Abstract
A class of noncoding RNAs, referred to as tsRNAs, is emerging with a potential to exert a new layer in gene regulation. These RNAs are breakdown products of tRNAs, either through active processing or passive cleavage or both. Since tRNAs are part of the general machinery for translation, their expression levels and activities are tightly controlled, raising the possibility that their breakdown products, tsRNAs, may provide a link between the overall translational status of a cell to specific changes in gene regulatory network. We hypothesize that Drosophila pupation, being a special developmental stage during which there is a global limitation of nutrients, represents a system in which such a link may readily reveal itself. We show that specific tsRNAs indeed show a dynamic accumulation upon entering the pupal stage. We describe experiments to characterize the mode of tsRNA action and, through the use of such gained knowledge, conduct a genome-wide analysis to assess the functions of dynamically expressed tsRNAs. Our results show that the predicted target genes are highly enriched in biological processes specific to this stage of development including metamorphosis. We further show that tsRNA action is required for successful pupation, providing direct support to the hypothesis that tsRNAs accumulated during this stage are critical to the gene expression program at this stage of development.
tRNA-derived small RNAs (tsRNAs, also known as tRNA-derived fragments) are among the most ancient and conserved RNA species across all organisms. Accumulating evidence suggests that, from bacteria to human, tsRNAs are not merely intermediates of the tRNA maturation process or residues of the tRNA degradation process as their biogenesis can show elaborate spatiotemporal patterns (Kumar et al. 2016; Sharma et al. 2016; Li et al. 2018; Dou et al. 2019; Krishna et al. 2021; Chen et al. 2021b). tsRNAs have a relatively wide range of size and abundant modifications, with a potential to interact with a variety of biomolecules to exert diverse functions (Haussecker et al. 2010; Schaefer et al. 2010; Tuorto et al. 2012; Hussain et al. 2013; Kumar et al. 2014; Guzzi et al. 2018; Luo et al. 2018). tsRNAs with a length of 20–24 nt can participate in post-transcriptional regulation through mRNA hybridization and Argonaute (AGO) engagement. In a way that resembles RNA interference (RNAi), tsRNAs could suppress the stability or translation of target mRNAs. For example, in mature B lymphocytes, a 3′ fragment derived from tRNAGlyGCC (referred to as 3′-tsRNAGlyGCC) has activities characteristic of a miRNA, including the capacity of binding to all four human AGOs and gene silencing via target sites in 3′ UTRs (Maute et al. 2013). In addition, tsRNAs can associate with RNA-binding proteins other than AGOs in a cell type– and status-specific manner. In breast cancer cells, four tsRNA species encompassing the anticodon loops (referred to as inter-tsRNAGluYUC, tsRNAAspGUC, tsRNAGlyUCC, and tsRNATyrGUA) have been reported to compete against oncogenic mRNAs for binding with YBX1 and suppress their stability (Goodarzi et al. 2015). Moreover, tsRNAs can frequently associate with translation machinery and related factors to participate in multiple layers of translation control, such as rRNA biogenesis, ribosome assembly, translation initiation, and translation elongation (Couvillion et al. 2010, 2012; Ivanov et al. 2011; Gebetsberger et al. 2012, 2017; Sobala and Hutvagner 2013; Keam et al. 2017; Kim et al. 2017). Finally, tsRNAs have been reported to bind to gene promoter regions to induce transcriptional reprogramming during early embryonic development of mouse and zebrafish (Chen et al. 2016, 2021a). These diverse and context-dependent functions of tsRNAs suggest a need for a comprehensive profiling and functional analysis of tsRNAs at the systems level within a developmental context.
Development is a process in which cells can adaptively respond to environmental fluctuations by rewiring gene regulatory networks. In particular, suppression on protein synthesis usually provides cells with a resource allocating solution within a few minutes of exposure to stress (López-Maury et al. 2008). As a class of key informational molecules in the process of translation, tRNAs may contribute to such a solution through noncanonical functions that are mediated by other forms, such as tsRNAs. Previous studies in cell cultures have shown that nutrient deprivation can cause tRNAs to break down, resulting in an accumulation of tsRNAs and down-regulation of translation (Lee and Collins 2005; Haussecker et al. 2010; Sobala and Hutvagner 2013; Luo et al. 2018). It remains unclear if such a scenario, or a varied version of it, could play out in a developmental context to contribute to the successful progression of developmental programs. Here we selected metamorphosis in Drosophila as a developmental window to probe this question aimed at gaining a systematic view of tsRNA action. Metamorphosis of insects is an intricate and deterministic developmental process. As larvae stop feeding to transition into the pupal stage, the nutritional input is cut off, forcing the animals to rely on the energy store accumulated by larvae to perform large-scale remodeling and complete the larva-to-adult transition (Merkey et al. 2011; Rolff et al. 2019; Li et al. 2022). A signature of the larva-to-pupa transition is a global decline in translation accompanied by a reduction in translation efficiency for proteins in the translation machinery itself (see Results), providing an excellent developmental system for a systematic analysis of tsRNA functions.
Results
A quantitative RNA-seq approach for analyzing tsRNAs during larva-to-pupa transition
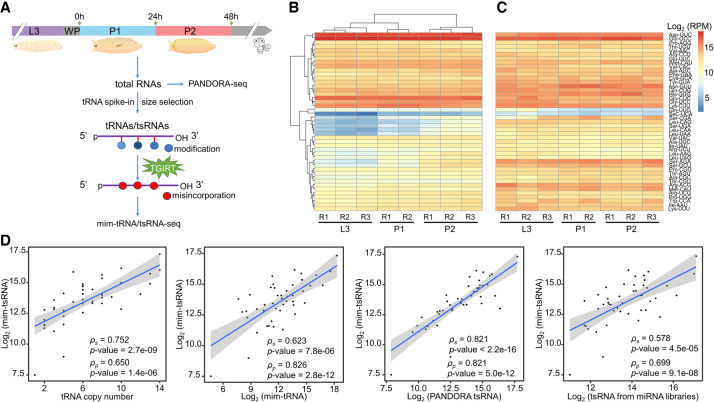
We used mim-tRNAseq (modification-induced misincorporation tRNA sequencing), an RNA-seq pipeline designed for tRNA analysis (Behrens et al. 2021), in our study of tsRNAs in Drosophila melanogaster. The library construction step of this pipeline includes necessary measures to minimize tRNA modifications that can hinder effective detection and quantitation in deep sequencing. In our experiments, we used size selection optimized specifically for detecting tsRNAs and tRNAs and adopted a “paired” library construction approach to permit direct comparisons between tRNA and tsRNA data for each RNA sample (Fig. 1A; Methods). We focused on three developmental stages spanning the larva-to-pupa transition in our study: third-instar larvae, first- and second-day pupae (L3, P1, and P2, respectively).
Figure 1.
Quantitative profiling of tRNA and tsRNA abundances during the larva-to-pupa transition of D. melanogaster. (A) Experimental design to co-profile tRNAs and tsRNAs in third-instar larvae (L3), first and second day pupae (P1 and P2). (B,C) Heatmaps showing the abundances (RPM, reads per million mapped reads) of tRNAs and tsRNAs from whole-body extracts of the examined developmental stages, respectively. Hierarchical clustering was performed on tRNA-seq libraries across different biological samples and across different tRNA genes (grouped according to the unique anticodons). (D) Scatterplots show that at the same or close stage, tsRNA abundances were correlated with tRNA gene copy numbers, tRNA abundances, tsRNA abundances measured by PANDORA-seq, and tsRNA abundances measured by miRNA-seq (SRP048223 and SRP000602), respectively. Solid lines: linear regression models; shades: 95% confidence interval (CI). Pearson's correlation and Spearman's correlation were labeled as ρp and ρs, respectively.
To evaluate the performance of mim-tRNAseq under our experimental setup, we analyzed different quantitative aspects of our data. Over 95% of the alignments with the genomic tRNA reference in both tRNA and tsRNA data sets are unambiguous (see Supplemental Table S1 for library information and mapping statistics). A nearly uniform pattern of read coverage along the length of nuclear genome-encoded tRNA with a modest weakening toward the 5′ end (Supplemental Fig. S1A) is generally consistent with published results (Behrens et al. 2021). In addition, the expression level of tRNAs showed a general correlation with the copy numbers of their corresponding genes (Pearson's correlation ρ ≈ 0.6 and P < 10−4 for all our data sets; Supplemental Fig. S1B). In particular, within each pair of isoacceptor tRNAs (tRNAGlyGCC/tRNAGlyUCC, tRNAGluCUC/tRNAGluUUC, and tRNALysCUU/tRNALysUUU), the relative difference in tRNA expression was also consistent with the difference in gene copy number (Supplemental Fig. S1C), suggesting single-nucleotide resolution of our data sets. The tRNA abundances (RPM, reads per million) displayed an excellent reproducibility between biological replicates and a relative stability from L3 to P2 (Fig. 1B). The overall stability of tRNA abundances during the larva-to-pupa transition was further supported by three additional analyses (Supplemental Fig. S1D–F): (1) absolute quantification of total tRNA concentration using a spike-in control (Escherichia coli tRNALysUUU); (2) quantification of total tRNAs using SYBR Gold staining; (3) northern blots against selected tRNAs (tRNAAspGUC, tRNAGlyGCC, and tRNAGluCUC). Together, these results document a suitability of the mim-tRNAseq pipeline in efficiently and accurately detecting tRNAs under our experimental setup.
