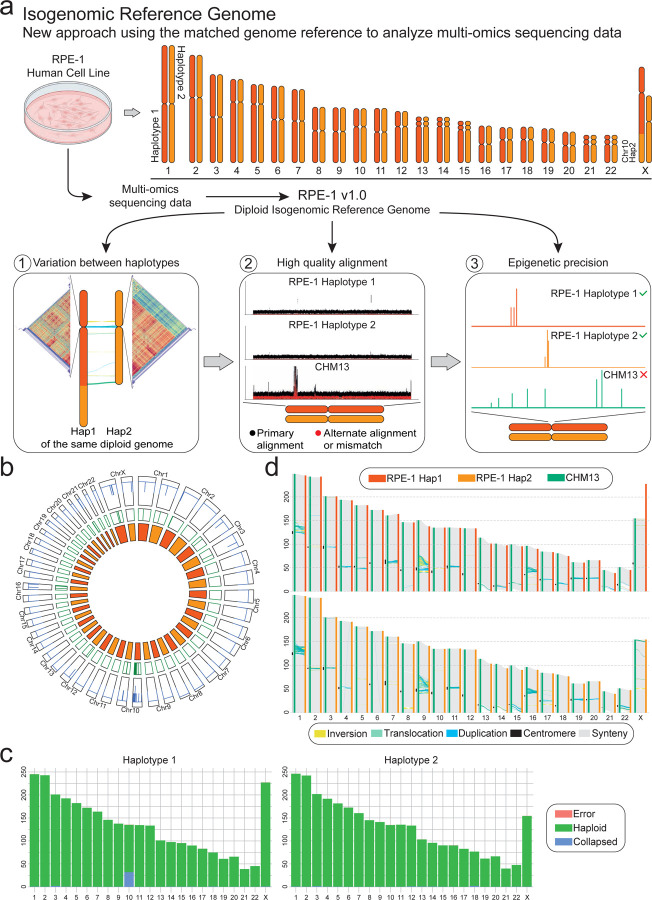
Figure 1. The diploid reference genome of RPE-1 enables faithful multi-omics data analysis.
(a) Schematic of the workflow from in vitro experiments to multi-omics data analyzed using the isogenomic reference genome (top). The advantages of this model are 1) the ability to assess the variation between haplotypes, 2) improved whole-genome alignment, and 3) faithful phased epigenomic peaks calling (bottom). (b) Regional and structural assembly errors identification using alignment Clipping information. Circos plot shows Assembly Quality Indicators (AQI), Clip-based Regional Errors (CREs) at precise location, for each chromosome of both haplotypes (from outside to inside). (c) Synteny between all chromosomes of RPE-1 haplotypes (Haplotype 1, Hap 1 and Haplotype 2, Hap 2) and CHM13 (haploid). RPE-1 shows comparable chromosome length to CHM13, with the majority of the variation in centromeric and pericentromeric regions. (d) Reliability of the RPE-1 chromosomes using read mapping. Regions flagged as ‘haploid’ are error-free (green).