Abstract
Changes in the amount of oligopeptide binding protein (OppA) in spontaneous kanamycin-resistant mutants of Escherichia coli were investigated. Among 20 colonies obtained from 108 cells cultured in the presence of 20 μg of kanamycin/ml, 1 colony had no detectable OppA and 7 colonies were mutants with reduced amounts of OppA. Sensitivity of wild-type cells to kanamycin increased slightly by transformation of the oppA gene, but the sensitivity of the mutants increased greatly by the transformation. A mutant with no OppA was found to be a nonsense mutant of the oppA gene at amino acid position 166. In a mutant having a reduced level of OppA, the reduction was due to the decrease in OppA synthesis at the translational level. These mutants were also resistant to other aminoglycoside antibiotics, including streptomycin, neomycin, and isepamicin. Isepamicin uptake activities decreased greatly in these two kinds of mutants. The results support the proposition that aminoglycoside antibiotics are transported into cells by the oligopeptide transport system, and that transport is an important factor for spontaneous resistance to aminoglycoside antibiotics.
Uptake of aminoglycoside antibiotics is a complex process that is still a matter of controversy (7, 21). Streptomycin is thought to be taken up by Escherichia coli by a process that may be subdivided into three consecutive phases: first, a rapid electrostatic binding to the cell; second, a slow rate of accumulation; and third, a much-enhanced rate of accumulation. Since there are several reports that aminoglycoside antibiotics are accumulated in E. coli by an active transport system (1, 2, 6), the second phase (a slow rate of accumulation) may involve the active transport system. The third phase, a much-enhanced rate of accumulation, may be explained by membrane permeabilization caused by the insertion of mistranslated proteins into the cytoplasmic membrane (3, 8). Another possibility has also been suggested for the third phase: the enhanced streptomycin uptake may involve the induction of a polyamine transport system by streptomycin, which can be utilized by streptomycin itself (10).
We isolated three clones carrying polyamine transport genes (pPT104, pPT79, and pPT71) and characterized them (9, 13, 16, 23). Using these three clones, we showed that aminoglycoside antibiotics do not up-regulate the polyamine transport system (15). We also proposed that the oligopeptide transport system is a candidate for the second phase, a slow rate of accumulation. This was based on the finding with E. coli DR112 (18) that sensitivity to aminoglycoside antibiotics increased due to the highly expressed oligopeptide binding protein (OppA), a component of the oligopeptide transport system, and decreased in cells lacking the oppA gene (15). To clarify whether the oligopeptide transport system is involved in the active transport of aminoglycoside antibiotics, we isolated spontaneous kanamycin-resistant mutants. Complete loss of or decrease in OppA was observed in 8 of 20 of these mutants. These results indicate that the oligopeptide transport system is involved in the uptake of aminoglycoside antibiotics, and that the system is down-regulated in some of the spontaneous kanamycin-resistant mutants.
Bacterial strains, plasmid, and culture conditions.
E. coli J53 (met pro thi) (5) was grown in Luria-Bertani medium (medium A) or M9 minimal medium (24) containing 100-μg/ml concentrations each of methionine and proline and 10 μg of thiamine/ml (medium B). Most of the experiments were performed with medium B. Spontaneous kanamycin-resistant mutants were isolated on 1.5% agar plates of Luria-Bertani medium containing 20 μg of kanamycin/ml by incubating the plates overnight at 37°C. The plasmid pPI5, containing the oppA gene located at 27 min of the E. coli chromosome, was prepared as described previously (17). E. coli cells containing pPI5 were cultured in the presence of 30 μg of chloramphenicol/ml to maintain the plasmid in E. coli cells. Cell growth was monitored by measuring the A540.
Determination of the nucleotide sequence of the oppA gene and measurement of the levels of OppA mRNA and OppA protein.
The oppA gene was amplified by PCR with 5′-GGGGAATTCGCCACATCATAATCC-3′ (sequence for positions −570 to −553 of the oppA gene, containing the EcoRI site) as 5′-end primer and 5′-GGGGTCGACACTCCTGCCCCACG-3′ (complementary sequence for positions 1657 to 1641 of the oppA gene, containing the SalI site) as 3′-end primer. The nucleotide sequence of the 2.2-kb oppA gene was determined by the dideoxy method of Sanger et al. (25). Determination of the transcription initiation site, dot blot analysis of OppA mRNA, and Western blot analysis of OppA were performed as described previously (12, 17) with E. coli cells harvested at an A540 of 0.3.
