Abstract
In previous studies, we have identified three promoters (P1, P2, and P3) in the regulatory region of the Escherichia coli aroP gene (P. Wang, J. Yang, and A. J. Pittard, J. Bacteriol. 179:4206–4212, 1997). Both P1 and P2 can direct mRNA synthesis for aroP expression, whereas P3 is a divergent promoter which overlaps with P1. The repression of transcription from the major promoter, P1, has been postulated to involve the activation of the divergent promoter, P3, by the TyrR protein (P. Wang, J. Yang, B. Lawley, and A. J. Pittard, J. Bacteriol. 179:4213–4218, 1997). In the present study, we confirmed the proposed mechanism of P3-mediated repression of P1 transcription by studying the binding of RNA polymerase to the promoters P1 and P3 in vitro in the presence and absence of TyrR protein and its cofactors. Our results show that (i) only one RNA polymerase molecule can bind to the DNA fragment carrying the aroP regulatory region, (ii) RNA polymerase has a higher affinity for P1 than for either P2 or P3 and binds to P1 in the absence of TyrR protein, (iii) in the presence of TyrR protein and its cofactor, phenylalanine or tyrosine, RNA polymerase preferentially binds to P3, and (iv) RNA polymerase does not respond to the activation-defective mutant TyrR protein TyrR-RQ10 and remains bound to P1 in the presence of TyrR-RQ10 and either of the cofactors.
The transcription of the aroP gene, which codes for a membrane protein responsible for the active transport of the three aromatic amino acids into cells, is repressed by the TyrR protein in the presence of any one of the three aromatic amino acids (12, 18). Previous genetic studies have identified the binding site for the TyrR protein, which comprises a strong and a weak TyrR box and is located downstream of the major promoter, P1, of the aroP gene (2). Recently, we have reported the identification of three promoters (P1, P2, and P3) in the upstream region of the aroP gene. Both P1 and P2 promoters can generate aroP mRNA, whereas the P3 promoter is a divergent promoter that overlaps P1 (Fig. 1) (16). We have shown that P2 is a minor promoter which contributes less than 20 percent of aroP expression in tyrR strains and that the expression of P2 transcription is almost totally repressed by TyrR and any of the cofactors.
FIG. 1.
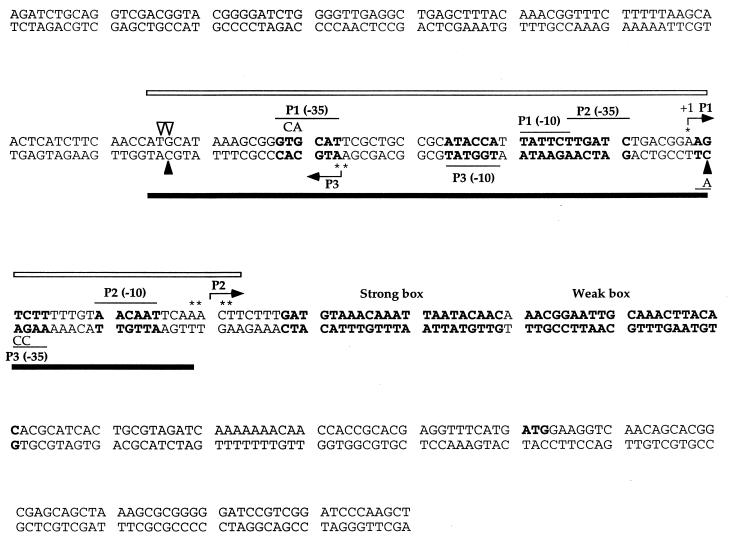
Nucleotide sequence of 320-bp DNA fragment containing the aroP regulatory region. The strong and weak TyrR boxes are shown in boldface. The −35 and −10 regions of the P1 and P2 promoters are overlined, and the −35 and −10 regions of the P3 promoter are underlined. The transcription start point for P1 is indicated with an asterisk and +1, and the transcription start points for P2 and P3 are marked with asterisks. The mutations in the −35 region of P1 (from GTGCAT to GCACAT) or P3 (from AAGACT to AACCAT) are also shown. The hypersensitive sites observed in DNase I footprinting in gel slice experiments using DNA fragments labelled in the top strand are indicated with triangles. The open triangles indicate the hypersensitive sites caused by the binding of RNA polymerase to the P1 promoter, and the filled triangles indicate the hypersensitive sites caused by the binding of RNA polymerase to the P3 promoter. The open bar shown above the DNA sequence represents the region protected by RNA polymerase when bound at the P1 promoter, and the filled bar shown below the DNA sequence represents the region protected by RNA polymerase when bound at the P3 promoter.
