Abstract
Naturally produced membrane vesicles (MVs), isolated from 15 strains of gram-negative bacteria (Citrobacter, Enterobacter, Escherichia, Klebsiella, Morganella, Proteus, Salmonella, and Shigella strains), lysed many gram-positive (including Mycobacterium) and gram-negative cultures. Peptidoglycan zymograms suggested that MVs contained peptidoglycan hydrolases, and electron microscopy revealed that the murein sacculi were digested, confirming a previous modus operandi (J. L. Kadurugamuwa and T. J. Beveridge, J. Bacteriol. 174:2767–2774, 1996). MV-sensitive bacteria possessed A1α, A4α, A1γ, A2α, and A4γ peptidoglycan chemotypes, whereas A3α, A3β, A3γ, A4β, B1α, and B1β chemotypes were not affected. Pseudomonas aeruginosa PAO1 vesicles possessed the most lytic activity.
Many gram-negative bacteria produce external membrane vesicles (MVs) during normal growth (3, 6, 7, 10, 14, 22, 24, 25). During their formation, MVs entrap several periplasmic components; for Pseudomonas aeruginosa these include alkaline phosphatase, phospholipase C, proelastase, protease, and peptidoglycan hydrolase (10, 11, 16). Because several of these components are virulence factors (including the lipopolysaccharide contained in the MV membrane), MVs may be important during the initial phases of infection, as they concentrate such factors and convey them to host tissue (10, 12). The partitioning of peptidoglycan hydrolase into MVs has led to another possibility. This cell wall-degrading enzyme could be used to lyse surrounding dissimilar bacteria in the donor bacterium’s environment, thereby releasing organic compounds for growth. An earlier study showed that MVs from P. aeruginosa PAO1 were capable of lysing Staphylococcus aureus, Escherichia coli, and another Pseudomonas strain (11).
For gram-positive bacteria, MVs attach to the cell wall surface where they break open, liberating the peptidoglycan hydrolase, which digests the underlying peptidoglycan of the wall; lysis ensues at the digestion site (11, 13). MVs lyse gram-negative bacteria by a subtly different mechanism. Here, the MV membrane fuses into the outer membrane of the host bacterium and, in so doing, introduces the MV luminal contents (containing peptidoglycan hydrolase) into the host’s periplasm. Once in the periplasm, the hydrolase is free to diffuse around the murein sacculus and can digest it at a number of different sites, causing multiple-site lysis (11). MVs are also able to adhere to S-layered gram-positive bacteria, where the peptidoglycan hydrolase migrates through the S-layer and attacks the underlying wall (13). S-layers composed of either protein or glycoprotein and which are arranged in oblique, square, or hexagonal lattice formats are unable to inhibit MV-mediated lysis (13).
Even though the evidence is growing that a number of different gram-negative bacteria produce MVs and that MVs are able to lyse other bacteria, most information on MVs other than those of P. aeruginosa is sketchy (12). It is now time to develop more information to increase the repertoire of MV-releasing bacteria and the lytic capability of their MVs. To this end, we have isolated MVs from 15 different strains of gram-negative bacteria and tested them for lytic behavior against 17 different gram-positive and gram-negative species possessing 11 different peptidoglycan chemotypes according to the classification of Schleifer and Kandler (20).
Bacterial strains.