To evaluate the feasibility of using the mim-tRNAseq pipeline to profile the dynamic expression of tsRNAs in our data sets, we generated tsRNA data (Fig. 1C) that were paired with the tRNA data (Fig. 1B). Figure 1D shows that, when grouped according to the anticodon, the tsRNA abundances showed significant correlations with the tRNA gene copy numbers, the tRNA abundances, the tsRNA abundances measured by PANDORA-seq (panoramic RNA display by overcoming RNA modification aborted sequencing) (Shi et al. 2021), and the tsRNA abundances obtained from published small RNA libraries (Pearson's correlation ρ = 0.65–0.82 and P < 10−5; see Supplemental Fig. S2 for comparisons of individual libraries). Furthermore, the changes of tsRNA abundances between different developmental stages revealed by our sequencing data were verified through the use of three high-abundance 5′-tsRNAs in northern blot analysis (see below and Fig. 2H for details). Together, these results supported a feasibility of using the mim-tRNAseq pipeline for quantitatively analyzing tsRNA data in our experimental setup. We referred to our adopted pipeline as mim-tRNA/tsRNA-seq.
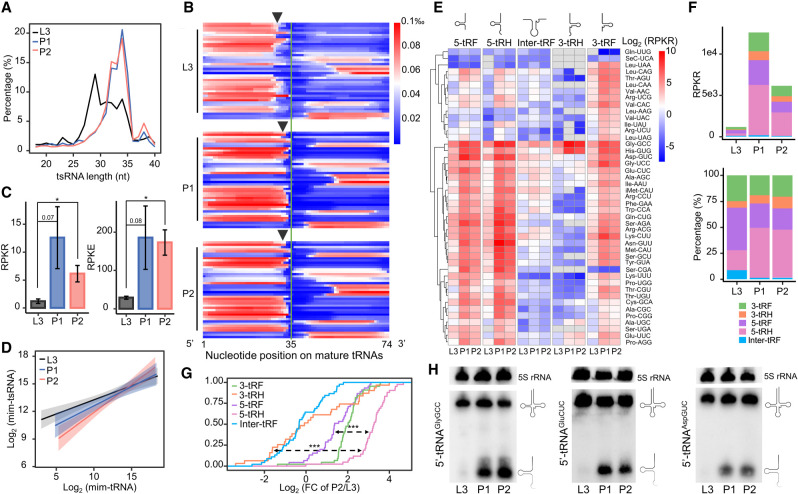
Figure 2.
Expression level of 5′-tsRNAs increases during the larva-to-pupa transition. (A) The length distributions of mim-tsRNA-seq reads in L3 (black), P1 (blue), and P2 (red). (B) A heatmap showing the normalized coverage of mim-tsRNA-seq reads at each nucleotide position along each mature tRNA. Each line presents one tRNA isoacceptor and they are ranked alphabetically. Note that the 3′ ends of 5′ tsRNAs in L3 are generally trimmed by ∼2 nt more than those in P1 and P2 (arrows marking the average positions of the 3′ ends at each stage). (C) Quantifications of total tsRNAs in different developmental stages, performed using either 2S rRNA as an internal control (RPKR, reads per kilo 2S rRNA reads) or E. coli tRNALysUUU as a spike-in control (RPKE, reads per kilo E. coli tRNA reads). Error bars represent SEM (standard error of the mean) computed from two or three biological replicates. Asterisks indicate significant differences (Student's t-test P < 0.01 denoted as **). (D) Linear models of the relationships between tsRNA and tRNA abundances in L3 (black), P1 (blue), and P2 (red). The log-log slopes are 0.308, 0.416, and 0.604 for L3, P1, and P2, respectively. (E) A heatmap showing the tsRNA abundances (RPKR) in L3, P1, and P2 stages. The mim-tsRNA-seq reads mapped to all cytoplasmic tRNA genes were grouped into five subclasses: 5-tRF, 5-tRH, Inter-tRF, 3-tRH, and 3-tRF. (F) Absolute (top) and relative (bottom) quantifications of total tsRNAs in each of the five subclasses. 3-tRF (green), 3-tRH (orange), 5-tRF (purple), 5-tRH (pink), and Inter-tRF (blue). (G) Empirical cumulative distribution function (ECDF) of the dynamic changes (P2/L3) in tsRNA levels (measured as RPKR) of different subclasses. Kolmogorov–Smirnov tests against the differences between 5-tRH versus 5-tRF and 5-tRH versus 3-tRH P < 0.001 (***). (H) Northern blots using probes against the 5′ ends of tRNAAspGUC, tRNAGlyGCC, and tRNAGluCUC. Each experiment has two independent replicates with consistent results.
Accumulation of 5′-tsRNAs during larva-to-pupa transition
According to our mim-tRNA/tsRNA-seq data, the size distribution of tsRNAs showed a sharper peak at ∼34 nt in early pupae than in larvae, with this peak (30–35 nt) accounting for 45.7%, 72.7%, and 71.2% of total tsRNAs in L3, P1, and P2, respectively (Fig. 2A). This could be explained by an active event of tRNAs being processed into tRNA halves during the larva-to-pupa transition. To infer this process, we calculated the relative coverage of tsRNA reads at each nucleotide position on the corresponding tRNAs in the three developmental stages. Figure 2B shows an uneven overall distribution of tsRNAs along the tRNA length, with a higher abundance in 5′-half than 3′-half for most tRNAs. A clear demarcation was detected near the middle of tRNAs separating the two halves, suggesting an overall preference of cleavage at the anticodon loop over D-loop or T-loop. This demarcation appeared sharper toward 5′-half than 3′-half, suggesting a relatively more subdued postcleavage processing for 5′-tsRNAs compared with 3′-tsRNAs. No such boundary was obvious at the position expected of T-loop cleavage, suggesting that further processing of 3′-tsRNAs was not a result of endonuclease cleavage at the T-loop.
The uneven distribution of tsRNA sequencing reads between 5′- and 3′-halves was independent of the developmental stages analyzed (Fig. 2B; see also Supplemental Fig. S3A for a similar pattern of PANDORA-seq data), suggesting an overall mechanistically similar pattern of cleavage within the anticodon loops at these stages. However, 5′-tsRNAs showed more degenerated 3′ ends in L3 than those in P1 and P2 (Fig. 2B, arrows), suggestive of de novo tRNA processing during the larva-to-pupa transition. To directly measure the change of tsRNA levels across different stages, we performed quantification by normalizing the tRNA-mapped reads to 2S rRNA (RPKR, reads per kilo 2S rRNA-mapped reads; see Supplemental Fig. S4A for northern blot against 2S rRNA documenting its stable expression during these stages) or to the E. coli spike-in (RPKE, reads per kilo E. coli tRNA-mapped reads). We observed a good correlation between the biological replicates of each developmental stage, indicating high reproducibility (Supplemental Fig. S4B). Both approaches supported a significant increase of total tsRNAs in P1 and P2 relative to L3, suggesting an overall accumulation of tsRNAs during the larva-to-pupa transition (Fig. 2C). Given the developmental stability of tRNA abundances, tsRNA accumulation also manifested itself as an increased slope of tsRNA abundance against their paired tRNA abundance (log-log slopes = 0.308, 0.416, and 0.604 arbitrary unit for L3, P1, and P2, respectively; Fig. 2D). Our results are in agreement with limited available findings reported previously (Luo et al. 2018) and, together, they documented an active accumulation of tsRNAs during the larva-to-pupa transition.
Based on tsRNA length and location in their corresponding tRNAs, we further grouped tsRNAs into five subclasses as described in Su et al. (2020): 5-tRFs, 5-tRHs, Inter-tRFs, 3-tRHs, and 3-tRFs (Fig. 2E, top; see also Supplemental Fig. S3B for PANDORA-seq data). We observed overall good correlations between tsRNAs in each of these subclasses and the corresponding tRNAs at each developmental stage, supportive of post-transcriptional biogenesis of all these tsRNAs (Supplemental Fig. S5A). We found that Inter-tsRNAs had the lowest overall levels (measured as RPKR) in all three stages (Fig. 2E, middle), further supporting a preference of tRNA cleavage at the anticodon loop. The overall accumulation in the pupal stages of 5-tRHs appeared slightly more robust than that of 3-tRHs (Fig. 2F), as further evidenced by a direct comparison of fold increases after pupation between these two tsRNA subclasses (Kolmogorov–Smirnov test P < 0.0001; Fig. 2G). Such asymmetric pattern of accumulation suggests an increased postcleavage stability of 5-tRHs relative to 3-tRHs. Together these results suggested a prominent contribution of 5′-tsRNAs, in particular 5-tRHs, to the overall increase in tsRNA levels during the larva-to-pupa transition.
To further confirm such an accumulation, we performed northern blot analyses on selected 5′-tsRNAs. Here we chose three with relatively high abundances: 5′-tsRNAAspGUC (RPKR = 52.9, 797.7, and 363.5 in L3, P1, and P2, respectively), 5′-tsRNAGlyGCC (RPKR = 98.8, 755.2, and 484.2), and 5′-tsRNAGluCUC (RPKR = 36.4, 249.7, and 123.4). We found that, although 5S rRNA and the corresponding tRNAs showed relatively stable levels of expression from L3 to P2, the selected 5′-tsRNAs all showed a clear increase after pupation (Fig. 2H). In fact, the expression of all the selected 5′-tsRNAs peaked in early pupae throughout the entire life cycle of D. melanogaster (Supplemental Fig. S5B). In contrast, 3′-tsRNAAspGUC (RPKR = 16.1, 85.6, and 60.2) was verified to have a consistently lower level of expression (Supplemental Fig. S5C). We also confirmed such an accumulation of selected 5′-tsRNAs in Drosophila simulans and Drosophila yakuba (Supplemental Fig. S5D). Altogether, these results suggested that 5′-tsRNA accumulation during the larva-to-pupa transition in Drosophila is a developmentally controlled and evolutionarily conserved event.
