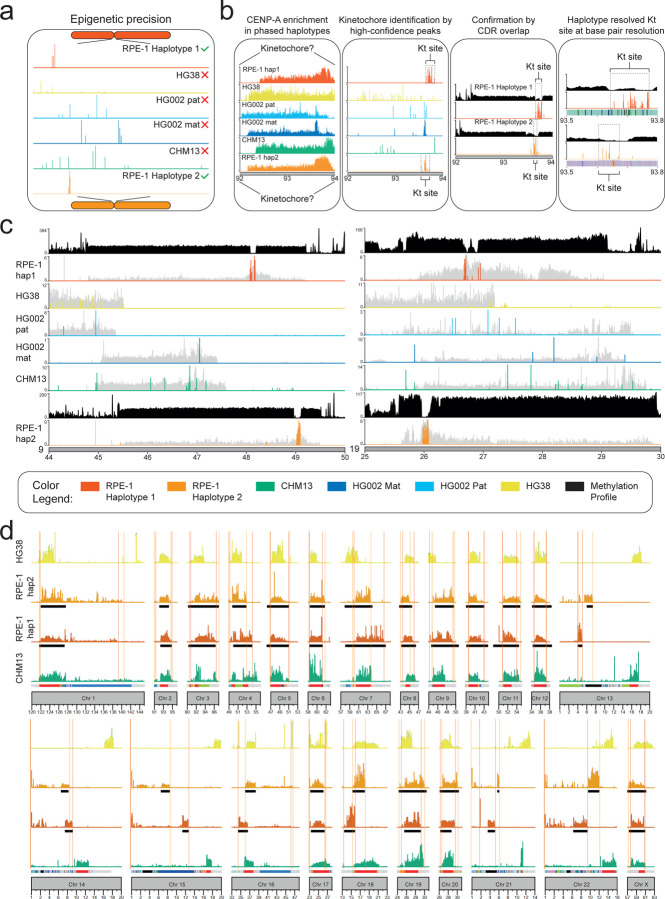
Figure 4. The isogenomic reference genome identifies high-confidence epigenomic landscapes.
(a) Phased peak calling improves when using the isogenomic genome as reference. (b) Pipeline describing the mapping of CENP-A CUT&RUN enrichment to reveals the precise kinetochore (KT) position with high confidence. High confidence CENP-A peak map to a single genomic location that differs between haplotypes defining the kinetochore site only when the isogenomic reference is used but not with any other reference tested, giving scattered or low significance peaks. This is confirmed by methylation analysis to overlap with the CDR. Kinetochore sequence and structure can be defined at base pairs resolution. (c) Example of methylation track (defining the CDR) and CENP-A high-confidence peaks (defining the kinetochore) for chromosomes 9 and 19 only using RPE-1 as reference but not with CHM13, HG38, HG002 mat or pat genomes. (d) The spread of CENP-A occupancy changes between haplotypes for all chromosomes, and when using different genomes raising questions on the validity of non-matched references for mapping reads to such polymorphic loci.