ABSTRACT
In the field of immunology, a systems biology approach is crucial to understanding the immune response to infection and vaccination considering the complex interplay between genetic, epigenetic, and environmental factors. Significant progress has been made in understanding the innate immune response, including cell players and critical signaling pathways, but many questions remain unanswered, including how the innate immune response dictates host/pathogen responses and responses to vaccines. To complicate things further, it is becoming increasingly clear that the innate immune response is not a linear pathway but is formed from complex networks and interactions. To further our understanding of the crosstalk and complexities, systems-level analyses and expanded experimental technologies are now needed. In this review, we discuss the most recent immunoprofiling techniques and discuss systems approaches to studying the global innate immune landscape which will inform on the development of personalized medicine and innovative vaccine strategies.
KEYWORDS: Systems immunology, innate immune profiling, innate immune cells, bioinformatics, RNA sequencing, O-Link, GWAS, metabolomics, multiplex cytokine profiling, luminex, mesoscale, multi-color flow cytometry, phospho-flow, CyTOF, MIBI, vaccinology, infectious disease
Background
Systems immunology utilizes multiple technical strategies to generate large amounts of data that, when integrated, can predict immunological responses and allow researchers to understand the complex interactions between the immune system’s cellular network and infectious organisms.1 Although there have been many advances in diagnostic testing for managing infectious diseases such as HIV, dengue and tuberculosis to facilitate early detection and more immediate care,2 personalized treatment options and treatment options for vulnerable populations such as the elderly are still needed. To this end, identification of novel biomarkers of immunity and of perturbations in immune responses remains critical.
With the immune system being made up of hundreds of cluster of differentiation (CD) antigens, cytokines, chemokines, specialized cell populations and thousands of genes, systems tools are required not only to study each component separately but also to combine these data. Combining these data allows researchers to understand the connections and associations that define the homeostatic activities of a healthy immune system and predict the response to changes in the environment, including exposure to harmful antigen and during vaccination against a large number of diseases.3 Systems immunology studies have been used to facilitate new hypotheses generation and inform mechanistic studies during the development of novel therapeutics and vaccines since the high-throughput methods used provide accurate and unbiased information in a timely fashion from large data sets.4
These large data sets are generated from multi-omics technologies including genomics, proteomics, metabolomics, microbiome profiling, and computational approaches.5 Together, these results form an integrated analysis of immune function at the molecular and cellular levels to establish predictive models of the networks and dynamic interactions between components of the immune system in health and disease. This will allow researchers to fully comprehend the value of new discoveries, particularly in the quest to manage infections and develop new vaccination strategies, and to define the worth of resulting data, its benefits, and risks, as well as its clinical usefulness, which is the key advantage of using the systems immunology approach.
Here we describe how systems immunology can be utilized for immune profiling with a particular focus on the innate immune system. To that end, we describe critical cells of the innate immune system, multiplexed technologies that can be used to generate large data sets, and how systems immunology studies have contributed to the advancements in the fields of vaccinology and infectious disease.
Cells of the innate immune system
Innate immune cells serve as a functional immune response to immediate pathogen threats, induced by an array of cytokine, receptor, and pattern recognition pathways (Figure 1). Such an activation allows for the stalling of infection as an immediate response, while a more specific and mounted immune response via adaptive immunity is generated. A key player in these roles is neutrophils, which are constantly circulating and are often the first to the source of invasion or inflammation.6 Neutrophils (PMN) function via cytokine production, phagocytosis, degranulation, and neutrophil extracellular traps formation (NETosis).6 Activation occurs shortly after engagement of specific pattern recognition receptors (PRRs) or with complement complexes that can induce phagocytosis. PMNs come in several types which have specific toll-like receptor (TLR) expression. There are two subsets of PMN: PMN-I and PMN-II. PMN-I expresses TLR-2, −4, −5, and −8, expresses IL-12 and CCL3, and activates M1 macrophages. PMN-II expresses TLR-2, −4, −7, and −9, allowing for release of IL-10 and CCL2, and activate alternative macrophages. PMNs will undergo apoptosis shortly after completing their response, a process that leads to the release of more granules and consequently more inflammation.
Figure 1.
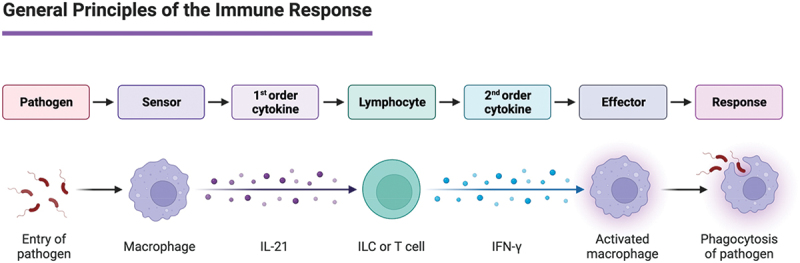
The immune system encompasses a unique population of cells and proteins that work together to protect the body from non-self-entities. The immune system in simple terms has two lines of defense: innate and adaptive immunity. The immune system has two basic lines of defense: innate and adaptive immunity. Innate immunity is antigen dependent, involving recognition of a non-self-antigen determined via unique structural or functional features of infectious agents. After recognition, the innate cells will initiate cytokine production to recruit more specific innate cells or initiate the antigen dependent adaptive immune system. Second order cytokines are produced from this interaction like IFN-γ leading to effector function like phagocytosis of the pathogen. Innate and adaptive immunity do not operate as separate mechanisms of host defense but rather complement each other.
Macrophages or developing monocytes form an additional component to the activation of innate immunity functioning via phagocytosis and additional cytokine and chemokine signaling during later stages after the onset of infection. Monocytes serve as the free roaming cell, which can differentiate into either pro-inflammatory (M1), anti-inflammatory (M2), or alternatively activated macrophages.7 Monocytes may not differentiate at all and remain active as phagocytic forms utilizing TLR4 and CD14 for gram negative bacterial identification and subsequent phagocytosis.8 Once activated, M1 macrophages will recruit more monocytes while inducing more MHC-II expression and maximizing their reactive oxygen species (ROS) and nitric oxide content to allow them to kill invasive pathogens more efficiently.7 Alternatively activated macrophages and M2 macrophages function in the recovery of inflammatory responses, modulating inflammation and inducing growth of cells and connective tissue to speed recovery after infection.7
Other cells are more capable of widespread damage via the expulsion of granules that can induce toxic effects against invasive forms and host cells alike. Such cells as eosinophils, basophils, and mast cells fill this role of granulocytes. Basophils have an expression of IgE that allows for specific targeting of pathogens as they are encountered.9 Eosinophils are more directed against parasitic infections, such as helminths, and their granules are cationic or histamine in nature. They also express IgE, which if an antigen interacts with the receptor or IL-5 is present, will induce degranulation.9 Mast cells predominately have granules of histamine, heparin, and serotonin, and will degranulate against a wide array of antigens, making them a culprit in allergic responses.10 Mast cells express TLR-1, −2, −4, and −6, and patrol many mucous membranes, allowing them to activate early if encountering a pathogen within these environments.
The main antigen presenting cells are dendritic cells (DCs), a cell that bridges innate and adaptive immunity. While other cells also serve as a function of antigen presentation, the DC has a dominant function in this role. Furthermore, their differentiation from hematopoiesis has arms in both the lymphoid and myeloid branches. DC has two subtypes, cDC1 and cDC2 that play the main role in antigen presentation to T-cells. Both classes up-regulate the expression of MHC-II, to improve antigen presentation.11 cDC1 has high functionality for cross presentation to both CD8+ and CD4+ T-cells, whereas cDC2 ismore attuned to CD4+ T cells. An additional subset of pDCs is more functional to the role of perpetuating the activation of cDC1, cDC2, and T cell activation via the secretion of Interferon alpha (IFN-α). IFN-α activates both cytotoxic and regulatory T cells (Tregs), which allows for a more substantial and coordinated immune response.
