Abstract
Tuberous sclerosis complex (TSC) is a multi-system genetic disease that causes benign tumors in the brain and other vital organs. The most debilitating symptoms result from involvement of the central nervous system and lead to a multitude of severe symptoms including seizures, intellectual disability, autism, and behavioral problems. TSC is caused by heterozygous mutations of either the TSC1 or TSC2 gene. Dysregulation of mTOR kinase with its multifaceted downstream signaling alterations is central to disease pathogenesis. Although the neurological sequelae of the disease are well established, little is known about how these mutations might affect cellular components and the function of the blood-brain barrier (BBB). We generated disease-specific cell models of the BBB by leveraging human induced pluripotent stem cell and microfluidic cell culture technologies. Using these microphysiological systems, we demonstrate that the BBB generated from TSC2 heterozygous mutant cells shows increased permeability which can be rescued by wild type astrocytes and with treatment with rapamycin, an mTOR kinase inhibitor. Our results further demonstrate the utility of microphysiological systems to study human neurological disorders and advance our knowledge of the cell lineages contributing to TSC pathogenesis.
Keywords: BBB, human stem cells, astrocytes, mTOR, rapamycin, microfluidics, tissue chips
Background
Neurogenetic disorders often present in young children due to their impact on the developing brain. The neurological manifestations of such disorders are typically severe and include epilepsy, intellectual disabilities, and autism (Moffat, Ka et al. 2015). Tuberous Sclerosis Complex (TSC) has been a prototypical model for neurogenetic disease for many years (Curatolo, Bombardieri and Jozwiak 2008). The study of TSC provides several advantages, including well-described clinical manifestations and a known genetic etiology from loss of TSC1 or TSC2 gene function with defined downstream signaling pathways (Ess 2006, Rosset, Netto and Ashton-Prolla 2017, Rosset, Vairo et al. 2017). In addition, affected downstream signaling pathways have been identified, with an ostensibly central role for dysregulation of mTOR kinase (Baybis, Yu et al. 2004, Kaper, Dornhoefer and Giaccia 2006, Lim, Gopalappa et al. 2017). Hamartin and tuberin, the TSC1 and TSC2 protein products respectively, normally inhibit mTOR signaling through an indirect pathway. TSC1 and TSC2-mutant cells thus have constitutively increased activity of mTOR kinase, which likely underlies the abnormal proliferation and differentiation of cells suspected to occur in multiple organs of patients with TSC (Ihrie and Henske 2022). This understanding has led to the development of mTOR inhibitors (“rapalogs”) that are FDA-approved and increasingly used for the treatment of several aspects of TSC (Krueger, Care et al. 2010, McCormack, Inoue et al. 2011, Bissler, Kingswood et al. 2013, Krueger, Capal et al. 2018).
While many animal models (mouse, rat, zebrafish, Drosophila) have been developed over the past 20 years (Uhlmann, Wong et al. 2002, Wong, Ess et al. 2003, Wenzel, Patel et al. 2004, Meikle, Talos et al. 2007, Kim, Speirs et al. 2011, Carson, Van Nielen et al. 2012, Tsai, Hull et al. 2012, Carson, Fu et al. 2013, Carson, Kelm et al. 2015), many key aspects of TSC pathogenesis remain poorly understood. These include species-specific impact of mTOR signaling as well as the requirements for heterozygous versus homozygous mutations of the TSC1/TSC2 genes. The advance of human induced pluripotent stem cell (iPSC) technology (Dolmetsch and Geschwind 2011, Higurashi, Uchida et al. 2013, Nityanandam and Baldwin 2015, Snow, Westlake et al. 2020) combined with tissue chip technology (Brown, Pensabene et al. 2015, Brown, Codreanu et al. 2016, Vernetti, Gough et al. 2017, Brown, Faley et al. 2020) has allowed the use of human tissue-based models to address these fundamental questions.
Although the neurological sequelae of TSC are well established, little is known about how TSC1 or TSC2 mutations affect different cellular and functional components of the brain. While expression of the astrocytic protein aquaporin-4, a component of the blood-brain barrier (BBB) is increased in epileptic cortex from patients with TSC and TSC mouse models (Short, Kozek et al. 2019), BBB function has not been well studied in TSC. We hypothesized that an abnormal BBB contributes to TSC pathogenesis. The study of the BBB in animal models of TSC although highly valuable also presents some challenges. Thus, species-specific differences in human versus mouse brain structure and cellular function have recently become more apparent. In addition, complicated genetic manipulations are needed to study the impact of Tsc2 mutations in specific cell types. Finally, the frequent occurrence of epilepsy or brain tumor phenotypes in animal models also represent potential confounding factors, as these symptoms could have secondary effects on the BBB. Therefore, to test our hypothesis and examine primary effects of TSC2 mutation on the BBB, we used an in vitro neurovascular unit (NVU) (Figure 1) to create TSC patient-specific brain tissue models that were generated by leveraging iPSC and microfluidic cell culture technologies (Brown, Pensabene et al. 2015, Brown, Codreanu et al. 2016, Brown, Faley et al. 2020). Using these microphysiological systems, we demonstrate that the TSC2 patient derived BBB shows increased permeability which can be rescued with rapamycin, an mTOR kinase inhibitor. Astrocytes appear to play a major role in this TSC BBB phenotype as replacement of TSC2-mutant astrocytes with wild type astrocytes also rescued the TSC2 patient derived BBB defect.
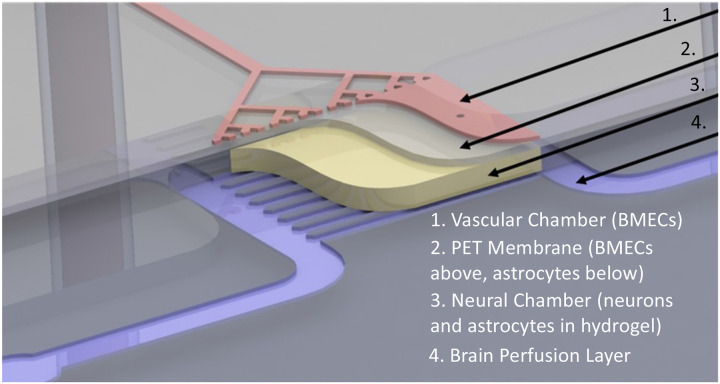
Figure 1. Schematic overview of NVU.
The vascular chamber (1, Pink) into which BMECs are loaded and the media for the vascular chamber is perfused. Porous PET membrane (2, Grey) with 3 μm pores supports a layer of BMECs on one side and a layer of astrocytes on the opposite “brain” neural cell chamber side. Neural chamber (4, Yellow) contains neurons and additional astrocytes within a hydrogel. Perfusion channels through the neural cell chamber indicated as Purple.
Our findings substantiate the use of microphysiological systems to study neurogenetic disorders. We interpret our results within what is known about the BBB in TSC and discuss how our findings expand this knowledge and support ongoing translational research related to TSC pathogenesis and treatment.
Methods
Derivation, validation, and differentiation of iPSCs
Three of the human induced pluripotent stem cell (iPSC) lines (control CC3, TSC patient TSP8-15, and TSP23-9) were derived at Vanderbilt University Medical Center and validated according to established protocols (Armstrong, Westlake et al. 2017, Neely, Davison et al. 2017). In brief, primary dermal fibroblasts were established from skin biopsies obtained after patients’ consent/assent under the Vanderbilt University Medical Center IRB protocol #080369. Fibroblasts were reprogrammed by electroporation with either CXLE plasmid vectors (Howden, Thomson and Little 2018) (lines CC3 and TSP8-15) using the Neon Transfection System (Life Technologies, Carlsbad, CA, USA) or CytoTune iPS 2.0 Sendai Reprogramming Kit (ThermoFisher) (line TSP23-9). Transfected fibroblasts were then plated at 5 x 104 cells/well into Matrigel-coated 6-well plates. Two days later, cells were transferred into TeSR-E7 medium and maintained until the emergence of iPSC colonies (about 4 weeks) which were then manually isolated and propagated in mTeSR medium (StemCell Technologies). Absence of plasmid integration and clearance of Sendai virus were confirmed, and normal karyotype verified using at least 20 metaphase spreads (Genetics Associates, Nashville, TN, USA) (Supplementary Figure 1A). Pluripotency markers were present as previously reported and pluripotency was further validated by Pluritest and/or the ability of the iPSC lines to differentiate into all three germline lineages using the hPSC TaqMan Scorecard (ThermoFisher A15870), and into neural lineages as we have previously described (Neely, Tidball et al. 2011, Brown, Codreanu et al. 2016, Tidball, Neely et al. 2016, Neely, Davison et al. 2017, Neely, Xie et al. 2021, Prince, Neely et al. 2021) (Neal, Marinelli et al. 2019). Isogenic iPSC lines TSP77+/+ and TSP77+/− were generated at Boston’s Children Hospital and previously validated as described (Sundberg, Tochitsky et al. 2018).
Cortical glutamatergic neuron differentiation
iPSCs were replated at a density of 2 x 104 cells/cm2 in mTeSR medium containing 10 μM Rho kinase inhibitor (Y-27632, Tocris #5849), which was removed after 24 hours. Once the cultures reached 100% confluency (day 0), neuralization was induced via an eleven-day dual-SMAD inhibition protocol using 0.4 μm LDN (Tocris # 6053) and 10 μM SB 431542 (Tocris #1614) as previously described (Chambers, Fasano et al. 2009, Neely, Litt et al. 2012). Starting on day 11, the neuronal cultures underwent further differentiation in cortical differentiation medium as previously reported (Neely, Xie et al. 2021, Prince, Neely et al. 2021). Around day 20-25, differentiating cells were passaged for the first time by incubating them with Accutase (StemCell Technologies, #01-0006) for 18-30 minutes and reseeding them into Matrigel (BD Bioscience #354277)-coated 6-well plates at 1 x 105 cells/cm2 in cortical differentiation medium containing 10 μM Rock-inhibitor (Tocris, Minneapolis, MN, USA #1254), which was removed after 24 hours. Cultures were maintained in cortical differentiation medium and replated at 3 x 105 cells/cm2 monthly. Neurons were harvested for seeding into the neurovascular units (NVUs) between days 80-120 of differentiation from iPSC stage. The cortical cultures derived from all iPSC lines contained abundant neurons with dense and complex neurite projections (Supplementary Figures 2A and B).
