Abstract
Patients with cystic fibrosis (CF) exhibit pronounced respiratory damage and were initially considered among those at highest risk for serious harm from SARS-CoV-2 infection. Numerous clinical studies have subsequently reported that individuals with CF in North America and Europe – while susceptible to severe COVID-19 – are often spared from the highest levels of virus-associated mortality. To understand features that might influence COVID-19 among patients with cystic fibrosis, we studied relationships between SARS-CoV-2 and the gene responsible for CF (i.e., the cystic fibrosis transmembrane conductance regulator, CFTR). In contrast to previous reports, we found no association between CFTR carrier status (mutation heterozygosity) and more severe COVID-19 clinical outcomes. We did observe an unexpected trend towards higher mortality among control individuals compared to silent carriers of the common F508del CFTR variant—a finding that will require further study. We next performed experiments to test the influence of homozygous CFTR deficiency on viral propagation, and showed that SARS-CoV-2 production in primary airway cells was not altered by absence of functional CFTR using two independent protocols. On the other hand, experiments performed in vitro strongly indicated that virus proliferation depended on features of the mucosal fluid layer known to be disrupted by absent CFTR in patients with CF, including both low pH and increased viscosity. These results point to the acidic, viscous, and mucus-obstructed airways in patients with cystic fibrosis as unfavorable for establishment of coronaviral infection. Our findings provide new and important information concerning relationships between the CF clinical phenotype and severity of COVID-19.
Keywords: SARS-CoV-2, Cystic fibrosis transmembrane conductance regulator, virus replication, mucus membrane, COVID-19, comorbidity
Graphical Abstract
• Several studies have suggested a mechanistic relationship between CFTR and COVID-19.
• In contrast to previous reports, we found no association between CF carrier status (mutational heterozygosity) and clinical severity of SARS-CoV-2 infection among nearly 3,000 patients with COVID-19 and ~6,000 controls.
• SARS-CoV-2 production in primary airway cells was not altered by homozygous absence of functional CFTR.
• Our results point to acidic, viscous, and mucus-obstructed airways in patients with cystic fibrosis as unfavorable for propagation of the coronavirus.
Introduction
Because cystic fibrosis (CF) is associated with severe respiratory insufficiency and pulmonary failure, patients homozygous for disease-associated cystic fibrosis transmembrane conductance regulator (CFTR) mutations were expected to be among the most seriously compromised due to COVID-19 (1–5). Early in the pandemic, a national patient registry in the United States described >1,250 individuals with CF infected by SARS-CoV-2, and reported a total of 11 deaths, more than half of which occurred among those with a previous lung transplant. In a global study of 149 SARS-CoV-2 infected patients with CF aged 0–74 years (without lung transplantation), three deaths due to SARS-CoV-2 were recorded. A recently published comprehensive study of 1542 CF patients infected with the coronavirus reported a mortality rate of 0.7% (6). These values were lower than case-fatality rates in the general population across various nations that averaged ~2–3% worldwide (7). Despite severely injured CF lungs, therefore, patients with CF who contract COVID-19 usually recover favorably (3, 8–20), although SARS-CoV-2 outcomes in CF could change for the worse at any time due to new viral variants or other factors.
The finding that individuals with CF may often be spared the most dire consequences of COVID-19 is not understood. A number of features have been suggested by others to help account for this clinical observation. Social distancing - to prevent transmission of bacterial or other respiratory pathogens - is routinely practiced by the CF patient community, and it has been proposed that SARS-CoV-2 might have been limited at least in part by diligent mask wearing or other health-related behavioral measures among patients with cystic fibrosis (13). However, since significant numbers of individuals with CF in North America and Europe have contracted COVID-19 (see above), clinical outcomes cannot simply be attributed to physical distancing without invoking other factors (e.g., diminished viral inoculum due to protective masks). It has also been speculated that because of chronically inflamed respiratory tissue, CF lungs might be less able to mount the cytokine storm or related pathogenic responses elicited by SARS-CoV-2. Other diseases characterized by chronic pulmonary inflammation, however, continue to be viewed as poor prognostic indicators for the virus (1, 21, 22). Individuals with CF are also younger than the general population, and infrequently exhibit SARS-CoV-2 risk factors such as marked obesity or coronary artery disease, although CF-related diabetes mellitus is a relatively common finding. Moreover, younger age at time of infection does not mitigate COVID-19 severity in chronic respiratory conditions such as bronchopulmonary dysplasia (21). While early in the pandemic, age did not appear to be a statistically significant indicator of CF hospitalization due to SARS-CoV-2 (3), subsequent studies have shown an association between older age and increased disease severity among patients with CF (6). Another potential consideration involves use of chronic CF medications (e.g., bronchodilators, anti-inflammatories, azithromycin, DNase, etc.) that in some cases might afford protection against coronaviral lung disease (for example (23–25)). The potential benefits of anti-inflammatory drugs have been supported in clinical studies of non-CF individuals infected with SARS-CoV-2 (26), although azithromycin in particular was not found to provide protection in CF (27, 28). In summary, while it is important to acknowledge that patients with CF have fared far better than originally envisioned, potential relationships between clinical CF, COVID-19, and CFTR function require further investigation.
CFTR is an epithelial chloride and bicarbonate ion channel that regulates composition and thickness of the airway surface fluid in human lungs (29–31). Specific features of CFTR cellular biology have been postulated as a link between CF and COVID-19 severity (32–39). For example, pre-pandemic studies of airway biology described CFTR localization in clathrin-coated pits (same location as the coronaviral receptor), debated CFTR as a regulator of endosomal pH (and therefore potentially contributing during viral proteolysis and/or uncoating), demonstrated CFTR regulation of airway mucosal pH, and showed local chloride concentration to be important during receptor attachment of other coronaviruses (40–55). Any of these factors might be taken to suggest mechanistic connections between CFTR and coronaviral pathogenesis.
In the present study, we investigated key aspects of F508del CFTR in relation to SARS-CoV-2 respiratory infection. We found higher CF carrier frequencies for a population of younger Italian males with severe COVID-19, but did not observe a similar association among Spanish males. We detected an overall trend towards protection from death in a large European cohort comprising heterozygotes for F508del CFTR, a finding that will require further study. These results are important for interpreting a number of earlier publications indicating that carrier status for CFTR mutations predisposes to more severe SARS-CoV-2 lung disease (56–58). We next tested whether homozygous CFTR defects would alter viral infectivity in vitro. We utilized primary human bronchial epithelial cell (hBEC) monolayers from individuals homozygous for F508del CFTR, and observed no statistically significant impact on virus replication , as judged by two virology laboratories employing independent protocols. Ionic composition, pH, viscosity, and protease balance are known to be strongly disrupted in airway fluids from patients with CF. We provide evidence that replacing airway epithelia-generated surface liquid in vitro with physiologic buffer leads to impairment of SARS-CoV-2 infection. Moreover, physicochemical properties that are well described features of CF respiratory secretions—low pH and elevated viscosity—inhibited viral production. Our findings support a model in which an acidic, viscous, and chronically disrupted CF pulmonary mucosal microenvironment may act to impair coronaviral propagation. The data are important for an improved mechanistic understanding of SARS-CoV-2 pathogenesis among a group of patients otherwise considered at high risk for life-threatening COVID-19.
Materials and Methods
Human DNA samples and genotyping
Two independent sets of patient samples were genotyped. In one study, blood was obtained from 424 individuals from the US hospitalized with severe COVID-19. The majority were from Northeast Georgia Medical Center, Gainesville, Georgia (295 samples) or Emory University Hospital, Atlanta, Georgia (119 samples), with ten samples from Elliot Hospital, Manchester, NH. Of the 424 samples, 296 came from patients that self-identified as “White - not Hispanic”, and 128 samples from individuals identified as “White” – which may include a subset of Hispanic ethnicity. A much larger study evaluated White subjects residing in Italy, Spain, or Norway, previously analyzed by Ellinghaus et al. and Degenhardt et al. (59, 60). Individuals from these cohorts were hospitalized with severe COVID-19 between February and September 2020 (European cohort), or between April and September 2020 (US cohort). SARS-CoV-2 positivity status was validated by RNA polymerase-chain-reaction (PCR) of nasopharyngeal swabs or other relevant biologic fluids and “severe” individuals were defined as SARS-CoV-2 positive, hospitalized patients requiring respiratory support ranging from nasal oxygen and mechanical ventilation to extracorporal membrane oxygenation (ECMO). In total, 9,545 European individuals with complete data on age and sex that passed quality control in a genome-wide association study (GWAS) by Degenhardt et al. (60) were evaluated for CFTR F508del. Twenty-eight samples failed to be typed for the CFTR variant, whereas 9,517 individuals were appropriate for analysis. Overall, 2,540 patients and 6,977 matched controls from the general population with unknown SARS-CoV-2 status were evaluated. General statistics regarding these individuals are shown in Supplementary Tables S2A–C. DNA in both studies was genotyped using a TaqMan SNP assay for the common F508del CFTR variant (rs113993960; ThermoFisher, MA, USA).
Cell lines
Vero E6 cells (American Type Culture Collection (ATCC), VA, USA) were propagated in Dulbecco’s Modified Eagle’s Medium (DMEM), with 10% v/v Serum Plus II Medium Supplement (Sigma-Aldrich, MO, USA), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine (GIBCO, ThermoFisher). Except where noted otherwise, Calu-3 pulmonary adenocarcinoma-derived cells (ATCC) were grown as submerged monolayers in Eagle’s Minimum Essential Medium (MEM), supplemented with 10% v/v fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, 1% minimum essential medium (MEM, GIBCO, ThermoFisher) non-essential amino acids, and 2 mM L-glutamine.