Innate Lymphoid Cells (ILCs) are a family of effector cells usually found on mucosal surfaces that rapidly secrete effector cytokines to help regulate the immune response to pathogens. ILCs do not possess rearranged antigen-specific cell receptors, but they mirror T helper cell function by the induction of signature cytokines and transcription factors. ILCs are divided into three groups: group 1 ILC, similar to Type 1 helper T cells (Th1) and includes natural killer cells (NK cells); group 2 ILCs, similar to Type 2 helper T cells (Th2); and group 3 ILC, similar to Type 17 and 22 helper T cells (Th17/22). These subtypes play an integral role in regulating adaptive Th1, Th2, and Th17/22 responses.12 Our group has also published on another important subtype of ILC, the ILCFR. ILCFR has been shown to inhibit the ability of T follicular helper (Tfh) cells to provide B cell help.13
Heterogeneity in the human innate immune system
The distribution of innate cells is dictated by their type, chemokines, and dynamics within the host, such as in infection or aging, which would manifest different localizations. Monocytes and granulocytes are often localized to the blood, or in the vicinity of highly vascularized tissue, where they remain throughout the lifetime of the host with minimal change with age.14 DCs follow a similar pattern, with localization to specific tissues that are notable to the different subsets of DCs. cDC1s are located in several systems, specifically the thymus and the T cell zone of the spleen, with functional variation depending on the difference between primary and secondary lymphoid tissue.10 In the thymus, cDC1s induce T-cell central tolerance and in the spleen, they induce T-cell cross tolerance.10 cDC2 is a mostly lymphoid-proximal localization, making it more functional to the surveillance of mucosal draining from the intestines and respiratory tract.15,16 However, while such patterns are present universally, some cDC2s are also found to be localized in the skin, but their frequencies are not consistent across individuals.15 pDCs are more consistent from person to person, following a cycle of peripheral development, and a subsequent migration to the thymus to develop immune tolerance.10 This suggests that the development of these innate cells may be variable across the population, and the mediated immune responses may be determined by these variations.16 Furthermore, ILCs, another regulator of inflammation and immunity, appear to show a similar heterogeneity as they also have both preserved mucosal tissues prevalence in certain ILC populations, whereas other subsets are more variable.17
Innate immune profiling using systems-based analyses
A key challenge in systems immunology is utilizing the correct bioinformatic tools to produce and integrate ‘omics datasets with each providing different insights into cellular processes. These processes are complemented by each other and can help give context to the magnitude of innate expression. The amount of raw data that can be produced by these analyses can be better complimented as a unit using systems immunology data integration using bioinformatics, which allows for patterning and subsequent predictions of the response of immune cells and has seen increasing use as more data are available to make stronger predictions. A system as complex as innate immunology requires these powerful and comprehensive approaches to investigate the systems as not merely a sum of its parts. Here we summarize how systems immunology characterization via genomics, transcriptomics, metabolomics, and multiplexed immunological assays can be used to create a large-scale comprehensive picture of vaccine immunology, infectious disease outcomes, and aging prediction (Figure 2).
Figure 2.
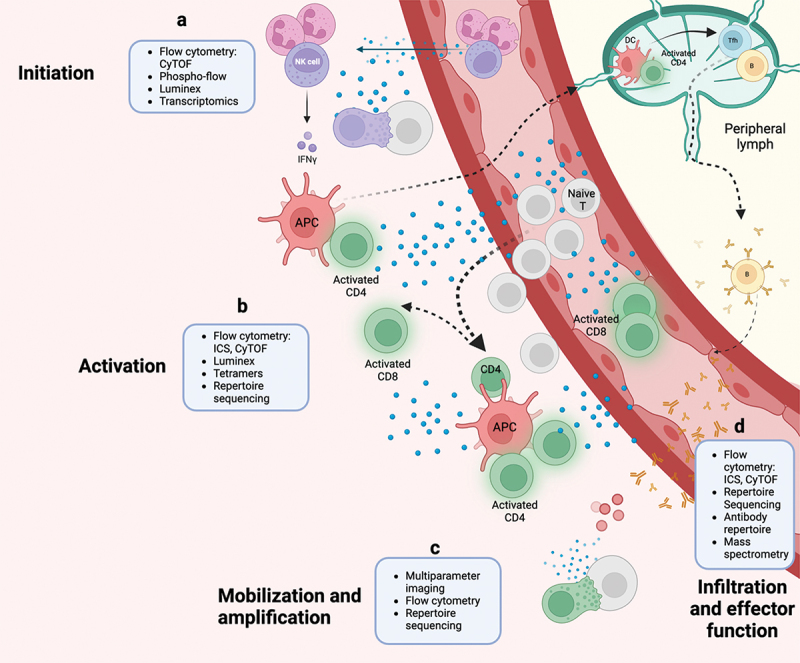
Systems innate immunology can be studied utilizing a variety of techniques combined with bioinformatic analyses. These analyses can not only be applied to different samples (ie peripheral blood, endotracheal aspirates, lymph node biopsies) but also to samples taken at different time points during infection, disease, or vaccination. (a) Innate cell subsets and their function have been characterized at early time points by flow cytometry, cytometry by time-of-flight (CyTOF), flow cytometry of phosphorylated proteins (phospho-flow), bead-based multiplex kit (Luminex) or other immunoassay like mesoscale discovery (MSD), and transcriptomics like RNA-seq to reveal inflammatory mediators and subsequent pathways in early infection or vaccination timepoints. (b) Innate initiation and activation of 1st order cytokines can be classified by phenotype and functional capability using flow cytometry for both surface and intracellular receptor expression along with effector proteins like cytokines and cytotoxic granules. Many of these effector proteins can also be detected by Luminex or MSD technology which can detect up to 80 analytes in a single sample. (c) Activated cells of the adaptive immune system can then be probed for antigen specificity by TCR or BCR repertoire sequencing, flow cytometry utilizing tetramer technology for peptide presentation, and multiparameter imaging. (d) These techniques are currently being expanded to cover tissue biopsies that will inform on the local microenvironment and infiltrating cell populations. APC, antigen-presenting cell; DC, dendritic cell; B, B cell; CD4, CD4+ T cell; NK, natural killer cell; T, T cell; ICS, intracellular cytokine staining; CD8, CD8+ T cell; Tfh, T follicular helper cell.
RNA sequencing
RNA-sequencing (RNA-seq) is a next-generation sequencing (NGS) approach that allows sequencing of RNA molecules. Bulk RNAseq is a valuable tool used to capture the expression profile of a large and diverse population of cells.18 RNAseq is commonly partnered with RT-qPCR to look at host mRNA targets involved with viral entry into host cells, the innate immune response including cytokine production, fundamental cellular processes, and interactions between miRNAs and mRNAs to alter host pathways.19 However, bulk RNAseq can mask which specific cell types are driving disease progressions or resistance. By contrast, single-cell RNAseq (sc-RNAseq) evaluates the individual expression of cells, and thousands at a time. When combined, bulk RNAseq and single-cell RNA seq are beneficial in establishing immune and inflammatory mechanisms of infection-induced organ damage.19,20 These tools have been incredibly useful during the SARS-CoV-2 pandemic. Both bulk and single cells are used to understand multivariant SARS-CoV-2 including viral immune evasion.19 Groups use this technology to understand the expression of mRNAs, proteins, posttranscriptional regulatory agents such as micro-RNAs (miRNA) and long noncoding RNA (lncRNA).19–21 Combining the expression of miRNAs with the upregulation of different genetic pathways which can be identified via the Kyoto Encyclopedia of Genes and Genomes database (KEGG) sheds light on which secretion and signaling pathways are altered during the course of SARS-CoV-2 infection in the lungs.19 In addition to peripheral samples, organ tissues can be directly sampled from patients to assess the severity of infection, upregulation of innate immune responses (including inflammatory genes IL-2, IL-6, IL-8, IL-17A, and NF-kB).19–22 Upregulation of these cytokines has downstream effects that can lead to organ injury and have therefore been valuable to understand the mechanisms during the pandemic.
OLINK
OLINK’s proteomic Proximity Extension Assay is a high throughput platform that measures thousands of proteins in blood with genomic data integration.23,24 OLINK is based on oligonucleotide linked antibody pairs that have slight affinity for different epitopes of the same protein. When these oligonucleotide linked antibodies are brought into proximity to each other on the same protein, the two unique oligonucleotides are extended by a DNA polymerase and amplified exponentially; quantitative real-time PCR (qPCR) is then used to amplify and quantify the oligonucleotides in the samples.23–25 Because of this technique’s specificity, small samples are needed per test (1uL) and previously frozen plasma samples that are decades old can be run with surprising accuracy, resulting in a high throughput and robust assay.23–25 OLINK has multiplex detection methods for inflammation and immunology, and targets 96 antigens in each test. Combining multiple panels allows users to look at thousands of proteins at once, painting an accurate depiction of the immune response.25 OLINK has been used in multiple fields of immunology, from cancer immunotherapy to infectious diseases. Recently during the COVID-19 pandemic, OLINK has been a value system biology approach for looking at proteins in infected persons.26,27 Differences in cytokine and chemokine profiles during different disease stages and disease severity are being evaluated, helping determine mechanisms underlying infection. When paired with other proteomic techniques such as RNAseq, OLINK data provide important insights into immune cell interaction, cell responses, and tissue/cell specific protein production during COVID-19 infection.26,27
GWAS
Individuals differ in their susceptibility to infectious diseases and in their ability to respond to vaccines. In fact, infectious variability and vaccine response heterogeneity among individuals are closely related to gene polymorphisms.28 Genome-wide association studies (GWAS) aim to identify associations of genotypes with phenotypes by testing for differences in the allele frequency of genetic variants between individuals who are ancestrally similar but differ phenotypically.29 GWAS reports blocks of correlated SNPs that all show a statistically significant association with the trait of interest, known as a genomic risk locus, which correlate with specific disease outcomes and play a regulatory function in gene expression.29,30 GWAS can be a beneficial tool in helping identify new targets for drugs as well as creating links between the immune system and other diseases.31 Most recently, GWAS has been instrumental in identifying host genetic factors that modulate the risk of infection and death from severe COVID-19 disease. A study by Neimi et al. looked at 49,562 individuals infected with SARS CoV-2 and identified 13 independent common risk variants, with many being in or near immune genes such as IFNAR2 and CXCR6.32 Several of these loci correspond to other lung or autoimmune inflammatory diseases as well.32 Additional studies also suggest a role in COVID for genes in the type I IFN pathway including TRL7.33 GWAS can help shed lights on biological insights of COVID-19 infection and help identify innovative treatment options with the hope of preventing severe disease and death.