Cortical astrocyte differentiation
We used a “spontaneous emergence approach” (Chandrasekaran, Avci et al. 2016) to make astrocyte cultures, as we have previously described (Miller, Schaffer et al. 2021). By around day 80 of cortical glutamatergic neuron differentiation, we begin to see astrocytes emerging alongside cortical glutamatergic neurons. By day 120-160 of differentiation, we passaged cells monthly at low density (0.5 x 105 cells/cm2), which leads to a continuous loss of neurons from the cultures and results in relatively pure astrocyte cultures after 3-4 passages. At that point astrocytes are transferred into astrocyte medium (ScienCell #1801) and replated at 0.4 x 105 cells/cm2 monthly until integration into the NVU. The large majority of cells in these astrocyte cultures express the astrocyte marker GFAP and/or S100B (Supplementary Figures 3).
BMEC-like differentiation
Brain microvascular endothelial-like cells (BMECs) were differentiated from iPSCs according to previously reported protocols, with minor modifications (Bosworth, Faley et al. 2017, Faley, Neal et al. 2019). Briefly, iPSCs were seeded at a concentration of 150,000 cells per well of a Matrigel-coated 6-well plate in E8 medium (ThermoFisher) containing Rock-inhibitor (10 μM; Y-27632). The following day, E8 medium was replaced with E6 medium (ThermoFisher Scientific) and changed daily for 4 days. On day 4, the medium was changed to Neurobasal (Gibco) medium supplemented with B27 (Gibco), 0.5 mM Glutamax (ThermoFisher Scientific), 20 ng/ml basic fibroblast growth factor (bFGF; PeproTech, Rocky Hill, NJ, USA), and 10 μM all-trans retinoic acid (RA; ThermoFisher Scientific) for 48 hours. BMECs were subcultured on day 6 in the same medium containing Rock-inhibitor (10 μM Y-27632). For traditional experiments, BMECs were subcultured either onto Polyester PET Transwells of 3 μm pore size or into 12-well tissue culture plates (Corning), both of which had been coated with a mixture of collagen IV (Sigma-Aldrich, 400 ng/ml) and fibronectin (Sigma-Aldrich, 100 ng/ml) in PBS for 24 hours at 37°C.
Transendothelial electrical resistance (TEER) Measurements
TEER was measured using an EVOM voltohmmeter with STX2 electrodes (World Precision Instruments) using standard protocols we have previously employed (Brown, Pensabene et al. 2015, Bosworth, Faley et al. 2017, Faley, Neal et al. 2019, Brown, Faley et al. 2020). Raw TEER values were adjusted through subtracting TEER values measured across an empty filter and then multiplied by filter surface area to yield TEER (Ωxcm2).
Transwell culture
Transwell cultures were seeded and cultured in parallel with the NVU cultures to allow for a direct comparison of cell phenotypes in these two different models. Thus, Transwell plates were coated with the same ECM components used in the vascular side of the NVU and seeded with the same BMECs suspension in the same medium. The BMECs were seeded onto the membrane of the apical side of the Transwell insert. Sample collection from the NVU and Transwell cultures was also performed in parallel.
NVU cell load and culture
NVU devices were coated with collagen IV and fibronectin as previously described (Brown, Pensabene et al. 2015). BMECs (control or TSC2-mutant) were seeded into the vascular chamber at a density of (8–10) × 106 cells/ml (day 0) (Brown, Pensabene et al. 2015, Brown, Codreanu et al. 2016). The device was oriented (neural cell “brain” chamber bottom, vascular chamber top) to allow BMEC attachment to the membrane overnight. On the following day, the astrocytes were loaded into the neural cell chamber at (2-5) × 106 cells/ml and the devices were inverted (neural cell chamber top, vascular chamber bottom) to allow for attachment of the astrocytes on the membrane side opposite to the seeded BMEC. Two hours after astrocyte loading, neurons (8–10) × 106 cells/ml suspended in a hydrogel (Mebiol Gel; Cosmo Bio Co. Ltd, #MBG-PMW20) were seeded into the neural cell chamber as previously described (Brown, Pensabene et al. 2015, Brown, Codreanu et al. 2016). Once the hydrogel had set, typically after 1 hour, media flow was started. The vascular chamber containing BMECs was perfused with Lonza EGM-2 medium and the neural chamber containing neurons and astrocytes was perfused with Neurobasal medium supplemented with B27 and Glutamax. Devices were maintained for a minimum of 24–48 hours before experiments were conducted.
NVU BBB permeability assay
Passive permeability measurements in NVUs
To measure BBB permeability in the NVUs we added 3kD (unless indicated otherwise) Alexa Fluor 680 dextran (excitation 680 and emission 706 nm; ThermoFisher) at a final concentration of 1 μM to the vascular compartment medium and perfused the NVU with this dextran solution for the entire duration of the culture (Brown, Faley et al. 2020). For passive permeability measurements, effluent was collected from the neural cell compartment over a fixed amount of time and the fluorescence intensity of the eluate quantified using a plate reader (Tecan M1000). Using the determined dye concentration in the brain compartment, we calculated the applied permeability coefficient, , using the standard equation:
where is the neural cell chamber volume in cm3, is the vascular chamber growth area in cm2, is the dextran concentration perfused into in the vascular compartment (μM), is the dextran concentration (μM) measured in the neural cell compartment eluate, and is the assay time in seconds (Frost, Jiang et al. 2019).
The effective permeability of the BMEC monolayer, , was calculated from the measured applied permeability by correcting for the permeability of a device without cells (but containing hydrogel and a membrane coated with collagen/fibronectin), , according to the equation:
Rapamycin treatment
Rapamycin (Thermo-Fisher) was reconstituted as a 10 μM stock solution in DMSO. 10 nM rapamycin in culture medium was then either perfused through the vascular chamber of the NVUs or added to the apical chamber of the Transwell cultures for 24 hours, after which BBB permeability measurements were carried out as described above.
Metabolomics
Neural cell and vascular chamber samples were stored at −80°C until analyzed via Liquid Chromatography-High Resolution Mass Spectrometry (LC-HRMS)-based metabolomics in the Vanderbilt Center for Innovative Technology (CIT) using previously described methods (Brown, Codreanu et al. 2016, Rogers, Sobolik et al. 2018, Mancini, Schrimpe-Rutledge et al. 2021) Briefly, effluent samples collected from both the vascular and brain chambers of the NVU were normalized by volume to 100 uL as previously reported (Brown, Codreanu et al. 2016). Metabolites were extracted with methanol/water 80:20. Heavy labeled phenylalanine-D8 and biotin-D2 were added to individual samples prior to protein precipitation. Following overnight incubation at −80°C, precipitated proteins were pelleted by centrifugation at 10,000 rpm for 10 min and metabolite extracts were dried down in vacuo and stored at −80°C.
Individual extracts were reconstituted in 50 μl of acetonitrile/water (3:97, v/v) with 0.1% formic acid containing heavy-labeled carnitine-D9, tryptophan-D3, valine-D8, and inosine-4N15, and centrifuged for 5 min at 10,000 rpm to remove insoluble material. A pooled quality control sample (QC) was prepared by pooling equal volumes of individual samples. The pooled QC sample was used for column conditioning (8 injections prior to sample analysis), retention time alignment and to assess mass spectrometry instrument reproducibility throughout the sample set.
Global, untargeted mass spectrometry analyses were performed on a high-resolution Q-Exactive HF hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a Vanquish UHPLC binary system (Thermo Fisher Scientific, Bremen, Germany). Extracts (5μL injection volume) were separated on a Hypersil Gold, 1.9 μm, 2.1 mm × 100 mm column (Thermo Fisher) held at 40 °C. LC was performed at 250 μL/min using solvent A (0.1% FA in water) and solvent B (0.1% FA in acetonitrile/water 80:20) with a gradient length of 30 min as previously described (Eberly, Beebout et al. 2020, Popay, Wang et al. 2021). Full MS analyses were acquired over the mass-to-charge ratio (m/z) range of 70-1,050 in positive ion mode. Full mass scan was acquired at 120,000 resolution with a scan rate of 3.5 Hz, automatic gain control (AGC) target of 1x106, and maximum ion injection time of 100 ms, and MS/MS spectra were collected at 15,000 resolution, AGC target of 2x105 ions, with a maximum ion injection time of 100 ms.
Metabolomics Data Processing and Pathway Analysis
Mass spectrometry raw data was imported, processed, normalized and reviewed using Progenesis QI v.3.0 (Non-linear Dynamics, Newcastle, UK). All MS and MS/MS sample runs were aligned against a pooled QC reference run. Unique ions (retention time and m/z pairs) were de-adducted and de-isotoped to generate unique “features” (retention time and m/z pairs). Data were normalized to all features and significance was assessed using p-values generated using ANOVA (analysis of variance) from normalized compound abundance data. Tentative and putative annotations were determined by using accurate mass measurements (<5 ppm error), isotope distribution similarity, and fragmentation spectrum matching (when applicable) by searching the Human Metabolome Database (Wishart, Jewison et al. 2013), METLIN (Smith, O'Maille et al. 2005), and the CIT’s in-house library. Annotations (Confidence Level 1-3 (Schrimpe-Rutledge, Codreanu et al. 2016) were determined for all compounds with a match to any of the searched libraries or databases. Metaboanalyst 5.0 (www.metaboanalyst.ca/) was used to perform pathway and metabolite enrichment analyses from annotated compounds with statistical significance (p-value ≤ 0.05) (Chong, Wishart and Xia 2019).