Measurement of OppA synthesis by an immunoprecipitation method.
E. coli cells were grown in medium B in which the methionine content was decreased from 100 to 10 μg/ml. When the A540 reached 0.2, [35S]methionine (1 MBq) was added to each 5-ml aliquot and cells were allowed to grow for 20 min. The amount of OppA synthesized was determined as described previously (12) by using 1,000,000 cpm of [35S]methionine-labeled protein and antibody against OppA. Radioactivity of labeled OppA was quantified with a Fujix Bas 2000 II imaging analyzer.
Measurement of isepamicin uptake and polyphenylalanine synthesis.
Determination of isepamicin uptake by intact cells was performed with 30 μM [14C]isepamicin (300 MBq/mmol) as described previously (15). Poly(U)-directed polyphenylalanine synthetic activity of ribosomes was measured in accordance with a previously published protocol (26).
Measurement of polyamine and amino acid contents.
Polyamine and amino acid contents were determined with a Toyo Soda high-performance liquid chromatography system (11) and a Hitachi 835-10 amino acid analyzer (14), using the trichloroacetic acid supernatant of E. coli. Protein levels were determined by the method of Lowry et al. (19).
Properties of spontaneous kanamycin-resistant mutants of E. coli.
Twenty spontaneous kanamycin-resistant colonies were isolated by culturing 108 E. coli J53 cells on a 1.5% agar plate containing 20 μg of kanamycin/ml. The amount of OppA was then determined by Western blot analysis using these 20 colonies. Colonies were classified into three groups: colonies having a normal amount of OppA (n = 12), colonies having 60 to 70% less OppA than the parent strain (n = 7), and colonies having no detectable OppA (n = 1). Since we were interested in the relationship between kanamycin resistance and its transport, the properties of mutants in the second and third groups were examined. The mutants from the second and third groups were termed m1 and m2, respectively.
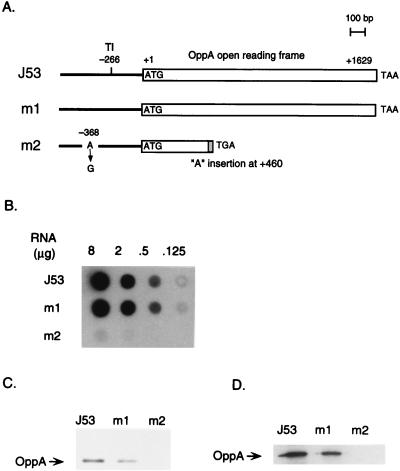
The structure of the oppA gene in the m1 and m2 mutants was determined from their nucleotide sequences (Fig. 1A). The synthesis of OppA mRNA in E. coli J53, m1, and m2 started at 266 nucleotides upstream from the initiation codon AUG. The sequence in the m1 mutant was the same as that in the parent strain, J53. Two mutations were observed in the nucleotide sequence of the oppA gene in the m2 mutant. Those were at positions −368 (A to G) and 460 (A insertion) (Fig. 1A). Thus, a termination codon appeared at amino acid position 166 of OppA.
FIG. 1.
Gene structure of oppA gene (A), OppA mRNA level (B), OppA protein level (C), and the rate of OppA synthesis (D) in E. coli J53, m1, and m2. The gene structure shown in panel A was constructed from the nucleotide sequences of oppA. TI, transcriptional initiation site. Shading in the open reading frame of the m2 mutant indicates encoded amino acids different from those of parent strain J53 due to the “A” insertion at position 460. Experiments were performed as described in the text.
The level of OppA mRNA was next measured by dot blot analysis. As shown in Fig. 1B, the level of OppA mRNA in the m1 mutant was the same as that of the parent strain, but only a small amount of OppA mRNA existed in the m2 mutant. The amount of OppA was measured by Western blot analysis. As shown in Fig. 1C, the amount of OppA in the m1 mutant was about one-third of that in the parent strain. OppA was not found in the m2 mutant. The amount of OppA was proportional to OppA synthesis (Fig. 1D).