Metabolomics
Metabolomics is a growing field of study, and it involves measuring small metabolic molecules including sugars, amino acids, bile acids, fatty acids, or lipids. Most commonly, metabolites are isolated via gas or liquid chromatography (GC or LC) and profiled using nuclear magnetic resonance (NMR) or mass spectrometry (MS).34 Metabolomics is frequently paired with other ‘omics such as genomics, transcriptomics, and proteomics with the intent of finding prognostic markers, predicting the evolution of disease, and understanding metabolic signatures of cells.34,35 Recently, metabolomics has been instrumental in mapping metabolic changes in innate immune cells during COVID-19 infection.35–37 Li et al. showed clear impairment of amino acid metabolism and energy metabolism in severe COVID-19 patients that correlates with inflammation.36 Specifically, the majority of these changes occurred with metabolites involved in the urea cycle, purine metabolism, arginine and proline metabolism, glutamate metabolism, and NAD+ synthesis.35–37 Metabolomic studies are also being used to link inflammation in COVID-19 with mitochondria-dependent energy metabolism, viral replication, coagulation, and fibrogenesis.36,37 This field can also help identify global molecular signatures during varying disease states and new targets for drug development.
Multiplex cytokine profiling
During an immune response to infection or vaccination where global tissue-level immune responses are needed, cell-to-cell communication is essential for immune cells to coordinate an efficient immune response. Cytokines and chemokines, which serve as mediators of intercellular communication during an immune response, play a crucial role in determining the path of an immune response to both infection and vaccination. As such, gaining an understanding of the dynamic changes to cytokine and chemokine levels throughout the immune response to infection and vaccination can provide major insights into understanding both the pathogenesis of a disease as well as the type of immune response that provides protection from infection. However, with over 100 cytokines known to exist and their levels being variable in different tissues and during an immune response, this is no small task.38 To help achieve an understanding of the role of cytokines and chemokines in shaping immune responses in a systemic manner, multiplexed cytokine and chemokine analysis through multiplexed bead-based immunoassays, such as Luminex® and Meso Scale Discovery (MSD) immunoassays, which can measure 60 or more proteins at once have now been developed.
Multiplexed cytokine and chemokine analysis has been utilized in several studies to help understand the immune response to both infection and vaccination. Examples of such a use include using multiplexed cytokine analysis to investigate the role of cytokines in the pathogenesis of Zika virus,39 gaining an understanding of differences in plasma inflammatory markers between severe and non-severe COVID-19 infections,27 identifying mycobacterium-specific cytokine biomarkers that distinguish latent and active tuberculosis (TB) infections,40 and measuring differences in cytokine levels in sera and semen to differentiate the different stages of HIV infection.41 In addition to being used to gain an understanding of immune responses to infection, multiplexed assays have also been used to gain an understanding of the immune response to vaccination. The use of multiplexed cytokine and chemokine assays has led to a greater understanding of the essential role of type-I and type-III IFN production by DCs for vaccine efficacy following influenza immunization,42 an understanding of how aging negatively impacts type-I IFN production in the elderly following stimulation by empty lipid nanoparticles (a component of current mRNA-based vaccines),43 suboptimal T cell responses in elderly individuals to COVID-19 vaccination,44 and to identify cytokines responsible for driving lymphoid tissue fibrosis-associated impaired vaccination responses.45 Importantly, while multiplexed protein assays can be utilized to gain an understanding of the immune response on a global scale, such as through serum and plasma analysis, multiplexed assays can also be run on samples taken directly from stimulated cells. Therefore, multiplexed protein assays provide a dynamic understanding of the cytokine and chemokine networks between immune cells that continually change during an immune response and offer the ability to gain deeper insights into immunological responses to infection and vaccination and novel therapeutic strategies.
Multicolor flow cytometry
Advances in flow cytometry have enabled high throughput multiparametric analyses of cellular phenotypes, biomarkers, and vaccine candidates46 based on fluorescent labeling. To date, multi-color panels of over 40 colors have been developed and executed.47 With the increasing complexity of flow cytometry panels, 2D manual gating is becoming the way of the past as automated analyses pave the way for improved reproducibility and data exploration. Validation of flow cytometry workflows for innate immunoprofiling of peripheral blood samples has enabled more efficient monitoring of innate responses within translational research and clinical settings.48 More recently, efforts to optimize workflows for high-dimensional analyses and multi-batch integration have expanded, improving visualization and clustering of flow cytometry data with algorithms including SPADE,49 FlowSOM,50 and UMAP.51 Furthermore, flow panels have evolved to include phospho-specific flow cytometry, or Phosphoflow, to examine single-cell phosphorylation events within signaling networks. Application of fluorescent cell barcoding (FCB), a mechanism to label cells with unique barcodes of fluorescence intensity and emission, with Phosphoflow has been previously utilized to screen a small-molecule library for T cell receptor and cytokine signaling inhibitors.52 Such analyses exemplify the high-dimensionality potential for flow cytometry analyses.
Cytometry by time of flight (CyTOF)
Traditional flow cytometry techniques remain limited in the number of potential parameters analyzed in a single sample. Recent developments have led to the generation of CyTOF, a mass cytometry technique enabling multiplexed analyses of limited samples with little signal overlap using a mass spectrometer. Unlike flow cytometry, which uses fluorescently labeled antibodies to label cellular markers, CyTOF offers unbiased high-dimensional characterization with the application of antibodies conjugated to rare heavy metal isotopes. An atomic mass cytometer can then detect the unique time-of-flight of each metal with detection overlaps limited to <2% compared to the 5–100% potential spectral overlap that occurs in flow cytometry.53 Additionally, CyTOF methods exhibit significantly lower background from autofluorescence, enabling better detection of low-expression markers even on myeloid cell populations with higher baseline autofluorescence.54 These capabilities have improved the output of large quantities of immunological data from small sample sizes to facilitate the understanding of complex biological systems, disease biomarkers, and therapeutic responses at a cellular level. For example, one study evaluated myeloid cell phenotypes in patients with progressive multiple sclerosis (MS), revealing an increased abundance of highly phagocytic and activated microglia and decreased tumor necrosis factor (TNF)Hi myeloid cells associated with chronic neuroinflammation and demyelination.55 A similar study further identified a novel CD56Hi NK cell signature in periventricular brain regions of MS patients.56 These investigations highlight the power of CyTOF to identify signatures of low and high-abundance immune cells in disease to set the stage for advanced targeted immunotherapies.
Multiplexed ion beam imaging (MIBI)
Like CyTOF, multiplexed ion beam imaging (MIBI) uses mass spectrometry to detect metal-tagged antibodies to analyze cellular protein markers. However, MIBI further enables analyses of 100 targets simultaneously while also providing details regarding cell morphology and localization through its application to whole tissue samples.57 This method overcomes the limitation of spectral and spatial overlap exhibited by immunohistochemistry, warranting its application in clinical diagnostics. Information produced using MIBI can be understood in a traditional imaging context, as well as through the application of high-dimensional analytics, to uncover discrete phenotypic and morphological characteristics of tissue samples including cancer biopsies. For example, a retrospective study used MIBI with Time-Of-Flight (MIBI-TOF) mass spectrometry to analyze 41 samples from triple-negative breast cancer (TNBC) patients. Data interpretation showed unique spatial immune composition and protein expression, including monocyte PD-L1 upregulation, in tumor microenvironments between patients, revealing hallmarks of tumor compartmentalization that correlated with overall survival.58 A similar study further demonstrated immune composition and protein co-expression patterns associated with TNBC survival and recurrence.59 These experiments illustrate the potential for MIBI to address complex immune state characterization in a variety of diseases to improve guidelines guiding therapeutic development.
Systems immunology data integration using bioinformatic platforms
Bioinformatics is the application of computational methods to gain additional insights from large datasets generated from high-throughput experiments.60 Reliable analysis pipelines have already been established for genomic and proteomic expression and networking, but with the increasing accessibility of high-performance computing, advanced techniques that rely on machine learning (ML) classification algorithms, such as random forest (RF), support vector machine (SVM), and K-nearest neighbor (KNN), can be used to identify patterns observed in specific groups of individuals exposed to certain conditions.60 Neural networks are another computational approach commonly used in the analysis of biological data. Unlike other approaches used for classifications such as RF, SVM, and KNN, neural networks can be used to provide predictions based on biological data. An example is training a model on protein data and providing accurate protein structure predictions.61 This application can aid in target identification by providing insight into exposed sites that can drive treatment development.