Our studies have also shown that the transcription of the major promoter, P1, is repressed as a result of activation of the P3 promoter by TyrR protein (15). Although, in the presence of 6 mM CaCl2, it is possible to demonstrate TyrR-mediated activation of transcription from P3 in vitro, no significant transcription from P3 can be demonstrated in vivo in the presence or absence of TyrR (16). Based on these results, it has been postulated that, in the presence of any of the cofactors, the TyrR protein binds and forms a transcriptionally nonproductive complex with RNA polymerase at the P3 promoter, thus inhibiting initiation of transcription from the P1 promoter (15). Because of the relative positions of each of the three promoters, it has also been suggested that the binding of one RNA polymerase molecule to one promoter excludes the binding of additional RNA polymerase molecules to other promoters in the aroP regulatory region (16).
To study the molecular mechanism of P3-mediated repression of P1 transcription, we have carried out experiments involving both a gel shift assay and DNase I footprinting in gel slices. Our results show that the TyrR protein inhibits the binding of RNA polymerase to the P1 promoter by recruiting RNA polymerase to the P3 promoter and offer confirmation for the previous model.
Analysis of protein-DNA complex formation by gel shift assay.
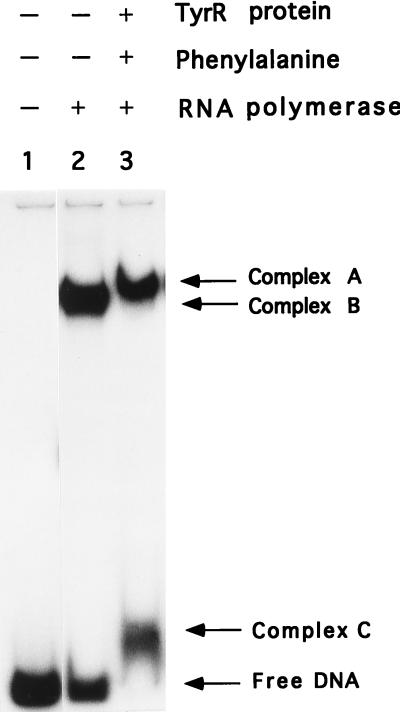
To test if the binding of an RNA polymerase molecule at one promoter prevents the access of a second RNA polymerase molecule to any one of the other promoters, we carried out a gel shift experiment. The 0.3-kb fragment containing the wild-type aroP regulatory region (P1+ P3+) (Fig. 1) was labelled with 32P by filling in the restriction end using [α-32P]dGTP, dATP, dCTP, dTTP, and Klenow enzyme. This aroP fragment was then incubated with RNA polymerase (400 nM) in the presence or absence of purified TyrR protein (200 nM) and phenylalanine (1 mM). The reaction mixtures were then analyzed by electrophoresis on a 5% native polyacrylamide gel.
