ml) were inoculated with individual colonies and grown overnight (30 °C, 14–16 h). Overnight cultures were diluted to OD600 = 0.2 (3 ml) and incubated until OD600 ≈ 1 (30 °C, 230 r.p.m., 4–5 h), corresponding to mid-log phase. Cultures were collected by centrifugation (4,000g, 5 min, r.t.), washed once with water, and transferred to a 1.5-ml microcentrifuge tube. Cells were centrifuged again (15,000g, 30 s, r.t.), snap-frozen in LN2, and stored at −80 °C until analysis. Lipids were extracted from cell pellets as described above and CoQ measurements were then performed using reverse-phase high-pressure liquid chromatography with electrochemical detection63 (HPLC-ECD), as described in the next section.
Targeted HPLC-ECD for CoQ
Extracted lipids were resuspended in 2-propanol (50 μl) and transferred to an amber vial. Sodium borohydride (2 μl of 10 mM in 2-propanol) was added to each vial, followed by brief vortexing and incubation (10 min, r.t.) to reduce the CoQ. Methanol (50 μl) was then added to each sample to remove excess sodium borohydride and the vials were flushed with argon gas. CoQ measurements were performed using reverse-phase HPLC-ECD66. Separation was performed using a C18 column (Thermo Scientific, Betasil C18, 100 × 2.1 mm, particle size 3 μm) at a flow rate of 0.3 ml min−1 with a mobile phase of 78% methanol, 10% 2-propanol, 10% acetonitrile and 2% ammonium acetate (1 M, pH 4.4). Electrochemical detection was performed using an ECD detector (Thermo Scientific ECD3000-RS) containing a 6020RS omni Coulometric Guarding Cell (E1) set to −200 mV and two 6011RS ultra Analytical Cells (E2 and E3) set to 600 mV. CoQ measurements were made on the analytical E2 electrode. For each experiment, CoQ6 and CoQ8 standards (Avanti) were prepared in the same manner as the experimental samples. Peak areas were quantified using Chromeleon 7.2.10 software (Thermo) and normalized to the CoQ8 internal standard. CoQ6 levels were further normalized to OD600 at collection.
Yeast growth assays
Starter cultures (SC, 2%D, 3 ml) were inoculated with individual colonies and incubated overnight (30 °C, 230 r.p.m., 14–16 h). Cells were pelleted and resuspended in respiratory medium (SC, 0.1%D, 3%G) containing the indicated additives at a density of 5 × 106 cells ml−1, then 100 μl of the resuspended cells were transferred to a sterile 96-well round-bottom plate (Thermo) with a Breathe-Easy cover seal (Diversified Biotech). Cultures were incubated (30 °C, 1,140 r.p.m.) in an Epoch2 plate reader (BioTek) with OD600 measured every 10 min.
Haem measurements
Yeast total haem measurements were performed using previously described methods with slight modifications67,68. Cells were grown under respiratory conditions as described previously and supplemented with glycine or ALA as indicated, then 1 × 108 cells were collected by centrifugation (4,000g, 5 min, r.t.), washed with water, and centrifuged again (15,000g, 30 s, r.t.). Pellets were snap-frozen in LN2 and stored at −80 °C. Frozen pellets were thawed on ice before being resuspended in 500 μl of oxalic acid (20 mM) and incubated in a closed box (16–24 h, 4 °C), then 500 μl of oxalic acid (2 M, preheated to 50 °C) was added to each sample, and each sample was divided in half into two 1.5-ml amber microcentrifuge tubes. One half of each sample was heated (95–100 °C, 30 min) while the other was incubated at r.t. Each tube was then centrifuged (16,000g, 2 min), and 200 μl of the supernatant was loaded into a 96-well black-bottom plate (Greiner). Haem fluorescence was measured on a Cytation 3 plate reader (BioTek) with excitation at 400 nm and emission at 620 nm. The fluorescence from the unheated sample was subtracted from the corresponding heated sample.
Quantitative PCR for COQ gene expression
Respiring yeast were cultured and harvested as described in the Yeast strains and cultures section for haem measurements. Total RNA was extracted using the MasterPure Yeast RNA Purification Kit (Lucigen). First-strand cDNA synthesis was performed using the EasyScript Plus cDNA Synthesis Kit (Lamda Biotech) using Oligo(dT) primers and 1 μg of RNA. Quantitative PCR (qPCR) was performed using the following reaction: 10 μl of Power SYBR Green PCR Master Mix (Thermo), 2.5 μl of cDNA (diluted 1:20) and 250 nM forward and reverse primers. Primers amplifying the yeast COQ2, COQ3, COQ4, COQ5, COQ6, COQ7, COQ8, COQ9 and ACT1 (reference) genes were used. For the qPCR cycle, an initial 2-min incubation at 50 °C was followed by 10-min denaturing at 95 °C, then 40 cycles of 95 °C for 15 s and 60 °C for 1 min were performed. qPCR data were collected using QuantStudio Real-Time PCR software v1.2 (Applied Biosciences). RNA abundance was calculated using the ΔΔCtmethod. The primers for qPCR are listed in Supplementary Table 1.
LC-MS/MS proteomics
Yeast growth
Yeast cultures were grown as described previously for respiration, then 1 × 108 cells were collected, snap-frozen in LN2, and stored at −80 °C.
Lysis and digestion
Yeast pellets were removed from −80 °C conditions and resuspended in lysis buffer (6 M guanidine hydrochloride, 100 mM Tris). The samples were then boiled at 100 °C for 5 min and sonicated in a bath sonicator (Qsonica) with a 5-min-long program of 20 s on, 10 s off. Methanol was added to each sample to 90% concentration, and the samples were centrifuged at 10,000g for 5 min to precipitate proteins. After precipitation, the supernatant was discarded from each sample, and the protein pellets were allowed to air-dry for 7 min. The dried pellets were resuspended in digestion solution (8 M urea, 10 mM TCEP, 40 mM CAA, 100 mM Tris), and the samples were sonicated in the bath sonicator with the same program as above to facilitate resolubilization. LysC (Wako Chemicals) was added to each resolubilized sample in an estimated 50:1 protein/enzyme ratio. The samples were incubated on a rocker at room temperature for 4 h before being diluted fourfold with 100 mM Tris. Trypsin (Promega) was added to each sample in an estimated 50:1 protein/enzyme ratio before they were incubated on a rocker at room temperature for 14 h. Each sample was finally acidified with TFA to a pH of 2, desalted by solid-phase extraction cartridges (Phenomenex) and dried under vacuum (Thermo Scientific).