In the context of immunology, these analyses can highlight the intricacies of interactions occurring within immune system pathways. Recently, these techniques have seen a drastic increase in utilization due to the prevalence of multiparametric assays and genomic sequencing in research projects that characterize protein expression, cytokine production, proliferation, and transcription markers, expanding our understanding of immune networks and their applications in the development of treatments for infectious diseases. A notable example is the recent SARS-CoV-2 pandemic, where bioinformatic pipelines were employed to characterize immune responses in individuals displaying different levels of severity.62,63 This type of analysis can be particularly effective in highlighting what immune interactions drive an observed response. Another application of these techniques is observing how innate signatures are affected in studies involving an aging population.64 Data availability for these techniques is made more accessible by data sharing platforms including NCBI Gene Expression Omnibus (GEO) for genomic data or Protein Data Bank (PDB) for data on protein structures.65,66 Databases like these allow for the efficient sharing and reuse of data that other labs can utilize to generate hypotheses and additional applications for these datasets.
Systems immunology utilization in vaccinology
Systems immunology has proven to be a powerful exploration tool which uses large, multiplexed data sets to gain a deep understanding of cellular and molecular partners orchestrating immune responses. Particularly in the context of vaccine-induced immune responses, systems immunology has aided in understanding how baseline, pre-vaccination immune landscapes shape post-vaccination responses, therefore gaining the ability to predict vaccine responses. To this end, characterization of immune signatures of vaccine responders has allowed for the rationale design of vaccine antigens and adjuvants to drive protective immune responses.67–70
Characterizing baseline immune landscapes to predict vaccine responses
Heterogeneity in vaccine responsiveness can be influenced by a variety of factors including age,71 sex,72 and ethnicity.73 Several studies using a systems approach have demonstrated that the innate immune landscape prior to vaccination has a profound effect on immune responses postvaccination. This has been demonstrated for several clinically approved vaccines such as the hepatitis B virus (HBV),74 and influenza75 vaccines.
Using transcriptional and cytometric profiling, the presence of inflammatory response transcripts and increased frequencies of pro-inflammatory innate cells, particularly CD40 expressing plasmacytoid DCs, pre-vaccination was found to correlate with weaker immune responses to the HBV vaccine.74 Similarly, increased frequencies of monocytes, increased transcription of innate sensing genes such as TLR4 and TLR8 and nucleotide-binding oligomerization domain containing protein 2 (NOD2) and increased transcription of inflammatory genes such as IFN-γ receptor, IL-13 receptor, and spleen tyrosine kinase (SYK) at baseline were found to negatively correlate with antibody responses post influenza vaccination.75 These findings suggest that inflammatory responses pre-vaccination may be detrimental to the induction of antibody responses.74,75
A more recent study has built on this work to identify a universal, baseline, pre-vaccination immune signature that is predictive of antibody responses across an array of vaccines.69 To do this, publicly available transcriptional data was integrated from different platforms of over 3,000 peripheral blood samples from 820 adults across 28 studies of 13 vaccines. Of the 13 vaccines, these spanned several vaccine platforms including live attenuated virus, inactivated virus, recombinant viral vector, recombinant protein, and bacterial glycoconjugate vaccines.69,70
Unsupervised clustering analysis of blood transcriptional modules was coupled to immunological function data to characterize pre-vaccination transcriptional profiles into three endotypes: high, mid, and low inflammatory. These endotypes were differentially defined by the expression of TLR genes, interferon stimulated genes (ISGs), and genes associated with cell metabolism downstream of the transcription factor NF-κB. These metabolic genes regulated by NF-κB are critical for innate and adaptive immune responses such as antiviral responses, antigen presentation, and B cell activation.69,70 Individuals in the high inflammatory endotype upregulated expression of transcriptomic markers of classical monocytes and DCs, which are associated with greater serum antibody responses. Contrary to the aforementioned studies,74,75 this suggests that baseline heightened innate immune activation mediated by monocytes and DCs favors vaccine-specific antibody production likely by aiding in germinal center reactions necessary for high-affinity antibodies.69,70 It is possible that pre-vaccination inflammation is a delicate balance where some can be beneficial to driving vaccine responses and excess can be detrimental. Nonetheless, these studies demonstrate that the innate immune landscape prior to vaccination affects vaccine induced immune responses. Using systems immunology to characterize these pre-vaccination immune landscapes aids in the ability to predict vaccine responders vs. non-responders as well as characterize and subsequently target non-responders through the precision design of vaccine antigens and adjuvants.
Characterizing immune signature following vaccination
In addition to aiding in the characterization of pre-vaccination immune signatures, systems immunology has also been used to characterize responses post-vaccination. This has enabled the identification of correlates of immune-mediated protection which can then be utilized to develop novel vaccines to currently incurable diseases.
Postvaccination immune signatures have been defined for several vaccines such as the yellow fever,76 influenza,75 and more recently, the SARS-CoV-2 mRNA vaccine.77 Similar to the findings in baseline immune signatures,69,70,74,75 postvaccination adaptive immune signatures were heavily dictated by early, innate immune responses.
Using functional genomics and multicolor flow cytometry, it was shown that the yellow fever vaccine 17D (YF17D) upregulated MyD88, a key adaptor protein needed for signaling, and TLR7. Genes downstream of MyD88 signaling such as the proinflammatory cytokines IL-6, IL-12, TNFα, and type I IFNs were also found to be upregulated postvaccination. Master transcription factors, such as NFκB and ETS2 (ETS proto-oncogene 2), were also found to be upregulated. Genes under the control of these master transcription factors are known to regulate the induction of several innate immune pathways such as type I IFNs, inflammasome activation, and complement. In line with this, YF17D vaccination led to increased caspase-1 and −5 which are known to be processed and activated by inflammasomes. Upregulation of components of the complement cascade, specifically C1qA and C1qB, were also found to be upregulated upon YF17D vaccination. Effector cells such as macrophages, NK cells, and DCs were also found to be critical in the early response to YF17D vaccination.76
Using microarray experiments, hemagglutinin inhibition (HAI) assays, and multicolor flow cytometry, vaccine responses to influenza vaccination across multiple seasons in both young and aged populations were evaluated.75 Compared to previous studies which evaluated a single influenza vaccine for one season in young individuals,78–83 evaluating responses in young and aged individuals to several influenza vaccines across multiple seasons addressed the issue of virus strains changing from year to year, what impact these variations may have on immune signatures, and how immune signatures in young individuals differ from those in aged individuals. Shared and consistent molecular signatures of influenza vaccine immunogenicity were seen across the five influenza vaccine seasons that were evaluated. Early after vaccination, strong innate responses were observed and characterized by the expression of IFNs and DC activation that positively correlated to later antibody response. TLR signaling and antigen presentation early postvaccination were also found to be enhanced. When comparing young and aged vaccination responses, aged responses were characterized by having enhanced frequencies of activated, cytotoxic NK cells and CD14+ CD16+ inflammatory monocytes with diminished expression of CD86. Prior to vaccination, monocytes were also increased in aged individuals who had a negative correlation to antibody response. These data suggest changes to the innate response in aged individuals result in diminished antibody responses to influenza vaccination.75 This study exemplifies how systems immunology can be used to understand what specific components of the immune system are responsible for the known suboptimal responses aged population have to many vaccines.75,84 Understanding the immune signature of aged vaccination responses allows for precision design of vaccine antigens and adjuvants that specifically modulate aged immune responses to increase overall vaccine efficacy.
Systems immunology was recently used to characterize immune responses to the novel mRNA vaccine platform used for the SARS-CoV-2 vaccine BNT162b2. Using Cytof, OLINK, and single-cell transcriptomics, it was found that BNT162b2 vaccination, specifically after the second immunization, enhanced innate immune responses. These enhanced innate immune responses were characterized by increased frequencies of CD14+ CD16+ inflammatory monocytes, higher concentrations of INF-γ in the plasma, and a transcriptional signature of innate antiviral immunity. This transcriptional signature in addition to monocyte-related signatures observed were associated with CD8+ T cells and neutralizing antibody responses. Although this study77 is important in understanding initial BNT162b2 vaccination responses, additional systems studies evaluating longitudinal mRNA vaccine responses are needed to understand the quickly waning immune responses against symptomatic COVID-19.85–88
Innate systems immunology in infection and immunity
Diseases are highly complex and exhibit great patient- to- patient variability. For medicine to evolve from an approach to one that considers therapies that are ‘suitable for all,’ high-throughput analysis technologies, mechanistic modeling, and big data generation are essential and will lead to more precise patient stratification and personalized treatment options. Dysregulation of the immune response during infection involves dynamic gene networks, immune signaling pathways, cellular networks, and host–pathogen interactions. Systems immunology approaches consider these complex immune pathways and apply a methodical analysis to each study to identify new predictors of disease and to accelerate the translation of this knowledge into therapies. Significant achievements in understanding immunological diseases using ‘omics and associated computational analysis methods have been made. However, gaps remain in data and connecting genetic variation, the impact of aging, and environmental factors to individual phenotypes and disease outcomes continue to elude researchers. Here, we present perspectives and discuss how recent advances in ‘omics technologies have aided in overcoming challenges toward complex disease.