Pulmonary cells grown at air-liquid interface (ALI)
Human bronchial epithelial cell models (i.e., airway epithelium) were obtained from commercial vendors as follows: three healthy (non-CF) donor cell lines (c2540s, Lonza, NC, USA; c12640, PromoCell, Heidelberg, Germany; FC0035, Lifeline Cell Technology, MA, USA) and three F508del/F508del CF donor cell lines (CF-AB-045–202 and CF-AB-060–901, Epithelix Sàrl, Switzerland; FC0103, Lifeline Cell Technology). Primary cells were propagated beginning at passage three on PureCol (Sigma-Aldrich) coated T75 flasks with Ex Plus expansion medium (Stemcell Technologies, Vancouver, Canada) at 37°C and 5% CO2/95% O2 until ~70–80% confluent. Monolayers were generated according to well established protocols (61–65) on 6.5 mm transwells (Corning) coated with collagen (type IV from human placenta, Sigma-Aldrich) by seeding at a density of 150,000 cells per insert, and maintained under submerged conditions in Ex Plus medium for three days. Apical and basolateral culture medium was aspirated and lower chamber fluid replaced with airway-liquid interface (ALI) medium (Stemcell Technologies). These conditions were maintained for 21–28 days until monolayers became fully differentiated. The respiratory epithelial model generated in this manner faithfully replicates in vivo characteristics of pulmonary mucosa such as basal, ciliated, and other cell types, junctional complexes, and SARS-CoV-2 propagation (61–73) (see also below).
Calu-3 cell air liquid interface cultures were expanded in MEM supplemented with 10% FBS and 1% MEM non-essential amino acids until 70–80% confluent. Approximately 150,000 cells were seeded on 6.5 mm transwells with culture medium present on both apical and basolateral surfaces. Medium from the apical chamber of each insert was removed three days after seeding, and monolayers maintained under ALI conditions an additional four days prior to analysis.
Production of virus stocks and studies of virus infection
SARS-CoV-2 isolate USA-WA1/2020 was obtained from BEI Resources (VA, USA) and propagated in Vero-E6 cell culture. Virus-containing medium was harvested two days post infection, when significant cytopathic effect was observed. Viral stocks were titered by serial dilution on Vero-E6 cells in a 96-well plate format, with seeding at 20,000 cells per well. The following day, infection for determining titer was performed with serially diluted virus. Cells were fixed 5–7 h post infection and stained for SARS-CoV-2 nucleocapsid (N) protein. This timing was chosen based on our observation that N protein expression can be detected as early as two hours post-infection (hpi), together with data showing by 10 hpi, infection will have begun to spread (74, 75). Viral titer was defined as the number of infectious units (IU, i.e., cells stained positive for N) per ml. Stocks used in these studies were between 2×104 and 2×106 IU/ml.
The mNeonGreen expressing reporter virus, icSARS-CoV-2_mNG, was acquired from World Reference Center for Emerging Viruses and Arboviruses (WRCEVA, TX, USA) in lyophilized culture medium (76). This virus is based on the USA-WA1/2020 isolate, but expresses mNeonGreen in place of Orf7a, allowing identification of infected cells by fluorescence microscopy. Virus was resuspended in phosphate buffered saline (PBS) and amplified by passage through Vero-E6 cells, followed by titering as above. Replication deficient adenovirus expressing GFP under control of a CMV promoter (AdV-GFP) was obtained from the Baylor University Gene Vector Core (Waco, TX, USA) and used as a control vehicle for infection, as well as one test of monolayer integrity following washing/replacement of overlying mucosal surface liquid.
SARS-CoV-2 replication kinetics in bronchial epithelial cells
Experiments to probe replication kinetics were performed at Rocky Mountain Laboratories, National Institutute of Allergy and Infectious Diseases (NIAID). Differentiated monolayers of human bronchial epithelial cells were grown in air-liquid interface transwells. Tissue samples from three CF and three non-CF donors were inoculated; four replicate wells were harvested per donor per timepoint. To assess virus replication kinetics, monolayers were gently washed with PBS twice; SARS-CoV-2 USA-WA1/2020 virus stock was diluted in growth media to achieve MOI 0.1 (based on stock virus titer in tissue culture infectious dose 50 (TCID50)), added to the apical surface of the monolayers, and incubated for 1 hr before removal and washing with PBS. To collect samples, 250 μl PBS was pipetted onto the apical surface of the monolayer, mixed and removed, at three, 12, 24, 48 and 72 hrs post inoculation. 140 μl of this supernatant was used for RNA extraction with the QiaAmp Viral RNA kit (Qiagen, MD, USA) according to manufacturer’s specifications. Five μl of RNA was used in a one-step real-time RT-PCR assay to detect E gRNA (77) with the QuantiFast probe kit (Qiagen, MD, USA) according to instructions of the manufacturer. In each run, standard dilutions of counted RNA standards were measured in parallel to calculate copy numbers in the samples. A single stock solution was used for viral inoculation at the t = 0 time point and verified for active virus.
Immunofluorescence microscopy
Cells were grown and infected in a 96-well plate format and fixed by removing medium and replacing with 100 μl 4% v/v paraformaldehyde in PBS. Following incubation at room temperature for 30–60 minutes, paraformaldehyde was replaced with 100 μl 0.1% v/v Triton X-100 in PBS to permeabilize cell membranes. After 10 minutes, Triton X-100 was aspirated and 100 μl blocking buffer (5% v/v FBS in PBS supplemented with 0.1% v/v Tween-20 (PBST)) incubated for one hour. Forty μl rabbit monoclonal anti-nucleocapsid antibody (SinoBiological #40143-R001) was diluted 1:5,000 in blocking buffer and added to plates overnight at 4°C. Samples were subsequently washed twice with 100 μl PBST and incubated with 40 μl goat anti-rabbit Alexa Fluor 647 (Invitrogen, #A-21244) and Hoechst-33342 (Invitrogen, ThermoFisher, MA, USA) diluted 1:2,000 and 1:10,000, respectively, in PBST for one hour at room temperature in the dark. Samples were then washed four times with 100 μl PBST and imaged using a Cytation 5 multi-mode microscope (Biotek, VT, USA). Total cell counts (nuclei) and infected cells were quantified with Gen5 software (Biotek).
Sputum Collection and Processing
CF hBECs in vitro do not consistently produce increased volumes of mucus (compared to wild-type) or exhibit the ‘plastering’ of viscous secretions to epithelial surfaces commonly observed in vivo. Moreover, cystic fibrosis hBECs fail to demonstrate key aspects of chronic CF inflammation, making it difficult to replicate certain hallmarks of in vivo sputum caused by homozygous loss of CFTR. That said, airway epithelial monolayers do mimic crucial elements of the in vivo mucosal surface, including transepithelial tissue resistance, membrane polarity, vectoral ion transport, cellular populations resembling those in vivo, and CF bioelectric abnormalities. In order to evaluate impact of in vivo airway secretions from patients with CF on viral production, freshly isolated sputum or processed sputum supernatants were obtained from the Biospecimen Registry at the Emory/Children’s Hospital Cystic Fibrosis Care Center in Atlanta. Sample collection was in accordance with institutional guidelines and all donors provided informed consent. Sputum samples were maintained under sterile conditions on ice at 4°C after expectoration. For some studies, supernatants were prepared as follows: 10 ml of PBS-EDTA was added per gram of sputum, followed by homogenization using multiple passages through a sterile 18-gauge needle. After two cycles of centrifugation (800g (10 min) and 3000g (20 min)) at 4°C, supernatants were collected. Complete protease inhibitor cocktail (Roche, Basel, Switzerland) was added with subsequent storage at (−80°C). If needed, a small amount of PBS was included to facilitate pipetting of viscous secretion.
Infection of submerged Calu-3 cells
Calu-3 cultures in 96-well plates were infected 24-hours post-seeding at near 100% confluency. Carboxymethylcellulose (CMC), a compound used to augment viscosity, was prepared at 2% w/v in PBS. After mixing to dissolve CMC powder, the solution was autoclaved to sterilize, and then diluted to various concentrations with PBS. CMC solutions were mixed 1:1 with EMEM culture medium and added to cell monolayers in a total volume of 100 μl. icSARS-CoV-2_mNG was added to cells at MOI of 0.01 in 20 μl EMEM. After 24 h, cells were fixed with 4% paraformaldehyde, and imaged using a Cytation 5 multi-mode microscope as above. A similar CMC protocol was applied to test hBEC monolayers.
Adjustment of ALI culture conditions
pH modification: Hank’s balanced salt solution (HBSS) was titrated to desired pH (between 6.5 and 8.0), and equilibrated at 37 °C, 5% CO2 for 30 minutes. Virus stocks were diluted 1:10 in modified HBSS prior to infection.
Mixing of sputum with SARS-CoV-2: virus stocks were diluted in sputum or supernatant prior to infection.
Viscosity: SARS-CoV-2 was diluted 1:10 in PBS or PBS supplemented with CMC (see above).
Washing: Two hundred μl of Pneumacult-ALI medium (Stemcell Technologies) was added apically to monolayers. After 20 minutes, medium was gently aspirated.
Virus recovery: ALI cultures were infected with 10 μl virus solution (SARS-CoV-2 or recombinant adenovirus) added to the apical surface of monolayers. Unless otherwise specified, cultures were not washed to remove airway surface liquid and mucus prior to infection. Following infection, RNA was harvested using Purelink RNA Mini Kit (Invitrogen, MA, USA).
qPCR to quantify SARS-CoV-2 RNA
For experiments performed at Emory University, RT-qPCR was conducted using the Center for Disease Control (Research Use Only) kit (IDT #10006713) and GoTaq® Probe 1-Step RT-qPCR (Promega, WI, USA). SARS-CoV-2 RNA quantification utilized a standard N1 primer and probe set, with host RNA levels measured by primers that amplify ribonuclease P (RNAseP). Changes in levels of SARS-CoV-2 RNA were assessed using the ΔΔCt method, where abundance of N RNA at 48 h post-infection was normalized first to cellular RNAseP from the corresponding sample, and subsequently to abundance at 0 h, i.e, input. This conventional approach to qPCR allows monitoring of relative levels of viral production but not absolute viral concentrations. Fold change in the level of SARS-CoV-2 N RNA was determined assuming a two-fold difference in RNA level per amplification cycle of qPCR.