LC-MS/MS proteomics data acquisition
Peptides were resuspended in 0.2% formic acid, and the concentration of each sample was determined using a NanoDrop One spectrophotometer (Thermo Scientific). The samples were then prepared in autosampler vials and loaded onto a 75-μm i.d. × 360-μm o.d. capillary column (New Objective) that was packed in-house69 with 1.7-μm BEH C18 particles and held at 50 °C throughout the analysis. Separations were performed with a Dionex UltiMate 3000 nano HPLC system (Thermo Scientific). The mobile phases were 0.2% formic acid in water (A) and 0.2% formic acid in 80% ACN (B). The peptides were analysed by an Orbitrap Eclipse system (Thermo Scientific) with the following parameters: Orbitrap MS1 resolution of 240,000; MS1 automatic gain control target of 1 × 106; MS1 maximum injection time of 50 ms; MS1 scan range of 300–1,500 m/z; dynamic exclusion of 20 ms; advanced peak determination70 toggled on; MS2 isolation window of 0.7 m/z; MS2 collision energy of 25%; ion trap MS2 resolution setting of turbo; MS2 automatic gain control target of 3 × 104; MS2 maximum injection time of 14 ms; MS2 scan range of 150–1,200 m/z.
Data analysis
MaxQuant71 (version 1.5.2.8) was used to process proteomics raw files against a database of reviewed yeast proteins plus isoforms from UniProt (downloaded 20 November 2019). Label-free quantification72 was used for relative quantification. Match-between-runs was enabled. Data were visualized and analysed using the Argonaut73 platform.
Targeted LC-MS/MS lipid measurements
Yeast growth
For measurements in respiration, yeast cells were grown and collected as described previously. Yeast pellets were snap-frozen in LN2 and stored at −80 °C.
For de novo PPHB and CoQ experiments, starter cultures (3 ml, SC, 2%D, 300 μM ALA) were inoculated with individual colonies and incubated (30 °C, 230 r.p.m., 18 h). OD600 was measured, and 2.5 × 107 cells were diluted into 50 ml of pABA− medium (2%D, 300 μM ALA) in a sterile 250-ml Erlenmeyer flask. Cells were incubated (30 °C, 230 r.p.m.) until OD600 ≈ 1.5–2 to deplete pABA in the cells, then 7.5 × 108 cells were centrifuged (3,000g, 5 min, r.t.) and resuspended in 50 ml of labelled pABA− medium (3%G, 300 μM ALA, 50 μM [phenyl-13C6]-4-HB). Immediately after the media swap, and at 2, 4 and 6 h after, 1 × 108 cells were collected by centrifugation (3,000g, 5 min, 4 °C) and washed with water. Cells were pelleted again (15,000g, 30 s, 4 °C), snap-frozen in LN2 and stored at −80 °C.
LC-MS/MS lipidomics data acquisition
Frozen pellets were thawed on ice and lipid extraction was performed as described previously. A Vanquish Horizon UHPLC system (Thermo Scientific) connected to an Exploris 240 Orbitrap mass spectrometer (Thermo Scientific) was used for targeted LC-MS analysis. A Waters Acquity CSH C18 column (100 mm × 2.1 mm, 1.7 μm) was held at 35 °C with the flow rate of 0.3 ml min−1 for lipid separation. A Vanquish binary pump system was employed to deliver mobile phase A, consisting of 5 mM ammonium acetate in ACN/H2O (70/30, vol/vol) containing 125 μl l−1 acetic acid, and mobile phase B consisting of 5 mM ammonium acetate in IPA/ACN (90/10, vol/vol) containing 125 μl l−1acetic acid. The gradient was set as follows: B was at 2% for 2 min and increased to 30% over the next 3 min, then further ramped up to 50% within 1 min and to 85% over the next 14 min, and then raised to 99% over 1 min and held for 4 min, before re-equilibration for 5 min at 2% B. Samples were ionized by a heated ESI source with a vaporizer temperature of 350 °C. The sheath gas was set to 50 units, auxiliary gas to 8 units and the sweep gas to 1 unit. The ion transfer tube temperature was kept at 325 °C with a 70% RF lens. The spray voltage was set to 3,500 V for positive mode and 2,500 V for negative mode. Targeted acquisition was performed with both tMS2 (targeted MS2) mode and tSIM (targeted selected ion monitoring) mode in the same injection: the tMS2 mode was for measuring CoQ6 (m/z 591.4408), [13]C6-CoQ6 (m/z 597.4609) and CoQ8 (m/z 727.5660) in positive polarity, and OPP (m/z 638.5432) and [13]C16-OPP (m/z 654.5968) in negative polarity at a resolution of 15,000, an isolation window of 2 m/z, normalized HCD collision energy of either 40% or stepped HCD energies of 30% and 50%, standard AGC target and auto maximum ion injection time; the tSIM mode was for measuring PPHB6 (m/z 545.4000), [13]C6-PPHB6 (m/z 551.4201) and [13]C12-PPHB6 (m/z 557.4403) in negative polarity at a resolution of 60,000, an isolation window of 2 m/z, standard AGC target and auto maximum ion injection time.
Data analysis
Targeted quantitative analysis of all acquired compounds was processed using TraceFinder 5.1 (Thermo Scientific) with a mass accuracy of 5 ppm. The result of peak integration was manually examined.
