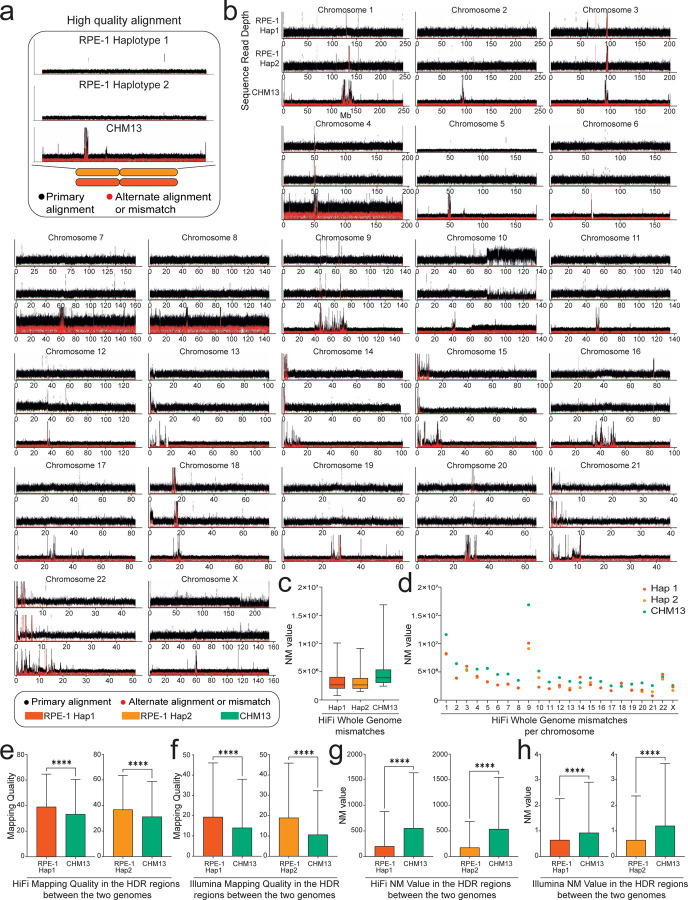
Figure 3. The isogenomic reference genome improves reads alignment.
(a) Read-depth profiles over RPE-1 Hap 1, RPE-1 Hap 2, and CHM13. The black and red dots represent, respectively, the coverage of the first and second most frequent alleles. (b) HiFi read-depth profile for all chromosomes. Using the RPE-1 haplotypes as reference improves the primary alignments compared to CHM13. The differences in coverage for long arm of chromosome 10 in RPE-1 genome are due to regional duplication (Methods). (c) Sequencing reads from a different batch of RPE-1 show lower edit distance (NM value) when aligned against the matched reference compared to CHM13, consistent across all chromosomes (d). HiFi and Illumina reads from High Diverged Regions (HDRs) show statistically significant increase in mapping quality (e-f), and lower NM value (g-h), when aligned against the RPE-1 haplotypes compared to other reference. p-values are from the student t-test (non-significant (ns) = p > 0.05, * p ≤ 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001).