Biological pathways
Arguably, the most important developments of system immunology have been the development and cataloging of databases. InnateDB and other International Molecular Exchange (IMEx) consortium databases provide systems-level analyses that give better insight into the complex networks of pathway interactions of the innate immune system.89,90 InnateDB is a systems biology database of mammalian and murine molecules and pathways involved in innate immunity. InnateDB is also a complete analysis platform that allows for annotation and database cross-referencing for each gene and protein in addition to a user-friendly bioinformatics interface.91,92 InnateDB provides a way to identify statistically significant pathways from over 3000 pathways that are stored and ontological terms from a list of user-selected genes. These terms describe the molecular function, biological process, and cellular compartment of the genes. Gene expression data can be included for up to 10 conditions at once and overlaid on pathways and networks of interest. Users can then construct interaction networks and visualize their data. Advances in mapping the interactomes are key to understanding systems innate immunology. Examples of comprehensive interaction databases included Pathguide,93 IntAct,94 MINT,95 and BioGRID.90,96 These databases store over 100,000 interactions across a range of species and include experimental types and publications. Network analysis has enabled a more complete picture of the relationships among genes, proteins, and other molecules and can include visualization of these interacting networks which might reveal unknown relationships in signaling cascades or pathways. This is evident when mapping mammalian TLR signaling pathways, which identified MyD88 as a gateway protein of which many pathways converge with positive and negative feedback and feedforward loops.97 These tools provide techniques for systems-level studies to provide novel insights into innate immunity, especially its context in infection.
Host–pathogen interactions and immune profiling
Most systems immunology approaches for studying correlates of protective immunity heavily focus on the adaptive system. Few studies focus on identifying the pre-infection environment and possible correlates of protection. However, there have been many studies using systems immunology to understand correlates of protection against Hepatitis C Virus (HCV).98,99 In these studies, published in the early 2000s, the researchers focused on identifying interferon signatures from peripheral blood mononuclear cells (PBMCs) and liver biopsies using microarray studies that correlated with HCV viral clearance. These studies provided snapshots of the overall immune response using acute HCV infection but fell far short of elucidating the entire immune microenvironment during infection.
However, with new advent of technologies, this is changing. Systems immunology has also been employed recently to study immune profiling. Early in the COVID-19 pandemic, one study used systems immunology to assess immunity to mild versus severed COVID-19 infection in humans.27 This study analyzed the immune responses in PBMCs of 76 COVID-19 patients and 69 healthy individuals from Hong Kong and Atlanta, GA, USA, to identify the underlying causes of immune response differences, especially between healthy young adults and adults with medical comorbidities including age, cardiovascular disease, cancer, or obesity. Understanding the immunological mechanisms of the diverse clinical presentations of diseases is a crucial step in designing therapeutics. Using mass cytometry and CITE-seq analysis, Arunachalam et al. analyzed PBMCs from COVID-19 patients. These techniques revealed common features of immune response induced in SARS-CoV-2 infection at a cellular level including decrease in plasmacytoid DC frequency and IFNα production. Additionally, monocytes and mDCs upon TLR stimulation have reduced proinflammatory cytokines IL-6, TNFα, IL-1β along with impaired rapamycin (mTOR) signaling. Using single-cell transcriptomics, the researchers saw an absence of type I IFNs and reduced HLA-DR in myeloid cells of patients with severe COVID-19. Analysis using systems immunology of the innate immune system helped reveal a spatial and temporal shift in COVID-19 patients.
Systems immunology has also been employed to understand the mechanisms behind the pathogenicity of the influenza virus including the known dysregulation of the host response with hopes that these network-based approaches could reveal novel therapeutic targets. Several studies have used transcriptional and proteomic analyses to determine the host immune response to influenza. A combination of whole tissue and single-cell transcriptomics has identified a feed-forward cytokine loop involving the recruitment of innate cells into infected tissue.100–103 Recent studies have also implicated post-transcriptional regulation in influenza pathogenesis. RNA seq analysis has identified miRNAs can modulate proinflammatory cytokine secretion thus affecting host response.104 Together, these studies provide insights into the programs that can modulate host immune response to pathogens.
Clinical research applications of systems immunology
Systems immunology and infection – improving patient stratification and therapeutic strategies
Significant achievements using ‘omics data have been made in personalized medicine, however the complexity of the immune system and the difficulties in delineating the networks that determine the response to infections/vaccines have now required the systems immunology approach to systematically characterize immune molecular and cellular networks in individual patients. Researchers must consider not only genetic factors and the formation of a memory response but also environmental factors. These factors will cause the manifestation of various clinical symptoms that can make it difficult for clinicians to identify patients infected with a particular disease. Hence, there is a need to identify reliable biomarkers and molecular signatures to precisely stratify patients for personalized treatment. Recent advances in immune subset deep phenotyping have used genomics, metagenomics, and metabolomics techniques to show that cytokine production is regulated by multiple genetic and nongenetic host factors and that cytokine production is relatively predictable with multiple baseline profiles. It has also been shown that there is a correlation of the immune response with genetic risk of disease.105 These studies have used mass cytometry to evaluate multiple cytotoxic molecules with detailed information of T and NK cell differentiation states, two key cell subsets involved in the elimination of infected cells.106
Monitoring and cure of viral diseases is particularly challenging as it requires comprehensive understanding of both the host and the pathogen’s individual features prior to demonstrating the outcomes of the infection in different individuals. System structural analysis of serotype DENV-specific Ig constant Fragment (Fc) has recently highlighted that progressive DHF/DSS patients develop an IgG1 humoral response which showed higher affinity for FcRγIIIA (CD16) due to afucosylated Fc glycans and therefore triggered platelet activation and the risk for thrombopenia and hemorrhagic damage.22 Similarly, metabolomics analysis of acute phase clinical sera samples from patients with dengue infection have identified 65 metabolites differentially expressed that predicted DHF/DSS progression, including α-linolenic acid, arachidonic acid, and docosahexaenoic acid.107
Schreiber et al. used immune transcriptomics to compare gene expression in HIV patients with and without severe non-nontyphoid Salmonella infections (iNTS).108 They found 1,200 genes that were upregulated in HIV patients with a Salmonella infection compared to patients without a bacterial infection. The genes that were upregulated in patients without bacterial infections were enriched in pathways associated with innate immunity/inflammation responses, whereas patients with concurrent bacterial infections had upregulation of genes associated with anti-inflammatory mediators. The authors speculate that this lack of innate immune signature could lead to utilization as a biomarker of poor prognosis in HIV patients with iNTS. These analyses can be used to investigate the manipulation of the immune system by pathogens. Published in 2007, Scott et al. engaged systems immunology to identify the local immune response in human monocytes and PBMCs to important Gram-positive and Gram-negative pathogens including methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus and Salmonella enterica serovar Typhimurium.109 Transcriptional changes were assessed to better understand the cellular cascade that occurs when specific peptides from these bacteria enter the cell.109 The biological connection of those gene expression changes was assessed using transcription factor binding site (TFBS) and network analysis with NetworkAnalyst revealing 11 pathways including the NFκB and Erk1/2 pathways.
Thus, screening patients for afucosylated Fc-IgG or identification of a metabolomics/transcriptomics signature of infection could aid in better targeting and triaging of ‘at risk’ patients, using acute phase clinical specimens and exemplifies the benefit of conducting systems immunology of infection disease. Seasonal influenza in young children can be dramatically severe due to a less developed immunological system. With the identical concept of improved prognostic criteria, a recent pilot study has shown that the nasopharyngeal microbial signature could predict severity of influenza in young children.110 The use of microbial signatures as prognostic biomarkers will also benefit translational and clinical settings in three ways: early recognition of a risky patient, a new therapeutic option to treat severe respiratory complication by manipulating microbiome; and mandatory vaccination/revised vaccine strategy for severe influenza-prone children.