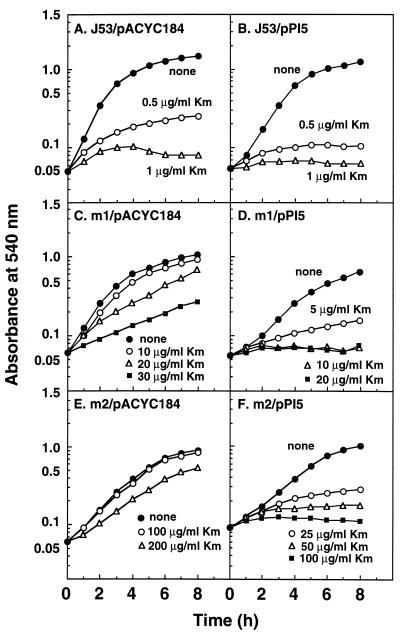
The degree of kanamycin resistance was determined by adding various concentrations of kanamycin to medium B. Cell growth was followed by measuring the A540. As shown in Fig. 2, the growth of the parent strain was inhibited strongly by 1 μg of kanamycin/ml, while the growth of the m1 and m2 mutants was resistant to 10- and 100-μg/ml concentrations of kanamycin, respectively (Fig. 2A, C, and E). The mutants transformed with the oppA gene exhibited greatly increased sensitivity, but the parent strain transformed with the oppA gene showed only slightly increased sensitivity (Fig. 2). The growth of the m1 and m2 mutants was completely inhibited by 10- and 100-μg/ml concentrations of kanamycin, respectively. These results indicate that OppA, a component of the active transport system of oligopeptides, is partly involved in the sensitivity to kanamycin.
FIG. 2.
Effect of kanamycin on growth of E. coli J53, m1, and m2. E. coli cells were transformed with either pACYC184 vector (A, C, E) or pPI5 containing the oppA gene (B, D, F). The concentrations of kanamycin (Km) used are shown in the figure. Each value is the average of duplicate determinations.
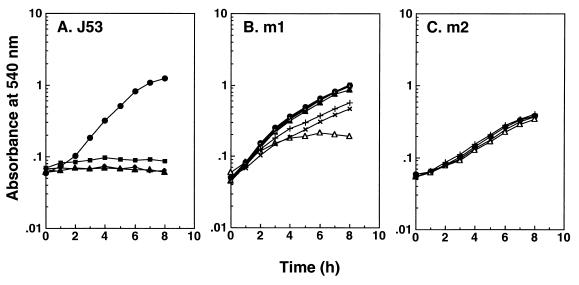
The sensitivity of the m1 and m2 mutants to other aminoglycoside antibiotics was next examined. As shown in Fig. 3, the m1 and m2 mutants were resistant to streptomycin, neomycin, and isepamicin. The degree of resistance was much greater in the m2 mutant than in the m1 mutant, similar to the degree of resistance seen with kanamycin. It has been reported that triornithine is taken up by the oligopeptide transport system and cell growth is inhibited due to its accumulation (20). Cell growth of the m1 and m2 mutants was resistant to triornithine by two- to threefold (data not shown).
FIG. 3.
Effect of aminoglycoside antibiotics on growth of E. coli J53 (A), m1 (B), and m2 (C). •, no antibiotic; ▴, streptomycin (10 μg/ml); ▵, streptomycin (30 μg/ml); ■, neomycin (3 μg/ml); □, neomycin (10 μg/ml); ×, neomycin (30 μg/ml); ⧫, isepamicin (3 μg/ml); ◊, isepamicin (10 μg/ml); +, isepamicin (30 μg/ml). Each value is the average of duplicate determinations.
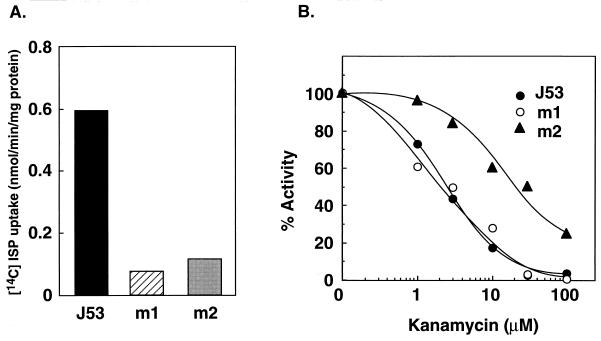
The aminoglycoside transport activity of the m1 and m2 mutants was measured with [14C]isepamicin. As shown in Fig. 4A, the activity of the two mutants was much lower than that of the parent strain. Protein synthetic activity of ribosomes from the mutants was then measured, since ribosomal mutation is also involved in the resistance to aminoglycoside antibiotics (4, 22). As shown in Fig. 4B, protein synthetic activity of ribosomes from the m2 mutant was more resistant to kanamycin than that of ribosomes from the parent strain and the m1 mutant. These results suggest that the m2 mutant is a double mutant of the oppA gene and a gene for ribosomal protein, which explains the strong resistance to aminoglycoside antibiotics.