Mitochondrial isolation
Starter cultures (3 ml, SC or SD, 2%D) were inoculated with individual colonies and incubated (30 °C, 230 r.p.m., 14–16 h), then 2 × 108 cells were diluted into 1 l of SC 2%D in a 5-l Erlenmeyer flask and incubated (30 °C, 230 r.p.m.) for 14–18 h to a final OD ≈ 6. Mitochondria were isolated as previously described74. Cells were collected by centrifugation (3,000g, 5 min, r.t.), washed with water, and centrifuged again (3,000g, 5 min, r.t.). The wet pellet weight of the cells was determined. Cells were resuspended in 2 ml g−1 dithiothreitol buffer (100 mM Tris-H2SO4, 10 mM dithiothreitol, pH 9.4) and shaken slowly (30 °C, 80 r.p.m., 20 min). Cells were pelleted, washed once with 7 ml g−1 Zymolyase buffer (1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4), and resuspended in 7 ml g−1 Zymolyase buffer with 3 mg g−1 Zymolyase 20T (Fisher) to generate spheroplasts. The yeast was shaken slowly (30 °C, 80 r.p.m.) for 30 min before being pelleted and washed with 7 ml g−1 Zymolyase buffer. Pellets were resuspended in 6.5 ml g−1 ice-cold homogenization buffer (0.6 M sorbitol, 10 mM Tris-HCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.2% (wt/vol) fatty-acid-free bovine serum albumin (BSA), pH 7.4). Spheroplasts were homogenized using 15 strokes of a tight-fitting glass-Teflon homogenizer and diluted twofold with homogenization buffer. The homogenate was centrifuged (1,500g, 5 min, 4 °C) to pellet cell debris and nuclei. The supernatant was centrifuged (4,000g, 5 min, 4 °C) to pellet additional debris, and the supernatant centrifuged again (4,000g, 5 min, 4 °C). Mitochondria were isolated by centrifuging the supernatant (12,000g, 15 min, 4 °C) and resuspended in SEM (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2) or SEP (0.6 M sorbitol, 1 mM EGTA, 50 mM potassium phosphate, pH 7.4), where indicated. Mitochondrial protein content was quantified by a BCA protein assay (Thermo).
Mitochondrial PPHB synthesis
Isolated mitochondria were resuspended in SEP at a concentration of 8 mg ml−1, kept on ice, and used within 4 h of isolation without freezing. PPHB synthesis was carried out using a modified protocol12. To begin the assay, 250 μl of mitochondria (2 mg per assay) were added to 750 μl of 1.3X substrate buffer (0.6 M sorbitol, 50 mM potassium phosphate, 1 mM EGTA, 9.1 mM MgCl2, 1.3 μM 4-HB, 13.3 μM [1,2-13C2]-IPP (Cambridge Isotope Laboratories), pH 7.4). The final concentrations in the reaction were as follows: 0.6 M sorbitol, 50 mM potassium phosphate, 1 mM EGTA, 7 mM MgCl2, 1 μM 4-HB, 10 μM [1,2-13C2]-IPP and 2 mg mitochondria, pH 7.4. Reactions were carried out at 30 °C in a 5-ml tube (Eppendorf) with constant shaking (500 r.p.m.). At the indicated time points, 300 μl of the reaction was removed and immediately extracted for lipids, then 300 μl of ice-cold methanol containing 1 μM CoQ8 as an internal standard was added to each sample, followed by 400 μl of ice-cold petroleum ether. The samples were vortexed (10 min, 4 °C) and centrifuged (3,000g, 3 min, 4 °C). The top petroleum ether layer was collected, and the extraction repeated again with another 400 μl of petroleum ether. The petroleum ether layers were pooled for each sample, dried under argon, and stored at −80 °C. Lipids were resuspended in 2-propanol (50 μl) and transferred to amber glass vials for targeted LC-MS/MS. Following the third time point, 50 μl of the remaining reaction was diluted with 2X sodium dodecyl sulfate (SDS) sample buffer for immunoblot analysis.
Bacterial uptake assays
Bacterial expression of mitochondrial carriers and whole-cell uptake assays were performed as described previously with slight modifications39,40. MBP-tagged fusion proteins were generated by combining the E. coli MBP containing the MalE signal peptide, a short linker containing a thrombin cleavage site, and the mitochondrial carrier gene. Fusion constructs were synthesized as gBlock gene fragments (IDT) and cloned into the pET-21b expression vector using the restriction enzymes NdeI and XhoI. Inserts were confirmed by sequencing. The plasmid map of pET-21b containing MBP-HEM25 is shown in Supplementary Fig. 1. The complete plasmid sequence and the sequence of the MBP-Hem25p fusion protein are provided in Supplementary Table 2. Mutants were generated by site-directed mutagenesis (SDM) (NEB; Supplementary Table 3).
Expression of MBP-tagged carriers was carried out in E. coli C43(DE3) cells (Biosearch Technologies)75. Single aliquots of commercially prepared cells were transformed according to the manufacturer’s instructions, and selected on LB agar plates containing 100 mg l−1 ampicillin (37 °C, overnight). Single colonies of freshly transformed cells were used to individually inoculate 4 ml of LB medium containing 100 mg l−1 ampicillin. Cultures were incubated overnight (37 °C, 230 r.p.m.) before being diluted 1:100 in 50 ml of LB medium containing 100 mg l−1 ampicillin. Refreshed cultures were incubated (37 °C, 230 r.p.m.) until OD600 = 0.5–0.6, at which point the cultures were removed and cooled on ice for 5–10 min. Protein expression was induced by adding 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) to the chilled cultures. Cells were induced overnight (20 °C, 230 r.p.m., 14–16 h). Single colonies were used to inoculate the starter cultures, with the subsequent refresh, culture and induction carried out independently.
Induced cells were collected by centrifugation (4,000g, 10 min, 4 °C), washed once with ice-cold KPi (50 mM potassium phosphate, pH 7.4), and centrifuged again (4,000g, 10 min, 4 °C). Cells were resuspended in ice-cold KPi to a cell density of OD600 = 10 and placed on ice until the start of the assay. OD600 was measured for each preparation, and adjusted accordingly to ensure the same number of cells in each assay.