Gene expression
Droplet digital PCR (ddPCR) employed the QX200 PCR System (Bio-Rad, CA, USA) according to manufacturer’s protocol. Total RNA was retrotranscribed in SuperScript IV VILO Master Mix (ThermoFisher Scientific), and cDNA products (10 ng) transferred to 96-well plates compatible with the droplet generator device. ddPCR master mix for gene expression was prepared by addition of two-fold diluted ddPCR supermix for probes (no dUTP) (Bio-Rad), with 0.9 μM and 0.25 μM final concentrations of primers and FAM probes, respectively. Target genes included: human-ACE2 (Hs01085333_m1), human-TMPRSS2 (Hs01122322_m1) (ThermoFisher Scientific), and CFTR (Hsa-CPE5056656) (Bio-Rad). Similar concentrations of reference human-TBP (Hsa-CIP0036255) or human-HPRT1 (Hsa-CPE5192872) primers and HEX probes (Bio-Rad) were studied as controls. Following PCR amplification of target and reference genes in droplets, reactions were evaluated using a QX200 Reader (Bio-Rad). QuantaSoftTM Software (Bio-Rad) determined the numbers of positive and negative droplets for each fluorophore in all samples (providing concentrations of target and reference DNA molecules in units of copies/μl input). Results were calculated as the ratio of copies of target per reference gene.
Statistics
F508del status and COVID-19 severity in human subjects
For evaluation of CFTR genotypes, we performed analyses in R version 3.6.1. Logistic regression based on COVID-19 case/control status was assessed, including age, sex, age*age, age2 and the first 10 principal components calculated from whole-genome genotypes post-GWAS quality control (59, 60). A meta-analysis across the different European studies (Italy, Spain and Norway) was conducted using an inverse-variance weighted fixed-effects approach and the R package metafor (doi: 10.18637/jss.v036.i03). We also stratified the analysis for sex and age, omitting age and sex covariates from the regression models, respectively. We selected age groups <=60 and >60 in correspondence to evaluation performed by the COVID-19 Human Genetics Initiative (HGI) and Degenhardt et al. (60). To test for possible correlation of a heterozygous (CF carrier status) CFTR genotype with disease severity, we grouped COVID-19 positive individuals into two cohorts: those who received nasal oxygen only were assigned the base “control” status, and individuals who received at least mechanical ventilation were assigned a “case” status. With mortality data available for a subset of individuals, we also performed a logistic regression analysis in the COVID-19 patients only with “survival at hospital release” coded as “control” and “death” coded as “case”. All analyses were conducted using an additive/dominant genetic model. For these studies, CFTR genotypes were coded as 0,1 with 0 representing wt/wt and 1 representing wt/F508del. We detected one F508del/F508del individual in the male Italian population with severe COVID-19 but excluded him from evaluation (any statistical significances remained when this individual was included in the analysis). Summary information describing patient demographics and treatment regimens is presented in Tables S1A, S1B, and S1C, with CF carrier frequencies shown in Tables 1 and 2. The corresponding allele frequencies are also summarized in Supplemental File S1. P-values of association were corrected for multiple testing using the Bonferroni-Holm correction method. Only studies with more than 50 affected and unaffected individuals within each stratum were considered (i.e. the Norwegian cohort was omitted for the substratified assessment).
Table 1.
Carrier frequencies of the CFTR F508del allele (i.e., F508del heterozygosity) in healthy individuals vs. individuals hospitalized with severe COVID-19 and meta-analysis for p-value of association with COVID-19 outcome. The p-value of association, corrected for multiple testing using the correction method of Benjamini-Holm (pFDR), was obtained from inverse-weighted fixed effects meta-analysis as detailed in Methods. Note: patient demographics and treatment regimens for study subjects are provided in Tables S1A, S1B, and S1C.
| Italy | Spain | Norway | ||||
|---|---|---|---|---|---|---|
| healthy | COVID-19 | healthy | COVID-19 | healthy | COVID-19 | |
| total | 3266 | 1251 | 3449 | 1226 | 262 | 62 |
| wt/wt | 3211 | 1228 | 3400 | 1206 | 259 | 61 |
| wt/F508del | 55 | 23 | 49 | 20 | 3 | 1 |
| carrier frequency, % | 1.68 | 1.84 | 1.42 | 1.63 | 1.15 | 1.61 |
| Meta-analysis | p = 0.205; pFDR = 1 | |||||
Table 2.
Carrier frequencies of the CFTR F508del allele in individuals with severe COVID-19 who survived at the time of hospital release vs. those who died with COVID-19, and meta-analysis p-value of association with COVID-19 outcome. The p-value of association, corrected for multiple testing using the correction method of Benjamini-Holm (pFDR), was obtained from inverse-weighted fixed effects meta-analysis as detailed in Methods.
| Italy | Spain | |||
|---|---|---|---|---|
| survived | died | survived | died | |
| total | 811 | 152 | 812 | 154 |
| wt/wt | 796 | 151 | 796 | 153 |
| wt/F508del5 | 15 | 1 | 16 | 1 |
| carrier frequency, % | 2.09 | 0.66 | 1.97 | 0.65 |
| Meta-analysis | p = 0.178; pFDR = 1 | |||
In vitro statistical analyses were performed using Graphpad Prism 9; tests included two-tailed t-tests (Fig. 2A–C); two-way ANOVA with Bonferroni’s post-hoc test (Fig. 1A, C; Fig, 4 A–C); repeated-measures ANOVA with Dunnett’s post-hoc test (Fig. 3A, C).
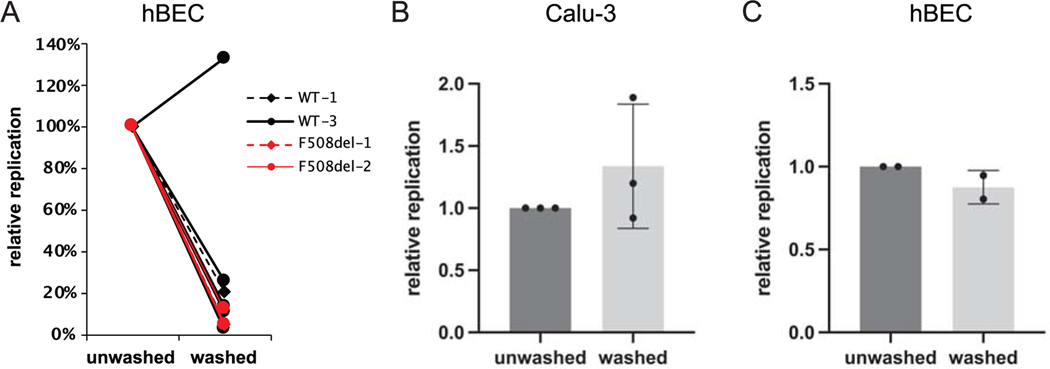
Figure 2.
Impact of washing mucosal fluid prior to infection by SARS-CoV-2. (A) Human primary airway (bronchial) epithelia (hBEC), or (B) Calu-3 cells were prepared at air-liquid interface, and treated with ~300 infectious units of SARS-CoV-2 in PBS. In A), two different WT hBEC cell models from different individuals were tested eight times, and two different CF hBEC models were tested three times, under both gently washed and unwashed conditions (11 experiments in total). Washing was conducted to remove periciliary fluid and mucus. Total RNA from cells was harvested at 48 h post infection and levels of SARS-CoV-2 RNA determined by RT-qPCR. Replication is expressed as relative viral RNA levels at 48 h, comparing unwashed vs. washed samples. The effects of washing were pronounced and, in many cases, essentially superimposable, making individual line graphs in Panel A difficult to distinguish. P value calculations by t-test included all 11 experiments (washed vs. unwashed, p < 0.0001). (C) Human primary bronchial epithelia were prepared as above at ALI (donor WT-1), then transduced with 1×108 infectious units (titered on 293T cells) of GFP-expressing recombinant adenovirus. After 48 h, GFP-positive cells were counted. Data in Panels (B) and (C) represent means and standard deviations. Overall viral production in Calu-3 cells was modestly (~2.5-fold) higher than in primary airway epithelial monolayers. Tissue origin from which cells are derived (airway glandular (Calu-3) versus pulmonary surface epithelial), growth conditions (including time required to achieve mature, polarizing cells), confluency, and other factors may contribute to differences in viral replication in hBEC versus Calu-3 model systems.
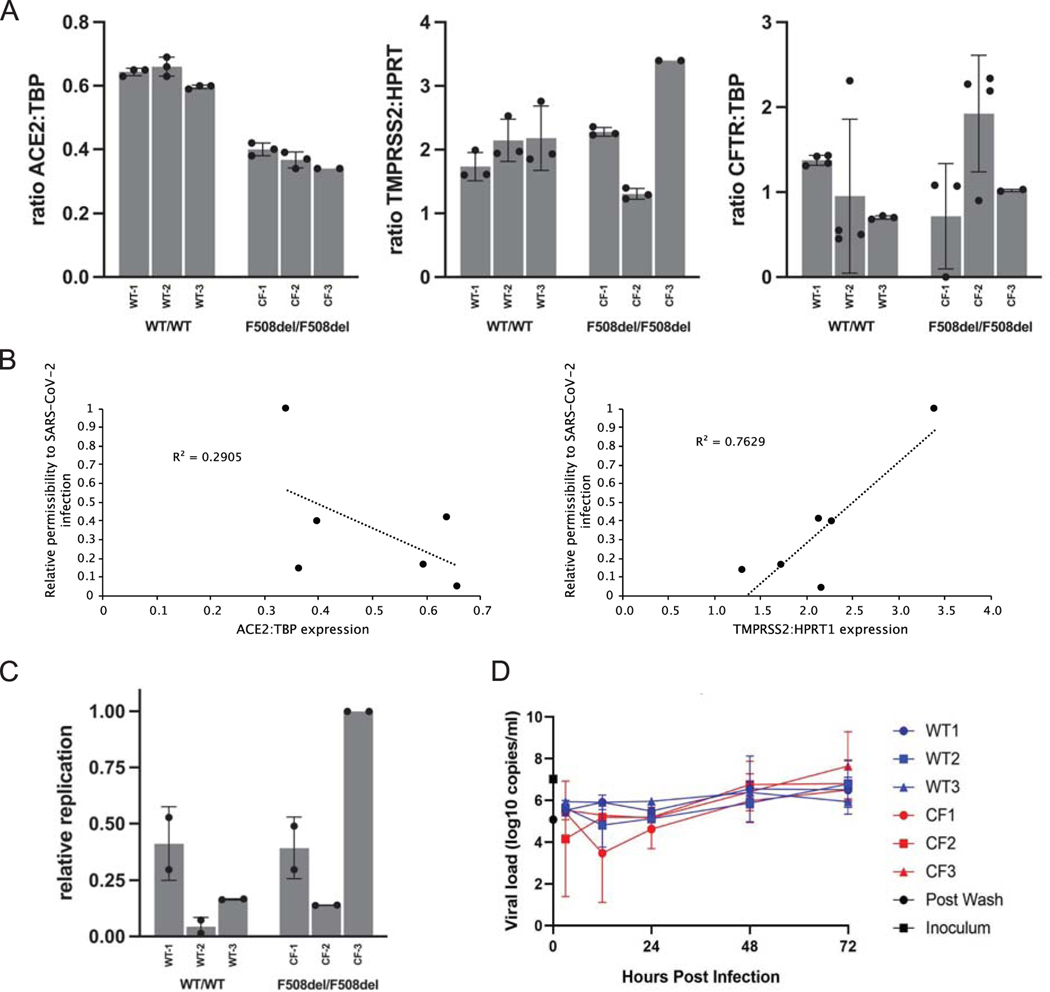
Figure 1.