Infectious disease and immunosenescence – new insights gained
Immunosenescence characterizes the decline of human immune system with aging. Older adults become more susceptible to infectious disease, cancer development, and less responsive to vaccinations. The progressive increased age of the population mandates new strategies to ensure sustained health and well-being. Novel approaches to counteract immunosenescence and understand the mechanism of age-related decline of the immune response to infection are required in basic and translational research. Work in the aging field has identified links between chronic inflammation in the immune system. For example, a longitudinal study of aging adults identified elevation of baseline phosphorylated STAT proteins that correlate with an elevation of inflammatory cytokines in the blood and the decreased ability of cells to respond to stimulus in vitro.111 Another metabolomic study linked this low-grade, chronic inflammation with specific inflammasome gene modules that classify older adults into either those with higher IL-1β expression and elevated oxidative stress and those without those characteristics. Elucidating these mechanisms show that targeting inflammasome components may abrogate chronic inflammation.112 We have also recently shown using a systems approach that the innate immune response is impaired in the elderly. In fact, monocytes and dendritic cells have reduced ability to respond to TLR ligands, and innate immune cells are unable to produce cytokines and express co-stimulatory factors that are needed for T and B cell help.64,113,114 We attributed this to the basal expression of increased pro-inflammatory cytokines that characterize immunosenescence. The aging response demands an interdisciplinary approach, one that ‘omics technologies create to facilitate the development of appropriate therapeutic strategies that will not only reduce infectious disease burden and risk but also improve life expectancy and quality. Learning from these findings may pave the way to developing novel and improved vaccines for older adults and the immunocompromised.
Concluding remarks
Systems immunology characterization via genomics, transcriptomics, metabolomics, and multiplexed immunological assays is a growing interest for scientists and clinicians and allows for a large-scale comprehensive picture of vaccine immunology, infectious disease outcomes, and aging prediction (Figure 2). Systems analyses promote the development of a personalized concept of medicine, thereby bringing personalized and precision medicine to the forefront of the clinic and revolutionizing the advancement of patient care and therapeutics. As this approach will tailor medical decisions and health care the level of individual, the need for better patient stratification and improvement of current diagnostic testing is crucial to select appropriate immune therapy based on individual biology. Technical improvement of biological markers measured in a lower-cost high-throughput format is appealing for routine care and will certainly contribute to making therapeutics more accessible to the majority.
Funding Statement
The author(s) reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Brodin P, Davis MM.. Human immune system variation. Nat Rev Immunol. 2017;17(1):21–14. doi: 10.1038/nri.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tay A, Pavesi A, Yazdi SR, Lim CT, Warkiani ME. Advances in microfluidics in combating infectious diseases. Biotechnol Adv. 2016;34(4):404–21. doi: 10.1016/j.biotechadv.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis MM, Tato CM, Furman D. Systems immunology: just getting started. Nat Immunol. 2017;18(7):725–32. doi: 10.1038/ni.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinform. 2018;19(6):1370–81. doi: 10.1093/bib/bbx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosales C. Neutrophil: a cell with many roles in inflammation or several cell types? Front Physiol. 2018;9. doi: 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yunna C, Mengru H, Lei W, Weidong C. Macrophage m1/m2 polarization. Eur J Pharmacol. 2020;877(173090):173090. doi: 10.1016/j.ejphar.2020.173090. [DOI] [PubMed] [Google Scholar]
- 8.Murray Peter J, Allen Judith E, Biswas Subhra K, Fisher Edward A, Gilroy Derek W, Goerdt S, Gordon S, Hamilton John A, Ivashkiv Lionel B, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chirumbolo S, Bjørklund G, Sboarina A, Vella A. The role of basophils as innate immune regulatory cells in allergy and immunotherapy. Hum Vaccin Immunother. 2018;14(4):815–31. doi: 10.1080/21645515.2017.1417711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchida AM, Ro G, Qiang L, Peterson KA, Round J, Dougan M, Dougan SK. Human differentiated eosinophils release il-13 in response to il-33 stimulation. Front Immunol. 2022;13. doi: 10.3389/fimmu.2022.946643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulfone-Paus S, Bahri R. Mast cells as regulators of t cell responses.Journal. Front Immunol. 2015;6. doi: 10.3389/fimmu.2015.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris ST, Durai V, Wu R, Theisen DJ, Ward JP, Bern MD, Davidson JT, Bagadia P, Liu T, Briseño CG, et al. Cdc1 prime and are licensed by cd4+ t cells to induce anti-tumour immunity. Nature. 2020;584(7822):624–9. doi: 10.1038/s41586-020-2611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Connor MH, Muir R, Chakhtoura M, Fang M, Moysi E, Moir S, Carey AJ, Terk A, Nichols CN, Metcalf T, et al. A follicular regulatory innate lymphoid cell population impairs interactions between germinal center tfh and b cells. Commun Biol. 2021;4(1):563. doi: 10.1038/s42003-021-02079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44(3):439–49. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 15.Gianchecchi E, Delfino DV, Fierabracci A. Natural killer cells: potential biomarkers and therapeutic target in autoimmune diseases? Front Immunol. 2021;12:12. doi: 10.3389/fimmu.2021.616853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audiger C, Rahman MJ, Yun TJ, Tarbell KV, Lesage S. The importance of dendritic cells in maintaining immune tolerance. J Immunol. 2017;198(6):2223–31. doi: 10.4049/jimmunol.1601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yudanin NA, Schmitz F, Flamar A-L, Thome JJC, Tait Wojno E, Moeller JB, Schirmer M, Latorre IJ, Xavier RJ, Farber DL, et al. Spatial and temporal mapping of human innate lymphoid cells reveals elements of tissue specificity. Immunity. 2019;50(2):505–19.e504. doi: 10.1016/j.immuni.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diallo I, Jacob RA, Vion E, Kozak RA, Mossman K, Provost P. Altered microRNA transcriptome in cultured human airway cells upon infection with sars-cov-2.Journal. Viruses. 2023;15(2):496. doi: 10.3390/v15020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Qiu H, Yang B, Chen Y, Zhu Q, Wen F, Peng M, Wang G, Guo G, Chen B, Maarouf M, et al. Influenza a virus-induced circrna circmertk negatively regulates innate antiviral responses. Microbiol Spectr. 2023;11(2):e03637–03622. doi: 10.1128/spectrum.03637-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang TT, Sewatanon J, Memoli MJ, Wrammert J, Bournazos S, Bhaumik SK, Pinsky BA, Chokephaibulkit K, Onlamoon N, Pattanapanyasat K, et al. Igg antibodies to dengue enhanced for fcγriiia binding determine disease severity. Science. 2017;355(6323):395–8. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods. 2007;4(4):327–9. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 24.Assarsson E, Lundberg M, Holmquist G, Björkesten J, Bucht Thorsen S, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, et al. Homogenous 96-plex pea immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PloS One. 2014;9(4):e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui M, Cheng C, Zhang L. High-throughput proteomics: a methodological mini-review. Lab Invest. 2022;102(11):1170–81. doi: 10.1038/s41374-022-00830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filbin MR, Mehta A, Schneider AM, Kays KR, Guess JR, Gentili M, Fenyves BG, Charland NC, Gonye ALK, Gushterova I, et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med. 2021;2(5):100287. doi: 10.1016/j.xcrm.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–20. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haralambieva IH, Ovsyannikova IG, Pankratz VS, Kennedy RB, Jacobson RM, Poland GA. The genetic basis for interindividual immune response variation to measles vaccine: new understanding and new vaccine approaches. Expert Rev Vaccines. 2013;12(1):57–70. doi: 10.1586/erv.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, Martin AR, Martin HC, Lappalainen T, Posthuma D. Genome-wide association studies. Nat Rev Methods Primers. 2021;1(1):59. doi: 10.1038/s43586-021-00056-9. [DOI] [Google Scholar]