5′-tsRNAs exert regulatory activities through target site recognition on mRNAs
To analyze the regulatory activities of tsRNAs during pupation, we performed tsRNA inhibition experiments in vivo using the “sponge” methodology. Here we expressed a silencing cassette with 10 repetitive sequences that are complementary to a 5′-tsRNA to be inhibited (Fig. 3A; Supplemental Table S2). This method follows Watson-Crick base pair complementarity, a method that has been successfully used in elucidating miRNA functions in a variety of species, both in vivo and in vitro (Ebert et al. 2007; Ebert and Sharp 2010a; Fulga et al. 2015). Five tsRNAs were selected for sponge analysis: 5′-tsRNAAspGUC, 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNAProUGG, and 5′-tsRNALysUUU. Among them, 5′-tsRNAAspGUC, 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, and 5′-tsRNAProUGG were selected because of their high abundances in early pupal stages according to our mim-tRNA/tsRNA-seq and PANDORA-seq data (Fig. 2E; Supplemental Fig. S3B). 5′-tsRNALysUUU was shown to be strongly induced under serum starvation in Drosophila S2 cells (Luo et al. 2018). Furthermore, we generated a sponge line against a sequence scrambled from 5′-tsRNAAspGUC (Supplemental Table S2), which represents a negative control in addition to no sponge (see below).
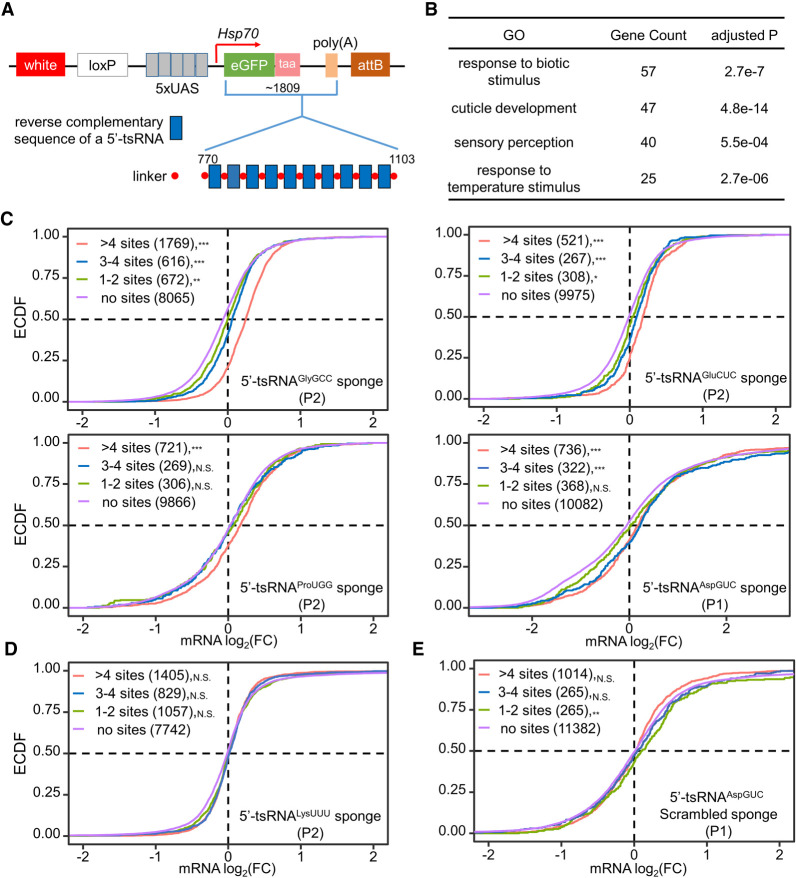
Figure 3.
Ubiquitous delivery of tsRNA sponges increases the levels of mRNAs with antisense target sites in early Drosophila pupae. (A) Construction of tsRNA sponges. The sponge element consists of 10 repetitive sequences (blue) that are complementary to a tsRNA of interest. It was placed within the 3′ UTR of eGFP under the control of five UAS sites in an attB vector. Six sponge elements were generated against, respectively, 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNAProUGG, 5′-tsRNAAspGUC, 5′-tsRNALysUUU, and a scrambled 5′-tsRNAAspGUC sequence. The expected sizes of the entire transcript and the sponge element for different tsRNA sponges are similar, and shown here are the numbers for 5′-tsRNAAspGUC. The transgenes were integrated into either attP40 (on second chromosome) or attP2 (on third chromosome) site via the phiC31 system. (B) Gene Ontology (GO) annotation reveals the enriched functional categories of 1097 genes with significantly higher mRNA expression (log2(fold change) > 1 and adjusted P < 0.05) in the five Tubulin-GAL4-driven sponge lines than in w1118. (C–E) ECDF plot of the fold differences in mRNA levels between each Tubulin-GAL4-driven sponge line and w1118 (DESeq2-calculated baseMean > 50). For 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNAProUGG, 5′-tsRNALysUUU (D) and their control, samples were collected from P2 stage. For 5′-tsRNAAspGUC (C), the 5′-tsRNAAspGUC scramble (E) and their control, samples were collected from P1 stage. Purple: genes without any predicted target sites; green: genes with 1–2 sites; blue: genes with 3–4 sites; red: genes with >4 sites.
To maximize the detection of changes in gene expression upon tsRNA inhibition, we used a ubiquitous driver, Tubulin-GAL4, to express each of the sponges and performed transcriptomic analyses in early pupae. For 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNAProUGG, and 5′-tsRNALysUUU, the samples were collected in P2; for 5′-tsRNAAspGUC, the samples were collected in P1 (because of lethality before P2; see Fig. 5B,C). To exclude the possibility that our sponges may affect tRNA levels or functions in either direct or indirect manners, we performed northern blot and qRT-PCR experiments. The quantification results showed that the expression levels of full-length tRNAs stayed mostly unaltered in Tubulin-GAL4 > 2 × sponge pupae (Supplemental Fig. S6A,B). In addition, by comparing the 5′-tsRNAAspGUC sponge line and its scramble control, our mRNA/Ribo-seq analysis did not detect a significant difference in either single-codon ribosome occupancies or translation efficiency (TE; see Methods) of genes enriched with codons GAC/U (Supplemental Fig. S6C,D). Furthermore, our PANDORA-seq analysis of the 5′-tsRNAAspGUC sponge and its scramble control showed that the levels of most of the detectable miRNAs and piRNAs were not significantly different (Supplemental Fig. S6E).
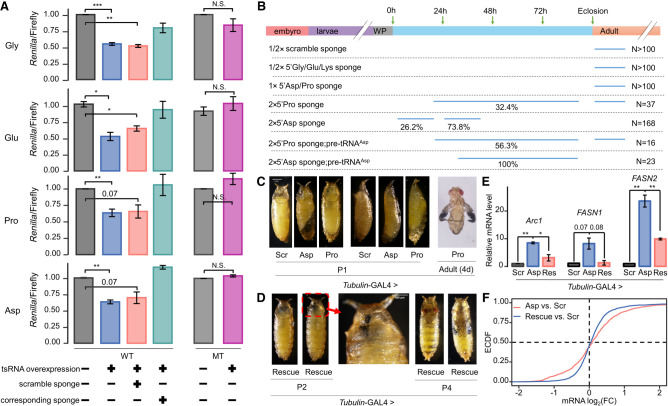
Figure 5.
Evaluation of regulatory activities of 5′-tsRNAs in vitro and during development. (A) Normalized Renilla/Firefly reporter activities to assay target site-dependent regulation by 5′-tsRNAs. WT: Renilla reporter gene carrying three target sites of each tsRNA. The sequences were cloned from native genes carrying predicted target sites (Supplemental Fig. S9B). MT: Renilla reporter gene carrying three sites with scrambled sequences in the seed match. For 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, and 5′-tsRNAProUGG, the sites were placed in 3′ UTR; for 5′-tsRNAAspGUC, the sites were placed in CDS with the consideration of their higher inhibitory effects (Supplemental Fig. S7C). tsRNA overexpression: synthetic tsRNA mimic and the pAWH vector to express the corresponding pre-tRNA. tsRNA sponge: synthetic RNA sequence that is reversely complementary to the tsRNA. Scramble sponge: synthetic RNA sequence that is randomly scrambled from the sponge. See Supplemental Tables S2–S5 for the sequences. (B) Summary of the estimated lethality time based on the stage at which pupae stopped developing. For 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNALysUUU, and the scramble 5′-tsRNAAspGUC, Tubulin-GAL4-driven one or two copies of sponge elements did not show any observable phenotypes (N > 100). For Tubulin-GAL4-driven one copy of sponge element against 5′-tsRNAAspGUC and 5′-tsRNAProUGG, some mild traits were observed but all pupae emerged (N > 100). (C) The pupal lethality phenotype of Tubulin-GAL4-driven two copies of sponge elements against 5′-tsRNAAspGUC and 5′-tsRNAProUGG. All 5′-tsRNAAspGUC-sponge pupae (“Asp”; N = 168) did not survive beyond P2. 26.2% of them had an abnormal body curvature and died during P1. 73.8% showed a failure in head inversion and an improper air bubble location during P2. The 5′-tsRNAProUGG-sponge pupae (“Pro”; N = 37) showed high phenotypic heterogeneity. 32.4% of them proceeded through P1 but failed to eclose. 67.6% emerged with various defects such as un-spread wings 4 d after emergence. The Tubulin-GAL4-driven two copies of scramble sponge (“Scr”) was used as control, and all tested pupae (N > 100) emerged without observable defects. (D) Phenotypic rescue with pre-tRNAAspGUC. All rescued pupae (“Rescue”; N = 23) proceeded through P1 with finished head evert process and correct air bubble location. 43.5% of them lived to P4 as late pharate adults, but had leg malformation and did not eclose. (E) qRT-PCR confirming that the mRNA levels of three selected 5′-tsRNAAspGUC target genes were restored by pre-tRNAAspGUC. All samples were collected from P1 pupae. Arc1, FASN1, and FASN2 were selected because of their known expression and function during the larva-to-pupa transition. rp49 was used as internal control. Each experiment had two independent replicates. Error bars represent SEM, and Student's t-tests were used. (F) mRNA-seq confirming that the mRNA levels of 5′-tsRNAAspGUC target genes (N = 1411) were restored by pre-tRNAAspGUC. Kolmogorov–Smirnov test P-value = 10−11.