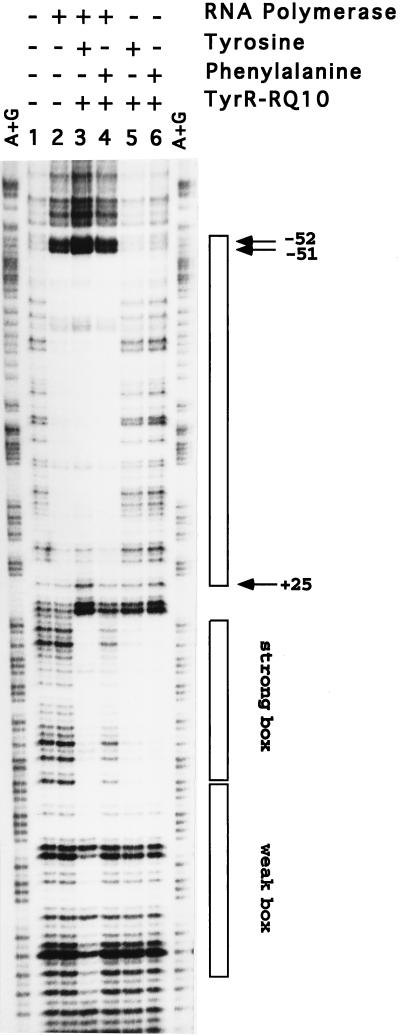
The results of these experiments are shown in Fig. 2. When only RNA polymerase was used, i.e., in the absence of TyrR, a single RNA polymerase-DNA complex (complex B) was observed. We have repeated this experiment using various concentrations of RNA polymerase (from 50 to 800 nM) and find that although the amount of the RNA polymerase-DNA complex increases with increasing amounts of RNA polymerase in the reaction mixture, in every case only one band is seen (results not shown), indicating that only one RNA polymerase molecule can bind to one aroP fragment. When the TyrR protein and phenylalanine were also added, two additional complexes were seen. One complex (complex A), which travelled a little slower than complex B, was shown to comprise DNA, TyrR protein, and RNA polymerase in DNase I footprinting in gel slice experiments (see the next section). The other complex (complex C), which travelled a little slower than native DNA, has been shown in separate experiments to comprise DNA and TyrR protein (results not shown). As expected, in both cases, only the strong TyrR box is protected, indicating that these two complexes contain a TyrR dimer. When tyrosine was substituted for phenylalanine, the same mobility shift occurred, indicating again the involvement of a dimer. It seems likely that the complex of DNA, RNA polymerase, and TyrR hexamer that would be expected to form in the presence of tyrosine and ATP (20) may have been too large to enter the gel. Similar experiments were carried out using DNA fragments in which either P1 or P3 had been inactivated by mutation, and equivalent complexes were identified and subsequently used in gel slice experiments.
FIG. 2.
Gel shift assay. The experiment was performed essentially as described by Taylor et al. (14). The 32P-labelled aroP fragment (0.5 nM) was incubated at 37°C for 25 min with RNA polymerase (400 nM) in the absence or presence of purified TyrR protein (200 nM) and phenylalanine (1 mM) in 30 μl of buffer which contained 5 mM Tris · Cl (pH 7.8), 3 mM magnesium acetate, 50 mM NaCl2, 6 mM CaCl2, 4% glycerol, 0.1 mM dithiothreitol, 0.1 mM EDTA, 0.2 mM ATP, and 0.25 mg of bovine serum albumin per ml. The incubation was allowed to proceed for 25 min at 37°C before addition of 5 μl of dye mix (30% glycerol, 60 mM EDTA, 0.15% xylene cyanol FF, 0.15% bromophenol blue). The samples were immediately loaded onto a 5% polyacrylamide gel (acrylamide-bisacrylamide, 37.5:1) containing 1 mM phenylalanine and 0.2 mM ATP.
Analysis of various protein-DNA complexes by DNase I footprinting in gel slices.
From our previous results, we would expect that when RNA polymerase alone binds to DNA or when it does so in the presence of TyrR protein but in the absence of the effector phenylalanine or tyrosine, it should bind predominantly to promoter P1. On the other hand, we would expect that the addition of TyrR protein plus tyrosine or phenylalanine should cause most of the RNA polymerase molecules to shift from P1 to P3. In order to test this hypothesis, the various protein-DNA complexes from the gel shift experiments were subjected to DNase I footprinting assays. In order to ascertain whether this technique would allow us to discriminate between RNA polymerase bound at promoter P1 or promoter P3, we first used mutant DNA fragments in which only P1 or P3 was active (these mutations are shown in Fig. 1).