RNA Seq
Total RNA was extracted from cells and purified with Rneasy mini kit (Qiagen) and quality was validated with an Agilent kit. RNA sequencing reads were adapter-trimmed and quality-filtered using Trimgalore v0.6.7 (Krueger et al., 2023). An alignment reference was generated from the GRCh38 human genome and GENCODE comprehensive gene annotations (Release 26), to which trimmed reads were aligned and counted using Spliced Transcripts Alignment to a Reference (STAR) v2.7.9a (Dobin et al., 2013) with quantMode GeneCounts parameter. Approximately 60 million uniquely mapped reads were acquired per sample. DESeq2 package v1.36.0 (Love et al. 2014) was used to perform sample-level quality control, low count filtering, normalization, and downstream differential gene expression analysis. Genomic features counted fewer than five times across at least three samples were removed. False discovery rate adjusted for multiple hypothesis testing with Benjamini-Hochberg (BH) procedure p value < 0.05 and log2 fold change >1 was used to define differentially expressed genes. Pairwise comparison with TSC2 heterozygous mutation (TSP8-15) versus control (CC3) was performed. Three replicates per condition were included for the differential gene expression analysis. Gene set enrichment analysis (GSEA) was performed using the R package Clusterprofiler (Yu, Wang et al. 2012) with gene sets from the Human MSigDB database v2022.1.Hs (Subramanian, Tamayo et al. 2005).
Statistics
Statistical significance was determined using a t test when changes were compared between two groups (GraphPad Prism 6). P< 0.05 was considered statistically significant unless otherwise stated. Values were expressed as means ± SEM. For comparisons of more than 2 groups, we used either one-way ANOVA (for normal distributions) with Bonferroni post hoc test or Kruska Wallis with Dunn’s post hoc test where a normal distribution cannot be confirmed. (p < 0.01 is indicated with ** in the figures throughout the manuscript).
Data Availability
The RNA-Seq data has been annotated and deposited to NCBI GEO with accession ID GSE235862.
Metabolomics data is in the process of being submitted to the NIH Common Fund's National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org. This work is supported by Metabolomics Workbench/National Metabolomics Data Repository (NMDR) (grant# U2C-DK119886), Common Fund Data Ecosystem (CFDE) (grant# 3OT2OD030544) and Metabolomics Consortium Coordinating Center (M3C) (grant# 1U2C-DK119889).
Results
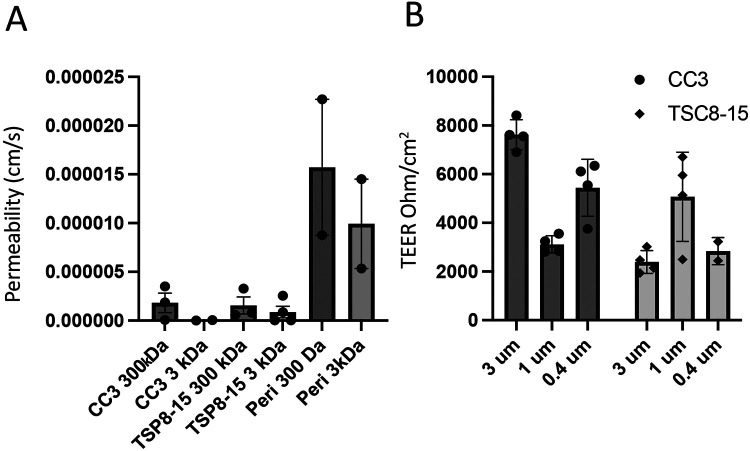
We initially used conventional monolayers of human iPSC-derived brain microvascular endothelial cells (BMECs) grown in a Transwell dish to evaluate barrier permeability of BMECs differentiated from control (CC3) and TSC2 heterozygous mutant (TSP8-15) iPSC lines. We delivered fluorescently labelled dextran of different sizes (300 or 3,000 Da) to the apical side of the barriers and quantified subsequent concentration in the basal side. The relative permeability of TSC mutant BMEC barriers showed a trend towards higher permeability than barriers from control BMECs for the 300 Da dextran, but the difference did not reach statistical significance (Figure 2A). As expected, pericytes, which do not form tight junctions (Bauer, Krizbai et al. 2014), cannot form a barrier other than the passive resistance created by the presence of cells in a monolayer on the filter membrane. Thus, the permeability for pericytes is greater than that of BMEC cultures (Figure 2A). We also looked at transendothelial electrical resistance (TEER) of BMECs cultured on membranes of varying pore sizes to evaluate the integrity of the endothelial barrier. A two-way ANOVA analysis showed that TEER was significantly affected by genotype, but not by membrane pore size (Figure 2B).
Figure 2. Comparison of barrier function of control and TSC2-mutant BMEC monocultures in Transwell plates.
A) TSC patient-derived (TSP8-15) BMEC transwell cultures showed a statistically non-significant trend towards larger permeability for the 3000 DA, but not 300 Da dextrans when compared to control (CC3) BMEC cultures. Pericytes (Peri) formed a minimal barrier (N = 2-4). B) Transendothelial electrical resistance (TEER) of CC3 and TSP8-15 BMECs cultured on membranes of varying pore sizes were measured. A two-way ANOVA analysis of the data indicated a genotype-dependent F (1,16) = 18.87 (P < 0.0005), but membrane pore-size independent F(2,16) = 1.91 (P = 0.1791) effect on TEER (overall P < 0.0001, N= 2-4).
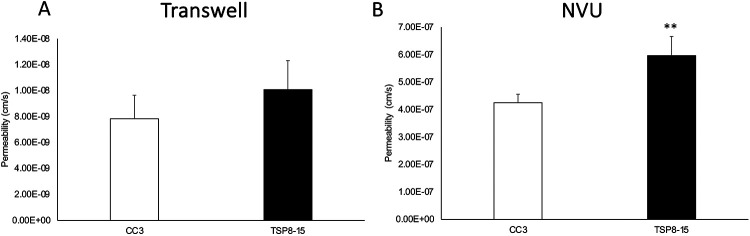
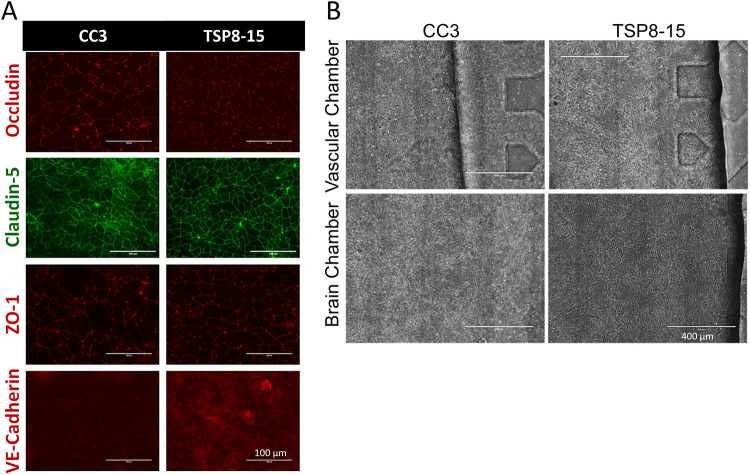
We next compared barrier permeabilities between transwell BMEC monocultures and the more complex multi-cell NVU assembly with “brain” (neural cell) and “vascular” sides (Figure 1) using 3 kD dextran. The difference between CC3 and TSP8-15 barrier function in the monoculture transwell model was not statistically significant, though TSP8-15 derived BMECs trended towards higher permeability (Figure 3A). In NVUs seeded with BMECs, astrocytes, and neurons we found a statistically significantly higher permeability in TSP8-15 than in control CC3-cell seeded NVUs (Figure 3B) (p < 0.01, N=5). The expression of the BMEC marker proteins occludin, claudin, and ZO-1 validated the BMEC-like nature of our CC3 and TSP8-15 cultures (Figure 4A). Figure 4B shows phase microscopy images of seeded vascular and neural compartments of CC3 and TSP8-15 NVUs.
Figure 3. Disruption of BBB in an NVU co-culture system.
(A) TSC patient-derived (TSP8-15) BMEC Transwell cultures 8 days in culture show a small, but not statistically significant higher permeability with 3 kD FITC dextran as compared to CC3 (p< 0.06, N=5). (B) In the NVU system, after 8 days co-culture with astrocytes and neurons of matching genotype, BBB permeability for the TSP8-15 was significantly increased compared to CC3 (p < 0.01, N=5).
Figure 4. Expression of junctional proteins in BMEC cultures.
(A) BMEC monolayers derived from CC3 and TSP8-15 iPSC lines express BMEC markers occludin, claudin-5, and ZO-1 and VE-cadherin. Scale bar for all images = 100 μm. (B) Phase images of seeded CC3 and TSP8-15 NVU vascular BMEC-containing compartments (top panels) and the neuron and astrocyte-containing neural cell chambers (bottom panels) are shown. Scale bar in all images = 400 μm.
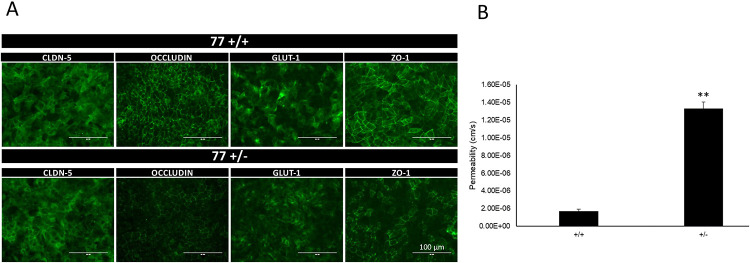
We extended our BBB permeability analyses to a set of isogenic iPSCs (TSP77; either wild type or heterozygous for a TSC2 mutation). As seen for CC3 control- (TSC2 wild type) and TSP8-15 TSC2 heterozygous mutant line BMECs, TSP77+/+ and TSP77+/− derived BMECs express the typical BMEC markers claudin-5, occluding, ZO-1 and Glut-1 (Figure 5A). Importantly, we observed a significantly higher permeability in TSP77+/− TSC2 heterozygous mutant cultures compared to TSP77+/+ isogenic wild type cultures (Figure 5B), thus confirming our permeability observations in the CC3 and TSP8-15 NVUs (Fig. 3B). A trend towards a higher difference in TSC (TSP8-15) versus control (CC3) BBB permeability was observed as early as day 2 after NVU seeding, and was significant by day 5 and thereafter (Fig. 6A and Fig. 7A).
Figure 5. Increased BBB permeability in NVU generated from TSC2 heterozygous mutant cells compared to isogenic control cells.
(A) TSP77+/+ and TSP77+/− derived BMECs express the BMEC marker proteins occluding, claudin-5, ZO-1 and Glut-1. Scale bar for all images 100 μm. (B) The BBB permeability in TSP77+/− NVUs is significantly increased compared to isogenic control TSP77 +/+ NVUs (p < 0.01, N = 5).