We used DESeq2 to conduct a two-factor analysis, accounting for both developmental stages (P1 vs. P2) and genotypes (Tubulin-GAL4 > 2 × sponge vs. no sponge). By contrasting the genotypes, we identified differentially expressed genes (DEGs) and performed Gene Ontology (GO) annotation. A total of 438 genes were identified with significantly lower mRNA expression [log2(fold change) < −1 and adjusted P < 0.05] in the five Tubulin-GAL4 > 2 × sponge lines, but without a significant clustering in any functional category. On the contrary, a total of 1097 genes with significantly higher mRNA expression in the sponge lines were enriched in a few key pathways for pupal development such as “response to biotic stimulus” and “cuticle development” (adjusted P = 10−7 and 10−14, respectively; Fig. 3B). Therefore, the pupa-accumulated 5′-tsRNA in our test could suppress specific gene activities that are crucial for pupal development.
The actions of tsRNAs on gene expression could depend on their recognition sites in targeted mRNAs (Maute et al. 2013; Luo et al. 2018). We combined three algorithms, TargetScan, phyloP, and miRanda, to predict mRNA target sites that could fulfill the following three criteria: (1) at least 7-bp perfect antisense match to the tsRNA sequence; (2) FDR-adjusted cross-species conservation score for the mRNA site < 0.05; (3) a minimum free energy (MFE) < −30 kCal/mol of local alignment. Based on the number of predicted target sites on each mRNA, we classified the expressed genes into four groups for each tsRNA sponge that we analyzed: no sites, 1–2 sites, 3–4 sites, and ≥5 sites. Figure 3C shows an mRNA level increase in response to the sponges against 5′-tsRNAAspGUC, 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, and 5′-tsRNAProUGG in a manner that is dependent on the number of predicted target sites. In contrast, when 5′-tsRNALysUUU was sponged, the mRNA levels of genes with or without putative target sites effectively had no difference (Fig. 3D). This tsRNA had the lowest abundance (RPKR = 89.6 in P2) among the tsRNAs tested, suggesting that the inhibitory effect detected in our sponge setup is dependent on tsRNA abundance. To further evaluate this point, we found that our data obtained with sponges against 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, and 5′-tsRNAProUGG each yielded similar results when either 5′-tsRNALysUUU sponge or no sponge was used as a negative control (Supplemental Fig. S7A). Furthermore, we found that the scramble sponge had no meaningful effect on mRNA levels irrespective of the number of “target” sites (Fig. 3E). In addition, our data obtained with the sponge against 5′-tsRNAAspGUC at P1 yielded similar results when either this scramble sponge or no sponge was used as a negative control (Supplemental Fig. S7B).
Effective mRNA-level inhibition has no strict requirements of either mRNA site location or tsRNA seed selection
To investigate whether tsRNAs may show a preference in target site location on mRNA, we selected genes containing predicted target sites in only one section of their transcripts and divided them into three classes based on target site positions on the transcript: 5′ UTR, CDS, and 3′ UTR. Supplemental Figure S7C shows that, among the 5′-tsRNAs analyzed, 5′-tsRNAAspGUC had a significant preference of target site location in CDS and 5′ UTR over 3′ UTR. However, 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, and 5′-tsRNAProUGG did not show such a preference, with varying degrees of inhibition when they were at different locations. These results suggested that the inhibitory effect exerted by tsRNAs through their presumed target genes is largely independent of tsRNA target site location on the transcript.
It is well documented that miRNA action mainly depends on the “seed” positioned at the nucleotide 2–8 at the 5′ end. To evaluate whether the mode of tsRNA action may share this feature, we analyzed the inhibitory effect of each 7-mer seed from 5′ to 3′ along 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNAProUGG, and 5′-tsRNAAspGUC. While the seeds at varied positions of 5′-tsRNAs did not show a gross bias in their ability to affect their presumed target mRNA levels, the majority of them showed clear derepression on target genes by the corresponding tsRNA sponge (Supplemental Fig. S7D). These results suggested that 5′-tsRNA recognition of their target sites was not restricted to the 5′ seed selection as seen for miRNAs.
Participation of accumulated tsRNAs in regulating the expression of metamorphosis genes
Our results described thus far suggested that tsRNAs could repress mRNA expression of target genes with an accommodating posture in terms of both seed selection and site location. Based on this mode of tsRNA action, we performed a genome-wide analysis to determine whether dynamically expressed tsRNAs may target genes that are relevant to developmental processes at early pupal stages. Here we analyzed tsRNAs that were expressed in P2 and compiled all mim-tsRNA-seq reads according to the functional unit of 7-mer seeds. This led to the identification of 2075 unique seeds. We selected the top 5% of these seeds with the highest abundance. At this abundance threshold, 104 seeds were captured, which contributed to 42% of the total tsRNA reads in P2 and had an increased RPKR by at least 2.1 fold in P2 than in L3 (Student's t-test P-value < 0.0001). Among them, only one was shared by known miRNA families: “GUAGAAU” as in miR-958, which was found to be adult gut-specific (Weigelt et al. 2019).
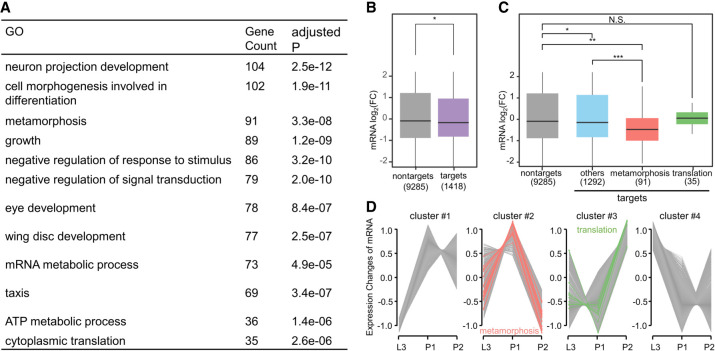
We used the 104 seeds to scan the genome for recognition sites (Methods). We calculated the density of these sites in the transcript of each protein-coding gene, and retrieved a total of 1418 genes with the top site density (9.1–55.8 sites per kb) and a detectable level of mRNA expression (DESeq2-calculated baseMean > 70). We considered them as putative target genes of highly expressed tsRNAs at P2 stage (also significantly up-regulated from L3 to P2 with fold change > 2.1 and Student's t-test P-value < 0.0001) for further analyses. GO annotation revealed that these genes were significantly enriched in essential biological processes including “metamorphosis” and “cytoplasmic translation” (Fig. 4A). Since this genome-wide screening was performed without a precondition of gene expression status, our results suggested that the accumulated tsRNAs at early pupal stages have a preference for targeting genes including those that are involved in biological processes taking place at this time.
Figure 4.
Identification of tsRNA function in early pupae. (A) GO enrichment of putative target genes (N = 1418) of highly expressed tsRNAs at P2 stage. (B) Shown are the fold differences between P2 and L3 in mRNA levels of putative tsRNA target genes and nontarget genes. Student's t-test P = 0.04. (C) Shown are the fold differences between P2 and L3 in mRNA levels of putative tsRNA target genes in three functional categories: GO:0007552 (“metamorphosis”), GO:0002181 (“translation”), and all other target genes (“others”). (D) Clustering of the time-series mRNA expression profiles from L3 to P2. A total of 6713 DEGs either between L3 and P1 or between P1 and P2 were used in the analysis. Hypergeometric test P = 10−8 and 10−7, for the metamorphosis group enrichment in cluster #2 (red) and the translation group enrichment in cluster #3 (green), respectively.
To examine whether the accumulated tsRNAs in early pupae exert regulatory activities on their putative target genes during normal development, we analyzed our mRNA-seq data of WT across different stages. Enriched GO groups of genes with an increased mRNA level include “mitochondrion organization” (GO:0007005), “cuticle development” (GO:0042335), and “protein targeting” (GO:0006605); those with a decreased level include “mitotic cell cycle” (GO:0000278), “metamorphosis” (GO:0007552), “lipid homeostasis” (GO:0055088), “alpha-amino acid metabolic process” (GO:1901605), and “proteolysis involved in cellular protein catabolic process” (GO:0051603). These results are in general agreement with existing knowledge (Zhang et al. 2020) about the developmental events during the larva-to-pupa transition. Figure 4B shows that the putative target genes had a significant overall reduction in mRNA levels (between P2 and L3). When these putative target genes were categorized according to GO annotation as “metamorphosis,” “cytoplasmic translation,” and “others,” they behaved differently from one another. Among the three groups, the “metamorphosis” group of tsRNA target genes showed the largest reduction (Fig. 4C, red). This effect was also observed for the “others” group but only in a modest manner (Fig. 4C, blue), but not for those in the “translation” group (Fig. 4C, green). In addition, a time-series profile analysis of the 6713 DEGs either between L3 and P1 or between P1 and P2 suggested that although the “metamorphosis” group of target genes was significantly enriched in the P2-downward cluster (Fig. 4D, red), the “translation” group was significantly enriched in the P2-upward cluster (Fig. 4D, green). We also examined specifically the changes of metamorphosis-associated gene product levels in response to the Tubulin-GAL4-driven sponges. Supplemental Figure S8 shows that mRNA levels of targets genes in the “metamorphosis” group were significantly higher than those of their nontarget counterparts (comparing red with green; Student's t-test P < 0.001) in 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, and 5′-tsRNAAspGUC sponge lines, respectively. Together, these results suggested a preferential impact of the accumulated tsRNAs at P2 stage on suppressing genes involved in metamorphosis.