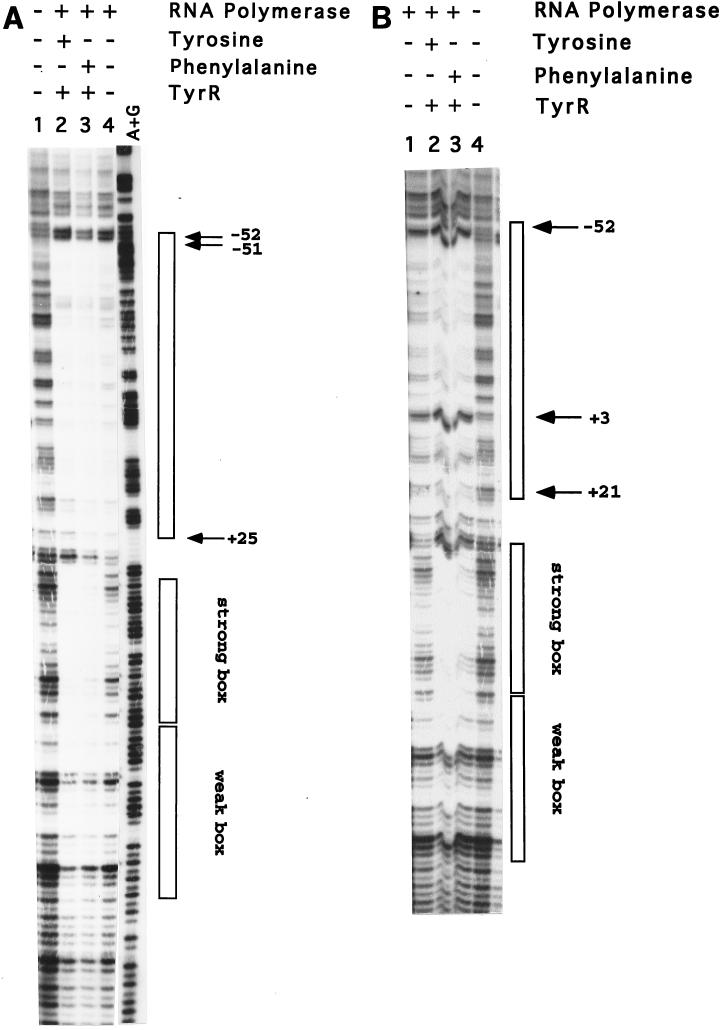
The results obtained with these mutant fragments (P1+ P3− and P1− P3+) are shown in Fig. 3. Whereas the region protected by RNA polymerase is fairly similar for each of the promoters, there are some important major differences. When the RNA polymerase binds to P1, the region from −53 to +25 (for simplicity, the numbering of positions in this paper are all relative to the +1 of P1) is protected and there are two distinctive hypersensitive bands at positions −51 and −52 (Fig. 3A). When the RNA polymerase binds to P3, it protects a region from −53 to +21 (Fig. 3B). Unlike the protection pattern for P1, there is only a single hypersensitive band in the −50 region, at −52 (Fig. 3B). Importantly there is an additional strong hypersensitive band at position +3 (Fig. 3B). Since in any one gel involving the wild-type fragment, we may have a mixture of some DNA molecules with RNA polymerase bound at P1 and others with RNA polymerase bound at P3, the existence of the P3-specific hypersensitive band should provide a dominant marker that can be used to measure binding to P3 in the presence or absence of binding to P1. Similarly, the presence of RNA polymerase molecules bound to P1 can be detected by the presence of the hypersensitive band at −51.
FIG. 3.