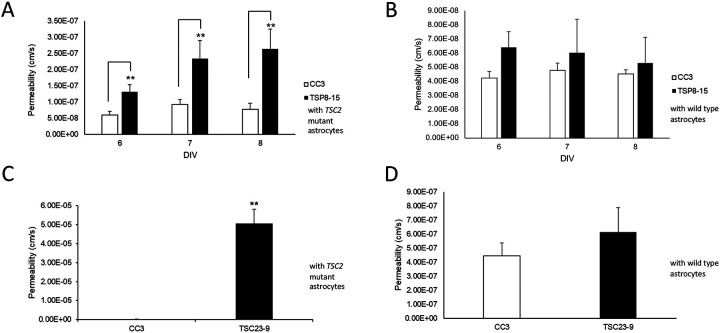
Figure 6. Control astrocytes rescue TSC BBB function.
A) The BBB permeability measured on days 6-8 in vitro (DIV) in TSC2 mutant (TSP8-15) NVUs is significantly higher than the one in control (CC3) NVUs (p < 0.01, N=5). B) The BBB permeability in TSC2 (TSP8-15) mutant NVUs seeded with control (CC3) astrocytes is not significantly different from the BBB permeability measured in control (CC3) NVUs (N=5, p>0.05). C) The BBB permeability in NVUs seeded with BMECs, neurons and astrocytes derived from hiPSC derived from a different TSC patient (TSP23-9) carrying a distinct TSC2 loss of function mutation is also significantly higher compared to control NVUs (CC3 = 2.95 x 10−7 cm/s). (p < 0.001, N=5). D) The presence of control (CC3) astrocytes in otherwise TSC2 mutant (TSP23-9) NVUs rescued BBB permeabilities to levels statistically indistinguishable from that measured in control (CC3) NVUs. (p= N.S., N=5).
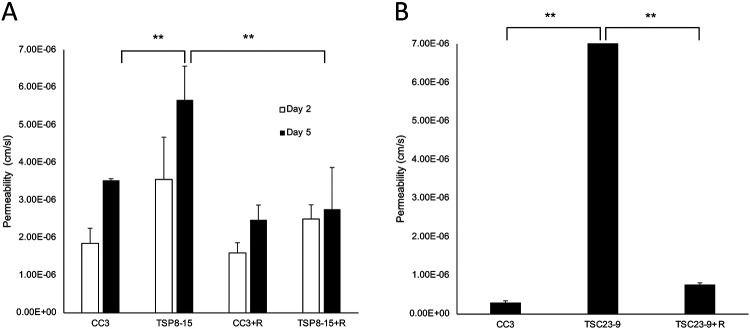
Figure 7. Rapamycin rescues TSC BBB permeability.
A) On day 5 in vitro, BBB permeability of TSP8-15 NVUs is significantly higher than that seen in CC3 NVUs (p< 0.01; N=5). Perfusion of the vascular compartment with rapamycin results in TSC BBB permeabilities statistically significantly reduced compared to vehicle-treated TSC NVUs and statistically indistinguishable from controls (CC3). B) As observed for the TSP8-15 NVUs, the BBB permeability of TSP23-9 NVUs is significantly higher than in CC3 NVUs on 5 day in culture (p < 0.001, N=5). BBB permeabilities were significantly smaller in rapamycin perfused TSP23-9 NVUs than their vehicle treated counterparts and at levels statistically indistinguishable from CC3 levels. The error bar for the TSP23-9 sample is too small to be resolved on this graph (p N.S., N=5).
Given the heightened difference in BBB permeability in NVUs (containing BMECs, neurons and astrocytes) compared to the transwell cultures (only containing BMECs), we hypothesized that a cell type other than BMECs, such as astrocytes contribute to BBB function either directly or through regulation of BMECs. A crucial strength of our experimental approach is the ability to independently generate cell lineages (BMECs, astrocytes, neurons) of different genotypes and then load them in desired combinations (TSC2-wild type or – heterozygous mutant) into individual NVUs. To test the role that astrocytes play in the TSC BBB phenotype, we compared BBB permeabilities in NVUs comprised of TSC2 mutant cells only and NVUs comprised of TSC2 mutant cells except for the astrocytes which were TSC2 wild type. This incorporation of control astrocytes into otherwise TSC2 mutant NVUs rescued BMEC barrier function to levels similar to those measured in control NVUs at all time points measured (Figure 6B). The same observation was made in NVUs populated with BMECs, astrocytes, and neurons differentiated from another human iPSC line (TSP23-9) derived from a different patient with the TSC2 gene harboring a distinct heterozygous TSC2 mutation. As observed for TSP8-15, BBB permeability in the TSP23-9 NVU was increased compared to control NVU (Figure 6C). Importantly, permeability was also rescued by substituting TSC2 wild type (CC3) astrocytes into otherwise TSP23-9 mutant NVU (Figure 6D). Collectively, these data indicate that control astrocytes are sufficient to rescue TSC2 mutant impaired BBB permeability.
A central impact of TSC2 gene mutations is dysregulation of mTOR kinase signaling. Application of rapamycin, an mTOR inhibitor, to the vascular compartment of the NVUs resulted in a significant decrease of the BMEC barrier permeabilities in TSC NVUs when compared to vehicle treated TSC NVUs, but did not affect BBB permeabilities in control NVUs (Figure 7A, 7B). We conclude that the impaired TSC2-mutant BBB function is reversible by a 24 hour treatment with rapamycin (10nM).
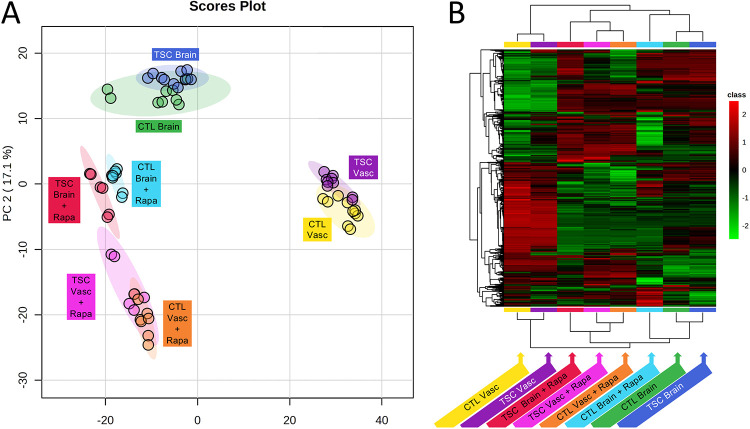
Given the impact of rapamycin on the TSC BBB phenotype, and the role mTOR signaling plays in the regulation of metabolism (Blenis 2017, Saxton and Sabatini 2017) we decided to investigate effects of genotype and rapamycin treatment on the exometabolome of the vascular and neural cell compartments of the NVU. We collected effluent from both the vascular and the neural cell side from control (CC3) and TSC2-mutant (TSP8-15) NVUs and used UPLC-IM-MS to perform unbiased metabolomics analysis. Not surprisingly we observed marked exometabolome differences between compartment types (neural cell versus vascular). Rapamycin treatment appeared to also cause clear differences in the metabolome, while the genotype (wild type versus heterozygous TSC2 mutation) had a smaller effect as shown in a principal component analysis (PCA, Figure 8A) and hierarchical metabolite heatmap (Figure 8B).
Figure 8. Effect of genotype and rapamycin on neural cell and BBB NVU exometabolomes.
A) Principal Component Analysis indicates a pronounced metabolic difference between tissue types (vascular versus brain). In addition, rapamycin treatment for brain and vascular compartments causes a shift in the metabolome. The genotype (wild type, CC3 versus TSC2 heterozygous mutation, TSP8-15) appeared to affect the metabolome to a much lesser degree. B) Hierarchical clustering maps provide global comparisons for differences of metabolite levels > 2-fold with a p value of less than 0.05. Different individual metabolite levels (rows) are clustered by genotype, tissue type and rapamycin treatment (columns). Metabolites are colored according to relative feature abundance across all samples ranging from low (green) to high (red).
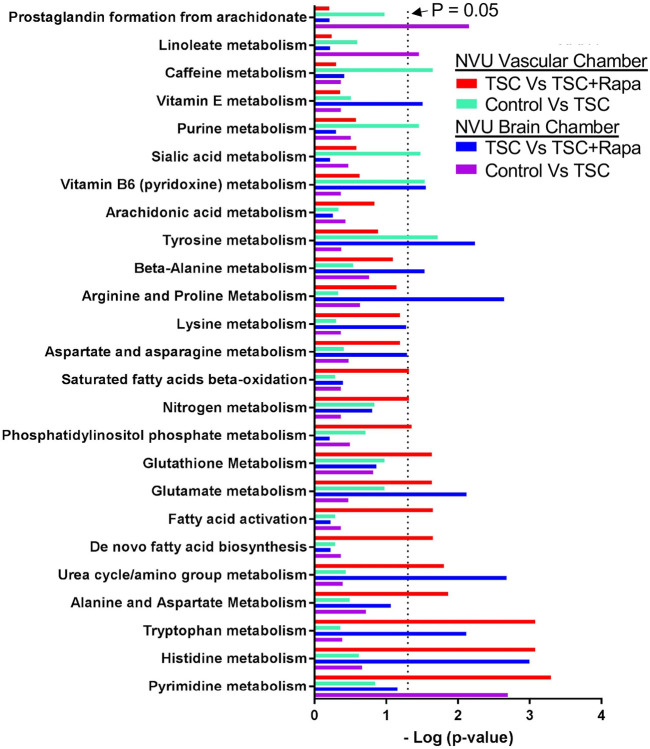
We performed a pathways analysis and show the top 25 pathways that differed between genotype (wild type versus TSC2 heterozygous mutant) and rapamycin treatment (Figure 9). We identified 5 pathways that differed between wild type and TSC2-mutant vascular tissue, these included caffeine-, purine-, sialic acid-, vitamin B6- and tyrosine metabolism. The same comparison between wild type and TSC2-mutated neural compartment revealed 3 different pathways (prostaglandin synthesis, linoleate- and pyrimidine metabolism). Rapamycin affected a total of 10 and 9 pathways in the vascular and neural compartments, respectively. Four pathways were affected in both tissues, they include urea cycle/amino group-, glutamate-, tryptophan- and histidine metabolism (Figure 9).