Experimental evaluation of tsRNA action and specificity
Our genome-wide, informatics-based analysis described thus far suggested a participation of pupa-enriched tsRNAs in regulating the mRNA levels of metamorphosis-related genes. To further evaluate tsRNA action and specificity, we performed gene-based analyses in both S2 cells and transgenic flies. Here we manipulated tsRNA activities through the uses of tsRNA sponges, tsRNA mimics, and pre-tRNA expression. In S2 cells, the expression of pre-tRNAs could moderately increase the intracellular levels of corresponding 5′-tsRNAs (see Supplemental Fig. S9A for northern blot analysis of pre-tRNAAspGUC expression as an example). Thus, we co-transfected a synthetic tsRNA mimic (Supplemental Table S3) and an expression vector of the corresponding pre-tRNA (Supplemental Table S4) to achieve overexpression. To examine whether the inhibitory effect of 5′-tsRNAs was dependent on specific target sites, we performed dual-luciferase reporter assays by inserting three native target sites (see Supplemental Table S5 and Supplemental Fig. S9B for sequences and local alignments, respectively) of each tested 5′-tsRNA into 3′ UTR or CDS of the Renilla gene. Figure 5A shows that the reporter activity was significantly reduced by overexpression of 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNAProUGG, and 5′-tsRNAAspGUC (blue), and that such an effect was blocked by their corresponding sponges (green) but not by the scramble sponge (pink). If the inserted sites were scrambled within the 7-mer seed match, tsRNA overexpression could not effectively repress the reporter activity (Fig. 5A, purple). In addition, we performed qRT-PCR in this system and confirmed similar results at the Renilla mRNA level (Supplemental Fig. S9C). These results document inhibitory activities of the predicted 5′-tsRNA target sites in a cell-based reporter system.
We also used our transgenic flies containing the sponge elements to evaluate the functional consequence and action specificity of 5′-tsRNAs in a developmental context. Driven by Tubulin-GAL4, two of the five sponges (5′-tsRNAAspGUC and 5′-tsRNAProUGG) showed pupal lethality when present at two copies (Fig. 5B). The pupal lethality phenotype caused by 5′-tsRNAAspGUC sponge was significantly rescued by a simultaneous expression of pre-tRNAAspGUC (Fig. 5C,D). This is a specific phenotypic rescue because it had no impact on improving the phenotype caused by another sponge against 5′-tsRNAProUGG. In particular, ∼90% pupae of Tubulin-GAL4 > 2 × 5′-tsRNAProUGG sponge were found to be insufficiently above the food surface and ∼32.4% pupae failed to eclose with varied defects. These traits could not be ameliorated by the expression of pre-tRNAAspGUC (Fig. 5B).
Furthermore, the observed phenotypic rescue by pre-tRNAAspGUC was accompanied by changes in mRNA levels of selected target genes. Arc1 encodes a retrovirus-like Gag protein that is required for regulation of neuronal synaptic plasticity, muscle development, and energy balance (Mosher et al. 2015; Ashley et al. 2018). According to our prediction, Arc1 transcript carries four conserved target sites of 5′-tsRNAAspGUC. qRT-PCR results showed that its mRNA level in P1 pupae of the Tubulin-GAL4-driven 5′-tsRNAAspGUC sponge was approximately eightfold higher than that in the scramble control, and such increase was effectively restored by one copy of UAS-pre-tRNAAspGUC (Fig. 5E). In fact, the Tubulin-GAL4-driven 5′-tsRNAAspGUC sponge had locomotion defects during the wondering stage (Supplemental Movie S1 in comparison to the scramble control shown in Supplemental Movie S2). FASN1 and FASN2 encode the two major fatty acid synthases in Drosophila (Chung et al. 2014; Garrido et al. 2015; Gramates et al. 2022). Fatty acids profiling (C14–C30) showed that heptadecanoic (17:0), stearic (18:0), and triacontanoic (30:0) acids were the most increased in 5′-tsRNAAspGUC sponge pupae (Supplemental Fig. S10A). qRT-PCR confirmed the rescue effect of pre-tRNAAspGUC on the mRNA levels of Arc1, FASN1, and FASN2 (Fig. 5E). mRNA-seq further confirmed that pre-tRNAAspGUC could restore the altered mRNA levels of 5′-tsRNAAspGUC target genes by 5′-tsRNAAspGUC sponge (Kolmogorov–Smirnov test P = 10−11; Fig. 5F). Together, these results documented the specificity of 5′-tsRNAAspGUC action on mRNA expression of predicted target genes in a developmental context.
We also used several other GAL4 lines to drive the 5′-tsRNAAspGUC sponge (listed in Methods). The ubiquitous Actin-GAL4 > 2 × sponge showed a similar pupal lethality phenotype with those driven by Tubulin-GAL4. Other tissue-specific drivers showed no obvious defects, except the oenocyte-specific Desat1-GAL4 (also known as PromE-GAL4). All Desat1-GAL4 > 1 × sponge pupae (N = 214) died before eclosion (Supplemental Fig. S10B). They showed misallocated bubbles in ventral abdomen at P2 and leg malformation at P4 (Supplemental Fig. S10C). 1 × pre-tRNAAspGUC rescued the abdominal trait in all P2 pupae (N = 9; Supplemental Fig. S10C), and three out of nine successfully emerged but died within the first day. Given the fact of the oenocyte-specific role of FASN2 during pupation (Billeter et al. 2009), we performed qRT-PCR and showed that, when driven by Desat1-GAL4, 1 × 5′-tsRNAAspGUC sponge elevated the mRNA level of FASN2, and such effect was restored by 1 × pre-tRNAAspGUC (Supplemental Fig. S10D). In addition, we used RiboTag (RPL13A-FLAG) to profile oenocyte-specific ribosome-associated mRNAs in the sponge pupae, and confirmed the increase of FASN2 expression (Supplemental Fig. S10E). These results further document that 5′-tsRNAAspGUC could act on specific target genes to achieve tissue-specific regulation during development.
tsRNAs contribute to the regulation of an overall decline in translation efficiency during the larva-to-pupa transition
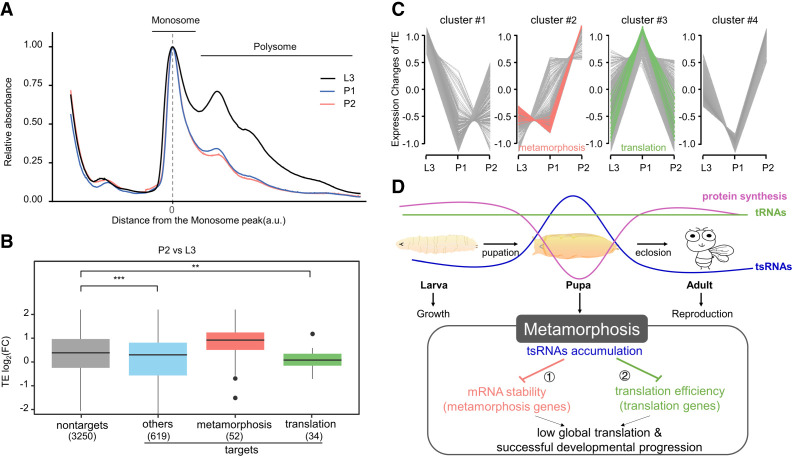
Previous studies in S2 cells suggested that tsRNAs contribute to serum starvation-induced reduction in global translation through preferentially down-regulating translation efficiency of translational machinery genes (Luo et al. 2018). As shown by polyribosome profiling (Fig. 6A), pupation is also accompanied by a decline in the overall translation. To investigate whether tsRNAs may be involved in such translation regulation with a developmental context, we performed Ribo-seq for estimating gene-specific TE. As detailed above for our analysis in Figure 4C, we similarly divided the predicted target genes of pupa-accumulated tsRNAs into three functional groups (metamorphosis, translation, and others). Figure 6B shows that, relative to the nontarget group, target genes in the translation group had significantly down-regulated TE during the larva-to-pupa transition, contrasting with an up-regulation in TE for genes in the metamorphosis group. Clustering of time-series TE profile from L3 to P2 further supported a discrepancy in the dynamic expression pattern between the two groups of tsRNA target genes (Fig. 6C). As shown in Figure 4C, genes in the translation group did not have a reduced mRNA expression level in P2. Therefore, tsRNAs accumulated in early pupae can operate through a distinct, translation-mediated mechanism in regulating the overall translation efficiency during pupation.
Figure 6.