The following strains were obtained from the American Type Culture Collection (ATCC), Rockville, Md.: Bacillus subtilis ATCC 6051, Deinococcus radiodurans ATCC 13939, Klebsiella pneumoniae ATCC 13883, Lactobacillus fermentum ATCC 11739, Lactococcus lactis ATCC 7962, Microbacterium imperiale ATCC 8365, Microbacterium laevaniformans ATCC 15953, S. aureus ATCC 25923, and Staphylococcus sp. strain ATCC 155. Two strains were obtained from the Czech Collection of Microorganisms, Brno, Czech Republic: Brachybacterium conglomeratum CCM2134 and Luteococcus japonicus CCM2142. Escherichia coli K-12 and K263, Enterobacter agglomerans UG-615, Citrobacter freundii UG-455, Klebsiella sp. strain UG-561, Micrococcus luteus UG-B32, Morganella morganii UG-326, Proteus vulgaris UG-355, P. aeruginosa PAO1, Pseudomonas trifolii UG-393, Salmonella arizonae UG-612, Salmonella cholerae-suis UG-316, Salmonella pullorum UG-147, Serratia marcescens UG-159, Shigella flexneri M90T, Streptococcus viridans UG-478, and Streptomyces griseus UG-A36 are from our departmental culture collection (UG strains), many of which were originally from the ATCC, the CCM, or the Ontario Veterinary College Animal Clinic. Mycobacterium kansasii ATCC 35775, Mycobacterium phlei 425, Mycobacterium smegmatis, and Mycobacterium fortuitum have been described in previous publications (17, 18). This group of gram-positive and gram-negative bacteria was chosen for two prime reasons: to be representative of (i) target cell peptidoglycan chemotypes in order to determine a bacteriolysis range and (ii) a broad spectrum of gram-negative genera in order to determine the MV producer range. The MV producer range is not meant to be all inclusive since other gram-negative bacteria are known to produce MVs (12). For the present study, Table 1 lists our strains, the peptidoglycan chemotypes, and the MV producers.
TABLE 1.
Lytic effects of MVs on bacterial species with different peptidoglycan chemotypes
| Strain | Peptidoglycan chemotypeb | Lytic effect of MVs froma:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli DH5α | E. coli K-12 | S. pullorum | S. arizonae | S. cholerae-suis | E. agglomerans | P. vulgaris | S. marcescens | Klebsiella strain UG-561 | K. pneumoniae | S. flexneri | C. freundii | M. morganii | P. trifolii | P. aeruginosa PAO1 | ||
| B. subtilis | A1α | + | + | + | ++ | ++ | + | + | ++ | ++ | ++ | ++ | ||||
| S. viridans | A1α | |||||||||||||||
| P. aeruginosa PAO1 | A1γ | +++ | +++ | ++ | ++ | ++ | ++ | +++ | ++ | + | ++ | ++++ | + | + | + | |
| E. coli K-12 | A1γ | ++ | + | +++ | ++ | ++ | ++ | ++ | ++ | +++ | ++ | +++ | ||||
| M. smegmatis | A1γ | ++ | ++ | ++ | +++ | ++ | +++ | +++ | +++ | ++ | ++ | ++++ | ||||
| M. phlei | A1γ | + | ++ | ++ | ++ | + | ++ | ++ | ++ | ++ | + | +++ | ||||
| M. luteus | A2α | + | + | ++ | ||||||||||||
| S. aureus | A3α | |||||||||||||||
| Staphylococcus sp. | A3α | |||||||||||||||
| D. radiodurans | A3β | |||||||||||||||
| S. griseus | A3γ | |||||||||||||||
| L. japonicus | A3γ | |||||||||||||||
| L. lactis | A4α | + | + | + | + | + | + | + | + | ++ | ||||||
| L. fermentum | A4β | |||||||||||||||
| B. conglomeratum | A4γ | + | + | ++ | ++ | + | ++ | ++ | ++ | ++ | +++ | |||||
| M. laevaniformans | B1α | |||||||||||||||
| M. imperiale | B1β | |||||||||||||||
Lytic effects were measured by diameters of the clear zones produced on agar plates: +, <1 cm; ++, 1 to 1.2 cm; +++, 1.2 to 1.4 cm; ++++, >1.4 cm. Negative results are not shown.
Classification of peptidoglycan chemotypes of bacteria is based on reference 20.
Occurrence of MVs.
All of the gram-negative bacteria used in our study produced MVs when grown in the appropriate broth medium (as listed in the ATCC culture directions) or in media typically used in our laboratory for nonfastidious cultures (e.g., Trypticase soy broth [Difco] is often used for the growth of E. coli, P. vulgaris, P. aeruginosa, and S. marcescens). These MVs could be isolated by a combination of differential centrifugation and filtration to separate them from the cells and ultracentrifugation to concentrate the MVs (10). Typically, 1 liter of broth produced approximately 0.5 to 1.0 mg (wet weight) of MVs derived from 150,000 × g pellets (10) of each bacterial strain.