Figure 9. Metabolic pathways altered by rapamycin, genotype, and tissue type in control (CC3) and TSC (TSP8-15) NVUs.
The top 25 significant response pathways are shown. In each pathway listed, at least one variable (rapamycin treatment, genotype, tissue type) showed a significant difference. Significance was defined as p-values less than or equal to 0.05 in pathways that had four or more metabolites with altered expression levels at a minimal two-fold change.
Finally, we used RNA Seq to look at differences in gene expression between control (CC3) TSC2 mutant (TSP8-15) neural cell compartments. In a comparison of TSP8-15 versus control (CC3), a total of 1143 up-regulated and 769 down-regulated genes were identified (log2foldchange >1 and padj<0.05). The top 100 most differentially expressed genes are listed in Supplementary Figure 4. GO pathways analysis identified multiple pathways pertaining to synapse function and formation, and glutamatergic synapses in particular. In addition, oxidative phosphorylation, ATP synthesis coupled electron chain transport, and respiratory electron chain transport, all pertaining to cellular respiration are also different in wild type and TSC2-heterozygus mutant neural compartments (Table 1).
Table 1. Top 10 up- and down regulated signaling pathways in TSC neural compartment when compared to control.
Based on the normalized enrichment score (NES) we analyzed RNA Seq data and rank the top 10 upregulated and the top 10 down regulated pathways in a comparison of TSC2 mutant versus control neural cell chambers.
| Pathway | setSize | % genes changed |
enrichment score | NES |
|---|---|---|---|---|
| GOCC_GLUTAMATERGIC_SYNAPSE | 306 | 33 | 0.537651966 | 2.32074029 |
| GOMF_EXTRACELLULAR_MATRIX_STRUCTURAL_CONSTITUENT | 140 | 34 | 0.586362661 | 2.3103844 |
| GOBP_LEARNING | 137 | 31 | 0.575619452 | 2.26138159 |
| HP_EPILEPTIC_ENCEPHALOPATHY | 105 | 36 | 0.592389496 | 2.24434551 |
| GOMF_EXTRACELLULAR_MATRIX_STRUCTURAL_CONSTITUENT_CONFERRING_TENSILE_STRENGTH | 40 | 38 | 0.700792853 | 2.23396967 |
| GOBP_PRESYNAPSE_ORGANIZATION | 48 | 38 | 0.666938819 | 2.2094885 |
| GOCC_SYNAPTIC_MEMBRANE | 343 | 33 | 0.504419524 | 2.20462272 |
| GOBP_MEMORY | 113 | 32 | 0.57033983 | 2.19403028 |
| GOBP_REGULATION_OF_SYNAPSE_ASSEMBLY | 90 | 38 | 0.591121008 | 2.19170979 |
| GOBP_COGNITION | 274 | 27 | 0.508160274 | 2.16550655 |
| GOCC_OXIDOREDUCTASE_COMPLEX | 112 | 63 | −0.540457606 | −2.1766616 |
| GOBP_OXIDATIVE_PHOSPHORYLATION | 128 | 62 | −0.525883212 | −2.1880898 |
| GOBP_ORGANIC_ACID_CATABOLIC_PROCESS | 215 | 47 | −0.495686186 | −2.21592 |
| GOBP_ATP_SYNTHESIS_COUPLED_ELECTRON_TRANSPORT | 84 | 70 | −0.576544217 | −2.227409 |
| GOBP_RESPIRATORY_ELECTRON_TRANSPORT_CHAIN | 103 | 61 | −0.555548849 | −2.2274795 |
| GOBP_ANTIGEN_PROCESSING_AND_PRESENTATION_OF_EXOGENOUS_PEPTIDE_ANTIGEN | 32 | 53 | −0.706102979 | −2.2758543 |
| GOCC_MHC_PROTEIN_COMPLEX | 20 | 85 | −0.804026119 | −2.2956252 |
| GOCC_MICROBODY_LUMEN | 44 | 52 | −0.669584925 | −2.326644 |
| GOBP_ANTIGEN_PROCESSING_AND_PRESENTATION_OF_EXOGENOUS_ANTIGEN | 36 | 53 | −0.70246313 | −2.3333633 |
| GOBP_ANTIGEN_PROCESSING_AND_PRESENTATION_OF_PEPTIDE_ANTIGEN | 52 | 42 | −0.672710779 | −2.4213981 |
Gene set analysis revealed an upregulation of K-ras signaling (Supplementary Figure 5A), glutamatergic synapse (Supplementary Figure 5B), and epileptic encephalopathy (Supplementary Figure 5C) when comparing TSC2 mutant (TSP8-15) versus control (CC3) neural cells for RNA expression differences.
Discussion
Neurogenetic disorders underlie many if not most causes of epilepsy, autism, and intellectual disabilities in children. TSC is a prototypical disease for understanding these severe manifestations due to known genetic causes, relevant downstream signaling pathways, and FDA-approved medications to treat some aspects of the disorder. TSC-associated epilepsy is one of the most debilitating aspects given the highly prevalent nature of seizures that are often very hard to control using standard approaches. Many patients in fact require multiple anti-seizure medications and consideration of dietary and surgical therapies.
Assessing mechanism underlying these pathologies has been done in animal model of TSC, and we have extensively modeled TSC using zebrafish and mice (Uhlmann, Wong et al. 2002, Wong, Ess et al. 2003, Kim, Speirs et al. 2011, Carson, Van Nielen et al. 2012, , Carson, Fu et al. 2013, Carson, Kelm et al. 2015). However, assessing cell-type specific contributions requires modifying the genotype of specific neural cell lineages which is very challenging. Here we use combinatorial human cell-based systems which present a unique opportunity to identify the contribution of multiple CNS cell lineages to the formation of the epileptic neural networks in TSC.
Genotype/phenotype relationships and gene dosage are key aspects in the pathogenesis of TSC. We chose to use TSC2 mutant iPSCs as the basis for all patient-derived cells in this study. This reflects the much more frequent occurrence of TSC2 versus TSC1 mutations in patients with TSC, as well as the generally more severe manifestations seen in patients with TSC2 mutations (Au, Williams et al. 2007). Importantly, all mutant cells we used here and showed a defect in the BBB were TSC2 heterozygous. This begins to address a central question in the field: is a homozygous mutation required for disease manifestations? TSC2 homozygous mutations were originally thought to be due to loss of heterozygosity (LOH) with “second hits” being acquired somatically during brain development and post-natal life. This model came from the initial assumption that the TSC1 and TSC2 gene would follow classic tumor suppressor paradigms including LOH. This concept was further supported by the many rodent and zebrafish models we and others have made over the past 10 years (Zeng, Rensing et al. 2011, Carson, Van Nielen et al. 2012, Fu and Ess 2013, Kim, Kowalski et al. 2013, Carson, Kelm et al. 2015). In these animal models, heterozygous loss of Tsc1 or Tsc2 genes generally have very mild or no phenotypes, whereas homozygous Tsc1 or Tsc2 mutant animals usually have severe manifestations. This contradiction between patient-based data from deep DNA sequencing of tubers as compared to animal models was a strong rationale for use of human iPSCs here. The minimal evidence for second hit homozygous mutant cells in the human brain has led to suggestions that heterozygous mutant cells are sufficient to cause disease in humans. More recent work in human brain and organoids (Giannikou, Lasseter et al. 2021, Eichmuller, Corsini et al. 2022) further suggests that LOH may not be required, especially in cells that natively have lower levels of hamartin or tuberin protein. Other groups have presented data that supports a role for heterozygous as well as homozygous TSC2 mutations in the pathogenesis of TSC (Blair, Hockemeyer and Bateup 2018, Winden, Sundberg et al. 2019).
Our studies show that astrocytes play a crucial role in the observed TSC BBB phenotype since control wild type astrocytes were able to rescue the TSC BBB dysfunction. We also recently reported impaired glutamate uptake by TSC as compared to wild type human astrocytes derived from the same iPSC cell lines used here (Miller, Schaffer et al. 2021). Thus, the findings reported here further highlight the critical role of astrocytes in TSC pathogenesis. Prior work in mouse models also implicated astrocyte dysfunction but mainly focused on homozygous mutant cells (Wong, Ess et al. 2003, Zeng, Ouyang et al. 2007). These findings are important, as they emphasize species-specific differences of genotype upon rodent versus human cell-based models of TSC and other related disorders. Another recent report details abnormalities of the mouse BBB and connection to seizures in an astrocyte restricted homozygous knockout of the Tsc1 gene (Guo, Zhang et al. 2023). Interestingly, treatment with RepSox, a TGF-β inhibitor that stabilizes the BBB decreased seizures and increased survival in this well-established mouse model of TSC.
Using metabolomics and RNA Seq, our results further detail changes in many pathways that are impacted by genotype and cell type. Notably, TSC2 heterozygous mutant BBB versus control BBB did not show increased mTOR signaling (data not shown). This is not unexpected, as human cells with heterozygous mutations generally do not show increased mTOR signaling using conventional immunoblotting, though homozygous mutant human cells usually do. Small differences in mTORC1 signaling from human TSC2 heterozygous cells with the very low amount of protein we can extract from an individual NVU however may not be detectable with the sensitivity of current methods. We did see an impact of genotype and rapamycin treatment on the metabolome of both the vascular and brain compartments of the NVU (Figures 8 and 9). Modest alterations in mTOR signaling then seem to be present given the impact of low dose rapamycin. This suggests that patients with TSC have diffusely abnormal BBB as all cells in patients are thought to be at least heterozygous mutant. This may be supported by clinical imaging studies in patients with TSC showing altered Apparent Diffusion Coefficient (ADC) brain imaging in TSC (Garaci, Floris et al. 2004). Other studies show increased inflammatory markers in the brains of patients with TSC, further supporting a potential mechanism of a “leaky” BBB allowing ingress of immune cells to the brain furthering dysfunction (Boer, Jansen et al. 2008, Boer, Troost et al. 2008). Alternatively, the abundance of these inflammatory markers may result from seizures secondarily causing BBB dysfunction. This further validates our approach with human iPSC and microphysiological systems as cell/cell functional interactions can be defined without confounding from seizures seen in most TSC animal models.