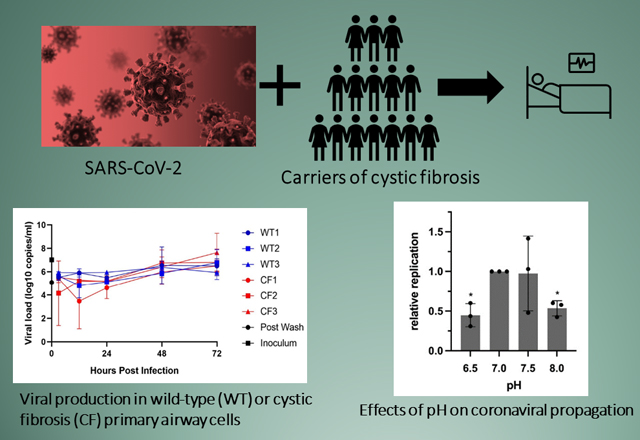
SARS-CoV-2 related features of primary human airway (bronchial) epithelium (hBEC) from three non-CF individuals or three patients with CF. Human primary cells were prepared as differentiated monolayers at air-liquid interface for each data point. (A) Quantitative analyses of ACE2, TMPRSS2, and CFTR expression in CF versus non-CF primary airway epithelia determined by ddPCR. Data is shown as the ratio of target gene expression to TBP or HPRT1 internal controls and calculated as mean +/− standard deviation (SD). Each bar represents 2–4 biological replicates per patient sample, with 2 ddPCR runs per replicate. (TMPRSS2 was normalized to HPRT1 due to expression at much higher levels than CFTR or ACE2 in primary airway epithelia.) For ACE2: CF vs. non-CF, p <0.0001; for TMPRSS2 and CFTR: CF vs. non-CF, p >0.05. (B) Correlation analysis between ACE2 and TMPRSS2 mRNA levels and SARS-CoV-2 replication. (C) hBEC monolayers were grown at air liquid interface and exposed to ~300 infectious units of SARS-CoV-2. Total RNA was harvested 48 h post infection and levels of SARS-CoV-2 RNA determined by RT-qPCR. Each bar represents a different individual with 2 biological replicates and 2 wells per repeat (4 wells total), with two duplicate qPCR samples per well (8 total qPCR values per bar). Propagation is expressed as viral RNA within cells, 48 h post infection, relative to sample CF-3 and normalized to RNaseP. Standard deviation bars are shown. Note that for certain points, error bars are shorter than height of symbols. (D) Virus replication kinetics in bronchial epithelial monolayers from three non-CF individuals and three individuals with CF were determined by inoculating separate monolayers with an MOI of 0.1 of SARS-CoV-2. Samples were collected from the apical surface at 3, 12, 24, and 48 h post infection. RNA was extracted and RT-qPCR performed to detect E gRNA. In each run, dilutions of counted RNA standards were run in parallel to calculate copy numbers in the samples. To confirm active virus, RT-qPCR was performed on the inoculum and immediately after removal of the wash. Data represent means and standard error of 4 replicate wells per donor per timepoint. Values of replication are variable between patient hBEC samples due to factors such as cell confluence, passage number, monolayer resistance, effects of genes other than CFTR, epistasis, etc. Differences between WT versus CF primary cells in (C) amount to modestly higher production of SARS-CoV-2 in the CF models, but overall (WT vs. CF) failed to reach statistical significance. CF samples in Figure 1D showed a maximal increase (peak value vs. 3-hour post inoculum timepoint) of 624-fold and non-CF samples showed a 370-fold increase.
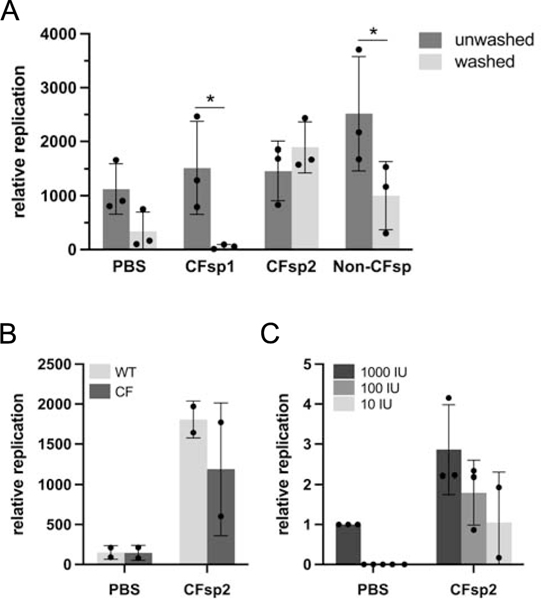
Figure 4.
Effect of CF or non-CF human sputum on SARS-CoV-2 infection. Primary bronchial epithelial cells were prepared as epithelial monolayers at air-liquid interface. (A) A subset of bronchial epithelial cell filters were washed to remove conditioned surface liquid and mucus. Prior to infection, 2,000 infectious units of SARS-CoV-2 were diluted 1:5 either in PBS or in freshly expectorated mucus collected from patients with cystic fibrosis or a healthy individual and added to cells in culture. Multiple comparison p values washed versus unwashed (Bonferroni): PBS: 0.57; CFsp1: 0.05; CFsp2: >0.99; Non-CFsp: 0.04. (B) Paired hBEC samples (WT/WT or F508del/F508del) were washed to remove surface liquid and mucus. SARS-CoV-2 viral stock was diluted 1:5 in PBS or sputum supernatant and 2,000 infectious units added to airway cell monolayers. PBS versus CFsp2, p = 0.01 (2-way ANOVA). (C) Prior to infection of unwashed non-CF epithelia, 1,000, 100 or 10 infectious units (IU) of SARS-CoV-2 were diluted in PBS or CF sputum supernatant 2. PBS versus CFsp2, p = 0.02; for virus dose effect, p = 0.03 (2-way ANOVA). Replication values in all panels represent viral RNA levels at 48 h. In (C), RNA levels are additionally normalized to the sample inoculated with 1,000 IU virus in PBS. Error bars in all panels represent standard deviations.
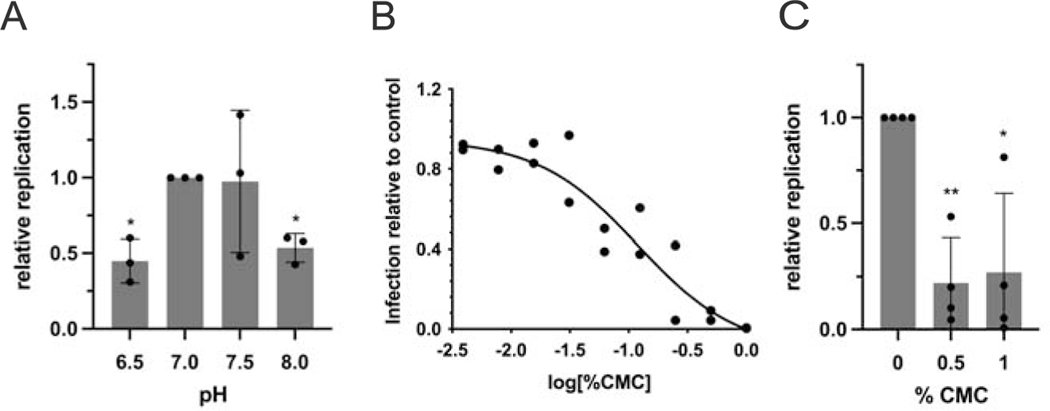
Figure 3.