FIG. 4.
Isepamicin (ISP) uptake activity (A) and polyphenylalanine synthetic activity of ribosomes (B) of E. coli J53, m1, and m2. Experiments were performed as described in the text with E. coli cells harvested at A540 = 0.3. Each value is the average of duplicate determinations.
We next investigated why the amount of OppA is smaller in the m1 mutant than in the parent strain. The amount of OppA mRNA in the m1 mutant was almost equal to that in the parent strain (Fig. 1B), suggesting that OppA synthesis is negatively regulated at the posttranscriptional level in the m1 mutant. Synthesis of OppA was measured by immunoprecipitation of [35S]methionine-labeled protein with antibody against OppA (12). The rate of OppA synthesis in the m1 mutant was about one-third of that in the parent strain (Fig. 1D), suggesting that decrease in OppA in the m1 mutant is due to a decreased rate of OppA synthesis.
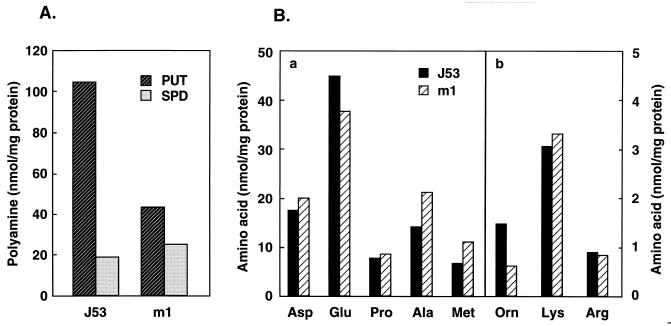
It has been reported that OppA synthesis is greatly stimulated by polyamines at the translational level (12, 17). Thus, the amounts of polyamines and amino acids were measured. As shown in Fig. 5A, the level of putrescine was lower in the m1 mutant than in the parent strain, J53. The levels of most amino acids were very similar in J53 and the m1 mutant, but the content of ornithine was much lower in the m1 mutant than in J53 (Fig. 5B). The results suggest that decrease in polyamines due to the change of ornithine metabolism may be one of the reasons for the decrease in OppA synthesis.
FIG. 5.
Polyamine (A) and amino acid (B) contents in E. coli J53 and m1. Experiments were performed as described in the text with E. coli cells harvested at A540 = 0.3. Each value is the average of duplicate determinations.
The results, taken together, indicate that the oligopeptide transport system is important for the uptake of aminoglycoside antibiotics, since a decrease in OppA was observed in about 40% of spontaneous kanamycin (aminoglycoside)-resistant mutants. However, it remains to be clarified which gene is mutated in the m1 mutant.
Acknowledgments
We thank A. J. Michael and K. Williams for their kind suggestions and help in preparing the manuscript. Thanks are also due to F. Ikegami and S. Yamaji for the operation of the amino acid analyzer and for kindly supplying isepamicin.