Our PCA analysis revealed greater metabolome differences between neural cell and vascular compartments and rapamycin treatment versus vehicle than between TSC2 genotypes. While there is chemical communication across the BBB, the major contributor to metabolic differences between the vascular and neural compartments are likely metabolic differences in cell lineages (BMECs versus neuron and astrocytes) involving many additional cellular processes aside from signaling pathways associated with TSC2. We identified a greater number of metabolic pathways changed by rapamycin than affected by the TSC2 genotype in the vascular- and neural compartments. Assessment of RNA transcriptional profiles of TSC2 wild type and TSC2-mutated neural compartments revealed differentially regulated pathways associated with synapse development and mitochondrial respiration, cellular functions intricately linked to CNS development.
Conclusions
In summary, we found altered BBB function within a human TSC2 heterozygous NVU microphysiological system. Replacement of TSC2-mutated astrocytes with TSC2 wild type astrocytes or treatment with rapamycin was sufficient to rescue the BBB phenotype. These findings have translational and clinical impact as impaired BBB may contribute to neurological disorders and future therapeutics for TSC may be designed to improve BBB function. As rapamycin and related compounds are being used clinically for some aspects of TSC, it is possible that alteration of the BBB permeability may be occurring and further focus on the BBB should yield important data and insights that may have clinical impact.
Supplementary Material
Supplementary Figure 1. iPSC Karyotypes. Karyotypes of all cell lines used were normal.
Supplementary Figure 2. Cortical neuronal cultures differentiated from control- and TSC- iPSC lines express the neuronal markers β3-tubulin and MAP2. Cortical neuronal cultures differentiated from control iPSC lines (A) and TSC iPSC lines (B) show a dense network of β3-tubulin- and MAP2-positive neurites. The fluorescence signals of the images in Fig. 2 were enhanced for optimal visualization of the markers; therefore, comparisons of expression levels between the different iPSC lines are not possible from this Figure. We have not observed any obvious and consistent genotype-dependent difference in the level or distribution of β3-tubulin or MAP2 expression. TSP77+/+ and TSP77+/− are isogenic TSC wild type and TSC2 heterozygous mutated iPSC lines, respectively. The neuronal cultures shown here were differentiated between 67-104 days before seeding into the NVUs. Scale bar is 100 μm.
Supplementary Figure 3. Astrocyte cultures differentiated from control and TSC- iPSC lines express the astrocytic markers GFAP and S100B. Astrocyte cultures differentiated from control iPSC lines and TSC iPSC lines show expression of GFAP and S100B. The expression levels of both GFAP and S100B varies from cell to cell within the same culture. A few cells express detectable levels of GFAP, but no S100B, some cells express S100B, but no GFAP and the majority of cells express both markers. Generally, the expression of S100B in control and TSC astrocytes co-expressing GFAP is lower than in cells expressing S100B only. The signals of the images were enhanced for optimal visualization of the markers and cell morphology; therefore, comparisons of expression levels between the lines are not possible from this Figure. However, we have not observed any obvious and consistent differences in GFAP or S100B expression between genotypes. TSP77+/+ and TSP77+/− are isogenic TSC2 wild type and TSC2 heterozygous mutated iPSC lines, respectively. The astrocyte cultures shown here were differentiated for >200 days. Scale bar is 100 μm.
Supplementary Figure 4. Genes expressed differentially in control (CC3) and TSPC-patient (TSP8-15) derived NVU neural chambers. The top 100 genes sorted by fold-difference with increased (blue-green) or decreased (orange) expression levels in the neural chamber of TSC2 mutant (TSP-15) compared to control (CC3) are shown.
Supplementary Figure 5. Gene set analysis of three gene pathways potentially relevant to TSC pathogenesis. We provide gene set analysis for three pathways that might be relevant to TSC CNS pathogenesis: K Ras signaling (A, NES = 1.69, setSize 178), Glutamatergic Synapse (B, NES = 2.32, setSize: 306) and Epileptic Encephalopathy (C, NES = 2.26, setSize 105).
Acknowledgements
This work was supported in part using the resources of the Center for Innovative Technology at Vanderbilt University. The microfluidic NVUs were fabricated by David K. Schaffer and Clayton M. Britt of the Vanderbilt Microfabricated Technologies Resource. The data that support the findings of this study are available from the corresponding author upon reasonable request. The Vanderbilt Creative Data Solutions Shared Resource (RRID:SCR_022366) performed the RNA-Seq data processing, differential gene expression analysis and Gene Set Enrichment Analysis.
Funding
Research reported in this publication was supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award No. UH3TR002097 and, through Vanderbilt University Medical Center, Award No. UL1TR002243 (K.C.E., J.P.W., M.D.N.). Also supported by the National Institutes of Health (R01NS118580 (R.A.I. and K.C.E.), and U54NS092090 (M.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- ANOVA
ANalysis Of VAriance
- BBB
Blood-Brain Barrier
- BMEC
Brain Microvascular Endothelial-like Cells
- iPSC
induced Pluripotent Stem Cell
- GSEA
Gene Set Enrichment Analysis
- LCHRMS
Liquid Chromatography-High Resolution Mass Spectrometry
- mTOR
mechanistic/mammalian Target Of Rapamycin
- NVU
NeuroVascular Unit
- PCA
Principal Component Analysis
- TEER
TransEndothelial Electrical Resistance
- TSC
Tuberous Sclerosis Complex
Footnotes
Ethics approval and consent to participate-all human samples were obtained under IRB approved studies at Vanderbilt University Medical Center (#080369) or Boston Children’s Hospital (#P00008224).
Availability of data and materials-available upon request
Competing interests-Dr. Mustafa Sahin reports grant support from Novartis, Biogen, Astellas, Aeovian, Bridgebio, and Aucta. He has served on Scientific Advisory Boards for Novartis, Roche, Regenxbio, SpringWorks Therapeutics, Jaguar Therapeutics, and Alkermes.
References
- Armstrong L. C., Westlake G., Snow J. P., Cawthon B., Armour E., Bowman A. B. and Ess K. C. (2017). "Heterozygous loss of TSC2 alters p53 signaling and human stem cell reprogramming." Hum Mol Genet 26(23): 4629–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K. S., Williams A. T., Roach E. S., Batchelor L., Sparagana S. P., Delgado M. R., Wheless J. W., Baumgartner J. E., Roa B. B., Wilson C. M., Smith-Knuppel T. K., Cheung M. Y., Whittemore V. H., King T. M. and Northrup H. (2007). "Genotype/phenotype correlation in 325 individuals referred for a diagnosis of tuberous sclerosis complex in the United States." Genet Med 9(2): 88–100. [DOI] [PubMed] [Google Scholar]
- Bauer H. C., Krizbai I. A., Bauer H. and Traweger A. (2014). ""You Shall Not Pass"-tight junctions of the blood brain barrier." Front Neurosci 8: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baybis M., Yu J., Lee A., Golden J. A., Weiner H., McKhann G. 2nd, Aronica E. and Crino P. B. (2004). "mTOR cascade activation distinguishes tubers from focal cortical dysplasia." Ann Neurol 56(4): 478–487. [DOI] [PubMed] [Google Scholar]
- Bissler J. J., Kingswood J. C., Radzikowska E., Zonnenberg B. A., Frost M., Belousova E., Sauter M., Nonomura N., Brakemeier S., de Vries P. J., Whittemore V. H., Chen D., Sahmoud T., Shah G., Lincy J., Lebwohl D. and Budde K. (2013). "Everolimus for angiomyolipoma associated with tuberous sclerosis complex or sporadic lymphangioleiomyomatosis (EXIST-2): a multicentre, randomised, double-blind, placebo-controlled trial." Lancet 381(9869): 817–824. [DOI] [PubMed] [Google Scholar]
- Blair J. D., Hockemeyer D. and Bateup H. S. (2018). "Genetically engineered human cortical spheroid models of tuberous sclerosis." Nat Med 24(10): 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenis J. (2017). "TOR, the Gateway to Cellular Metabolism, Cell Growth, and Disease." Cell 171(1): 10–13. [DOI] [PubMed] [Google Scholar]
- Boer K., Jansen F., Nellist M., Redeker S., van den Ouweland A. M., Spliet W. G., van Nieuwenhuizen O., Troost D., Crino P. B. and Aronica E. (2008). "Inflammatory processes in cortical tubers and subependymal giant cell tumors of tuberous sclerosis complex." Epilepsy Res 78(1): 7–21. [DOI] [PubMed] [Google Scholar]
- Boer K., Troost D., Jansen F., Nellist M., van den Ouweland A. M., Geurts J. J., Spliet W. G., Crino P. and Aronica E. (2008). "Clinicopathological and immunohistochemical findings in an autopsy case of tuberous sclerosis complex." Neuropathology 28(6): 577–590. [DOI] [PubMed] [Google Scholar]