Genes in the translation group that are predicted targets of pupa-accumulated tsRNAs show a reduction in translation efficiency during pupation. (A) Absorbance profiles at 254 nm of RNA samples from L3 (gray), P1 (blue), and P2 (green). For each experiment, both the absorbance and the position were normalized to the 80S monosome peak. (B) Shown are the fold differences between P2 and L3 in TE of putative tsRNA target genes in different functional categories. Protein-coding genes that had mRNA-seq FPKM > 10 in both L3 and P2 were included. (C) Clustering of the time-series TE profiles from L3 to P2. A total of 3380 genes (mRNA-seq FPKM > 5) with TE that differed by at least twofold either between L3 and P1 or between P1 and P2 were used in the analysis. Hypergeometric test P = 10−4 and 10−6, for the metamorphosis group enrichment in cluster #2 (red) and the translation group enrichment in cluster #3 (green), respectively. (D) A graphic illustration of tsRNA regulatory activities during the larva-to-pupa transition of Drosophila. Despite the stable level of tRNAs (green), this transition is marked with an accumulation of tsRNAs (blue) and a decline of overall protein synthesis (purple), providing a special developmental context for understanding of tsRNA action. In a target site-dependent manner, the early pupa-accumulated tsRNAs specifically down-regulate gene expression in the metamorphosis group at the mRNA level (suppression arrow 1) and genes in the translation group at the translation level (suppression arrow 2). Subsequently, the global translation activity is reduced. Under this framework, the accumulation of functionally active tsRNAs instruct Drosophila metamorphosis to proceed through the larva-to-pupa-to-adult transition.
Discussion
This study is designed to advance our understanding of tsRNA function in a developmental context in Drosophila. It focuses on a developmental transition from an active growth and feeding state of larvae to a nonfeeding and starved state of pupae in preparation of metamorphosis (Fig. 6D). This transition is marked with a decline in ATP level (Yamada et al. 2020), overall translation (Fig. 6A), and an accumulation of tsRNAs (Fig. 2), providing a special developmental context for understanding of tsRNA action. Here, we take a genome-wide, informatics-based approach toward establishing the mode of tsRNA action through the use of sponges for highly accumulated tsRNAs during this developmental transition. Our results suggest that tsRNAs can regulate gene expression through two distinct mechanisms acting on either mRNA levels or translation efficiency. tsRNAs accumulated in pupae exert preferential inhibitory effects on mRNA levels of genes involved in metamorphosis and on translation efficiency of genes encoding components of the translation machinery. These results document that tsRNAs accumulated at a developmental time exert regulatory activities at that time, suggesting that accumulation and action of tsRNAs are an integral part of the regulatory network instructing developmental progression (Fig. 6D).
tsRNA biogenesis is an inseparable aspect of tsRNA biology and our study provides a first comprehensive atlas of tsRNAs during the larva-to-pupa transition in Drosophila. Here we adopted the mim-tRNAseq method to simultaneously quantitate tRNAs and tsRNAs, through the use of optimized size ranges in library construction and enzymatic treatments, primarily including T4PNK and TGIRT (Behrens et al. 2021). The length distribution of tsRNAs in our mim-tRNA/tsRNA-seq libraries peaks around 34 nt, which is generally consistent with a previous report using PANDORA-seq and small RNA-seq (Isakova et al. 2020; Shi et al. 2021). This suggests that our approach provides a more reliable detection of tsRNAs than approaches using miRNA libraries. The clear demarcation detected at the anticodon loop and an inferred active processing of tRNAs into tsRNAs stem from our method that was tailored specifically for tsRNAs. As shown in the quantification results of different tsRNA subclasses (Fig. 2E; Supplemental Fig. S3B), mim-tRNA/tsRNA-seq showed an advantage in precisely detecting the 5′-tsRNA accumulation in early pupae. Such accumulation was further verified by northern blots in the cases of 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, and 5′-tsRNAAspGUC. However, unlike the significant increase in 5′-tsRNA levels during the larva-to-pupa transition, tRNA levels appeared to be relatively stable. In addition, our analysis of TGIRT-introduced misincorporation identified little difference in tRNA modification profiles between larvae and early pupae (Supplemental Fig. S11). Because accumulation of 5′-tsRNAs takes place without significant changes in tRNA modifications analyzed, potential differences across developmental stages influencing such an accumulation likely come from other means, such as undetected modifications, the activity of tRNA cleavage enzymes, or the stability of tsRNAs. A prominent contribution of 5-tRHs to the tsRNA accumulation (Fig. 2G) supports a role of postcleavage stability of tsRNAs in regulating tRNA/tsRNA homeostasis during the larva-to-pupa transition.
We have taken an informatics approach—complemented by gene-based analyses—toward an understanding of tsRNA action and specificity in a developmental system. Here we use two algorithms, TargetScan and miRanda, to probe the mode of tsRNA-mRNA interaction that can best explain our mRNA-seq data in response to sponge perturbations. These software had been used to predict the post-transcriptional regulation of specific tsRNAs on putative target genes in human and Drosophila cells (Luo et al. 2018; Chen et al. 2022; Tu et al. 2023). We note that the utilization of these software as a tool in our study is not meant to be evidence for a strictly miRNA-like or seed-dependent mode of target recognition. To the contrary, based on these bioinformatics tools, our results suggest a reduced dependence on both selection of seed positions and site locations relative to miRNAs. This distinction may be reflective of the differences in size, abundance, modifications, and associated Argonaute proteins between tsRNAs and miRNAs. In Drosophila, miRNAs, siRNAs, and piRNAs execute post-transcriptional regulation by primarily binding to AGO1, AGO2, and AGO3/PIWI/AUB, respectively (Okamura et al. 2004; Tomari et al. 2007; Iwasaki et al. 2015). While AGO3/PIWI/AUB are predominantly expressed in germline cells and early embryos, both AGO1 and AGO2 are expressed throughout the life cycle. According to our western blot analysis (Supplemental Fig. S12), AGO2 might be the only Argonaute protein expressed at a relatively high level in P2 pupae. It is possible that the presence of different AGO proteins at different developmental stages may contribute to the noncanonical target recognition mode of early pupae-accumulated tsRNAs.
Several miRNAs, including bantam and miR-14, have been reported to influence the fitness during this developmental transition by specifically targeting and tuning key hormonal pathways (Varghese and Cohen 2007; Boulan et al. 2013). In our study, the pupal lethality phenotype caused by tsRNA sponges suggests that 5′-tsRNAProUGG and 5′-tsRNAAspGUC are required for successful completion of metamorphosis. The timing of lethality coincides with that of 5′-tsRNA accumulation to reach their peak expression levels. As shown in Figures 4C and 6B, these tsRNAs act on a wide range of genes by regulating either the mRNA level or translation efficiency. The metamorphosis defects caused by tsRNA sponges are likely because of dysregulation of a collection of genes including, but not limited to, Arc1 and FASN2. Accordingly, our rescue experiments to document the specificity of sponge-induced phenotype were performed by overexpressing full-length tRNAs, an approach that has been successfully used previously (Kuscu et al. 2018). Through the use of gene-based analyses, we have verified such specificity in both reporter assays and in a tissue-specific manner with the use of FASN2 as a molecular marker. In our experiments, not all tsRNA sponges could cause a phenotype and, additionally, the defects were moderate for Tubulin-GAL4-driven one copy of the sponge transgenes. These results suggest that, similar to what has been observed with miRNA sponges (Davis et al. 2006; Esau 2008; Fulga et al. 2015), tsRNA sponges need to be present at a sufficient level to exert inhibitory activities.
The sponge approach has been widely used in studying miRNA functions. We adopted this approach to facilitate our analysis of tsRNAs that show a newly uncovered property of accumulation at the larva-to-pupa transition. While this approach has provided useful insights into the action of these tsRNAs in our study, there are limitations inherent to all sponge-related approaches, especially when used in the tsRNA study. In particular, the gold standard of verifying the specificity of the phenotype induced by a miRNA sponge is through editing the miRNA gene itself (Cohen 2009), but this is not feasible for tsRNA sponges. In evaluating miRNA sponge specificity, another test is to compare the phenotype and gene expression profiles not only between a sponge and its negative control but also between the negative control and no treatment (Ebert and Sharp 2010b). We have taken a similar test in our study and our results showed that, just like the wild-type line, the negative control lines showed neither the observed phenotypes nor specific effects on tsRNA target genes (Figs. 3D,E, 5B). In our analysis, putative tsRNA target genes that had elevated mRNA levels in sponge lines did not show such an increase in either the wild-type line or in the negative control lines (Fig. 3; Supplemental Fig. S7). Furthermore, we used two algorithms, miRanda and miRNAsong (Barta et al. 2016), to examine the interactions between our tsRNA sponges and their corresponding tsRNAs. The miRanda calculation suggested a highly stable structure (MFE < −59 kCal/mol) for all the five sponge-tsRNA pairs in this study, but the interactions between the tsRNA sponges and all known D. melanogaster miRNAs were much less stable (MFE > −17 kCal/mol). The miRNAsong calculation (MFE cutoff: −25 kCal/mol and seed type: canonical 7mer) predicted four potential off-target miRNAs: miR-9378-5p, miR-4949-5p, miR-1003-5p, and miR-966-5p, but they all had a negligible expression in early pupal stages (PANDORA-seq: no detection; miRNA-seq: 0–44 RPM). With respect to the phenotypes detected in two of the sponge lines, we used full-length tRNA expression in a rescue experiment. Our results showed that pre-tRNAAspGUC had a rescue effect in the 5′-tsRNAAspGUC sponge line, but not in the 5′-tsRNAProUGG sponge line, a finding supportive of sponge specificity. However, this rescue remained only partial, which underscores the limitations of both the perturbation and rescue approaches available to us in studying tsRNA action. Taken together, our results described in this report are supportive of the utility and specificity of tsRNA sponges despite a current insufficiency to formally rule out unintended effects including off-targets.