To start the assay, 200 μl of cells were added to 200 μl of 2X substrate buffer (2 mM MgCl2 and radiolabelled substrate at double the final concentration in KPi) and mixed briefly by pipetting. Assays were carried out at room temperature in a 1.5-ml microcentrifuge tube. [1-14C]-IPP and [phenyl-14C6]-4-HB were purchased from American Radiolabeled Chemicals as lyophilized solids. Radiolabelled compounds were resuspended in KPi to ~10 mM, and the exact concentration determined using scintillation counting and the supplier’s provided specific activity. At each time point, 100 μl of the assay mixture was removed from the reaction tube and filtered under vacuum to separate the cells from the incubation medium. The filtration apparatus consisted of a 25 mm filter placed atop a glass frit membrane support base (Millipore). The filter and membrane support were attached, using a stopper, to a side-arm flask connected to a house vacuum line. To reduce non-specific binding of the substrate to the filter, filtration was carried out using 0.22-μm mixed cellulose ester (MCE) filters (Millipore) for IPP uptake assays and 0.2-μm Whatman Nuclepore track-etched hydrophilic membrane filters (Cytiva) for 4-HB uptake assays42. Immediately following filtration, the filters were washed with 5 ml of ice-cold KPi. Once washed, the filters were removed from the support and placed in a 7-ml scintillation vial (Fisher), then 5 ml of Ultima Gold MV liquid scintillation cocktail (PerkinElmer) was added to each vial before analysis by liquid scintillation counting. Counts per minute were converted to disintegrations per minute (DPM) using the counting efficiency of the counter. IPP and 4-HB were quantified using a standard curve.
To determine the total bacterial protein content, protein from a known amount of bacteria were extracted using the B-PER Bacterial Protein Extraction Reagent (Thermo). The concentration of extracted proteins was determined using a BCA assay (Thermo), which was then used to calculate the amount of protein per data point.
Bacterial OPP synthesis
The culturing, expression and preparation of E. coli C43(DE3) cells were performed exactly as described in the Bacterial uptake assays section. Following preparation, cells were resuspended to a cell density of OD600 = 10 in ice-cold KPi and kept on ice until the start of the assay.
To start the assay, 400 μl of cells were added to 400 μl of 2X substrate buffer (2 mM MgCl2 and 100 μM [1,2-13C2]-IPP in KPi, pre-warmed to 37 °C for 5 min) in a 1.5-ml microcentrifuge tube. The assay mixture was rapidly mixed by pipetting before being placed in a heat block set to 37 °C. At 1, 5, 15 and 30 min following the start of the assay, 200 μl of the assay mixture was removed and added to 800 μl of ice-cold KPi in a separate tube. Cells were pelleted by centrifugation (10,000g, 1 min, 4 °C) and the supernatant was removed by aspiration. Cell pellets were snap-frozen in LN2, and stored at −80 °C until analysis.
Lipid extractions were performed as described above with the notable exception of 1 μM CoQ6 as the internal standard (CoQ8 was not used as it is present in bacteria). LC-MS/MS lipidomics was performed as described above. The MS was operated in positive and negative parallel reaction monitoring (PRM) mode, acquiring targeted scans to detect CoQ6, CoQ8, [13C16]-CoQ8, OPP and [13C16]-OPP.
CoQ rescue experiments
The yeast expression vector p416 GPD containing the constitutive glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter was modified to contain the endogenous HEM25 promoter. PCR was used to amplify a 744-bp segment directly upstream of the HEM25 gene from yeast genomic DNA (Supplementary Table 4). This segment, containing the endogenous promoter, was then cloned into the SacI-BamHI sites of a pre-digested p416 GPD vector, yielding a p416 vector containing the HEM25Endogenous Promoter (p416 HEP). The HEM25 gene was amplified from yeast genomic DNA and cloned into the BamHI-XhoI sites of the p416 GPD or the p416 HEP vectors. The PCR primers were designed so that a short linker and a FLAG tag were added to the end of the open reading frame (Supplementary Table 4). HEM25-FLAG mutants were synthesized as gBlocks (IDT) and cloned into the BamHI-XhoI sites of the p416 GPD or p416 HEP vectors. We were not able to PCR-amplify p416, so SDM could not be performed. The sequences of the cloned HEM25 promoter and the inserts were confirmed by sequencing. Verified constructs were transformed into hem25Δ yeast using the LiAc/SS carrier DNA/PEG method76. For controls, empty vectors were also transformed into WT W303, coq6Δ and hem25Δ yeast. Respiratory growth and targeted lipidomics were performed as described above. For immunoblotting, 1 × 108 cells were collected for whole-cell lysates. To check mitochondrial localization, mitochondria were isolated as described above, with the exception that the cultures were collected during respiratory growth. Cultures for mitochondrial isolation were scaled up to 1 l to produce enough mitochondria for immunoblotting. Cell numbers and culture volumes were scaled appropriately to keep the conditions consistent.
HEM25 orthologues were synthesized as codon-optimized gBlocks (IDT) and cloned into the BamHI-XhoI sites of the p416 ADH vector. Constructs were sequenced and transformed into hem25Δ yeast. Fermentative growth and targeted lipidomics were performed as described above.
Sequence alignments and bioinformatics analysis
Hem25p orthologues were curated from the PANTHER database (release 17.0)47. Sequences were aligned using Clustal Omega in Jalview 2.10.377,78. The phylogenetic tree was generated using Jalview 2.10.3 and FigTree v1.4.4. The structure of Hem25p was predicted using AlphaFold (accessed 1 September 2022) and visualized using PyMol 2.079,80.
Immunoblotting
Antibodies
Primary antibodies for this study include anti-FLAG (Sigma F1804, 1:2,500–1:5,000), anti-FLAG (Cell Signaling Technology #14793, 1:1,000), anti-Act1 (Abcam ab8224, 1:5,000), anti-Coq1 (custom made at Genscript, 1:2,000), anti-Vdac1 (Abcam ab110326, 1:4,000–1:5,000) and anti-MBP (NEB E8032S, 1:10,000). Secondary antibodies include IRDye 680RD goat anti-rabbit (LI-COR 926-68071, 1:15,000), IRDye 680RD goat anti-mouse (LI-COR 926-68070, 1:15,000), IRDye 800CW (LI-COR 926-32210, 1:15,000), anti-mouse immunoglobulin-G (IgG) horseradish peroxidase (HRP)-linked (Cell Signaling Technology #7076, 1:2,000-1:100,000) and anti-rabbit IgG HRP-linked (Cell Signaling Technology #7074, 1:20,000–1:100,000).