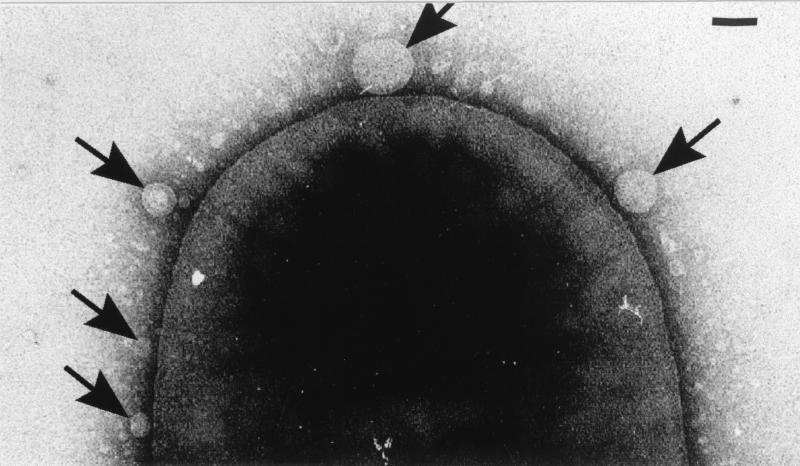
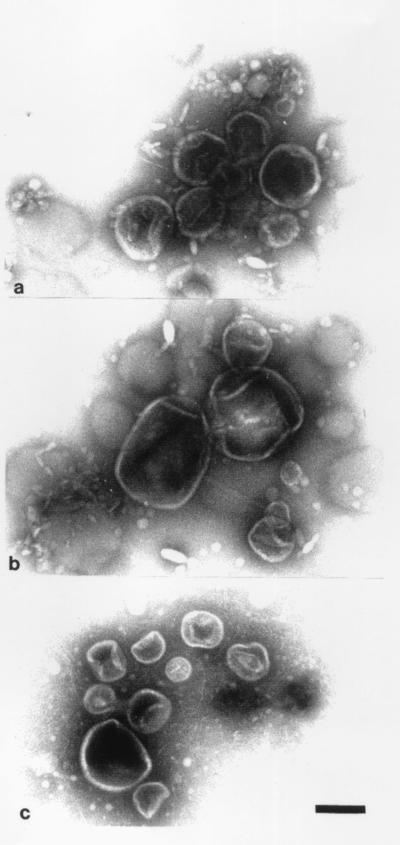
MV formation could be monitored by electron microscopy and was similar to that previously seen with P. aeruginosa (10, 12). By negative staining, it could be seen that the outer membrane blebbed outwards (Fig. 1) until an MV had separated from the cell. The isolated MVs from the various strains revealed pliable spherically shaped vesicles about 50 to 250 nm in diameter which were easily deformed by surface tension during drying in the negative stain (Fig. 2). Yet, even with this distortion, it was apparent that MVs from each strain possessed a relatively constant diameter (those of E. coli [Fig. 2a] and P. aeruginosa [Fig. 2c] were ∼100 to 150 nm, whereas those of S. marcescens were ∼150 to 250 nm [Fig. 2b] and were amongst the largest of all MV preparations). Freeze-substitution and conventional embeddings (8, 10) confirmed that the MVs were bilayered structures and that during their formation, periplasm was entrapped (10, 12). Immunogold labelling for the major 26-kDa peptidoglycan hydrolase in the MVs produced by P. aeruginosa confirmed its presence, as was first suggested by Li et al. (16). It therefore appeared that MV formation in all strains used in this study was consistent with that in earlier studies (10, 12, 16).
FIG. 1.
Transmission electron micrograph of MVs forming on the surface of S. marcescens (arrows). The cell has been negatively stained with 2% (wt/vol) uranyl acetate and imaged in a Philips EM300 electron microscope operating under standard conditions at 60 kV. Bar = 100 nm.
FIG. 2.
MVs isolated from E. coli K-12 (a), S. marcescens (b), and P. aeruginosa PAO1 (c) which have been negatively stained and imaged as outlined in the legend to Fig. 1. All panels are the same magnification. Bar = 100 nm.
Killing of bacteria by MVs.
