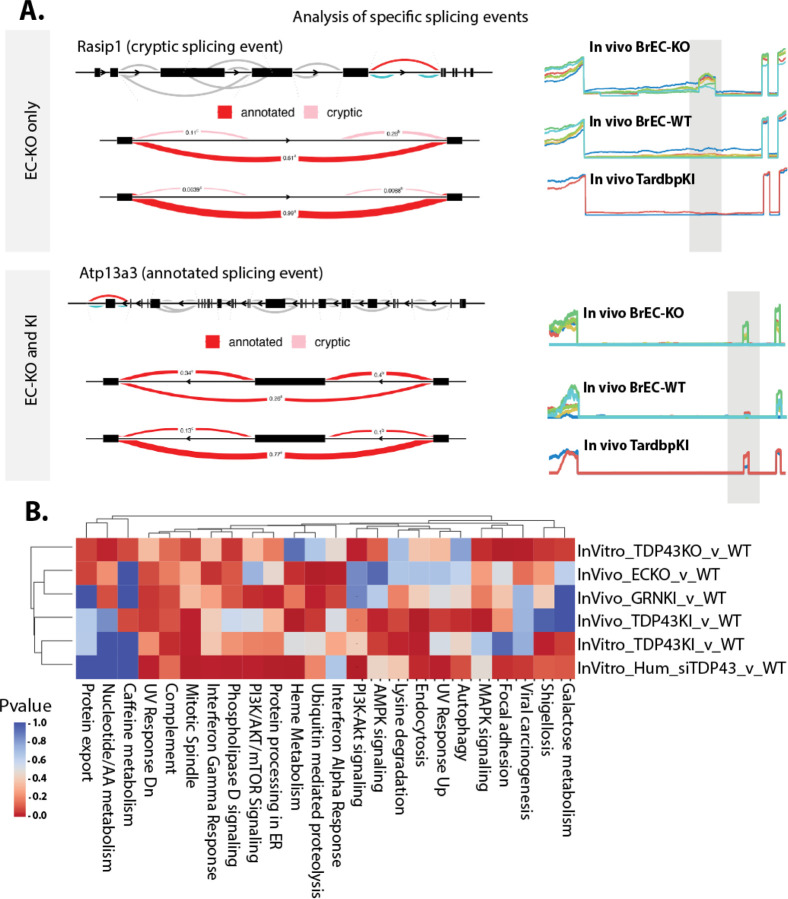
Figure 7. Splicing alterations associated with the loss of nuclear TDP-43 affect proteins in pathways enriched in altered mRNA transcripts.
(A) Leafcutter (Leafviz) analysis of example splicing events differentially regulated in BrEC-KO cells in vivo, some of which are also seen in KI cells in vivo. Line plots to the right show read density in each of the replicates across the alternatively spliced region. (B) Heat map and clustering (UPGMA algorithm) of the most consistently affected KEGG and Hallmark pathways, and determine by GseaPy ENRICHR. (B) FDR value derived from GSEA analysis is shown.