CoQ rescue experiments
Protein lysates from whole yeast cells were prepared81 by adding 150 μl of lysis buffer (2 M NaOH, 1 M β-mercaptoethanol) to cell pellets and incubating them for 10 min on ice with periodic vortexing, then 150 μl of 50% trichloroacetic acid (TCA) was added, and the samples were incubated on ice for 10 min with periodic vortexing to precipitate proteins. The samples were centrifuged (14,000g, 2 min, r.t.) and the supernatant discarded. The remaining pellet was washed with 1 ml of acetone before being pelleted again (14,000g, 2 min, r.t.). The pellet was left to air-dry before being resuspended in 120 μl of 0.1 M NaOH. Protein concentrations were determined by BCA protein assay (Thermo) and diluted twofold with 2X SDS sample buffer.
Protein (30 μg) was loaded onto NuPAGE 4–12% Bis-Tris gels (Thermo) and separated (200 V, 35 min). Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Sigma) and blocked with 5% non-fat dry milk (NFDM) in tris-buffered saline with 0.1% Tween 20 (TBST) (1 h, r.t.). The membranes were then probed with primary anti-FLAG (Sigma F1804, 1:2,500) antibodies diluted in 5% NFDM in TBST (overnight, 4 °C). Membranes were washed three times with TBST and then probed with secondary antibodies diluted in 5% NFDM in TBST (1 h, r.t.). Membranes were washed three times with TBST and developed by enhanced chemiluminescence (ECL). For the blotting of cells harbouring constructs containing the endogenous HEM25 promoter, the SuperSignal West Atto substrate (Thermo, A38554) was used. Blotting for cells harbouring constructs containing the constitutive GPD promoter used the SuperSignal West Dura substrate (Thermo, 34075). Developed membranes were imaged on a ChemiDoc system running Image Lab Touch Software 3.0.1.14 (Bio-Rad) before being stripped (Thermo, 46430). Stripped membranes were blocked in 5% NFDM in TBST (1 h, r.t.) before being probed with anti-Act1 (Abcam ab8224, 1:5,000) loading control antibody (1 h, r.t.). Membranes were washed three times with TBST before being probed with the secondary antibody diluted in 5% NFDM (1 h, r.t.). Membranes were washed three times before being developed using the SuperSignal West Dura substrate. Membranes then were imaged using the ChemiDoc (Bio-Rad).
For immunoblots of isolated mitochondria, mitochondria were prepared as described above. Mitochondrial protein content was quantified by BCA assay (Thermo) before being diluted twofold with 2X SDS sample buffer. For conditions that used the constitutive GPD promoter, 15 μg of the respective mitochondrial protein sample was loaded onto NuPAGE 4–12% Bis-Tris gels (Thermo). This amount was increased to 20 μg for samples using the endogenous HEM25. Proteins were separated (200 V, 35 min), transferred to a PVDF membrane (Sigma), and blocked with 5% NFDM (1 h, r.t.). Membranes were then probed with primary anti-FLAG (Cell Signaling Technology #14793, 1:1,000) antibodies diluted in 5% (NFDM) in TBST (1 h, r.t.). Membranes were washed three times with TBST and then probed with secondary antibodies diluted in 5% NFDM in TBST (1 h, r.t.). Membranes were washed six times with TBST and developed using ECL. Like the whole-cell immunoblots above, samples using the endogenous HEM25 promoter were developed with the SuperSignal West Atto substrate (Thermo, A38554). Those using the constitutive GPD promoter used the SuperSignal West Dura substrate (Thermo, 34075). Developed membranes were imaged on a ChemiDoc system (Bio-Rad) before being stripped (Thermo, 46430). Stripped membranes were blocked in 5% NFDM in TBST (1 h, r.t.) before being probed with anti-Vdac1 (Abcam ab110326, 1:5,000) mitochondrial loading control antibody (overnight, 4 °C). Membranes were washed three times with TBST before being probed with secondary antibodies (1 h, r.t.). Membranes were washed three times before being developed with SuperSignal West Dura substrate and imaged on the ChemiDoc system (Bio-Rad).
Mitochondrial PPHB synthesis
Samples were collected and prepared as described in the Mitochondrial PPHB synthesis section. Protein (15 μg) was loaded onto a NuPAGE 4–12% Bis-Tris gel (Thermo) and separated (200 V, 35 min). Proteins were transferred to a PVDF membrane (Sigma), cut between the expected sizes of Coq1p and VDAC, and blocked with 4% NFDM in TBST (1 h, r.t.). The membranes were then probed with primary antibodies diluted in 4% NFDM (overnight, 4 °C). The membranes were washed three times with TBST before being probed with secondary antibodies (1 h, r.t.). Membranes were washed three times with TBST before being imaged on a LI-COR Odyssey CLx Imaging System and analysed with LI-COR Image Studio software (version 5.2.5).
E. coli membranes
Approximately 40 ml*OD units (e.g. 4 mL of a culture with OD600 = 10) of induced cells were collected (4,000g, 15 min, 4 °C), the supernatant was removed, snap-frozen in LN2, and stored at −80 °C until blotting. Frozen pellets were thawed on ice and resuspended in 2 ml of ice-cold lysis buffer (10 mM Tris pH 7 containing protease inhibitors). Cells were sonicated until clear (Branson Ultrasonics, 45% amplitude, 5-s pulse, 50% duty cycle, microtip). The lysed cells were diluted with ice-cold lysis buffer to a final volume of 9 ml. Unlysed cells and insoluble material were pelleted by centrifugation (27,000g, 15 min, 4 °C). The supernatant was transferred to a 14-ml tube (Beckman 344060) and ultracentrifuged to pellet membranes (150,000g, 1 h, 4 °C). The supernatant was removed, and the membrane pellet was resuspended in 75 μl of ice-cold lysis buffer. Protein concentrations were determined by BCA protein assay (Thermo) and the sample diluted twofold in 2X SDS sample buffer.
Protein (5 μg) was loaded onto a NuPAGE 4–12% Bis-Tris gel (Thermo) and separated (150 V, 1 h). Proteins were transferred to a PVDF membrane (Sigma) and blocked with 5% NFDM in TBST. The membrane was then probed with the primary anti-MBP antibody (NEB E8032S, 1:10,000) diluted in 5% NFDM (overnight, 4 °C). The membrane was washed three times with TBST and probed with secondary antibodies (1 h, r.t.) diluted in 5% NFDM in TBST. The membrane was washed three times with TBST before being imaged on a LI-COR Odyssey CLx Imaging System.