Effect of mucosal pH and viscosity on SARS-CoV-2 infection. (A) Human primary airway cells were prepared at air-liquid interface, and treated with approximately 2,000 infectious units of SARS-CoV-2 in HBSS at the indicated pH. Total RNA was harvested 48 h post infection and levels of SARS-CoV-2 RNA determined by RT-qPCR. Replication is expressed as viral RNA levels, relative to results at pH 7 (*p < 0.05). (B) Calu-3 cells were seeded in 96-well plates. Prior to infection, cultures were switched to medium adjusted to contain the indicated percentage (v/v) of carboxymethylcellulose. Cells were infected at an MOI of 0.01 with icSARS-CoV-2-mNG. After 48 h, monolayers were fixed and number of infected cells per well determined by fluorescence microscopy. (C) Human bronchial epithelial monolayers at air-liquid interface were treated with approximately 1,000 infectious units of SARS-CoV-2 diluted in PBS containing the indicated percentage (w/v) of carboxymethylcellulose. Error bars in (A) and (C) represent standard deviations. (**p = 0.009 at 0.5% CMC and *p = 0.048 at 1% CMC compared to control)
Results
CFTR F508del carrier status does not impact likelihood of hospitalization from severe COVID-19 (first pandemic wave)
Mutant CFTR carrier status is associated with a number of non-CF respiratory diseases, including bronchiectasis, sinusitis, and bronchitis (78). Earlier studies have reported that CFTR mutation carrier status predisposes to more severe SARS-CoV-2 infection (56–58). Among 424 samples from U.S. patients hospitalized with severe COVID-19, we detected eight individuals heterozygous for CFTR F508del (1.9%). The expected carrier frequency among the Caucasian population in the United States is ~2–3%. This finding suggested carriers of F508del exhibit similar susceptibility to severe COVID-19 compared to the overall U.S. population (p > 0.05, binominal test, observed vs. expected distribution). In order to extend the analysis in a much larger number of subjects, we determined F508del heterozygous status in 2,540 patients hospitalized with COVID-19 and 6,977 controls from a European cohort in Italy, Spain, and Norway. Although we detected slightly higher frequencies of F508del carriers in patients exhibiting severe COVID-19 compared to control individuals (frequencies, controls/COVID-19): Italy - 1.68%/1.84%; Spain - 1.42%/1.63%; Norway - 1.15%/1.61%), these differences were not statistically significant (all p values > 0.05; meta-analysis: p = 0.205; Table 1 and Supplemental File S1).
Younger age as a COVID-19 risk factor among male Italian F508del carriers
We next analyzed F508del CFTR carrier status in the European cohort for the following population categories: male or female gender; age 60 or under; age over 60; and disease severity (requiring nasal oxygen vs. mechanical ventilation). Italian males under 60 requiring COVID-19 hospitalization had higher CFTR F508del carriership than in the healthy Italian population (3.73% vs. 1.74%; p = 0.014). The same did not hold among Spanish males (1.92% vs. 1.54%; p = 0.471; Supplemental File S1). In an attempt to explain this difference, we examined potential confounders (disease severity and comorbidities) in both the Italian and Spanish cohorts. The younger Italian COVID-19 patients, on average, received ventilation more often than the Spanish group (87.9% vs. 45.1%, respectively; Supplemental Table S1C). However, the level of respiratory support could not account for differences in F508del carrier frequency for the two populations (p = 0.793; Supplemental Fig. S1A). Hypertension, coronary artery disease, and diabetes were also comparably represented in the younger male COVID-19 patients (Supplemental Table S1C), and no differences were observed in carriership of F508del with regard to hypertension or coronary artery disease (p = 0.707 and p = 0.893, respectively; Supplemental Fig. S1A).
F508del CFTR and a trend towards protection from COVID-19 related death
Analysis of survival was possible in a total of 1,930 individuals from Italy and Spain. F508del carrier frequencies among patients who survived were three times higher than those who died (~2.0% vs. ~0.65%). An OR of 0.36 (0.08 – 1.59) indicated a trend towards protective effects of carrier status (p = 0.178; Table 2; Supplemental File S1). While a tendency for protection of CF carriers from COVID-19 mortality could be of interest from an evolutionary standpoint (see below), the data indicate larger numbers of study subjects will be necessary to determine the significance of this finding in detail.
ACE2, TMPRSS2, and CFTR mRNA expression in CF and non-CF airway epithelia
Because no robust effects on COVID-19 outcomes could be ascribed to F508del CF carrier (heterozygous) status, we next evaluated the homozygous CF genotype – which leads to a very different set of clinical findings than mutant CF heterozyosity. Patients homozygous for CFTR variants exhibit mucus and airway surface fluid acidification, hyperviscosity, blanketing of the airways by CF secretions, and occlusion of access to the mucosal surface. As noted above, homozygosity for CFTR mutations in vivo has also elicited patient outcomes considerably less severe than anticipated. We therefore performed studies directed towards impact of homozygous CFTR variants in relation to features of SARS-CoV-2 pathogenesis. This included potential mechanistic relationships between complete CFTR deficiency in vitro and aspects of SARS-CoV-2 infection. We began by testing expression of two proteins important for viral entry, ACE2 and TMPRSS2 (79), in CF-derived F508del/F508del primary airway epithelial monolayers and in matched airway cells from healthy donors expressing wild-type CFTR. ACE2 mRNA expression was reduced by approximately 30% in CF cells (p <0.0001). This result is similar to a previous report in which Bezzerri et al. showed ACE2 was deficient in CF primary airway epithelia as judged by quantitative immunofluorescence. The same report indicated decreased SARS-CoV-2 replication and diminished IL6 expression in the CF samples (80). TMPRSS2 RNA expression was variable (Fig. 1A), but (unlike ACE2) demonstrated moderate correlation with SARS-CoV-2 replication (Fig. 1B, R2 = 0.76), compatible with importance of secreted proteases during the viral infectious cycle (30, 81). Protease/anti-protease imbalance is well established in CF airways in vivo (82–91), and the possibility that clinical features of CF lung disease related to protease activity might influence SARS-CoV-2 infection has been suggested previously (32, 39). As expected, F508del CFTR mRNA levels were similar to wild-type (p = 0.440), i.e., the F508del variant elicits a protein folding abnormality without markedly altering CFTR mRNA abundance (Fig. 1A). Comparable levels of KRT5 mRNA, a pulmonary basal cell marker, indicated that monolayers contained similar airway progenitor cell differentiation (92). (CF and non-CF samples differed for KRT5 mRNA by 1.16%; p = 0.48.)
Absence of CFTR and effects on viral propagation in primary airway epithelial monolayers
To test whether lack of functional CFTR can impact infection by SARS-CoV-2, we examined primary human bronchial epithelial cells from healthy controls and patients with CF encoding the F508del/F508del genotype. Air liquid interface epithelial monolayers were infected and virus replication monitored. Using two independent protocols at two virology laboratories, we observed donor-to-donor differences, but comparable replication levels of SARS-CoV-2 in CF versus non-CF monolayers under conditions studied here (Fig. 1C and D). This result contasts findings from others (80).
Apical surface fluid replacement impacts viral infectivity
CFTR regulates depth and composition of the thin (<10 micron) fluid layer bathing mucosal surfaces of human airways in vivo (31). In order to examine the potential influence of conditioned ASL during SARS-CoV-2 infection, we studied replication in washed (using fresh PneumaCult-ALI medium at pH = 7.4) or unwashed cultures – i.e., without or with conditioned secretions produced by airway cells. We found that replacement of conditioned surface fluid with physiologic buffer dramatically reduced infectivity (p<0.0001; Fig. 2A). When we repeated the same experiment using ALI cultures from Calu-3 (airway serous glandular) cells, no significant differences were observed, indicating specificity for primary airway epithelium (Fig. 2B). To determine whether replacement of conditioned surface fluid is a general phenomenon that could suppress transduction by other viral constructs, we tested hBEC ALI cultures using recombinant (replication deficient) adenovirus, and found no impact on viral transgene expression (Fig. 2C). The adenovirus result also provides an indication of monolayer integrity and viability during a standard washing protocol, since no decrement of adenoviral gene transfer due to washing was observed. These findings suggest constituent(s) of mucosal secretion from primary human bronchial cells may help promote SARS-CoV-2 replication, although other potential contributors (e.g., mild or transient distortion of monolayers during washing, with a particularly strong effect on SARS-CoV-2) might also be considered as contributory. In either case, because hBECs are commonly used for mechanistic and other studies of COVID-19 pathogenesis—and since ALI monolayers are typically washed to remove mucus in experiments of this type—findings shown in Figure 2A should be prominently considered when using the hBEC system.
Impact of features that contribute to CF airway disease in vivo
CFTR at the plasma membrane mediates HCO3− secretion, and absent CFTR has been shown to confer low pH of the in vivo periciliary fluid layer, which alters host defense mechanisms, protease/antiprotease balance, and innate immunity (81, 93–99). Because CF animal models, as well as patients with cystic fibrosis, exhibit acidic airway surface fluid – and since chronically inflamed/infected CF mucus itself is strongly acidified – we tested viral infection of hBEC ALI cultures at pH between 6.5 and 8. Significantly diminished SARS-CoV-2 replication was observed at either low or high pH values (p = 0.043 for pH = 6.5, p = 0.027 for pH 8.0; Fig. 3A). In this context, it is important to note that CF airways in vivo are also characterized by copious (infected/inflamed) mucinous secretions that adhere to the epithelial surface and range in pH between 2.9 and 6.5 (100). Diminished pH of ASL and acidic mucus in vivo might therefore be expected to reduce SARS-CoV-2 propagation.
Defective ion transport caused by absence of functional CFTR leads to depletion of airway surface liquid and dramatically increases mucus viscosity among individuals with cystic fibrosis (for review, see (29)). Unlike studies of epithelial monolayers in cell culture, years of CF lung disease in vivo result in a dense mucus matrix that blankets portions of the airway surface. As a model for viscosity, we added carboxymethylcellulose (CMC) to periciliary fluid in vitro, and observed robust, dose-dependent blunting of viral infection (Fig. 3B). Viscous media significantly inhibited viral production by either hBEC or Calu-3 ALI cultures (Fig. 3B and C).
CF airway secretions variably influence SARS-CoV-2 replication
Finally, we tested whether CF respiratory secretions, themselves, might impact SARS-CoV-2 propagation. We performed infections of washed monolayers in the presence of freshly isolated CF mucus. Likely because CF sputum is highly variable in terms of microflora, protease/antiprotease balance, viscosity, pH, and other features, results from these studies were patient dependent. In some cases, CF sputum strongly activated viral production (compare CF mucus sample 1 and sample 2, Fig. 4A). Sputum constituents from certain individuals were found to partially or fully complement loss of viral replication after washing (Fig. 4B). Interestingly, human sputum samples that permitted viral propagation despite removal of conditioned ASL remained active at very low viral innocula (Figure 4C), suggesting that mucosal fluid composition may be especially important when minimal virus is deposited at the apical cell surface (as often occurs during infection in vivo). In either case, these findings provide a means by which constituents of airway fluid that impact SARS-CoV-2 might be investigated in the future.