- Bosworth A. M., Faley S. L., Bellan L. M. and Lippmann E. S. (2017). "Modeling Neurovascular Disorders and Therapeutic Outcomes with Human-Induced Pluripotent Stem Cells." Front Bioeng Biotechnol 5: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. A., Codreanu S. G., Shi M., Sherrod S. D., Markov D. A., Neely M. D., Britt C. M., Hoilett O. S., Reiserer R. S., Samson P. C., McCawley L. J., Webb D. J., Bowman A. B., McLean J. A. and Wikswo J. P. (2016). "Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit." J Neuroinflammation 13(1): 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. A., Faley S. L., Shi Y., Hillgren K. M., Sawada G. A., Baker T. K., Wikswo J. P. and Lippmann E. S. (2020). "Advances in blood-brain barrier modeling in microphysiological systems highlight critical differences in opioid transport due to cortisol exposure." Fluids Barriers CNS 17(1): 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. A., Pensabene V., Markov D. A., Allwardt V., Neely M. D., Shi M., Britt C. M., Hoilett O. S., Yang Q., Brewer B. M., Samson P. C., McCawley L. J., May J. M., Webb D. J., Li D., Bowman A. B., Reiserer R. S. and Wikswo J. P. (2015). "Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor." Biomicrofluidics 9(5): 054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson R. P., Fu C., Winzenburger P. and Ess K. C. (2013). "Deletion of Rictor in neural progenitor cells reveals contributions of mTORC2 signaling to tuberous sclerosis complex." Hum Mol Genet 22(1): 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson R. P., Kelm N. D., West K. L., Does M. D., Fu C., Weaver G., McBrier E., Parker B., Grier M. D. and Ess K. C. (2015). "Hypomyelination following deletion of Tsc2 in oligodendrocyte precursors." Ann Clin Transl Neurol 2(12): 1041–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson R. P., Van Nielen D. L., Winzenburger P. A. and Ess K. C. (2012). "Neuronal and glia abnormalities in Tsc1-deficient forebrain and partial rescue by rapamycin." Neurobiol Dis 45(1): 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M. and Studer L. (2009). "Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling." Nat Biotechnol 27(3): 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran A., Avci H. X., Leist M., Kobolak J. and Dinnyes A. (2016). "Astrocyte Differentiation of Human Pluripotent Stem Cells: New Tools for Neurological Disorder Research." Front Cell Neurosci 10: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Wishart D. S. and Xia J. (2019). "Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis." Curr Protoc Bioinformatics 68(1): e86. [DOI] [PubMed] [Google Scholar]
- Curatolo P., Bombardieri R. and Jozwiak S. (2008). "Tuberous sclerosis." Lancet 372(9639): 657–668. [DOI] [PubMed] [Google Scholar]
- Dolmetsch R. and Geschwind D. H. (2011). "The human brain in a dish: the promise of iPSC-derived neurons." Cell 145(6): 831–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberly A. R., Beebout C. J., Tong C. M. C., Van Horn G. T., Green H. D., Fitzgerald M. J., De S., Apple E. K., Schrimpe-Rutledge A. C., Codreanu S. G., Sherrod S. D., McLean J. A., Clayton D. B., Stratton C. W., Schmitz J. E. and Hadjifrangiskou M. (2020). "Data highlighting phenotypic diversity of urine-associated Escherichia coli isolates." Data Brief 31: 105811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmuller O. L., Corsini N. S., Vertesy A., Morassut I., Scholl T., Gruber V. E., Peer A. M., Chu J., Novatchkova M., Hainfellner J. A., Paredes M. F., Feucht M. and Knoblich J. A. (2022). "Amplification of human interneuron progenitors promotes brain tumors and neurological defects." Science 375(6579): eabf5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ess K. C. (2006). "The neurobiology of tuberous sclerosis complex." Semin Pediatr Neurol 13(1): 37–42. [DOI] [PubMed] [Google Scholar]
- Faley S. L., Neal E. H., Wang J. X., Bosworth A. M., Weber C. M., Balotin K. M., Lippmann E. S. and Bellan L. M. (2019). "iPSC-Derived Brain Endothelium Exhibits Stable, Long-Term Barrier Function in Perfused Hydrogel Scaffolds." Stem Cell Reports 12(3): 474–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost T. S., Jiang L., Lynch R. M. and Zohar Y. (2019). "Permeability of Epithelial/Endothelial Barriers in Transwells and Microfluidic Bilayer Devices." Micromachines (Basel) 10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C. and Ess K. C. (2013). "Conditional and domain-specific inactivation of the Tsc2 gene in neural progenitor cells." Genesis 51(4): 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaci F. G., Floris R., Bozzao A., Manenti G., Simonetti A., Lupattelli T., Curatolo P. and Simonetti G. (2004). "Increased brain apparent diffusion coefficient in tuberous sclerosis." Radiology 232(2): 461–465. [DOI] [PubMed] [Google Scholar]
- Giannikou K., Lasseter K. D., Grevelink J. M., Tyburczy M. E., Dies K. A., Zhu Z., Hamieh L., Wollison B. M., Thorner A. R., Ruoss S. J., Thiele E. A., Sahin M. and Kwiatkowski D. J. (2021). "Correction: Low-level mosaicism in tuberous sclerosis complex: prevalence, clinical features, and risk of disease transmission." Genet Med 23(10): 2022. [DOI] [PubMed] [Google Scholar]
- Guo D., Zhang B., Han L., Rensing N. R. and Wong M. (2023). "Cerebral vascular and blood brain-barrier abnormalities in a mouse model of epilepsy and tuberous sclerosis complex." Epilepsia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi N., Uchida T., Lossin C., Misumi Y., Okada Y., Akamatsu W., Imaizumi Y., Zhang B., Nabeshima K., Mori M. X., Katsurabayashi S., Shirasaka Y., Okano H. and Hirose S. (2013). "A human Dravet syndrome model from patient induced pluripotent stem cells." Mol Brain 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden S. E., Thomson J. A. and Little M. H. (2018). "Simultaneous reprogramming and gene editing of human fibroblasts." Nat Protoc 13(5): 875–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie R. A. and Henske E. P. (2022). "Modeling tuberous sclerosis with organoids." Science 375(6579): 382–383. [DOI] [PubMed] [Google Scholar]
- Kaper F., Dornhoefer N. and Giaccia A. J. (2006). "Mutations in the PI3K/PTEN/TSC2 pathway contribute to mammalian target of rapamycin activity and increased translation under hypoxic conditions." Cancer Res 66(3): 1561–1569. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Kowalski M. L., Carson R. P., Bridges L. R. and Ess K. C. (2013). "Heterozygous inactivation of tsc2 enhances tumorigenesis in p53 mutant zebrafish." Dis Model Mech. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Speirs C. K., Solnica-Krezel L. and Ess K. C. (2011). "Zebrafish model of tuberous sclerosis complex reveals cell-autonomous and non-cell-autonomous functions of mutant tuberin." Dis Model Mech 4(2): 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger D. A., Capal J. K., Curatolo P., Devinsky O., Ess K., Tzadok M., Koenig M. K., Narayanan V., Ramos F., Jozwiak S., de Vries P., Jansen A. C., Wong M., Mowat D., Lawson J., Bruns S., Franz D. N. and Group T. S. R. (2018). "Short-term safety of mTOR inhibitors in infants and very young children with tuberous sclerosis complex (TSC): Multicentre clinical experience." Eur J Paediatr Neurol 22(6): 1066–1073. [DOI] [PubMed] [Google Scholar]
- Krueger D. A., Care M. M., Holland K., Agricola K., Tudor C., Mangeshkar P., Wilson K. A., Byars A., Sahmoud T. and Franz D. N. (2010). "Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis." N Engl J Med 363(19): 1801–1811. [DOI] [PubMed] [Google Scholar]
- Lim J. S., Gopalappa R., Kim S. H., Ramakrishna S., Lee M., Kim W. I., Kim J., Park S. M., Lee J., Oh J. H., Kim H. D., Park C. H., Lee J. S., Kim S., Kim D. S., Han J. M., Kang H. C., Kim H. H. and Lee J. H. (2017). "Somatic Mutations in TSC1 and TSC2 Cause Focal Cortical Dysplasia." Am J Hum Genet 100(3): 454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini V., Schrimpe-Rutledge A. C., Codreanu S. G., Sherrod S. D., McLean J. A., Picton H. M. and Pensabene V. (2021). "Metabolomic Analysis Evidences That Uterine Epithelial Cells Enhance Blastocyst Development in a Microfluidic Device." Cells 10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack F. X., Inoue Y., Moss J., Singer L. G., Strange C., Nakata K., Barker A. F., Chapman J. T., Brantly M. L., Stocks J. M., Brown K. K., Lynch J. P. 3rd, Goldberg H. J., Young L. R., Kinder B. W., Downey G. P., Sullivan E. J., Colby T. V., McKay R. T., Cohen M. M., Korbee L., Taveira-DaSilva A. M., Lee H. S., Krischer J. P., Trapnell B. C., National C. Institutes of Health Rare Lung Diseases and M. T. Group (2011). "Efficacy and safety of sirolimus in lymphangioleiomyomatosis." N Engl J Med 364(17): 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L., Talos D. M., Onda H., Pollizzi K., Rotenberg A., Sahin M., Jensen F. E. and Kwiatkowski D. J. (2007). "A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival." J Neurosci 27(21): 5546–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. R., Schaffer D. K., Neely M. D., McClain E. S., Travis A. R., Block F. E. 3rd, McKenzie J., Werner E. M., Armstrong L., Markov D. A., Bowman A. B., Ess K. C., Cliffel D. E. and Wikswo J. P. (2021). "A bistable, multiport valve enables microformulators creating microclinical analyzers that reveal aberrant glutamate metabolism in astrocytes derived from a tuberous sclerosis patient." Sens Actuators B Chem 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J. J., Ka M., Jung E. M. and Kim W. Y. (2015). "Genes and brain malformations associated with abnormal neuron positioning." Mol Brain 8(1): 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal E. H., Marinelli N. A., Shi Y., McClatchey P. M., Balotin K. M., Gullett D. R., Hagerla K. A., Bowman A. B., Ess K. C., Wikswo J. P. and Lippmann E. S. (2019). "A Simplified, Fully Defined Differentiation Scheme for Producing Blood-Brain Barrier Endothelial Cells from Human iPSCs." Stem Cell Reports 12(6): 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely M. D., Davison C. A., Aschner M. and Bowman A. B. (2017). "From the Cover: Manganese and Rotenone-Induced Oxidative Stress Signatures Differ in iPSC-Derived Human Dopamine Neurons." Toxicol Sci 159(2): 366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely M. D., Litt M. J., Tidball A. M., Li G. G., Aboud A. A., Hopkins C. R., Chamberlin R., Hong C. C., Ess K. C. and Bowman A. B. (2012). "DMH1, a highly selective small molecule BMP inhibitor promotes neurogenesis of hiPSCs: comparison of PAX6 and SOX1 expression during neural induction." ACS Chem Neurosci 3(6): 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely M. D., Xie S., Prince L. M., Kim H., Tukker A. M., Aschner M., Thimmapuram J. and Bowman A. B. (2021). "Single cell RNA sequencing detects persistent cell type- and methylmercury exposure paradigm-specific effects in a human cortical neurodevelopmental model." Food Chem Toxicol 154: 112288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely N. A. Tidball K. C. Ess and Bowman A., Eds. (2011). Induced pluripotent stem cells (iPSCs) - an emerging model system for the study of human neurotoxicology. Neuromethods: Cell Culture techniques. New York, Humana press. [Google Scholar]