To determine the amount of MBP-Hem25p in each sample, isolated bacterial membranes were run alongside a standard curve of recombinant, purified MBP (Abcam). Proteins were separated on a gel, transferred to a PVDF membrane, blocked and incubated with primary anti-MBP antibody as described above. The membrane was washed three times with TBST and probed with secondary anti-mouse IgG HRP-linked antibody (Cell Signaling Technology #7076, 1:20,000, 1 h, r.t.) diluted in 5% NFDM in TBST. The membrane was washed three times with TBST before being developed using the SuperSignal West Dura substrate. The membrane was subsequently imaged on a ChemiDoc system (Bio-Rad). Densitometry was performed using Image Lab software 6.1 (Bio-Rad). The determined values were then used to calculate the amount of MBP-Hem25p in each data point.
Statistics and reproducibility
All experiments were performed in at least biological triplicate, unless otherwise stated. No statistical method was used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. All results are presented as the arithmetic mean ± s.d. No data were excluded from the analyses. Statistical analysis were performed in Prism 9.0 or 9.3.1 (GraphPad Software) or in Microsoft Excel. P values were calculated using unpaired, two-sided Student’s t-test with values less than 0.05 considered significant. For fold changes, the average of the control or WT replicates was set to 1, with the other samples normalized accordingly.
Extended Data
Extended Data Fig. 1. Hem25p drives CoQ production independently of its role in heme biosynthesis.
a, Relative protein abundances of hem25Δ cells compared to WT cells versus statistical significance. CoQ biosynthetic proteins and Complex Q proteins are highlighted. Raw data from the Y3K dataset33(respiration-RDR condition) are displayed as the mean from three biologically independent samples with two-sided Student’s t-test used. b, Relative transcript abundances of CoQ biosynthetic genes from hem25Δ with ALA supplementation compared to WT cells without ALA supplementation (mean ± SD, n = 3 biologically independent samples). c, Relative CoQ6 abundances in WT and hem25Δ yeast. Cells were treated with either vehicle (ethanol) or 10 μM CoQ6. CoQ6 supplementation raised the cellular CoQ levels of WT and hem25Δ yeast to similar levels (p = 0.3642 WT+CoQ6 vs hem25Δ+CoQ6, mean ± SD, n = 3 biologically independent samples). d, Relative protein abundances of WT cells with CoQ6, hem25Δ cells without CoQ6, and hem25Δ cells with CoQ6 compared to WT cells without CoQ6(mean, n = 3 biologically independent samples, two-sided Student’s t-test). e, Relative abundance of unlabeled PPHB and CoQ in WT, hem25Δ, coq6Δ, and coq6Δhem25Δ yeast from the experiment in Fig. 2d (mean ± SD, n = 3 biologically independent samples); ND, not detected. f, Normalized abundance of unlabeled PPHB levels in isolated coq6Δ and coq6Δhem25Δ mitochondria (mean ± SD, n = 3 biologically independent samples). g, Coq1p levels in isolated mitochondria from each replicate, assessed by immunoblotting. Source numerical data and unprocessed blots are available in source data.
Extended Data Fig. 2. 4-HB import by pcaK and unadjusted IPP uptake kinetics.
a, Immunoblot for MBP-Hem25p, MBP-SLC25A38, and MBP-Yhm2p in E. coli membranes preparations. b, Steady-state kinetics of [1-14C]-IPP uptake in cells expressing MBP-Hem25p or the empty vector. Initial rates (nmol/min) values represent the mean ± SD of three biologically independent samples. c, Time course of 50 μM [phenyl-14C6]-4-HB uptake by E. coli cells expressing the Pseudomonas putida PcaK or the empty vector (*p = 0.0002, **p = 0.0042, ***p = 0.0006, ****p = 0.0011 empty vector vs pcaK, mean ± SD, n = 3 biologically independent samples, two-sided Student’s t-test). Source numerical data and unprocessed blots are available in source data.
Extended Data Fig. 3. Hem25p function is required for CoQ biosynthesis.
a, Relative PPHB abundances in hem25Δ yeast carrying WT and mutant HEM25FLAG constructs under the control of the endogenous HEM25 promoter. Levels are relative to WT yeast carrying the empty expression vector (*p = 0.0011, **p = 0.0006, ***p = 0.0116, ****p = 0.0009 HEM25FLAG vs mutants or empty vector, mean ± SD, n = 3 biologically independent samples, two-sided Student’s t-test). b, c, Immunoblots of whole cell lysates (b) or isolated mitochondria (c) from WT, coq6Δ, or hem25Δ cells expressing various EV and Hem25p-FLAG constructs under the control of the endogenous HEM25promoter. d, e, Relative CoQ (d) and PPHB (e) abundances in hem25Δ yeast carrying WT and mutant HEM25FLAG constructs under the control of the constitutive GPD promoter. Levels are relative to WT yeast carrying the empty expression vector (*p = 0.0006, **p = 0.0005, ***p = 0.0009, ****p = 0.0019, #p = 0.0012, ##p = 0.0014, ###p = 0.0012, ####p = 0.0012 HEM25FLAG vs mutants or empty vector, mean ± SD, n = 3 biologically independent samples, two-sided Student’s t-test). f, g, Immunoblots of whole cell lysates (f) or isolated mitochondria (g) from WT, coq6Δ, or hem25Δ cells expressing various EV and Hem25p-FLAG constructs under the control of the constitutive GPD promoter. h, Time course of 50 μM [1-14C]-IPP uptake by E. coli cells expressing WT MBP-Hem25p, mutant MBP-Hem25p, or the empty vector (*p = 0.0494, **p = 0.0023, ***p = 0.0136, ****p = 0.0063 empty vector vs WT, mean ± SD, n = 3 biologically independent samples, two-sided Student’s t-test). i, Immunoblot for WT and mutant MBP-Hem25p in E. coli membranes preparations. j, k Normalized abundance of unlabeled OPP (j) and CoQ8 (k) in E. coli cells expressing the EV, WT MBP-Hem25p, or the R181P MBP-Hem25p mutant (mean ± SD, n = 3 biologically independent samples). Source numerical data and unprocessed blots are available in source data.