DNase I footprinting in gel slices. The experiments were performed essentially as described by Straney et al. (13) and Hanamura and Aiba (3). Gel shift experiments were carried out as described in the legend to Fig. 2. The free DNA band and the bands containing protein-DNA complexes were excised from the polyacrylamide gel, and the resulting gel slices (about 30 μl) were each incubated at room temperature for 15 min in a solution (10 μl) containing 0.1 μg of DNase I per ml, 10 mM Tris · Cl (pH 8.0), 2 mM dithiothreitol, 5% glycerol, and 0.5 mg bovine serum albumin per ml. The DNase I cleavage reaction was initiated by adding 5 μl of starting solution containing 50 mM MgCl2 and 50 mM CaCl2. After incubation at 37°C for 2 min, the reaction was terminated by the addition of 30 μl of stop buffer containing 0.1 M EDTA and 0.15% SDS. The resulting DNA fragments were eluted from the gel slices and analyzed on a 6% sequencing gel against an A+G ladder produced by the Maxam and Gilbert method (8). (A) DNase I footprinting in gel slice experiments carried out using the 0.3-kb DNA fragment (P1+ P3−) containing only the functional aroP P1 promoter. (B) DNase I footprinting in gel slice experiments carried out using the 0.3-kb DNA fragment (P1− P3+) containing only the intact aroP P3 promoter.
We next carried out the same footprinting experiments using a wild-type fragment (P1+ P3+) and RNA polymerase in the presence and absence of TyrR protein and one or the other of its cofactors. These results are shown in Fig. 4. As can be clearly seen, in the absence of TyrR protein, the protection pattern is identical to that observed for P1. However, in the presence of TyrR protein and tyrosine or phenylalanine, the appearance of the strong hypersensitive band at +3 clearly indicates that a significant number of molecules of RNA polymerase are now binding to P3. By observing the loss of the hypersensitive band at −51 we can conclude that in the presence of tyrosine nearly all of the RNA polymerase molecules have moved to P3, whereas in the presence of phenylalanine, although there is a clear indication of binding of RNA polymerase to P3, one can also detect some molecules binding to P1. A cartoon which summarizes the results of these experiments is shown in Fig. 5.
FIG. 4.
DNase I footprinting in gel slice experiments carried out using the 0.3-kb DNA fragment containing the wild-type aroP regulatory region (P1+ P3+). The experimental conditions are as described in the legend to Fig. 3.
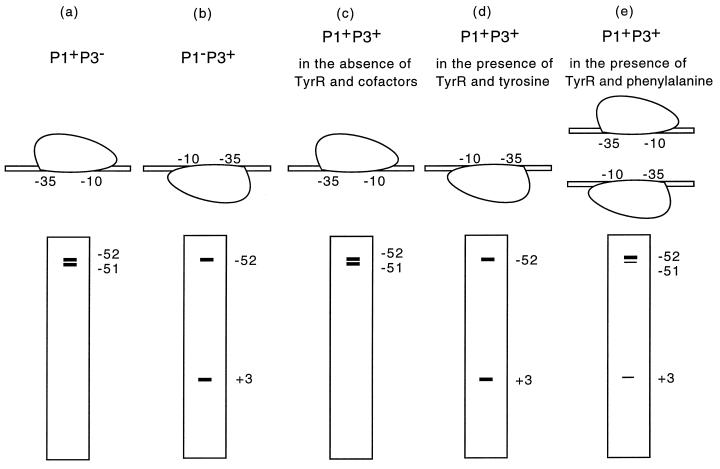
FIG. 5.
Cartoon depicting results from the DNase I footprinting in gel slice experiments. (a) RNA polymerase binds only to the P1 promoter (P1+ P3−). The protection pattern shows two hypersensitive bands at −52 and −51. (b) RNA polymerase binds only to the P3 promoter (P1− P3+). The protection pattern shows the loss of the hypersensitive band at −51 and the gain of a new strong hypersensitive band at +3. (c) In the absence of TyrR, RNA polymerase binds selectively to the P1 promoter on a wild-type aroP fragment (P1+ P3+) (same pattern as for panel a). (d) In the presence of TyrR and tyrosine, RNA polymerase binds selectively to the P3 promoter on a wild-type aroP fragment (P1+ P3+) (same pattern as for panel b). (e) Mixture of molecules with RNA polymerase bound either to P1 or to P3 on a wild-type aroP fragment (P1+ P3+) in the presence of TyrR and phenylalanine (combined patterns of panels a and b).