Discussion
Less-severe-than-projected outcomes among individuals with CF suggest that partial or complete deficiency of CFTR might play a role during pathogenesis of COVID-19 (3, 8–18). In this report, we describe studies to evaluate relationships between CFTR genotype and SARS-CoV-2 infection. Based on initial experiments in a group of 424 White individuals from the United States hospitalized for treatment of COVID-19 (Host Genetics Initiative [HGI] classification 2A) at both a major tertiary center and regional care facilities, we observed no evidence that F508del heterozygosity influenced likelihood of COVID-19 hospital admission. Next, we evaluated a much larger number of individuals from Italy, Spain, and Norway and again found F508del carrier frequency to be similar between those hospitalized with SARS-CoV-2 infection (HGI classification 2B; Table 1) and non-hospitalized controls. Our data did suggest younger (60 years or under) Italian male CF carriers were more frequently at risk for hospitalization due to COVID-19. This finding, however, was not reproduced among a greater number of European males. In contrast to earlier reports, therefore, we did not observe a correlation between COVID-19 severity and F508del carrier status. Moreover, comorbidities such as CAD, hypertension, or diabetes could not account for differences observed in young Italian males versus the population as a whole. Other modifying features should therefore be considered in studies that propose an association between CF-carrier status and SARS-CoV-2 lung disease (56–58). This is in addition to ways distinct viral strains (such as those appearing later in the pandemic) might influence susceptibility to coronaviral infection, immune response, or particular aspects of a specific study population (e.g., Spanish, Italian, Norwegan, or American).
The observation that COVID-19 mortality among F508del carriers was approximately one third that of individuals without the CFTR mutation is compatible with lower case-fatality rates in the CF patient population at large (6). While analyses of heterozygous carrier status did not reach statistical significance, the tendency towards protection from death is of potential interest. Natural selective pressure, such as an unknown lethal infectious disease over millennia of human history, is believed to have caused the high prevalence of F508del CFTR among individuals of European descent (101–105). Although the etiologic pathogen has never been well established, a variety of epidemics have been implicated. Very little has been reported regarding widespread historical infections by coronavirus in Europe (106, 107). Whether certain SARS-CoV-2 variants or other coronaviral pathogens may exhibit greater sensitivity to CFTR function will require further study.
Because partial CFTR deficiency (i.e., CF carrier status) was not found to correlate with COVID-19 clinical findings, we next investigated the extent to which complete loss of CFTR function (F508del homozygosity) might influence SARS-CoV-2 infection in vitro. In these studies, we tested virus replication using an airway model shown by numerous laboratories to recapitulate features of coronaviral pathogenesis (for examples, see (62, 63, 66–73)). In contrast to other studies , we found no decrease of SARS-CoV-2 replication due to F508del CFTR homozygosity. Our data indicate that under standard conditions tested here, presence or absence of functional CFTR does not alter the intracellular environment (lysosomal uncoating, post-endoplasmic reticulum viral assembly, etc.) in ways that affect virus propagation.
Previous findings in animal models and humans have shown that CF airway surface fluid is strongly acidified, a result in agreement with function of CFTR as a pathway for mucosal bicarbonate secretion (81, 93–95, 108). pH has been reported as 5.2 +/− 0.3 in CF airway surface liquid from neonates, versus 6.85–7.65 within control newborn lungs. Moreover, in chronic mucopurulent (reduced) sputum samples taken from patients with CF, very strong acidification (pH 2.9 – 6.5) has been demonstrated (81, 93–96, 100). Delivery of virus to hBECs utilizes a comparatively large volume of buffered solution (versus the 10 μm airway surface liquid depth), making determination of pH-related effects on native ASL in vitro problematic. For example, studies using SARS-CoV-2 infection of airway monolayers typically apply large inocula compared to much smaller volumes of conditioned ASL present in a tissue culture well (e.g., 100 μl inoculated viral volume versus 1.9 μl ASL in a single well of a 24-well plate). Accordingly, viral addition in vitro dilutes the 10-micron ASL depth (by ~50-fold) and resets pH to match the buffer in which virus is being added. In the present experiments, we tested controlled alterations of mucosal acidity, and found that mucosal fluid pH below 7.0 inhibits viral infection (Fig. 3).
Besides regulating pH, ion transport by CFTR plays an essential role maintaining proper viscosity of human respiratory secretions. In a prevailing model, pH and/or ionic composition govern expansion and elasticity of mucins produced by surface epithelium or submucosal glands (109–112). Since a conventional CF lung phenotype involving hyperviscous mucus and chronic infection/inflammation can require years to develop (and cannot be adequately reproduced by the hBEC system), we modified airway liquid viscosity using carboxymethylcellulose – and found robust suppression of SARS-CoV-2 (Fig. 3). Mucosal surfaces of chronically diseased lungs from individuals homozygous for mutations in CFTR are blanketed by thick and hyperviscous secretion. Based on findings presented here, it is reasonable to imagine that among patients with CF, SARS-CoV-2 might be less able to reach susceptible lung cells – or that spread from an initial focus of infection to adjoining respiratory tissue could be vitiated by an acidic, viscous, and otherwise imbalanced CF micro-environment that is less favorable to viral propagation.
While investigating a possible role for CF human respiratory secretion during SARS-CoV-2 replication, we found that washing of conditioned medium from airway epithelial monolayers conferred a marked decrease in virus infectivity (Fig. 2). Primary airway cells have been used extensively during mechanistic and pharmaco-therapeutic characterization of COVID-19, and such monolayers are routinely washed for removal of mucus just prior to infection. Impaired production of coronavirus after removing conditioned medium should therefore be considered in experiments of this type, particularly when designing protocols to test SARS-CoV-2 pathogenesis and/or anti-viral drug development. Results in Figure 2 are important from an investigative perspective, since the findings establish that a standard protocol for washing airway monolayers can inhibit viral replication.
Care was taken in our studies to select in vitro conditions that – to the extent possible – faithfully replicate aspects of in vivo airway mucosa. Polarizing respiratory monolayers, studies of freshly isolated mucus from individuals with CF, and direct modulation of apical surface pH were employed. Newer methods for in vivo analysis of viral infection are emerging. For example, recent progress with non-invasive approaches to testing mucus viscosity by optical coherence tomography and particle tracking microrheology may allow airway mucus viscoelastic properties to be directly examined in vitro or in vivo – although additional work will be needed before the technology can be applied to experiments using COVID-19 (113). In the same context, methods for directly monitoring ASL pH are technically demanding, but aerosolization of SARS-CoV-2 in CF animals or human pulmonary tissue explants (as a test of CF in vivo susceptibility to viral pathogenesis) could represent a “physiologic” protocol for evaluating transmission. However, conditions that approximate infection of human subjects (site of inoculum, droplet deposition and composition, anatomic compartment most likely to be involved, relevance of cough/respiration, or other features) remain difficult to mimic. That being said, the current findings delineate a novel mechanistic model involving CF and coronavirus – and point to the need for additional studies of this type.
In summary, while there is no question that individuals with CF, as well as obligate heterozygotes who carry a single CFTR mutation remain at substantial risk from COVID-19, numerous reports have unexpectedly shown that patients with CF experience clinical outcomes no worse than the general population. In contrast to other findings, the data presented here fail to demonstrate a prominent association between heterozygous loss of CFTR and susceptibility to severe COVID-19. Our results instead emphasize the importance of homozygous CFTR deficiency in vivo and resulting chronic changes of mucosal fluid as features that influence coronavirus infectivity. In particular, the data point to physicochemical properties of pulmonary secretion among individuals with CF (acidic pH, altered composition, viscosity) that may limit SARS-CoV-2 replication. These findings provide a framework by which studies to test relationships between ASL, CFTR and COVID-19 can be better understood in the future.
Study approval
For the Italian, Spanish and Norwegian studies, information on recruiting centers and Ethics Committee Approval IDs are detailed in Supplementary Table 1a of Degenhardt et al. (60). In short, all centers from which individuals were recruited obtained ethics approval from their local ethics committee. Subjects were recruited from four Italian, seven Spanish, and eight Norwegian centers. All US samples were evaluated from remnant blood derived from anonymized individuals hospitalized for COVID-19 without patient identifiers. These samples were designated “not human subjects research” by institutional ethics panels.
Supplementary Material
Acknowledgements
We are grateful to Jan Tindall for editing the manuscript. We thank Samuel Smoot (Elliot Hospital, Manchester, NH) and members of the Northeast Georgia Medical Center and of the Emory University Clinical Laboratory for help preparing blood samples. We also thank the Severe COVID-19 GWAS Consortium for contribution of their data. A full list of all members is shown in Supplemental File S2. Full original study acknowledegements are presented in Degenhardt et al. (60). This study was carried out in part using anonymized data provided by the Catalan Agency for Quality and Health Assessment, within the framework of the PADRIS Program. The authors of the study would like to acknowledge all GCAT project investigators who contributed to the generation of the GCAT data. A full list of the investigators is available from www.genomesforlife.com. We also thank the Blood and Tissue Bank from Catalonia (BST) and all the GCAT volunteers that participated in the study. SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281 was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH. icSARS-CoV-2_mNeonGreen was deposited by Pei-Yong Shi and obtained through the World Reference Center for Emerging Viruses and Arboviruses, NIAID, NIH.