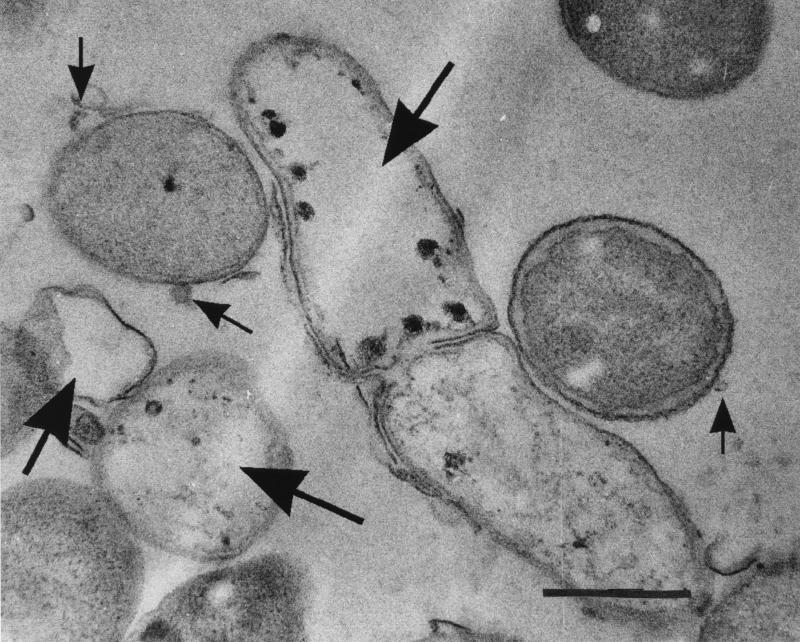
Killing was measured by using an agar plate assay in which suspensions of the various host bacteria were mixed with molten agar (45°C) and poured into plastic petri dishes. For this assay, it was important that the host bacteria were not actively growing and dividing (i.e., peptidoglycan turnover was greatly reduced so that lesions in cell walls produced by MVs could not be readily repaired) but that the bacteria were still alive and healthy. For this reason, no nutrients were added to the agar but only 20 mM sodium phosphate buffered saline, pH 7.0, was used. Under these conditions at 37°C, bacteriolysis in control plates was not seen after overnight incubation and the addition of a nutrient broth to the plate induced growth. When 10 μg of MVs was added (this volume contained 10 μg of protein) to the plates, a circular zone of lysis was often observed (Table 1). Electron microscopy of bacteria in these zones of lysis supported the view that the bacteria which were subjected to the MVs had been killed (Fig. 3). In addition when the Gram stain was performed on gram-positive bacteria in these zones, these bacteria had become gram negative. Attempts to culture bacteria (both gram positive and gram negative) were negative and proved that rampant lysis had occurred.
FIG. 3.
Cell of E. coli K-12 which was lysed by MVs from P. aeruginosa PAO1 during the agar plate assay. This preparation was fixed with 2% (wt/vol) glutaraldehyde followed by 2% (wt/vol) osmium tetroxide, embedded in LR white, and thin sectioned. The sections were stained with uranyl acetate and lead citrate and imaged as outlined in the legend to Fig. 1. The large arrows point to lysed cells, whereas the small arrows point to MVs that have the dimensions of those from P. aeruginosa. Bar = 1 μm.
Electron microscopy was used in an effort to determine (in a physical sense) how the MVs killed the bacteria. In all observations, the claims of Kadurugamuwa and Beveridge (11) were supported. For gram-positive cells, the cell wall immediately underlying an attached MV was digested and these “single-hit” sites lysed the cell in a manner similar to that detected by Kadurugamuwa et al. (13). For gram-negative bacteria, MVs attached to and fused into the outer membrane, liberating their luminal contents into the periplasm of the targeted cells. For this reason, multiple sites on the murein sacculus (as seen in reference 10) were digested and multisite lysis occurred.
Lytic effect of MVs on bacteria possessing different peptidoglycan chemotypes.
The lytic effects of various MVs according to our agar plate assay are shown in Table 1. Notably, all A1γ strains (including those with cell walls containing such complicated secondary polymers as mycolic acids, lipoarabinomannan, and arabinogalactan [e.g., Mycobacterium spp.]) were significantly lysed by MVs isolated from most gram-negative strains. In addition, A1α (B. subtilis ATCC 6051), A4α (L. lactis ATCC 7962), and A4γ (B. conglomeratum CCM2134) bacteria were also lysed. There was slight lysis of M. luteus (A2α) by MVs from S. marcescens, P. aeruginosa, and P. trifolii. Overall, MVs from P. aeruginosa appeared to be the most potent and had the widest killing spectrum. MVs from E. agglomerans, Klebsiella strain 561, C. freundii, and M. morganii had the least activity. As expected, when donor bacteria were subjected to their own MVs in the agar plate assay, there was very little lysis (see E. coli K-12 and P. aeruginosa PAO1 in Table 1). A previous study, using a cell suspension system, showed similar trends in that MVs from P. aeruginosa PAO1 did not lyse the donor strain (11). Interestingly, a dissimilar strain of P. aeruginosa was more sensitive to the PAO1 MVs in the 1996 study, which emphasizes just how stringent autolysin regulation must be (11).