This work was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Andry K, Bockrath R C. Dihydrostreptomycin accumulation in E. coli. Nature. 1974;251:534–536. doi: 10.1038/251534a0. [DOI] [PubMed] [Google Scholar]
- 2.Bryan L E, Van den Elzen H M. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob Agents Chemother. 1977;12:163–177. doi: 10.1128/aac.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busse H-J, Wöstmann C, Bakker E P. The bactericidal action of streptomycin: membrane permeabilization caused by the insertion of mistranslated proteins into the cytoplasmic membrane of Escherichia coli and subsequent caging of the antibiotic inside the cells due to degradation of these proteins. J Gen Microbiol. 1992;138:551–561. doi: 10.1099/00221287-138-3-551. [DOI] [PubMed] [Google Scholar]
- 4.Choi E C, Nishimura T, Tanaka N. Mutational alterations of either large or small ribosomal subunit for the kanamycin resistance. Biochem Biophys Res Commun. 1980;94:755–762. doi: 10.1016/0006-291x(80)91299-1. [DOI] [PubMed] [Google Scholar]
- 5.Clowes R C, Rowley D. Some observations on linkage effects in genetic recombination in Escherichia coli K-12. J Gen Microbiol. 1954;11:250–260. doi: 10.1099/00221287-11-2-250. [DOI] [PubMed] [Google Scholar]
- 6.Damper P D, Epstein W. Role of the membrane potential in bacterial resistance to aminoglycoside antibiotics. Antimicrob Agents Chemother. 1981;20:803–808. doi: 10.1128/aac.20.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David B D. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev. 1987;51:341–350. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David B D, Chen L, Tai P C. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci USA. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuchi T, Kashiwagi K, Kobayashi H, Igarashi K. Characteristics of the gene for spermidine and putrescine transport system mapped at 15 min on Escherichia coli chromosome. J Biol Chem. 1991;266:20928–20933. [PubMed] [Google Scholar]
- 10.Höltje J-V. Streptomycin uptake via an inducible polyamine transport system in Escherichia coli. Eur J Biochem. 1978;86:345–351. doi: 10.1111/j.1432-1033.1978.tb12316.x. [DOI] [PubMed] [Google Scholar]
- 11.Igarashi K, Kashiwagi K, Hamasaki H, Miura A, Kakegawa T, Hirose S, Matsuzaki S. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol. 1986;166:128–134. doi: 10.1128/jb.166.1.128-134.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi K, Saisho T, Yuguchi M, Kashiwagi K. Molecular mechanism of polyamine stimulation of the synthesis of oligopeptide-binding protein. J Biol Chem. 1997;272:4058–4064. doi: 10.1074/jbc.272.7.4058. [DOI] [PubMed] [Google Scholar]
- 13.Kashiwagi K, Hosokawa N, Furuchi T, Kobayashi H, Sasakawa C, Yoshikawa M, Igarashi K. Isolation of polyamine transport-deficient mutants of Escherichia coli and cloning of the genes for polyamine transport proteins. J Biol Chem. 1990;265:20893–20897. [PubMed] [Google Scholar]
- 14.Kashiwagi K, Igarashi K. Adjustment of polyamine contents in Escherichia coli. J Bacteriol. 1988;170:3131–3135. doi: 10.1128/jb.170.7.3131-3135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashiwagi K, Miyaji A, Ideka S, Tobe T, Sasakawa C, Igarashi K. Increase of sensitivity to aminoglycoside antibiotics by polyamine-induced protein (oligopeptide-binding protein) in Escherichia coli. J Bacteriol. 1992;174:4331–4337. doi: 10.1128/jb.174.13.4331-4337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991;266:20922–20927. [PubMed] [Google Scholar]
- 17.Kashiwagi K, Yamaguchi Y, Sakai Y, Kobayashi H, Igarashi K. Identification of the polyamine-induced protein as a periplasmic oligopeptide binding protein. J Biol Chem. 1990;265:8387–8391. [PubMed] [Google Scholar]
- 18.Linderoth N, Morris D R. Structural specificity of the triamines sym-homospermidine and aminopropylcadaverine in stimulating growth of spermidine auxotrophs of Escherichia coli. Biochem Biophys Res Commun. 1983;117:616–622. doi: 10.1016/0006-291x(83)91245-7. [DOI] [PubMed] [Google Scholar]
- 19.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Naider F, Becker J M. Multiplicity of oligopeptide transport system in Escherichia coli. J Bacteriol. 1975;122:1208–1215. doi: 10.1128/jb.122.3.1208-1215.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichols W W, Young S N. Respiration-dependent uptake of dihydrostreptomycin by Escherichia coli. Its irreversible nature and lack of evidence for a uniport process. Biochem J. 1985;228:505–512. doi: 10.1042/bj2280505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozaki M, Mizushima S, Nomura M. Identification and functional characterization of the protein controlled by the streptomycin-resistant locus in E. coli. Nature. 1969;222:333–339. doi: 10.1038/222333a0. [DOI] [PubMed] [Google Scholar]
- 23.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem. 1993;268:146–152. [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor: stationary phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]