Funding
This work was supported by the Italian Ministry of Health (Ministero della Salute) [Ricerca Finalizzata RF-2016-02364358]; Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Ricerca corrente; Fondazione IRCCS Ca’ Granda core COVID-19 Biobank [RC100017A]; “Liver BIBLE” [PR-0391]; and Innovative Medicines Initiative 2 joint undertaking of the European Union (EU) Horizon 2020 research and innovation programme and European Federation of Pharmaceutical Industries and Associations Programme Horizon 2020 [under grant agreement No. 777377] for the project LITMUS and the European Union, programme “Photonics” [under grant agreement 101016726] to LV. JCH was funded by the Research Council of Norway [grant no 312780] and a philanthropic donation from Vivaldi Invest A/S owned by Jon Stephenson von Tetzchner. The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the report. AF was supported by a grant from the German Federal Ministry of Education and Research [01KI20197]. This work was also funded by a generous philanthropic donation from Banca Intesa San Paolo to RA. This study makes use of data generated by the GCAT-Genomes for Life cohort study of the Genomes of Catalonia, Fundacio IGTP; IGTP is part of the CERCA Program/Generalitat de Catalunya, and GCAT was funded by Acción de Dinamización del ISCIII-MINECO and the Ministry of Health of the Generalitat of Catalunya [ADE 10/00026]; with additional support by the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) [2017-SGR 529], and National Grant [PI18/01512] and VEIS project [001-P-001647], co-funded by European Regional Development Fund (ERDF), “A way to build Europe”. EdW and BNW were supported by the Intramural Research Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations
- ACE2
Angiotensin Converting Enzyme 2
- ALI
Air-liquid interface
- ASL
Airway surface liquid
- CAD
Coronary artery disease
- CF
Cystic Fibrosis
- CFTR
Cystic Fibrosis Transmembrane Conductance Regulator
- CI
Confidence interval
- CMC
Carboxymethylcellulose
- COVID-19
Coronavirus Disease 2019
- ddPCR
droplet digital polymerase chain reaction
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
Fetal Bovine Serum
- GFP
Green fluorescent proein
- GWAS
Genome-Wide Association Studies
- hBEC
human bronchial epithelial cells
- HGI
Host Genetics Initiative
- MEM
Minimum Essential Medium
- MOI
Multiplicity of infection
- OR
Odds Ratio
- pFDR
positive false discovery rate
- PBS
phosphate-buffered saline
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- TMPRSS2
Transmembrane Serine Protease 2
- WT
wild-type
Footnotes
Conflict of interest statement
The authors declare no competing interest.
Data availability
All statistical data are publicly available as Supplemental Material.
References
- 1.CDC, People with Certain Medical Conditions. (May 13, 2021). [Google Scholar]
- 2.CysticFibrosisFoundation (2021) COVID-19 [video]. (https://www.youtube.com/watch?v=8BV2JH2wRFM). [Google Scholar]
- 3.McClenaghan E. et al. , The global impact of SARS-CoV-2 in 181 people with cystic fibrosis. J Cyst Fibros 19, 868–871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goyal P. et al. , Clinical characteristics of Covid-19 in New York City. N Engl J Med 382, 2372–2374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustamante L. (2021) Cystic fibrosis patients fight for higher spot in coronavirus vaccine line [video]. (NBC10 Philadelphia, https://www.nbcphiladelphia.com/news/coronavirus/cystic-fibrosis-patients-fight-for-higher-spot-in-coronavirus-vaccine-line/2699345/?amp). [Google Scholar]
- 6.Carr SB et al. , Factors associated with clinical progression to severe COVID-19 in people with cystic fibrosis: A global observational study. J Cyst Fibros 10.1016/j.jcf.2022.06.006 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y, Hiyoshi A, Montgomery S, COVID-19 case-fatality rate and demographic and socioeconomic influencers: worldwide spatial regression analysis based on country-level data. BMJ Open 10, e043560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fainardi V, Longo F, Chetta A, Esposito S, Pisi G, Sars-CoV-2 infection in patients with cystic fibrosis. An overview. Acta Biomed 91, e2020035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bezzerri V, Lucca F, Volpi S, Cipolli M, Does cystic fibrosis constitute an advantage in COVID-19 infection? Ital J Pediatr 46, 143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corvol H. et al. , First wave of COVID-19 in French patients with cystic fibrosis. J Clin Med 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathew HR, Choi MY, Parkins MD, Fritzler MJ, Systematic review: cystic fibrosis in the SARS-CoV-2/COVID-19 pandemic. BMC Pulm Med 21, 173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosgriff R. et al. , A multinational report to characterise SARS-CoV-2 infection in people with cystic fibrosis. J Cyst Fibros 19, 355–358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bain R. et al. , Clinical characteristics of SARS-CoV-2 infection in children with cystic fibrosis: An international observational study. J Cyst Fibros 20, 25–30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beltramo G. et al. , Chronic respiratory diseases are predictors of severe outcome in COVID-19 hospitalised patients: a nationwide study. Eur Respir J 10.1183/13993003.04474-2020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombo C. et al. , SARS-CoV-2 infection in cystic fibrosis: A multicentre prospective study with a control group, Italy, February-July 2020. PLoS One 16, e0251527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naehrlich L. et al. , Incidence of SARS-CoV-2 in people with cystic fibrosis in Europe between February and June 2020. J Cyst Fibros 10.1016/j.jcf.2021.03.017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mondejar-Lopez P. et al. , Impact of SARS-CoV-2 infection in patients with cystic fibrosis in Spain: Incidence and results of the national CF-COVID19-Spain survey. Respir Med 170, 106062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray DM et al. , COVID-19 and pediatric lung disease: a South African tertiary center experience. Front Pediatr 8, 614076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason K, Hasan S, Darukhanavala A, Kutney K, COVID-19: Pathophysiology and implications for cystic fibrosis, diabetes and cystic fibrosis-related diabetes. J Clin Transl Endocrinol 26, 100268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CysticFibrosisFoundationPatientRegistry (2020) Annual Data Report. (Bethesda, Maryland: ). [Google Scholar]
- 21.Moeller A. et al. , COVID-19 in children with underlying chronic respiratory diseases: survey results from 174 centres. ERJ Open Res 6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang T, Tran Thi Anh T, Comparison of Comorbidities in Relation to Critical Conditions among Coronavirus Disease 2019 Patients: A Network Meta-Analysis. Infect Chemother 53, 13–28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber AG, Chau AS, Egeblad M, Barnes BJ, Janowitz T, Nebulized in-line endotracheal dornase alfa and albuterol administered to mechanically ventilated COVID-19 patients: a case series. Mol Med 26, 91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desilles JP et al. , Efficacy and safety of aerosolized intra-tracheal dornase alfa administration in patients with SARS-CoV-2-induced acute respiratory distress syndrome (ARDS): a structured summary of a study protocol for a randomised controlled trial. Trials 21, 548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okur HK et al. , Preliminary report of in vitro and in vivo effectiveness of dornase alfa on SARS-CoV-2 infection. New Microbes New Infect 37, 100756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horby P. et al. , Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med 384, 693–704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldenburg CE et al. , Effect of Oral Azithromycin vs Placebo on COVID-19 Symptoms in Outpatients With SARS-CoV-2 Infection: A Randomized Clinical Trial. Jama 326, 490–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anonymous, Azithromycin for community treatment of suspected COVID-19 in people at increased risk of an adverse clinical course in the UK (PRINCIPLE): a randomised, controlled, open-label, adaptive platform trial. Lancet 397, 1063–1074 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowe SM, Miller S, Sorscher EJ, Cystic fibrosis. N Engl J Med 352, 1992–2001 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Manfredi C, Tindall JM, Hong JS, Sorscher EJ, Making precision medicine personal for cystic fibrosis. Science 365, 220–221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorscher EJ, “Cystic fibrosis” in Harrison’s Principles of Internal Medicine, Jameson JL, et al. , Ed. (McGraw-Hill Education/Medical, New York, NY, 2021). [Google Scholar]
- 32.Stanton BA, Hampton TH, Ashare A, SARS-CoV-2 (COVID-19) and cystic fibrosis. Am J Physiol Lung Cell Mol Physiol 319, L408–l415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peckham D, McDermott MF, Savic S, Mehta A, COVID-19 meets cystic fibrosis: for better or worse? Genes Immun 21, 260–262 (2020). [DOI] [PubMed] [Google Scholar]
- 34.McElvaney OJ et al. , Alpha-1 antitrypsin for cystic fibrosis complicated by severe cytokinemic COVID-19. J Cyst Fibros 20, 31–35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdel Hameid R, Cormet-Boyaka E, Kuebler WM, Uddin M, Berdiev BK, SARS-CoV-2 may hijack GPCR signaling pathways to dysregulate lung ion and fluid transport. Am J Physiol Lung Cell Mol Physiol 320, L430–l435 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Recchiuti A. et al. , Resolvin D1 and D2 reduce SARS-CoV-2-induced inflammatory responses in cystic fibrosis macrophages. Faseb j 35, e21441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenhut M, Shin JI, Pathways in the pathophysiology of coronavirus 19 lung disease accessible to prevention and treatment. Front Physiol 11, 872 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abraham EH et al. , Cystic fibrosis improves COVID-19 survival and provides clues for treatment of SARS-CoV-2. Purinergic Signal 10.1007/s11302-021-09771-0, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bitossi C. et al. , SARS-CoV-2 Entry Genes Expression in Relation with Interferon Response in Cystic Fibrosis Patients. Microorganisms 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biwersi J, Verkman AS, Functional CFTR in endosomal compartment of CFTR-expressing fibroblasts and T84 cells. Am J Physiol 266, C149–156 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Haggie PM, Verkman AS, Defective organellar acidification as a cause of cystic fibrosis lung disease: reexamination of a recurring hypothesis. Am J Physiol Lung Cell Mol Physiol 296, L859–867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C. et al. , Staphylococcus aureus survives in cystic fibrosis macrophages, forming a reservoir for chronic pneumonia. Infect Immun 85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukacs GL et al. , The cystic fibrosis transmembrane regulator is present and functional in endosomes. Role as a determinant of endosomal pH. J Biol Chem 267, 14568–14572 (1992). [PubMed] [Google Scholar]
- 44.Dunn KW et al. , Regulation of endocytic trafficking and acidification are independent of the cystic fibrosis transmembrane regulator. J Biol Chem 269, 5336–5345 (1994). [PubMed] [Google Scholar]