In the above experiments, the DNA fragments had been labelled at the 3′ end of the top strand. The experiments were also done using fragments labelled in the bottom strand. In the experiment involving the P1− P3+ fragment and RNA polymerase, two distinct hypersensitive bands at −3 and +10 were observed (data not shown). These two P3-specific hypersensitive bands were not present when the P1+ P3− fragment was used (data not shown). With the wild-type fragment (P1+ P3+), neither band was present in complexes involving RNA polymerase alone but both were present in complexes involving RNA polymerase and TyrR protein with either phenylalanine or tyrosine (data not shown). These results add further confirmation for the postulated shift from P1 to P3 in the presence of TyrR protein and phenylalanine or tyrosine.
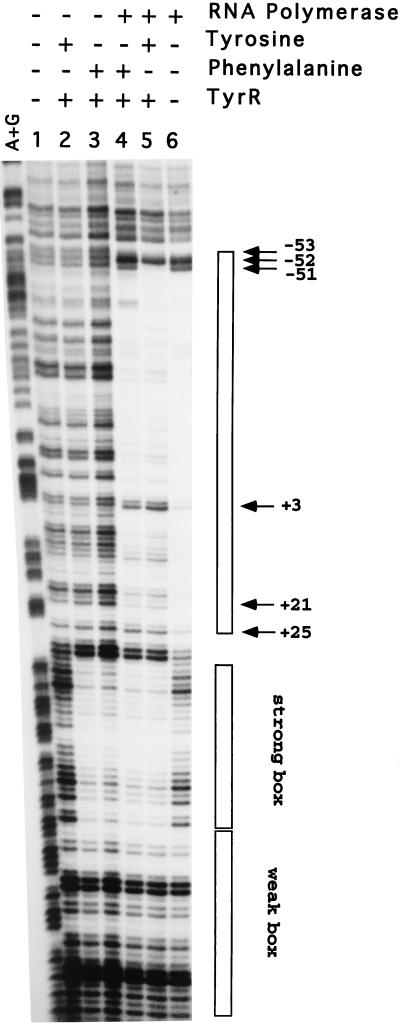
A tyrR mutant, which codes for an altered TyrR protein containing an arginine-to-glutamine change at position 10, has previously been isolated (21). This mutant TyrR protein, TyrR-RQ10, is completely defective in activation of transcription from both the mtr and tyrP+3 promoters (21, 22) and is also unable to repress transcription from the aroP P1 promoter (15). To see if the inability of TyrR-RQ10 to repress P1 results from the failure of this mutant protein to recruit RNA polymerase to P3, we repeated the DNase I footprinting in gel slice experiment using the purified TyrR-RQ10 protein and the DNA fragment carrying the wild-type aroP regulatory region. The results shown in Fig. 6 clearly show that, while TyrR-RQ10 maintains the ability to bind to the TyrR box region, it fails to recruit RNA polymerase to the divergent P3 promoter.
FIG. 6.
DNase I footprinting in gel slice experiments carried out with the 0.3-kb DNA fragment containing the wild-type aroP regulatory region (P1+ P3+) in the presence of TyrR-RQ10 (200 nM). The experimental conditions are as described in the legend to Fig. 3.
Discussion.
The results from the gel shift assay show that only one RNA polymerase molecule is able to bind to one DNA fragment carrying the upstream region of the aroP gene. In vitro analysis of the protein-DNA complexes, using the technique of DNase I footprinting in gel slices, reveals that RNA polymerase has a higher affinity for P1 than for P2 and P3 and occupies the P1 promoter region in the absence of TyrR protein. However, in the presence of TyrR protein and cofactors, RNA polymerase binds to the P3 region by cooperative interaction with TyrR molecules bound at the TyrR boxes. As the complexes used in these experiments appear to have involved TyrR dimers rather than hexamers, the general very weak protection of the downstream weak TyrR box is not unexpected.