- 30.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22(9):1748–59. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cano-Gamez E, Trynka G. From gwas to function: using functional genomics to identify the mechanisms underlying complex diseases. Front Genet. 2020;11. doi: 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemi MEK, Karjalainen J, Liao RG, Neale BM, Daly M, Ganna A, Pathak GA, Andrews SJ, Kanai M, Veerapen K, et al. Mapping the human genetic architecture of covid-19. Nature. 2021;600(7889):472–7. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, et al. Inborn errors of type i ifn immunity in patients with life-threatening covid-19. Science. 2020;370(6515):eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araújo R, Bento LFN, Fonseca TAH, Von Rekowski CP, da Cunha BR, Calado CRC. Infection biomarkers based on metabolomics. Metabolites. 2022;12(2):92. doi: 10.3390/metabo12020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costanzo M, Caterino M, Fedele R, Cevenini A, Pontillo M, Barra L, Ruoppolo M. Covidomics: the proteomic and metabolomic signatures of covid-19. Int J Mol Sci. 2022;23(5):2414. doi: 10.3390/ijms23052414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Ning N, Li B, Luo D, Qin E, Yu W, Wang J, Yang G, Nan N, He Z, et al. Longitudinal metabolomics reveals ornithine cycle dysregulation correlates with inflammation and coagulation in COVID-19 severe patients. Front Microbiol. 2021;12. doi: 10.3389/fmicb.2021.723818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buyukozkan M, Alvarez-Mulett S, Racanelli AC, Schmidt F, Batra R, Hoffman KL, Sarwath H, Engelke R, Gomez-Escobar L, Simmons W, et al. Integrative metabolomic and proteomic signatures define clinical outcomes in severe covid-19. iScience. 2022;25(7):104612. doi: 10.1016/j.isci.2022.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 2014;5:5. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tappe D, Pérez-Girón JV, Zammarchi L, Rissland J, Ferreira DF, Jaenisch T, Gómez-Medina S, Günther S, Bartoloni A, Muñoz-Fontela C, et al. Cytokine kinetics of zika virus-infected patients from acute to reconvalescent phase. Med Microbiol Immunol. 2016;205(3):269–73. doi: 10.1007/s00430-015-0445-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tebruegge M, Dutta B, Donath S, Ritz N, Forbes B, Camacho-Badilla K, Clifford V, Zufferey C, Robins-Browne R, Hanekom W, et al. Mycobacteria-specific cytokine responses detect tuberculosis infection and distinguish latent from active tuberculosis. Am J Respir Crit Care Med. 2015;192(4):485–99. doi: 10.1164/rccm.201501-0059OC. [DOI] [PubMed] [Google Scholar]
- 41.Vanpouille C, Introini A, Morris SR, Margolis L, Daar ES, Dube MP, Little SJ, Smith DM, Lisco A, Gianella S. Distinct cytokine/chemokine network in semen and blood characterize different stages of hiv infection. AIDS. 2016;30(2):193–201. doi: 10.1097/QAD.0000000000000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Athale S, Banchereau R, Thompson-Snipes L, Wang Y, Palucka K, Pascual V, Banchereau J. Influenza vaccines differentially regulate the interferon response in human dendritic cell subsets. Sci Transl Med. 2017;9(382):eaaf9194. doi: 10.1126/scitranslmed.aaf9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connors J, Joyner D, Mege NJ, Cusimano GM, Bell MR, Marcy J, Taramangalam B, Kim KM, Lin PJC, Tam YK, et al. Lipid nanoparticles (lnp) induce activation and maturation of antigen presenting cells in young and aged individuals. Commun Biol. 2023;6(1):188. doi: 10.1038/s42003-023-04555-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tut G, Lancaster T, Sylla P, Butler MS, Kaur N, Spalkova E, Bentley C, Amin U, Jadir A, Hulme S, et al. Antibody and cellular immune responses following dual COVID-19 vaccination within infection-naive residents of long-term care facilities: an observational cohort study. Lancet Healthy Longev. 2022;3(7):e461–e9. doi: 10.1016/S2666-7568(22)00118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kityo C, Makamdop KN, Rothenberger M, Chipman JG, Hoskuldsson T, Beilman GJ, Grzywacz B, Mugyenyi P, Ssali F, Akondy RS, et al. Lymphoid tissue fibrosis is associated with impaired vaccine responses. J Clin Invest. 2018;128(7):2763–73. doi: 10.1172/JCI97377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellebedy AH, Jackson KJL, Kissick HT, Nakaya HI, Davis CW, Roskin KM, McElroy AK, Oshansky CM, Elbein R, Thomas S, et al. Defining antigen-specific plasmablast and memory b cell subsets in human blood after viral infection or vaccination. Nat Immunol. 2016;17(10):1226–34. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park LM, Lannigan J, Jaimes MC. Omip-069: forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytometry Part A. 2020;97(10):1044–51. doi: 10.1002/cyto.a.24213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Pan K, de Bruin-Versteeg S, Damasceno D, Hernández-Delgado A, van der Sluijs-Gelling AJ, van den Bossche WBL, de Laat IF, Díez P, Naber BAE, Diks AM, et al. Development of a standardized and validated flow cytometry approach for monitoring of innate myeloid immune cells in human blood. Front Immunol. 2022;13:13. doi: 10.3389/fimmu.2022.935879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with spade. Nat Biotechnol. 2011;29(10):886–91. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y. Flowsom: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry Part A. 2015;87(7):636–45. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 51.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using umap. Nat Biotechnol. 2019;37(1):38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 52.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3(5):361–8. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 53.Leipold MD, Newell EW, Maecker HT. Multiparameter phenotyping of human pbmcs using mass cytometry. In: Shaw A, editor. Multiparameter phenotyping of human PBMCs using mass cytometry. Immunosenescence: methods and protocols. New York (NY): Springer New York; 2015. p. 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tighe RM, Redente EF, Yu Y-R, Herold S, Sperling AI, Curtis JL, Duggan R, Swaminathan S, Nakano H, Zacharias WJ, et al. Improving the quality and reproducibility of flow cytometry in the lung. An official American thoracic society workshop report. Am J Respir Cell Mol Biol. 2019;61(2):150–61. doi: 10.1165/rcmb.2019-0191ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Böttcher C, van der Poel M, Fernández-Zapata C, Schlickeiser S, Leman JKH, Hsiao C-C, Mizee MR, Adelia, Vincenten MCJ, Kunkel D, et al. Single-cell mass cytometry reveals complex myeloid cell composition in active lesions of progressive multiple sclerosis. Acta Neuropathol Commun. 2020;8(1):136. doi: 10.1186/s40478-020-01010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodríguez-Lorenzo S, van Olst L, Rodriguez-Mogeda C, Kamermans A, van der Pol SMA, Rodríguez E, Kooij G, de Vries HE, van der Pol SM. Single-cell profiling reveals periventricular cd56bright nk cell accumulation in multiple sclerosis. eLife. 2022;11:11(e73849. doi: 10.7554/eLife.73849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–42. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, Yang S-R, Kurian A, Van Valen D, West R, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174(6):1373–87.e1319. doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patwa A, Yamashita R, Long J, Risom T, Angelo M, Keren L, Rubin DL. Multiplexed imaging analysis of the tumor-immune microenvironment reveals predictors of outcome in triple-negative breast cancer. Commun Biol. 2021;4(1):852. doi: 10.1038/s42003-021-02361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park JJ, Lee KAV, Lam SZ, Moon KS, Fang Z, Chen S. Machine learning identifies t cell receptor repertoire signatures associated with COVID-19 severity. Commun Biol. 2023;6(1):76. doi: 10.1038/s42003-023-04447-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, Wang J, Sattar A, Yang Y, Zhou Y. Improving prediction of secondary structure, local backbone angles and solvent accessible surface area of proteins by iterative deep learning. Sci Rep. 2015;5(1):11476. doi: 10.1038/srep11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garg M, Li X, Moreno P, Papatheodorou I, Shu Y, Brazma A, Miao Z. Meta-analysis of COVID-19 single-cell studies confirms eight key immune responses. Sci Rep. 2021;11(1):20833. doi: 10.1038/s41598-021-00121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, Bach K, Sungnak W, Worlock KB, Yoshida M, et al. Single-cell multi-omics analysis of the immune response in covid-19. Nat Med. 2021;27(5):904–16. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metcalf TU, Wilkinson PA, Cameron MJ, Ghneim K, Chiang C, Wertheimer AM, Hiscott JB, Nikolich-Zugich J, Haddad EK. Human monocyte subsets are transcriptionally and functionally altered in aging in response to pattern recognition receptor agonists. J Immunol. 2017;199(4):1405–17. doi: 10.4049/jimmunol.1700148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Costanzo LD, Christie C, Duarte JM, Dutta S, Feng Z. Ww PDBc. Protein data bank: the single global archive for 3d macromolecular structure data. Nucleic Acids Res. 2019;47(D1):D520–D8. doi: 10.1093/nar/gky949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Tilbeurgh M, Maisonnasse P, Palgen J-L, Tolazzi M, Aldon Y, Dereuddre-Bosquet N, Cavarelli M, Beignon A-S, Marcos-Lopez E, Gallouet A-S, et al. Innate cell markers that predict anti-hiv neutralizing antibody titers in vaccinated macaques. Cell Rep Med. 2022;3(10):100751. doi: 10.1016/j.xcrm.2022.100751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fourati S, Tomalin LE, Mulè MP, Chawla DG, Gerritsen B, Rychkov D, Henrich E, Miller HER, Hagan T, Diray-Arce J, et al. Pan-vaccine analysis reveals innate immune endotypes predictive of antibody responses to vaccination. Nat Immunol. 2022;23(12):1777–87. doi: 10.1038/s41590-022-01329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pedroza-Pacheco I, McMichael AJ. Immune signature atlas of vaccines: learning from the good responders. Nat Immunol. 2022;23(12):1654–6. doi: 10.1038/s41590-022-01361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24(8):1159–69. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 72.Chambers C, Skowronski DM, Rose C, Serres GD, Winter A-L, Dickinson JA, Jassem A, Gubbay JB, Fonseca K, Drews SJ, et al. Should sex be considered an effect modifier in the evaluation of influenza vaccine effectiveness? Open Forum Infect Dis. 2018;5(9):ofy211. doi: 10.1093/ofid/ofy211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kurupati R, Kossenkov A, Haut L, Kannan S, Xiang Z, Li Y, Doyle S, Liu Q, Schmader K, Showe L, et al. Race-related differences in antibody responses to the inactivated influenza vaccine are linked to distinct pre-vaccination gene expression profiles in blood. Oncotarget. 