- 45.Jouret F, Devuyst O, CFTR and defective endocytosis: new insights in the renal phenotype of cystic fibrosis. Pflugers Arch 457, 1227–1236 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Lukacs GL, Segal G, Kartner N, Grinstein S, Zhang F, Constitutive internalization of cystic fibrosis transmembrane conductance regulator occurs via clathrin-dependent endocytosis and is regulated by protein phosphorylation. Biochem J 328 (Pt 2), 353–361 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poschet JF et al. , Hyperacidification of cellubrevin endocytic compartments and defective endosomal recycling in cystic fibrosis respiratory epithelial cells. J Biol Chem 277, 13959–13965 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Poschet JF et al. , Endosomal hyperacidification in cystic fibrosis is due to defective nitric oxide-cylic GMP signalling cascade. EMBO Rep 7, 553–559 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biwersi J, Emans N, Verkman AS, Cystic fibrosis transmembrane conductance regulator activation stimulates endosome fusion in vivo. Proc Natl Acad Sci U S A 93, 12484–12489 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jouret F. et al. , Cystic fibrosis is associated with a defect in apical receptor-mediated endocytosis in mouse and human kidney. J Am Soc Nephrol 18, 707–718 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Bradbury NA et al. , Characterization of the internalization pathways for the cystic fibrosis transmembrane conductance regulator. Am J Physiol 276, L659–668 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Barasch J. et al. , Defective acidification of intracellular organelles in cystic fibrosis. Nature 352, 70–73 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Dho S, Grinstein S, Foskett JK, Plasma membrane recycling in CFTR-expressing CHO cells. Biochim Biophys Acta 1225, 78–82 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Guy JL et al. , Angiotensin-converting enzyme-2 (ACE2): comparative modeling of the active site, specificity requirements, and chloride dependence. Biochemistry 42, 13185–13192 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Rushworth CA, Guy JL, Turner AJ, Residues affecting the chloride regulation and substrate selectivity of the angiotensin-converting enzymes (ACE and ACE2) identified by site-directed mutagenesis. Febs j 275, 6033–6042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baldassarri M. et al. , Severe COVID-19 in Hospitalized Carriers of Single CFTR Pathogenic Variants. J Pers Med 11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delanghe JR, De Buyzere ML, Speeckaert MM, Genetic polymorphisms in the host and COVID-19 infection. Adv Exp Med Biol 1318, 109–118 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Sarantis P, Koustas E, Papavassiliou AG, Karamouzis MV, Are cystic fibrosis mutation carriers a potentially highly vulnerable group to COVID-19? J Cell Mol Med 24, 13542–13545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellinghaus D. et al. , Genomewide Association Study of severe Covid-19 with respiratory failure. N Engl J Med 383, 1522–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Degenhardt F. et al. , Detailed stratified GWAS analysis for severe COVID-19 in four European populations. Hum Mol Genet 10.1093/hmg/ddac158 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgan R. et al. , A medium composition containing normal resting glucose that supports differentiation of primary human airway cells. Sci Rep 12, 1540 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chhetri BK et al. , Marine Natural Products as Leads against SARS-CoV-2 Infection. J Nat Prod 85, 657–665 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanderheiden A. et al. , Type I and Type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J Virol 94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung WJ et al. , Increasing the endoplasmic reticulum pool of the F508del allele of the cystic fibrosis transmembrane conductance regulator leads to greater folding correction by small molecule therapeutics. PLoS One 11, e0163615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birket SE et al. , Combination therapy with cystic fibrosis transmembrane conductance regulator modulators augment the airway functional microanatomy. Am J Physiol Lung Cell Mol Physiol 310, L928–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hou YJ et al. , SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446.e414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sungnak W. et al. , SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26, 681–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y. et al. , SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proceedings of the National Academy of Sciences 118, e2022643118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryu G, Shin HW, SARS-CoV-2 Infection of Airway Epithelial Cells. Immune Netw 21, e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mulay A. et al. , SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep 35, 109055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morrison CB et al. , SARS-CoV-2 infection of airway cells causes intense viral and cell shedding, two spreading mechanisms affected by IL-13. Proc Natl Acad Sci U S A 119, e2119680119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Broadbent L. et al. , An endogenously activated antiviral state restricts SARS-CoV-2 infection in differentiated primary airway epithelial cells. PLoS One 17, e0266412 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Subramaniyan B. et al. , Characterization of the SARS-CoV-2 Host Response in Primary Human Airway Epithelial Cells from Aged Individuals. Viruses 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ogando NS et al. , SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol 101, 925–940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nyayanit DA et al. , Transcriptome & viral growth analysis of SARS-CoV-2-infected Vero CCL-81 cells. Indian J Med Res 152, 70–76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie X. et al. , An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848.e843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corman VM et al. , Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller AC et al. , Cystic fibrosis carriers are at increased risk for a wide range of cystic fibrosis-related conditions. Proc Natl Acad Sci U S A 117, 1621–1627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoffmann M. et al. , SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bezzerri V. et al. , SARS-CoV-2 viral entry and replication is impaired in Cystic Fibrosis airways due to ACE2 downregulation. Nat Commun 14, 132 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Massip-Copiz MM, Santa-Coloma TA, Extracellular pH and lung infections in cystic fibrosis. Eur J Cell Biol 97, 402–410 (2018). [DOI] [PubMed] [Google Scholar]
- 82.McKelvey MC, Weldon S, McAuley DF, Mall MA, Taggart CC, Targeting Proteases in Cystic Fibrosis Lung Disease. Paradigms, Progress, and Potential. Am J Respir Crit Care Med 201, 141–147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Margaroli C. et al. , Elastase Exocytosis by Airway Neutrophils Is Associated with Early Lung Damage in Children with Cystic Fibrosis. Am J Respir Crit Care Med 199, 873–881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liou TG et al. , Sputum biomarkers and the prediction of clinical outcomes in patients with cystic fibrosis. PLoS One 7, e42748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gray RD et al. , Delayed neutrophil apoptosis enhances NET formation in cystic fibrosis. Thorax 73, 134–144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dubois AV et al. , Influence of DNA on the activities and inhibition of neutrophil serine proteases in cystic fibrosis sputum. Am J Respir Cell Mol Biol 47, 80–86 (2012). [DOI] [PubMed] [Google Scholar]
- 87.Dittrich AS et al. , Elastase activity on sputum neutrophils correlates with severity of lung disease in cystic fibrosis. Eur Respir J 51 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Gehrig S. et al. , Lack of neutrophil elastase reduces inflammation, mucus hypersecretion, and emphysema, but not mucus obstruction, in mice with cystic fibrosis-like lung disease. Am J Respir Crit Care Med 189, 1082–1092 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Weldon S. et al. , miR-31 dysregulation in cystic fibrosis airways contributes to increased pulmonary cathepsin S production. Am J Respir Crit Care Med 190, 165–174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gaggar A. et al. , The role of matrix metalloproteinases in cystic fibrosis lung disease. Eur Respir J 38, 721–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turnbull AR et al. , Abnormal pro-gly-pro pathway and airway neutrophilia in pediatric cystic fibrosis. J Cyst Fibros 19, 40–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smirnova NF et al. , Detection and quantification of epithelial progenitor cell populations in human healthy and IPF lungs. Respir Res 17, 83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abou Alaiwa MH et al. , Neonates with cystic fibrosis have a reduced nasal liquid pH; a small pilot study. J Cyst Fibros 13, 373–377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coakley RD et al. , Abnormal surface liquid pH regulation by cultured cystic fibrosis bronchial epithelium. Proc Natl Acad Sci U S A 100, 16083–16088 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ojoo JC, Mulrennan SA, Kastelik JA, Morice AH, Redington AE, Exhaled breath condensate pH and exhaled nitric oxide in allergic asthma and in cystic fibrosis. Thorax 60, 22–26 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tate S, MacGregor G, Davis M, Innes JA, Greening AP, Airways in cystic fibrosis are acidified: detection by exhaled breath condensate. Thorax 57, 926–929 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pezzulo AA et al. , Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature 487, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abou Alaiwa MH et al. , pH modulates the activity and synergism of the airway surface liquid antimicrobials β-defensin-3 and LL-37. Proc Natl Acad Sci U S A 111, 18703–18708 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shah VS et al. , Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science 351, 503–507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK, Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio 6, e00767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Poolman EM, Galvani AP, Evaluating candidate agents of selective pressure for cystic fibrosis. J R Soc Interface 4, 91–98 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gabriel SE, Brigman KN, Koller BH, Boucher RC, Stutts MJ, Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science 266, 107–109 (1994). [DOI] [PubMed] [Google Scholar]
- 103.Pier GB et al. , Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature 393, 79–82 (1998). [DOI] [PubMed] [Google Scholar]
- 104.Bosch L. et al. , Cystic fibrosis carriership and tuberculosis: hints toward an evolutionary selective advantage based on data from the Brazilian territory. BMC Infect Dis 17, 340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cuthbert AW, Halstead J, Ratcliff R, Colledge WH, Evans MJ, The genetic advantage hypothesis in cystic fibrosis heterozygotes: a murine study. J Physiol 482 (Pt 2), 449–454 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Piret J, Boivin G, Pandemics throughout history. Front Microbiol 11, 631736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.NIH (https://www.niaid.nih.gov/diseases-conditions/coronaviruses (last accessed 10/13/3021)) Coronaviruses. (National Institute of Allergy and Infectious Diseases; ). [Google Scholar]
- 108.Li X. et al. , Electrolyte transport properties in distal small airways from cystic fibrosis pigs with implications for host defense. Am J Physiol Lung Cell Mol Physiol 310, L670–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xie Y. et al. , Acidic submucosal gland pH and elevated protein concentration produce abnormal cystic fibrosis mucus. Dev Cell 54, 488–500.e485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tang XX et al. , Acidic pH increases airway surface liquid viscosity in cystic fibrosis. J Clin Invest 126, 879–891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abou Alaiwa MH et al. , Repurposing tromethamine as inhaled therapy to treat CF airway disease. JCI Insight 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoegger MJ et al. , Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science 345, 818–822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Birket SE et al. , Ivacaftor Reverses Airway Mucus Abnormalities in a Rat Model Harboring a Humanized G551D-CFTR. Am J Respir Crit Care Med 202, 1271–1282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All statistical data are publicly available as Supplemental Material.