Extended Data Fig. 4. SLC25A38 does not contribute to CoQ biosynthesis.
a, Normalized PPHB abundance in hem25Δ cells expressing Hem25p-FLAG or SLC25A38. (mean ± SD, n = 3 biologically independent samples) b, Relative lipid abundances in hem25Δ yeast compared to WT versus statistical significance with CoQ10 and the CoQ10 biosynthetic intermediate demethoxy-coenzyme Q (DMQ10,) highlighted. c, Relative protein abundance SLC25A38KO cells compared to WT cells versus statistical significance with CoQ-related (COQ3-COQ9) and OxPhos-related proteins highlighted. For panels (b) and (c), raw lipidomic and proteomic data, respectively, from the MITOMICS resource46 are displayed as the mean from three independent samples with two-sided Welch’s t-test used. Source numerical data are available in source data.
Supplementary Material
Acknowledgements
We thank members of the Pagliarini laboratory and K. Henzler-Wildman for helpful discussions and insights throughout this project. This work was supported by NIH R35 GM131795 (D.J.P.), P41 GM108538 (J.J.C. and D.J.P.), F31 AG064891 (J.T.), T32 GM140935 (J.T.), T32 DK007120 (S.W.R.) and the BJC Investigator Program (D.J.P.).
Footnotes
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this Article.
Data availability
Proteomics raw files have been deposited in the MassIVE repository under accession nos. MSV000089127 and MSV000092359. The lipidomics datasets, including raw files and quantified peak areas, have been deposited in the MassIVE repository and can be accessed with accession no. MSV000092744. Data supporting the findings of this study and materials used in this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
- 1.Lester RL & Crane FL The natural occurrence of coenzyme Q and related compounds. J. Biol. Chem 234, 2169–2175 (1959). [PubMed] [Google Scholar]
- 2.Jones ME Pyrimidine nucleotide biosynthesis in animals: genes, enzymes and regulation of UMP biosynthesis. Annu. Rev. Biochem 49, 253–279 (1980). [DOI] [PubMed] [Google Scholar]
- 3.Frerman FE Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase. Biochem. Soc. Trans 16, 416–418 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Guerra RM & Pagliarini DJ Coenzyme Q biochemistry and biosynthesis. Trends Biochem. Sci 48, 463–476 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emmanuele V. et al. Heterogeneity of coenzyme Q10 deficiency: patient study and literature review. Arch. Neurol 69, 978–983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y & Hekimi S The efficacy of coenzyme Q10 treatment in alleviating the symptoms of primary coenzyme Q10 deficiency: a systematic review. J. Cell. Mol. Med 26, 4635–4644 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefely JA & Pagliarini DJ Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem. Sci 42, 824–843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awad AM et al. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 62, 361–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y & Hekimi S Understanding ubiquinone. Trends Cell Biol. 26, 367–378 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Gold PH & Olson RE Studies on coenzyme Q. The biosynthesis of coenzyme Q9 in rat tissue slices. J. Biol. Chem 241, 3507–3516 (1966). [PubMed] [Google Scholar]
- 11.Payet LA et al. Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem. Biol 23, 1241–1250 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Casey J & Threlfall DR Synthesis of 5-demethoxyubiquinone-6 and ubiquinone-6 from 3-hexaprenyl-4-hydroxybenzoate in yeast mitochondria. FEBS Lett. 85, 249–253 (1978). [DOI] [PubMed] [Google Scholar]
- 13.Momose K & Rudney H 3-Polyprenyl-4-hydroxybenzoate synthesis in the inner membrane of mitochondria from p-hydroxybenzoate and isopentenylpyrophosphate. A demonstration of isoprenoid synthesis in rat liver mitochondria. J. Biol. Chem 247, 3930–3940 (1972). [PubMed] [Google Scholar]
- 14.Tzagoloff A & Dieckmann CL PET genes of Saccharomyces cerevisiae. Microbiol. Rev 54, 211–225 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson KP et al. Defining intermediates and redundancies in coenzyme Q precursor biosynthesis. J. Biol. Chem 296, 100643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor EB Functional properties of the mitochondrial carrier system. Trends Cell Biol. 27, 633–644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmieri F & Monne M Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim. Biophys. Acta 1863, 2362–2378 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Vogtle FN et al. Landscape of submitochondrial protein distribution. Nat. Commun 8, 290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgenstern M. et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19, 2836–2852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozeir M. et al. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol 18, 1134–1142 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Marobbio CMT, Agrimi G, Lasorsa FM & Palmieri F Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22, 5975–5982 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barkovich RJ et al. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem 272, 9182–9188 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Jonassen T & Clarke CF Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J. Biol. Chem 275, 12381–12387 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Wang Y. et al. SLC25A39 is necessary for mitochondrial glutathione import in mammalian cells. Nature 599, 136–140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi X. et al. Combinatorial GxGxE CRISPR screen identifies SLC25A39 in mitochondrial glutathione transport linking iron homeostasis to OXPHOS. Nat. Commun 13, 2483 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson R. et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 10, 119–130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luk E, Carroll M, Baker M & Culotta VC Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl Acad. Sci. USA 100, 10353–10357 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diessl J. et al. Manganese-driven CoQ deficiency. Nat. Commun 13, 6061 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naranuntarat A, Jensen LT, Pazicni S, Penner-Hahn JE & Culotta VC The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem 284, 22633–22640 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lunetti P. et al. Characterization of human and yeast mitochondrial glycine carriers with implications for heme biosynthesis and anemia. J. Biol. Chem 291, 19746–19759 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Murray JP et al. Glycine and folate ameliorate models of congenital sideroblastic anemia. PLoS Genet. 12, e1005783 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson A. et al. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae*. J. Biol. Chem 280, 31397–31404 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Stefely JA et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol 34, 1191–1197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee AY et al. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science 344, 208–211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desbats MA et al. The COQ2 genotype predicts the severity of coenzyme Q10 deficiency. Hum. Mol. Genet 25, 4256–4265 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Allan CM et al. Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J. Biol. Chem 290, 7517–7534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh EJ et al. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys 463, 19–26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dufay JN, Fernández-Murray JP & McMaster CR SLC25 family member genetic interactions identify a role for HEM25 in yeast electron transport chain stability. G3 Genes Genomes Genet. 