The results of these in vitro experiments provide direct evidence in support of the model positing that the TyrR protein represses transcription initiation of the aroP P1 promoter by recruiting RNA polymerase to the divergent P3 promoter, thereby excluding the binding of RNA polymerase to P1. Although both tyrosine and phenylalanine were shown to be involved in TyrR-mediated recruitment of RNA polymerase to the P3 promoter, tyrosine had a greater effect than phenylalanine. This observation agrees with previous findings obtained from in vivo experiments, that the level of TyrR-mediated repression of P1 transcription by tyrosine is greater than that by phenylalanine (16). The failure of RNA polymerase to be activated to bind to P3 in response to the mutant TyrR protein, TyrR-RQ10, indicates that the same amino acid residue (arginine-10) which plays a critical role in TyrR-mediated activation of the mtr and tyrP+3 promoters (21, 22) is also involved in the recruitment of RNA polymerase to the P3 promoter of the aroP gene.
In these results, we do not detect any involvement of the P2 promoter. Since it is located 21 bases downstream of P1, RNA polymerase binding to P2 would give a quite distinctive pattern of protection. Although in the absence of TyrR protein, P2 is transcribed as efficiently as P1 from supercoiled templates in vitro (16), the same is not true from linear templates where transcription from P2 is only about 1/10 of that from P1 (unpublished results). Since linear DNA fragments were used in these experiments, the failure to detect binding of RNA polymerase to P2 agrees with the earlier observations. It should also be noted that P2 is a much weaker promoter than P1 in vivo (16).
Repression of transcription from procaryotic promoters can be achieved via different mechanisms. In many cases, repression involves direct competition between a repressor and RNA polymerase for access to a promoter. More recently, it has been demonstrated in several systems such as those involving GalR, KorB, and the P4 protein of phage φ29 that a repressor and RNA polymerase can bind simultaneously to a promoter (1, 9, 19). In these cases, the repressors act to inhibit either the isomerization of closed-to-open complex or promoter clearance. The negative regulation of some other procaryotic transcription systems has been shown to involve a divergent promoter (3–7, 17). In these systems, the molecular mechanism of autoregulation of the Escherichia coli crp gene is most analogous to that of the TyrR-mediated repression of the aroP P1 promoter (3). By using a variety of in vitro approaches, Hanamura and Aiba (3) have demonstrated that the cyclic AMP (cAMP)-cAMP receptor protein complex bound at a site downstream of the crp promoter activates transcription of a divergent promoter and this stimulation of transcription results in a blockage of transcription initiation from the crp promoter. The divergent promoter of crp is transcriptionally productive both in vivo and in vitro upon stimulation by the cAMP receptor protein and cAMP (3, 10, 11), whereas the divergent aroP promoter is virtually nonproductive in vivo in both the presence and absence of TyrR protein and its cofactors (16). Preliminary results suggest that the transcriptional nonproductivity of the aroP P3 promoter is important for effective repression of P1 transcription (data not shown).
Acknowledgments
This work was supported by a collaborative research grant from the Australian Research Council and the Japan Society for Promotion of Science. P. Wang is the recipient of an Australian Agency for International Development Scholarship.
We thank H. Camakaris and J. Gowrishankar for their comments on the manuscript.