While this study suggests a role of tsRNAs in regulating both mRNA levels and translation efficiency during pupation, our current understanding remains incomplete. For example, according to our model (Fig. 6D), tsRNA target genes in the translation group are preferentially down-regulated through a translation-mediated mechanism, whereas those in the metamorphosis group are down-regulated at their mRNA levels. Although both mechanisms have been suggested for tsRNA-mediated regulation (Goodarzi et al. 2015; Huang et al. 2017; Luo et al. 2018), it remains elusive how tsRNAs select different sets of target genes and act on them via different mechanisms within a single developmental context. In addition, tsRNAs could regulate developmental programs in ways that are distinct from the RNAi-like mechanism (Chen et al. 2016, 2021a; Sharma et al. 2016). Given the above-mentioned caveats about the use of tsRNA sponge and tRNA overexpression in vivo, it is possible that alternative or additional functioning modes of tsRNAs may be operative during the larva-to-pupa transition. For example, although the pupa-accumulated tsRNAs do not appear to have an impact on either the levels or the activities of full-length tRNAs (Supplemental Fig. S6), they could act as aptamers for other gene regulatory machineries possibly via interacting with different biomolecules (Chen and Zhou 2023).
It has been well documented that tsRNAs derived from cleavage at the anticodon loop are induced by serum starvation, UV, hypoxia, and other stresses (Lee and Collins 2005; Yamasaki et al. 2009; Emara et al. 2010; Sobala and Hutvagner 2013; Luo et al. 2018). Pupation represents a developmental stage that has an “internally starved” state with limited energy production. Thus, a decline in global translation during the larva-to-pupa transition as seen in Figure 6A is a direct and effective solution to a cost-benefit optimization problem because of the huge cost of amino acid synthesis and polypeptide assembly (Yamada et al. 2020). Such a decline is accompanied by tsRNA accumulation without an apparent tRNA level reduction (Supplemental Fig. S1). Therefore, the performance objective of tsRNA accumulation here is not to deplete genuine tRNAs but to confine gene regulatory activities in the specific developmental context. As in serum-deprived S2 cells (Luo et al. 2018), tsRNAs accumulated in pupae preferentially reduce the translation efficiency of mRNAs coding for translation-related genes, thus allowing normal translation of other genes on demand. These results thus suggest a two-pronged design of embedding tsRNAs in the gene regulatory network instructing metamorphosis: translation-mediated action to influence overall translation and regulation of mRNA levels of metamorphosis-related genes. These mechanisms together confer an ability to pupa-accumulated 5′-tsRNAs in safeguarding the progression of metamorphosis.
Methods
Fly husbandry, stocks, transgenic strains, and classification of pupal stages
All flies were reared and crossed on standard corn medium at 25°C and 60% humidity. Drosophila stocks used in this study include w1118, Actin-GAL4, Tubulin-GAL4, Eyeless-GAL4, Elav-GAL4, GMR-GAL4, Hh-GAL4, Ptc-GAL4, Vg-GAL4, MHC-GAL4, Dilp2-GAL4, Cg-GAL4, Phm-GAL4, Desat1-GAL4.E800 (BDSC 65405), UAS-RPL13A-FLAG (BDSC 83684), Drosophila simulans (DSSC 14021-0251.195), and Drosophila yakuba (DSSC 14021-0261.01).
For each tsRNA sponge (against 5′-tsRNAGlyGCC, 5′-tsRNAGluCUC, 5′-tsRNAAspGUC, 5′-tsRNAProUGG, or 5′-tsRNALysUUU), 10 repeats of the sequence that is reversely complementary to 5′ half of the corresponding mature tRNA with 4-bp spacers (Supplemental Table S2) were incorporated into the multiple cloning site (XholI/XbalI), 34 bp downstream to the stop codon of eGFP (Fig. 3A). As a negative control for 5′-tsRNAAsp sponge, a scrambled version of the reversely complementary sequence was generated using GenScript (Supplemental Table S2). For expression of pre-tRNAAspGUC, the genomic fragment of tRNA:Asp-GTC-1-6 (FBgn0051602), from the upstream 223 bp to the downstream 228 bp, was amplified (see Supplemental Table S4 for PCR primers) and then inserted into the XholI/XbalI sites of pUAST-attB. All the transgenic strains were created by integrating the pUAST-attB vector with the donor transgenes to cytological positions 25C6, 68A4, and 75B1 via the phiC31 system. Microinjection services were provided by the Drosophila Resource and Technology facility, Center for Excellence in Molecular Cell Science, CAS.
The chronological age after pupation was determined as hours because the white pupa completely stopped wriggling. After that, 0–24 h, 24–48 h, 48–72 h, and >72 h were labeled as P1–P4, respectively. According to the morphological markers, the P1–P4 ages of our w1118 and the other control strains roughly correspond to Bownes's developmental stages p1–p5, p5–p6, p7–p9, and p10–p15 (Bainbridge and Bownes 1981), respectively.
S2 cell culture, transfection, and luciferase reporter assays
Drosophila S2 cells were cultured in Schneider's Insect Medium (Gibco) containing 10% FBS and Mycoplasma Prevention Reagent (TransGen FM501) without CO2 to reach 2 × 106 to 4 × 106 cells/mL. Transfection was performed in ∼5 × 105 cells per well using transfection reagent according to the manufacturer's instructions (JetPRIME).
We used Dual-Luciferase Reporter Assay System (Promega) by cloning three tandem repeats of an endogenous target site (a perfect 7-mer match to the nucleotides 2–8 of 5′-tsRNA + upstream 15 bp + downstream 8 bp; Supplemental Table S5) into the multiple cloning site (XholI/NotI) of psiCHECK-2. For 5′-tsRNAGlu, 5′-tsRNAGly, and 5′-tsRNAPro, the target sites were placed in the 3′ UTR of Renilla; for 5′-tsRNAAsp, the target sites were incorporated before the stop codon of Renilla. To contrast with the endogenous target sites, the mutated sites were created by scrambling the 7-mer seed match (Supplemental Table S5).
To examine the tsRNA actions and specificity, we performed co-transfection to quantify the effects of overexpression and sponge of tsRNAs on Renilla expression. For overexpression, cells were bathed with synthetic single-stranded RNA mimics of each 5′-tsRNA (Supplemental Table S3; Azenta) at a final concentration of 20 nM, along with the pAWH vector expressing the corresponding full-length tRNA (Supplemental Table S4). After transfection for 48 h, the luciferase activities of Firefly and Renilla were measured according to the manufacturer's instructions (Promega).
RNA isolation and qRT-PCR
Total RNA was isolated from tissue samples or S2 cell culture using RNAiso Plus (Takara 9108), and the concentration was measured by NanoDrop. For tRNA qRT-PCR, before reverse transcription, RNAs were pretreated by rtStar™ tRF&tiRNA Pretreatment Kit (Arraystar AS-FS-005) to remove internal m1A, m1G, and m3C. Reverse transcription was performed using HiScript III first Strand cDNA Synthesis Kit (Vazyme R312-01). Quantitative PCR was performed using 2× Universal SYBR Green Fast qPCR Mix (ABclonal RK21203) and quantified using LineGene 9600plus (Bioer Technology). Data from tissue samples were all normalized to rp49; Renilla data from Dual-Luciferase Reporter Assay in S2 cells were normalized to Firefly. All RT-PCR primers are listed in Supplemental Table S6.
Northern blot analysis and total RNA staining
Briefly, 7–10 μg of total RNA was routinely run in 10 mL of 10% PAGE gel with 7 M urea at 130 V for about 1 h. Then, for detection of total tRNAs, SYBR Gold (Invitrogen S11494) staining was performed. For northern blot analyses, RNA was transferred to Nylon membranes (Millipore INYC00010) for 1 h with 0.5× TBE, cross-linked (UVP Crosslinker CX-2000UV), and hybridized at 55°C overnight with 5′ Digoxigenin-labeled probes (Supplemental Table S7) in hybridization buffer with 5× SSC, 1% SDS, and 1× Denhardt's reagent (Sangon Biotech B548209). After blocking in maleic acid buffer with 1% block reagent (Roche 11096176001) at room temperature for 1 h, the membrane was incubated in blocking buffer with 1:10000 anti-DIG-AP antibody (Roche 11093274910). The membrane was washed in maleic acid buffer with Tween 20 for 15 min twice, incubated with pH 9.5 detection buffer, reacted with CDP-Star (Roche 12041677001) at 37°C for 15 min, and then exposed. Imaging was performed using Chemilluminescence Imaging System (Clinx), and quantification was performed using ImageJ.
cDNA library construction for tRNAs and tsRNAs
A synthetic mimic of E. coli tRNALysUUU (sequence in Supplemental Table S3; Azenta) was added to the total RNAs as a spike-in at 0.06 pmol/μg. Then the RNA samples were analyzed on 15% TBE-Urea gels alongside Low Range ssRNA Ladder (NEB N0364), and the 60–100 nt (tRNA fraction) and 20–40 nt (tsRNA fraction) bands were respectively excised (for PANDORA-seq in L3 samples, 15–35 nt bands were excised).
For mim-tRNA/tsRNA-seq, RNAs from both fractions were treated with deacylation buffer (pH = 9.0) and T4 PNK (NEB M021) to generate terminal 3′-OH and 5′-P ends. The resolved RNAs were sequentially ligated with 3′-adaptor and 5′-adaptor by T4 RNA ligase 2 truncated (NEB M0242) and T4 RNA ligase 1 (NEB M0204), respectively. Reverse transcription was performed with TGIRT-III (HaiGene D0310) in low salt buffer (50 mM Tris-HCl pH = 8.3, 75 mM KCl, 3 mM MgCl2) at 42°C for 16 h. For PANDORA-seq (Shi et al. 2021), the tsRNA fraction was treated with tRF&tiRNA Pretreatment Kit (Arraystar AS-FS-005) to generate terminal 3′-OH and 5′-P ends and to remove internal m1A, m1G, and m3C. The resolved RNAs were sequentially ligated with 3′- and 5′-adaptors, and then reverse-transcribed with SuperScript IV (Thermo Fisher Scientific 18090010).