7, 1861–1873 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravaud S. et al. Impaired transport of nucleotides in a mitochondrial carrier explains severe human genetic diseases. ACS Chem. Biol 7, 1164–1169 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Mifsud J. et al. The substrate specificity of the human ADP/ATP carrier AAC1. Mol. Membr. Biol 30, 160–168 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Castegna A. et al. Identification and functional characterization of a novel mitochondrial carrier for citrate and oxoglutarate in Saccharomyces cerevisiae. J. Biol. Chem 285, 17359–17370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nichols NN & Harwood CS PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol 179, 5056–5061 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson AJ, Overy C & Kunji ER The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl Acad. Sci. USA 105, 17766–17771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guernsey DL et al. Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat. Genet 41, 651–653 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Heeney MM et al. SLC25A38 congenital sideroblastic anemia: phenotypes and genotypes of 31 individuals from 24 families, including 11 novel mutations, and a review of the literature. Hum. Mutat 42, 1367–1383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rensvold JW et al. Defining mitochondrial protein functions through deep multiomic profiling. Nature 606, 382–388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas PD et al. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian K. et al. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. J. Cell Biol 218, 1353–1369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eisenberg-Bord M. et al. The endoplasmic reticulum-mitochondria encounter structure complex coordinates coenzyme Q biosynthesis. Contact 2, 2515256418825409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vest KE, Leary SC, Winge DR & Cobine PA Copper import into the mitochondrial matrix in Saccharomyces cerevisiae is mediated by Pic2, a mitochondrial carrier family protein. J. Biol. Chem 288, 23884–23892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boulet A. et al. The mammalian phosphate carrier SLC25A3 is a mitochondrial copper transporter required for cytochrome c oxidase biogenesis. J. Biol. Chem 293, 1887–1896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casey J & Threlfall DR Formation of 3-hexaprenyl- 4-hydroxybenzoate by matrix-free mitochondrial membrane-rich preparations of yeast. Biochim. Biophys. Acta 530, 487–502 (1978). [DOI] [PubMed] [Google Scholar]
- 53.Runquist M, Ericsson J, Thelin A, Chojnacki T & Dallner G Isoprenoid biosynthesis in rat liver mitochondria. Studies on farnesyl pyrophosphate synthase and trans-prenyltransferase. J. Biol. Chem 269, 5804–5809 (1994). [PubMed] [Google Scholar]
- 54.Grünler J, Ericsson J & Dallner G Branch-point reactions in the biosynthesis of cholesterol, dolichol, ubiquinone and prenylated proteins. Biochim. Biophys. Acta 1212, 259–277 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Keller RK Squalene synthase inhibition alters metabolism of nonsterols in rat liver. Biochim. Biophys. Acta 1303, 169–179 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Kardon JR et al. Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell 161, 858–867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rondelli CM et al. The ubiquitous mitochondrial protein unfoldase CLPX regulates erythroid heme synthesis by control of iron utilization and heme synthesis enzyme activation and turnover. J. Biol. Chem 297, 100972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stefely JA et al. Cerebellar ataxia and coenzyme Q deficiency through loss of unorthodox kinase activity. Mol. Cell 63, 608–620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziegler M. et al. Welcome to the family: identification of the NAD+ transporter of animal mitochondria as member of the solute carrier family SLC25. Biomolecules 11, 880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luongo TS et al. SLC25A51 is a mammalian mitochondrial NAD+ transporter. Nature 588, 174–179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kory N. et al. MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci. Adv 6, eabe5310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Girardi E. et al. Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import. Nat. Commun 11, 6145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 63.Baudin A,Ozier-Kalogeropoulos O,Denouel A,Lacroute F& Cullin C A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329–3330 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Longtine MS et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Wach A, Brachat A, Alberti-Segui C, Rebischung C & Philippsen P Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065–1075 (1997). [DOI] [PubMed] [Google Scholar]
- 66.Ismail A. et al. Coenzyme Q biosynthesis: evidence for a substrate access channel in the FAD-dependent monooxygenase Coq6. PLoS Comput. Biol 12, e1004690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sassa S. Sequential induction of heme pathway enzymes during erythroid differentiation of mouse Friend leukemia virus-infected cells. J. Exp. Med 143, 305–315 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sinclair PR, Gorman N & Jacobs JM Measurement of heme concentration. Curr. Protoc. Toxicol 00, 8.3.1–8.3.7 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Shishkova E, Hebert AS, Westphall MS & Coon JJ Ultra-high pressure (>30,000 psi) packing of capillary columns enhancing depth of shotgun proteomic analyses. Anal. Chem 90, 11503–11508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hebert AS et al. Improved precursor characterization for data-dependent mass spectrometry. Anal. Chem 90, 2333–2340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tyanova S, Temu T & Cox J The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Cox J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brademan DR et al. Argonaut: a web platform for collaborative multi-omic data visualization and exploration. Patterns 1, 100122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meisinger C, Pfanner N & Truscott KN Isolation of yeast mitochondria. Methods Mol. Biol 313, 33–39 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Miroux B & Walker JE Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol 260, 289–298 (1996). [DOI] [PubMed] [Google Scholar]
- 76.Gietz RD & Schiestl RH High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc 2, 31–34 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Waterhouse AM, Procter JB, Martin DMA, Clamp M & Barton GJ Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sievers F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539–539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varadi M. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Veling MT et al. Multi-omic mitoprotease profiling defines a role for Oct1p in coenzyme Q production. Mol. Cell 68, 970–977 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics raw files have been deposited in the MassIVE repository under accession nos. MSV000089127 and MSV000092359. The lipidomics datasets, including raw files and quantified peak areas, have been deposited in the MassIVE repository and can be accessed with accession no. MSV000092744. Data supporting the findings of this study and materials used in this study are available from the corresponding author on reasonable request. Source data are provided with this paper.