It was clear that the MVs used in our present study did not lyse bacteria possessing peptidoglycan chemotypes far removed from the A1γ chemotype of the donor strains. Of all the chemotypes attacked by the MVs, A1γ was by far the most easily attacked. This is not surprising since MVs encapsulate peptidoglycan hydrolases, which would (presumably) be components of the donor bacterium’s autolysin system. In fact, a small 22-kDa endotransglycosylase which is bound to the outer membrane by means of a lipid substituent is part of E. coli’s autolysin system (15). Autolysins are used during peptidoglycan metabolism as the parent cell grows and divides. It is possible that MVs could contain small quantities of peptidoglycan digestion products as well as the autolysins, although these products were not detected in a separate study of MVs (13). When MVs attach to foreign cells and liberate their luminal constituents to a substrate, the substrate must be recognizable to the released enzymes and A1γ peptidoglycan would be the closest fit. The bacteria with A2, A3, A4, and B1 chemotypes would have dissimilar peptidoglycans which may not be recognized by the MV enzymes; in this case, there would be no cell wall digestion and no lysis.
Some lysis was seen with A1α and A2α strains (Table 1). These are the chemotypes used in our study which are most similar to A1γ; in A1α, an l-lysine replaces meso-diaminopimelic acid at position 3 of the peptide stem and is directly linked to the terminal d-alanine at position 4 on the adjacent peptide stem. The A2α chemotype frequently uses the l-lysine and d-alanine at the same positions for cross-linking, but a five-amino-acid linkage unit is used to join the two stems together (e.g., M. luteus has a —d-Ala←l-Lys(Gly)←d-Glu←l-Ala— linkage unit [20]). These differences in peptidoglycan structure reduced the lytic power of MVs but did not entirely retard it.
Chemotypes A3α to B1β are very different from A1γ (20). For example, cross-bridging can occur at position 2 instead of position 3 of the peptide stem, peptide stem chemistry can be different, cross-bridging linker units are common, and cross-linkage percentages vary (20). It was not surprising that cells possessing these “foreign” peptidoglycan chemotypes were resistant to MVs from A1γ-chemotype donor bacteria (Table 1). These results point (indirectly) to the conclusion that distinct peptidoglycan hydrolases from donor autolysin systems are responsible for the killing action on targeted cells and that their most effective action is on bacteria with similar peptidoglycans.
The lysis of L. lactis (A4α) and B. conglomeratum (A4γ) was unexpected and surprising (Table 1). Although the two chemotypes have meso-diaminopimelic acid at position 3 in the peptide stems (as do A1γ strains), both chemotypes also possess quite dissimilar interpeptide cross-linking units (20). It is possible that other hydrolytic enzymes from the MVs also came into play during lysis of these bacteria. For example, protease, lipase, or phospholipase C (10) could have directly affected the underlying plasma membrane of these susceptible bacteria or could have digested essential secondary polymers in the cell wall (some secondary polymers [e.g., those in Enterococcus hirae and Streptococcus pneumoniae] [4, 5, 9, 21] regulate autolytic activity).
Detection of hydrolysis of peptidoglycan by MV peptidoglycan hydrolases by using an SDS-PAGE zymogram system.