REFERENCES
- 1.Choy H E, Park S W, Aki T, Parrack P, Fujita N, Ishihama A, Adhya S. Repression and activation of transcription by Gal and Lac repressors: involvement of alpha subunit of RNA polymerase. EMBO J. 1995;14:4523–4529. doi: 10.1002/j.1460-2075.1995.tb00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chye M-L, Pittard J. Transcription control of the aroP gene in Escherichia coli K-12: analysis of operator mutants. J Bacteriol. 1987;169:386–393. doi: 10.1128/jb.169.1.386-393.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanamura A, Aiba H. Molecular mechanism of negative autoregulation of Escherichia coli crp gene. Nucleic Acids Res. 1991;19:4413–4419. doi: 10.1093/nar/19.16.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershberger P A, Mita B C, Tripatara A, deHaseth P L. Interference by Pr-bound RNA polymerase with Prm function in vitro: modulation by the bacteriophage λ cI protein. J Biol Chem. 1993;268:8943–8948. [PubMed] [Google Scholar]
- 5.Hershberger P A, deHaseth P L. RNA polymerase bound to the Pr promoter of bacteriophage λ inhibits open complex formation at the divergently transcribed Prm promoter: implications for an indirect mechanism of transcriptional activation by λ repressor. J Mol Biol. 1991;222:479–494. doi: 10.1016/0022-2836(91)90491-n. [DOI] [PubMed] [Google Scholar]
- 6.Krause H M, Higgins N P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986;261:3744–3752. [PubMed] [Google Scholar]
- 7.Livrelli V, Lee I W, Summers A O. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. I. Metalloregulatory protein MerR mutants. J Biol Chem. 1993;268:2623–2631. [PubMed] [Google Scholar]
- 8.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 9.Monsalve M, Mencía M, Rojo F, Salas M. Activation and repression of transcription at two different phage φ29 promoters are mediated by interaction of the same residues of regulatory protein P4 with RNA polymerase. EMBO J. 1996;15:383–391. [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto K, Hara S, Bhasin R, Freundlich M. Evidence in vivo for autogenous control of cyclic AMP receptor protein gene (crp) in Escherichia coli by divergent RNA. J Bacteriol. 1988;170:5076–5079. doi: 10.1128/jb.170.11.5076-5079.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto K, Freundlich M. Mechanism for the autogenous control of the crp operon: transcriptional inhibition by a divergent RNA transcript. Proc Natl Acad Sci USA. 1986;83:5000–5004. doi: 10.1073/pnas.83.14.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 458–484. [Google Scholar]
- 13.Straney D C, Straney S B, Crothers D M. Synergy between Escherichia coli CAP protein and RNA polymerase in the lac promoter open complex. J Mol Biol. 1989;206:41–57. doi: 10.1016/0022-2836(89)90522-6. [DOI] [PubMed] [Google Scholar]
- 14.Taylor J D, Ackroyd A J, Halford S E. The gel shift assay for the analysis of DNA-protein interactions. Methods Mol Biol. 1994;30:263–279. doi: 10.1385/0-89603-256-6:263. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Yang J, Lawley B, Pittard A J. Repression of the aroP gene of Escherichia coli involves activation of a divergent promoter. J Bacteriol. 1997;179:4213–4218. doi: 10.1128/jb.179.13.4213-4218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Yang J, Pittard A J. Promoters and transcripts associated with the aroP gene of Escherichia coli. J Bacteriol. 1997;179:4206–4212. doi: 10.1128/jb.179.13.4206-4212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wek R C, Hatfield G W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988;203:643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]
- 18.Whipp M J, Pittard A J. Regulation of aromatic amino acid transport systems in Escherichia coli K-12. J Bacteriol. 1977;132:453–461. doi: 10.1128/jb.132.2.453-461.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams D R, Motallebi-Veshareh M, Thomas C M. Multifunctional repressor KorB can block transcription by preventing isomerization of RNA polymerase-promoter complexes. Nucleic Acids Res. 1993;21:1141–1148. doi: 10.1093/nar/21.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson T J, Maroudas P, Howlett G J, Davidson B E. Ligand-induced self-association of the Escherichia coli regulatory protein TyrR. J Mol Biol. 1994;238:309–318. doi: 10.1006/jmbi.1994.1294. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Camakaris H, Pittard A J. Further genetic analysis of the activation function of the TyrR regulatory protein. J Bacteriol. 1996;178:1120–1125. doi: 10.1128/jb.178.4.1120-1125.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Camakaris H, Pittard A J. In vitro transcriptional analysis of TyrR-mediated activation of the mtr and tyrP+3 promoters of Escherichia coli. J Bacteriol. 1996;178:6389–6393. doi: 10.1128/jb.178.21.6389-6393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]