2016;7:1949–2553. (Electronic). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fourati S, Cristescu R, Loboda A, Talla A, Filali A, Railkar R, Schaeffer AK, Favre D, Gagnon D, Peretz Y, et al. Pre-vaccination inflammation and b-cell signalling predict age-related hyporesponse to hepatitis b vaccination. Nat Commun. 2016;7(1):10369. doi: 10.1038/ncomms10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43:1097–4180. (Electronic). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR III, Castro E, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205(13):3119–31. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, Grigoryan L, Trisal M, Edara VV, Lai L, et al. Systems vaccinology of the bnt162b2 mRNA vaccine in humans. Nature. 2021;596(7872):410–6. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:1097–4172. (Electronic). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bucasas KL, Franco Lm Fau - Shaw CA, Shaw Ca Fau - Bray MS, Bray Ms Fau - Wells JM, Wells Jm Fau - Niño D, Niño D, Fau-Arden N, Arden N, Fau - Quarles JM, Quarles J, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203:1537–6613. (Electronic). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao RG, Suarez NM, Obermoser G, Lopez SMC, Flano E, Mertz SE, Albrecht RA, García-Sastre A, Mejias A, Xu H, et al. Differences in antibody responses between trivalent inactivated influenza vaccine and live attenuated influenza vaccine correlate with the kinetics and magnitude of interferon signaling in children. J Infect Dis. 2014;210(2):224–33. doi: 10.1093/infdis/jiu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Franco LM, Bucasas KL, Wells JM, Niño D, Wang X, Zapata GE, Arden N, Renwick A, Yu P, Quarles JM, et al. Integrative genomic analysis of the human immune response to influenza vaccination. ELife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9(1):659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li G-M, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boraschi D, Italiani P. Immunosenescence and vaccine failure in the elderly: strategies for improving response. Immunol Lett. 2014;162(1, Part B):346–53. doi: 10.1016/j.imlet.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 85.Pilishvili T, Gierke R, Fleming-Dutra KE, Farrar JL, Mohr NM, Talan DA, Krishnadasan A, Harland KK, Smithline HA, Hou PC, et al. Effectiveness of mRNA COVID-19 vaccine among US Health care personnel. N Engl J Med. 2021;385(25):e90. doi: 10.1056/NEJMoa2106599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin D-Y, Gu Y, Wheeler B, Young H, Holloway S, Sunny S-K, Moore Z, Zeng D. Effectiveness of COVID-19 vaccines over a 9-month period in north carolina. N Engl J Med. 2022;386(10):933–41. doi: 10.1056/NEJMoa2117128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rosenberg ES, Dorabawila V, Easton D, Bauer UE, Kumar J, Hoen R, Hoefer D, Wu M, Lutterloh E, Conroy MB, et al. Covid-19 vaccine effectiveness in new york state. N Engl J Med. 2021;386(2):116–27. doi: 10.1056/NEJMoa2116063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tartof SY, Slezak JM, Fischer H, Hong V, Ackerson BK, Ranasinghe ON, Frankland TB, Ogun OA, Zamparo JM, Gray S, et al. Effectiveness of mRNA bnt162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021;398(10309):1407–16. doi: 10.1016/S0140-6736(21)02183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FSL, Cesareni G, et al. Protein interaction data curation: the international molecular exchange (imex) consortium. Nat Methods. 2012;9(4):345–50. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–45. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan THW, Shah N, et al. Innatedb: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4(1):218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL, Lynn DJ. Innatedb: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41(D1):D1228–D33. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34(suppl_1):D504–D6. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. Intact—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35(suppl_1):D561–D5. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chatr-Aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. Mint: the molecular interaction database. Nucleic Acids Res. 2007;35(suppl_1):D572–D4. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stark C, Breitkreutz B-J, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The biogrid interaction database: 2011 update. Nucleic Acids Res. 2011;39(suppl_1):D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol. 2006;2(1):2006.0015. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, et al. Genomic analysis of the host response to hepatitis c virus infection. Proc Natl Acad Sci USA. 2002;99(24):15669–74. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bigger Catherine B, Brasky Kathleen M, Lanford Robert E. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis c virus infection. J Virol. 2001;75(15):7059–66. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brandes M, Klauschen F, Kuchen S, Germain Ronald N. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154(1):197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Askovich PS, Sanders CJ, Rosenberger CM, Diercks AH, Dash P, Navarro G, Vogel P, Doherty PC, Thomas PG, Aderem A, et al. Differential host response, rather than early viral replication efficiency, correlates with pathogenicity caused by influenza viruses. PloS One. 2013;8(9):e74863. doi: 10.1371/journal.pone.0074863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheung CY, Chan EY, Krasnoselsky A, Purdy D, Navare AT, Bryan JT, Leung CKL, Hui KPY, Peiris JSM, Katze MG. H5n1 virus causes significant perturbations in host proteome very early in influenza virus-infected primary human monocyte-derived macrophages. J Infect Dis. 2012;206(5):640–5. doi: 10.1093/infdis/jis423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lietzén N, Öhman T, Rintahaka J, Julkunen I, Aittokallio T, Matikainen S, Nyman TA, Iwasaki A. Quantitative subcellular proteome and secretome profiling of influenza a virus-infected human primary macrophages. PLoS Pathog. 2011;7(5):e1001340. doi: 10.1371/journal.ppat.1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Y, Chan Eric Y, Li J, Ni C, Peng X, Rosenzweig E, Tumpey Terrence M, Katze Michael G. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol. 2010;84(6):3023–32. doi: 10.1128/JVI.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bakker OB, Aguirre-Gamboa R, Sanna S, Oosting M, Smeekens SP, Jaeger M, Zorro M, Võsa U, Withoff S, Netea-Maier RT, et al. Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses. Nat Immunol. 2018;19(7):776–86. doi: 10.1038/s41590-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bengsch B, Ohtani T, Herati RS, Bovenschen N, Chang K-M, Wherry EJ. Deep immune profiling by mass cytometry links human t and nk cell differentiation and cytotoxic molecule expression patterns. J Immunol Methods. 2018;453:453(3–10. doi: 10.1016/j.jim.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Voge NV, Perera R, Mahapatra S, Gresh L, Balmaseda A, Loroño-Pino MA, Hopf-Jannasch AS, Belisle JT, Harris E, Blair CD, et al. Metabolomics-based discovery of small molecule biomarkers in serum associated with dengue virus infections and disease outcomes. PLoS Negl Trop Dis. 2016;10(2):e0004449. doi: 10.1371/journal.pntd.0004449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schreiber F, Lynn DJ, Houston A, Peters J, Mwafulirwa G, Finlay BB, Brinkman FSL, Hancock REW, Heyderman RS, Dougan G, et al. The human transcriptome during nontyphoid salmonella and hiv coinfection reveals attenuated nfκb-mediated inflammation and persistent cell cycle disruption. J Infect Dis. 2011;204(8):1237–45. doi: 10.1093/infdis/jir512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scott MG, Dullaghan E, Mookherjee N, Glavas N, Waldbrook M, Thompson A, Wang A, Lee K, Doria S, Hamill P, et al. An anti-infective peptide that selectively modulates the innate immune response. Nat Biotechnol. 2007;25(4):465–72. doi: 10.1038/nbt1288. [DOI] [PubMed] [Google Scholar]
- 110.Stanley Langevin MP, Smith E, Morrison J, Bent Z, Green R, Barker K, Solberg O, Gillet Y, Javouhey E, Lina B, et al. Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J Gener Virol. 2017;98(10):2425–37. doi: 10.1099/jgv.0.000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shen-Orr Shai S, Furman D, Kidd Brian A, Hadad F, Lovelace P, Huang Y-W, Rosenberg-Hasson Y, Mackey S, Grisar Fatemeh AG, Pickman Y, et al. Defective signaling in the jak-stat pathway tracks with chronic inflammation and cardiovascular risk in aging humans. Cell Systems. 2016;3(4):374–84.e374. doi: 10.1016/j.cels.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Furman D, Chang J, Lartigue L, Bolen CR, Haddad F, Gaudilliere B, Ganio EA, Fragiadakis GK, Spitzer MH, Douchet I, et al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat Med. 2017;23(2):174–84. doi: 10.1038/nm.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Metcalf TU, Cubas RA, Ghneim K, Cartwright MJ, Grevenynghe JV, Richner JM, Olagnier DP, Wilkinson PA, Cameron MJ, Park BS, et al. Global analyses revealed age-related alterations in innate immune responses after stimulation of pathogen recognition receptors. Aging Cell. 2015;14(3):421–32. doi: 10.1111/acel.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Connors J, Taramangalam B, Cusimano G, Bell MR, Matt SM, Runner K, Gaskill PJ, DeFilippis V, Nikolich-Žugich J, Kutzler MA, et al. Aging alters antiviral signaling pathways resulting in functional impairment in innate immunity in response to pattern recognition receptor agonists. GeroScience. 2022;44(5):2555–72. doi: 10.1007/s11357-022-00612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