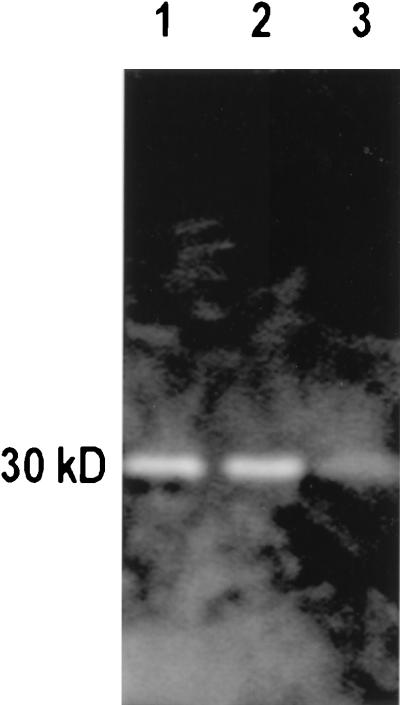
In previous publications we have used a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) zymogram system to detect and separate peptidoglycan hydrolases (1, 11, 16, 23). When used for MVs, the MVs are solubilized in boiling 2% (wt/vol) SDS and run on SDS-PAGE gels containing isolated peptidoglycan sacculi. After the hydrolases have been separated into distinct bands in the gel by electrophoresis, they are renatured and allowed to digest the incorporated sacculi; the gels are then stained to emphasize the regions of clearing, which are due to peptidoglycan hydrolysis (1, 16, 23). When MVs from each of the donor bacteria were run in gels containing A1γ peptidoglycan sacculi from P. aeruginosa, one or more zymogram bands were seen (Fig. 4). This helps confirm that the MVs contain peptidoglycan hydrolases.
FIG. 4.
Typical zymogram showing how peptidoglycan hydrolases can be separated by SDS-PAGE and identified by their digestion of murein sacculi within the gel. In this case, sacculi from E. coli K-12 are the substrates in the gel and the SDS-soluble components of MVs from S. marcescens UG-159 (lane 1), P. aeruginosa PAO1 (lane 2), and S. pullorum UG-147 (lane 3) have been run into the gel. Once separated from one another, the peptidoglycan hydrolases were renatured and the gels were stained according to Bernadsky et al. (1). All of the MVs from the three species possessed ∼30-kDa hydrolases that digested the E. coli murein sacculi.
Why do bacteria secrete MVs into their surroundings?
This study suggests that the production of MVs by gram-negative bacteria is a natural and common phenomenon. Although the present data is derived from cultures grown in flasks under laboratory conditions, we commonly see MVs blebbing from bacteria taken directly from field sites when soil, sediment, animal model, freshwater, and marine systems are sampled or when laboratory biofilm simulations are developed (2) and electron microscopy is performed. The present study suggests that most MVs contain hydrolytic enzymes and that these include peptidoglycan hydrolases. Previous studies have shown that, as MVs bleb from the cell surface, they entrap periplasmic constituents within their lumen. Taken together, it is likely that MVs are structures which gram-negative bacteria use to partition and concentrate periplasmic components so that they can be sent (and maybe even targeted) to the environment to perform particular functions. Certainly, bacteria secrete a number of soluble extracellular enzymes by various secretion routes (19), but diffusion must quickly dilute enzyme concentrations as they move from the cell. Those enzymes contained in MVs are concentrated and remain so until they reach a particulate substrate. It is possible that MV systems have evolved over time to increase delivery efficiency. In the specific case of our study, it is probable that peptidoglycan hydrolases are included as MV constituents for the purpose of lysing neighboring dissimilar cells in the environment so as to increase the nutrient load for the donor cell. Since these peptidoglycan hydrolases appear to be normal components of the donor cell’s autolysin system (16), their prime extracellular activity is on neighboring bacteria possessing A1γ or closely related peptidoglycans. In the harsh reality of the microbial world, little quarter is given to neighboring cells and “predatory MVs” may give certain gram-negative bacteria an advantage over their non-MV-producing counterparts.
Acknowledgments
We thank Zdena Pácová for providing CCM strains, N. Allen for help with Fig. 1, and Diane Moyles and Robert Harris of our laboratory for their technical assistance.
This work was supported by operating grants to T.J.B. and A.J.C. from the Canadian Bacterial Diseases Network, which is funded as a National Center of Excellence. The electron microscopy was performed in the NSERC Guelph Regional STEM Facility, which is partially funded by a Natural Sciences and Engineering Research Council of Canada Major Facility Access grant to T.J.B.