- Nityanandam A. and Baldwin K. K. (2015). "Advances in reprogramming-based study of neurologic disorders." Stem Cells Dev 24(11): 1265–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popay T. M., Wang J., Adams C. M., Howard G. C., Codreanu S. G., Sherrod S. D., McLean J. A., Thomas L. R., Lorey S. L., Machida Y. J., Weissmiller A. M., Eischen C. M., Liu Q. and Tansey W. P. (2021). "MYC regulates ribosome biogenesis and mitochondrial gene expression programs through its interaction with host cell factor-1." Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince L. M., Neely M. D., Warren E. B., Thomas M. G., Henley M. R., Smith K. K., Aschner M. and Bowman A. B. (2021). "Environmentally relevant developmental methylmercury exposures alter neuronal differentiation in a human-induced pluripotent stem cell model." Food Chem Toxicol 152: 112178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M., Sobolik T., Schaffer D. K., Samson P. C., Johnson A. C., Owens P., Codreanu S. G., Sherrod S. D., McLean J. A., Wikswo J. P. and Richmond A. (2018). "Engineered microfluidic bioreactor for examining the three-dimensional breast tumor microenvironment." Biomicrofluidics 12(3): 034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset C., Netto C. B. O. and Ashton-Prolla P. (2017). "TSC1 and TSC2 gene mutations and their implications for treatment in Tuberous Sclerosis Complex: a review." Genet Mol Biol 40(1): 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset C., Vairo F., Bandeira I. C., Correia R. L., de Goes F. V., da Silva R. T. B., Bueno L. S. M., de Miranda Gomes M. C. S., Galvao H. C. R., Neri J., Achatz M. I., Netto C. B. O. and Ashton-Prolla P. (2017). "Molecular analysis of TSC1 and TSC2 genes and phenotypic correlations in Brazilian families with tuberous sclerosis." PLoS One 12(10): e0185713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R. A. and Sabatini D. M. (2017). "mTOR Signaling in Growth, Metabolism, and Disease." Cell 169(2): 361–371. [DOI] [PubMed] [Google Scholar]
- Schrimpe-Rutledge A. C., Codreanu S. G., Sherrod S. D. and McLean J. A. (2016). "Untargeted Metabolomics Strategies-Challenges and Emerging Directions." J Am Soc Mass Spectrom 27(12): 1897–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short B., Kozek L., Harmsen H., Zhang B., Wong M., Ess K. C., Fu C., Naftel R., Pearson M. M. and Carson R. P. (2019). "Cerebral aquaporin-4 expression is independent of seizures in tuberous sclerosis complex." Neurobiol Dis 129: 93–101. [DOI] [PubMed] [Google Scholar]
- Smith C. A., O'Maille G., Want E. J., Qin C., Trauger S. A., Brandon T. R., Custodio D. E., Abagyan R. and Siuzdak G. (2005). "METLIN: a metabolite mass spectral database." Ther Drug Monit 27(6): 747–751. [DOI] [PubMed] [Google Scholar]
- Snow J. P., Westlake G., Klofas L. K., Jeon S., Armstrong L. C., Swoboda K. J., George A. L. Jr. and Ess K. C. (2020). "Neuronal modeling of alternating hemiplegia of childhood reveals transcriptional compensation and replicates a trigger-induced phenotype." Neurobiol Dis 141: 104881. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. and Mesirov J. P. (2005). "Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles." Proc Natl Acad Sci U S A 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg M., Tochitsky I., Buchholz D. E., Winden K., Kujala V., Kapur K., Cataltepe D., Turner D., Han M. J., Woolf C. J., Hatten M. E. and Sahin M. (2018). "Purkinje cells derived from TSC patients display hypoexcitability and synaptic deficits associated with reduced FMRP levels and reversed by rapamycin." Mol Psychiatry 23(11): 2167–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball A. M., Neely M. D., Chamberlin R., Aboud A. A., Kumar K. K., Han B., Bryan M. R., Aschner M., Ess K. C. and Bowman A. B. (2016). "Genomic instability associated with p53 knockdown in the generation of Huntington's disease human induced pluripotent stem cells." PLoS.ONE . 11(3): e0150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P. T., Hull C., Chu Y., Greene-Colozzi E., Sadowski A. R., Leech J. M., Steinberg J., Crawley J. N., Regehr W. G. and Sahin M. (2012). "Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice." Nature 488(7413): 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann E. J., Wong M., Baldwin R. L., Bajenaru M. L., Onda H., Kwiatkowski D. J., Yamada K. and Gutmann D. H. (2002). "Astrocyte-specific TSC1 conditional knockout mice exhibit abnormal neuronal organization and seizures." Ann Neurol 52(3): 285–296. [DOI] [PubMed] [Google Scholar]
- Vernetti L., Gough A., Baetz N., Blutt S., Broughman J. R., Brown J. A., Foulke-Abel J., Hasan N., In J., Kelly E., Kovbasnjuk O., Repper J., Senutovitch N., Stabb J., Yeung C., Zachos N. C., Donowitz M., Estes M., Himmelfarb J., Truskey G., Wikswo J. P. and Taylor D. L. (2017). "Functional Coupling of Human Microphysiology Systems: Intestine, Liver, Kidney Proximal Tubule, Blood-Brain Barrier and Skeletal Muscle." Sci Rep 7: 42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel H. J., Patel L. S., Robbins C. A., Emmi A., Yeung R. S. and Schwartzkroin P. A. (2004). "Morphology of cerebral lesions in the Eker rat model of tuberous sclerosis." Acta Neuropathol 108(2): 97–108. [DOI] [PubMed] [Google Scholar]
- Winden K. D., Sundberg M., Yang C., Wafa S. M. A., Dwyer S., Chen P. F., Buttermore E. D. and Sahin M. (2019). "Biallelic Mutations in TSC2 Lead to Abnormalities Associated with Cortical Tubers in Human iPSC-Derived Neurons." J Neurosci 39(47): 9294–9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S., Jewison T., Guo A. C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., Bouatra S., Sinelnikov I., Arndt D., Xia J., Liu P., Yallou F., Bjorndahl T., Perez-Pineiro R., Eisner R., Allen F., Neveu V., Greiner R. and Scalbert A. (2013). "HMDB 3.0--The Human Metabolome Database in 2013." Nucleic Acids Res 41 (Database issue): D801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M., Ess K. C., Uhlmann E. J., Jansen L. A., Li W., Crino P. B., Mennerick S., Yamada K. A. and Gutmann D. H. (2003). "Impaired glial glutamate transport in a mouse tuberous sclerosis epilepsy model." Ann Neurol 54(2): 251–256. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L. G., Han Y. and He Q. Y. (2012). "clusterProfiler: an R package for comparing biological themes among gene clusters." OMICS 16(5): 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L. H., Ouyang Y., Gazit V., Cirrito J. R., Jansen L. A., Ess K. C., Yamada K. A., Wozniak D. F., Holtzman D. M., Gutmann D. H. and Wong M. (2007). "Abnormal glutamate homeostasis and impaired synaptic plasticity and learning in a mouse model of tuberous sclerosis complex." Neurobiol Dis 28(2): 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L. H., Rensing N. R., Zhang B., Gutmann D. H., Gambello M. J. and Wong M. (2011). "Tsc2 gene inactivation causes a more severe epilepsy phenotype than Tsc1 inactivation in a mouse model of tuberous sclerosis complex." Hum Mol Genet 20(3): 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. iPSC Karyotypes. Karyotypes of all cell lines used were normal.
Supplementary Figure 2. Cortical neuronal cultures differentiated from control- and TSC- iPSC lines express the neuronal markers β3-tubulin and MAP2. Cortical neuronal cultures differentiated from control iPSC lines (A) and TSC iPSC lines (B) show a dense network of β3-tubulin- and MAP2-positive neurites. The fluorescence signals of the images in Fig. 2 were enhanced for optimal visualization of the markers; therefore, comparisons of expression levels between the different iPSC lines are not possible from this Figure. We have not observed any obvious and consistent genotype-dependent difference in the level or distribution of β3-tubulin or MAP2 expression. TSP77+/+ and TSP77+/− are isogenic TSC wild type and TSC2 heterozygous mutated iPSC lines, respectively. The neuronal cultures shown here were differentiated between 67-104 days before seeding into the NVUs. Scale bar is 100 μm.
Supplementary Figure 3. Astrocyte cultures differentiated from control and TSC- iPSC lines express the astrocytic markers GFAP and S100B. Astrocyte cultures differentiated from control iPSC lines and TSC iPSC lines show expression of GFAP and S100B. The expression levels of both GFAP and S100B varies from cell to cell within the same culture. A few cells express detectable levels of GFAP, but no S100B, some cells express S100B, but no GFAP and the majority of cells express both markers. Generally, the expression of S100B in control and TSC astrocytes co-expressing GFAP is lower than in cells expressing S100B only. The signals of the images were enhanced for optimal visualization of the markers and cell morphology; therefore, comparisons of expression levels between the lines are not possible from this Figure. However, we have not observed any obvious and consistent differences in GFAP or S100B expression between genotypes. TSP77+/+ and TSP77+/− are isogenic TSC2 wild type and TSC2 heterozygous mutated iPSC lines, respectively. The astrocyte cultures shown here were differentiated for >200 days. Scale bar is 100 μm.
Supplementary Figure 4. Genes expressed differentially in control (CC3) and TSPC-patient (TSP8-15) derived NVU neural chambers. The top 100 genes sorted by fold-difference with increased (blue-green) or decreased (orange) expression levels in the neural chamber of TSC2 mutant (TSP-15) compared to control (CC3) are shown.
Supplementary Figure 5. Gene set analysis of three gene pathways potentially relevant to TSC pathogenesis. We provide gene set analysis for three pathways that might be relevant to TSC CNS pathogenesis: K Ras signaling (A, NES = 1.69, setSize 178), Glutamatergic Synapse (B, NES = 2.32, setSize: 306) and Epileptic Encephalopathy (C, NES = 2.26, setSize 105).
Data Availability Statement
The RNA-Seq data has been annotated and deposited to NCBI GEO with accession ID GSE235862.
Metabolomics data is in the process of being submitted to the NIH Common Fund's National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org. This work is supported by Metabolomics Workbench/National Metabolomics Data Repository (NMDR) (grant# U2C-DK119886), Common Fund Data Ecosystem (CFDE) (grant# 3OT2OD030544) and Metabolomics Consortium Coordinating Center (M3C) (grant# 1U2C-DK119889).