REFERENCES
- 1.Bernadsky G, Beveridge T J, Clarke A J. Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of select gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1994;176:5225–5232. doi: 10.1128/jb.176.17.5225-5232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beveridge T J, Makin S A, Kadurugamuwa J L, Li Z S. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20:291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee S N, Das J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J Gen Microbiol. 1967;49:1–11. doi: 10.1099/00221287-49-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland R F, Daneo-Moore L, Wicken A J, Shockman G D. Effect of lipoteichoic acid and lipids on lysis of intact cells of Streptococcus faecalis. J Bacteriol. 1976;127:1582–1584. doi: 10.1128/jb.127.3.1582-1584.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleveland R F, Höltje J-V, Wicken A J, Tomasz A, Daneo-Moore L, Shockman G D. Inhibition of bacterial wall lysis by lipoteichoic acids and related compounds. Biochem Biophys Res Commun. 1975;67:1128–1135. doi: 10.1016/0006-291x(75)90791-3. [DOI] [PubMed] [Google Scholar]
- 6.Devoe I W, Gilchrist J E. Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J Exp Med. 1973;138:1156–1166. doi: 10.1084/jem.138.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorward D E, Garon C F, Judd R C. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham L L, Harris R, Villiger W, Beveridge T J. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J Bacteriol. 1991;172:1623–1633. doi: 10.1128/jb.173.5.1623-1633.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höltje J-V, Tomasz A. Lipoteichoic acid: a specific inhibitor of autolysin activity in Pneumococcus. Proc Natl Acad Sci USA. 1975;72:1690–1694. doi: 10.1073/pnas.72.5.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadurugamuwa J L, Beveridge T J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadurugamuwa J L, Beveridge T J. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadurugamuwa J L, Beveridge T J. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother. 1997;40:615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- 13.Kadurugamuwa J L, Mayer A, Messner P, Sára M, Sleytr U B, Beveridge T J. S-layered Aneurinibacillus and Bacillus spp. are susceptible to the lytic action of Pseudomonas aeruginosa membrane vesicles. J Bacteriol. 1998;180:2306–2311. doi: 10.1128/jb.180.9.2306-2311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo K, Takade A, Amako K. Release of outer membrane vesicles from Vibrio cholerae and Vibrio parahaemolyticus. Microbiol Immunol. 1993;37:149–152. doi: 10.1111/j.1348-0421.1993.tb03192.x. [DOI] [PubMed] [Google Scholar]
- 15.Kraft A R, Templin M, Höltje J-V. Membrane-bound lytic endotransglycosylase in Escherichia coli. J Bacteriol. 1998;180:3441–3447. doi: 10.1128/jb.180.13.3441-3447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z S, Clarke A J, Beveridge T J. A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division, and secretion in surface membrane vesicles. J Bacteriol. 1996;178:2479–2488. doi: 10.1128/jb.178.9.2479-2488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul T R, Beveridge T J. Reevaluation of envelope profiles and cytoplasmic ultrastructure of mycobacteria processed by conventional embedding and freeze-substitution protocols. J Bacteriol. 1992;174:6508–6517. doi: 10.1128/jb.174.20.6508-6517.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul T R, Beveridge T J. Preservation of surface lipids and ultrastructure of Mycobacterium kansasii using freeze-substitution. Infect Immun. 1994;62:1542–1550. doi: 10.1128/iai.62.5.1542-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–471. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 131–166. [Google Scholar]
- 22.Wai S M, Takade A, Amako K. The release of outer membrane vesicles from the strains of enterotoxigenic Escherichia coli. Microbiol Immunol. 1995;39:451–456. doi: 10.1111/j.1348-0421.1995.tb02228.x. [DOI] [PubMed] [Google Scholar]
- 23.Watt S R, Clarke A J. Initial characterization of two extracellular autolysins from Pseudomonas aeruginosa PAO1. J Bacteriol. 1994;176:4784–4789. doi: 10.1128/jb.176.15.4784-4789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitmire W M, Garon C F. Specific and nonspecific responses of murine B cells to membrane blebs of Borrelia burgdorferi. Infect Immun. 1993;61:1460–1467. doi: 10.1128/iai.61.4.1460-1467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wispelwey B, Hansen E J, Scheld M. Haemophilus influenzae outer membrane vesicles induced blood-brain barrier permeability during experimental meningitis. Infect Immun. 1989;57:2559–2562. doi: 10.1128/iai.57.8.2559-2562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]