All cDNAs were amplified by Phusion DNA Polymerase (NEB M0530) through 14 PCR cycles, and the products within correct size ranges were collected for sequencing (Illumina NovaSeq 6000 PE150 for mim-tRNA/tsRNA-seq and PANDORA-seq; Illumina NovaSeq 6000 SE50 PANDORA-seq in L3 samples).
Ribosome profiling, Ribo-seq, RiboTag-seq, and mRNA-seq
Ribosome profiling and Ribo-seq library construction for larvae and pupae were performed as previously described (Zhang et al. 2018). Oenocyte-specific RiboTag experiments were performed on P2 pupae of Desat1-GAL4 > UAS-RPL13A-FLAG,UAS-5′-tsRNAAspGUC sponge or Desat1-GAL4 > UAS-RPL13A-FLAG,UAS-scramble as previously described (Huang et al. 2019). Anti-FLAG antibody (Proteintech 66008-4-Ig) was used. Library construction for mRNA-seq was performed by VAHTS Universal V8 RNA-seq Library Prep Kit for Illumina (Vazyme NR605) or Biopharmaceutical Public Service Platform. Sequencing was performed with PE150 chemistry on Illumina NovaSeq 6000.
Sequencing read processing, alignment, and quantification
For mRNA-seq and RiboTag-seq, reads were mapped to the D. melanogaster genome (FlyBase r6.36) using HISAT2 v2.2.1 (Kim et al. 2019), and then summarized using featureCounts v2.0.1 (Liao et al. 2014). Fragments per kilobase of exon per million mapped (FPKM) was then calculated to quantify each mRNA-coding gene. For the differential expression analysis, the count data were normalized and analyzed using DESeq2 (Love et al. 2014).
For Ribo-seq, after clipping for the adaptor sequences using cutadapt v3.4 (Martin 2011), reads with a remaining size of 26–34 nt were kept, and then sequentially mapped to rRNAs, tRNAs, and the transcript with the longest CDS for each protein-coding gene using Bowtie 2 v2.2.5 (Langmead and Salzberg 2012) with default parameters. Ribosome protected fragment (RPF) was measured as the counts of reads uniquely mapped to the CDS (excluding the first five and the last five codons) and then normalized to per million total CDS-mapped reads. TE was calculated as RPF/FPKM. At the single-codon resolution (Supplemental Fig. S6C), for each RPF read with a length of 30–32 nt, we determined the codon that occupies a ribosomal A-site as the 15th–17th trinucleotides from the 5′ end and in the same frame of CDS. All the other reads (∼50%) were discarded to ensure the accuracy. For the genes enriched with codons matching with tRNAAspGUC (Supplemental Fig. S6D), we selected 1112 genes with the top density of codons GAC/U for 5′-tsRNAAspGUC and mRNA-seq detection in all examined libraries.
For PANDORA-seq, reads within 20–40 nt were kept after clipping adaptors, and then mapped to tRNAs using Bowtie 2 v2.2.5 with default parameters. For mim-tRNA/tsRNA-seq, reads were processed and analyzed using mim-tRNAseq v1.2 (Behrens et al. 2021) with the default parameters for D. melanogaster.
Differential expression analysis, functional annotation, and time-series profile clustering
DEGs were considered with a significant change when FDR-adjusted P-value < 0.05 and |log2(fold change)| > 1 in a DESeq2 analysis. For functional annotation, AnnotationDbi and clusterProfiler (Yu et al. 2012) were used with pvalueCutoff = 0.01, qvalueCutoff = 0.05. For time-series analyses, Mfuzz (Kumar and Futschik 2007) was used to perform soft clustering on the dynamic profiles of DEGs.
Prediction of tsRNA target genes
To predict the target genes of tsRNAs that were accumulated in early pupae, we obtained all possible 7-mers from the tsRNA reads of mim-tRNA/tsRNA-seq data at the P2 stage. These 7-mers comprised a total of 2075 unique seeds, from which 104 highly abundant seeds (top 5%) were retrieved. Then, TargetScan 7.0 (Agarwal et al. 2015) was used to predict antisense perfect match in mRNA transcripts for the 104 seeds, and phyloP (Pollard et al. 2010) was used to compute the conservation score of each putative target site. The densities of conserved sites (FDR-adjusted phyloP score < 0.001) in the mRNA transcripts of each protein-coding gene were summarized. Finally, we retrieved a total of 1418 genes with high site density (top 15%) and reliable mRNA expression (DESeq2-calculated baseMean > 70), and considered them as the target genes of P2-stage tsRNAs. For the five 5′-tsRNAs and antisense-scramble tsRNA, whose regulatory activities were further experimentally evaluated, we used TargetScan7.0 predicted conserved sites (FDR-adjusted phyloP score < 0.05) and an additional criterion of minimum free energy < −30 kcal/mol for the local alignment computed with miRanda (Enright et al. 2003).
Free fatty acid profiling
Lipids were extracted from 30 mg of frozen tissues using a modified version of the Bligh and Dyer's method as described previously (Song et al. 2020). For each experimental setting, four independent samples were prepared and stored at −80°C until further analysis. Lipidomic analyses were conducted at LipidALL Technologies using a Shimadzu Nexera 20AD HPLCcoupled with Sciex QTRAP 6500 PLUS as reported previously (Lam et al. 2021). Individual lipid species were quantified by referencing to spiked internal standards. Free fatty acids were quantitated using d31-16:0 (Sigma-Aldrich) and d8-20:4 (Cayman Chemicals).
Western blot analysis
Whole larvae or pupae at L3, P1, and P2 stages were lysed in RIPA lysis buffer (FUDE, FD009) with PMSF (Beyotime; ST506) and PhosStop phosphatase inhibitor cocktail (Beyotime; P1045). Rabbit anti-AGO1 (Abcam ab5070; 1:1000), Rabbit anti-AGO2 (Abcam ab5072; 1:1000), HRP-conjugated β-Tubulin Mouse mAb (ABclonal AC030; 1:5000), and anti-rabbit HRP-conjugated secondary antibodies (FUDE FDR007) were used.
Statistical significance
For each sample subjected to deep sequencing in this study, the number of biological replicates and the mapping statistics were summarized in Supplemental Table S1. For northern blot, western blot, RT-qPCR, and luciferase reporter assay, respectively, each setting had at least two independent sample preparations and experiments. All statistical tests used in this study have been indicated in the legends to the related figures.
Data access
All raw RNA sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA957228. All custom scripts required to reproduce the work are available at GitHub (https://github.com/junlingshi/tsRNA_Drosophila_metamorphosis/) and as Supplemental Code.
Supplementary Material
Acknowledgments
This study was supported in part by the National Key Research and Development Program of China (2018YFA0800102 and 2021YFC2700403) and the National Natural Science Foundation of China (31871249 and 31871452). We acknowledge support of Zhejiang University (ZJU) School of Medicine affiliated Women's Hospital and Children's Hospital. We thank Hai Huang, JianHua Huang, Qi Zhou, and Xiaohang Yang of ZJU for fly strains.
Author contributions: J.S., J.M., and F.H. conceived the study and designed the experiments; J.S. and J.X. performed experiments and generated data; J.S. analyzed the data and generated all figures; J.M. and F.H. acquired funding; J.S., J.M., and F.H. wrote the paper, and all approved the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278128.123.
Competing interest statement
The authors declare no competing interests.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T. 2018. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172: 262–274.e11. 10.1016/j.cell.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge SP, Bownes M. 1981. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol 66: 57–80. [PubMed] [Google Scholar]
- Barta T, Peskova L, Hampl A. 2016. miRNAsong: a web-based tool for generation and testing of miRNA sponge constructs in silico. Sci Rep 6: 36625. 10.1038/srep36625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A, Rodschinka G, Nedialkova DD. 2021. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell 81: 1802–1815.e7. 10.1016/j.molcel.2021.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. 2009. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature 461: 987–991. 10.1038/nature08495 [DOI] [PubMed] [Google Scholar]
- Boulan L, Martin D, Milán M. 2013. Bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol 23: 473–478. 10.1016/j.cub.2013.01.072 [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhou T. 2023. Emerging functional principles of tRNA-derived small RNAs and other regulatory small RNAs. J Biol Chem 299: 105225. 10.1016/j.jbc.2023.105225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. 2016. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351: 397–400. 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- Chen L, Xu W, Liu K, Jiang Z, Han Y, Jin H, Zhang L, Shen W, Jia S, Sun Q, et al. 2021a. 5′ half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci Adv 7: eabh0494. 10.1126/sciadv.abh0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang X, Shi J, Yan M, Zhou T. 2021b. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem Sci 46: 790–804. 10.1016/j.tibs.2021.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu Z, Cai H, Peng Y, Yang L, Wang Z. 2022. Identifying differentially expressed tRNA-derived small fragments as a biomarker for the progression and metastasis of colorectal cancer. Dis Markers 2022: 2646173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Loehlin DW, Dufour HD, Vaccarro K, Millar JG, Carroll SB. 2014. A single gene affects both ecological divergence and mate choice in Drosophila. Science 343: 1148–1151. 10.1126/science.1249998 [DOI] [PubMed] [Google Scholar]
- Cohen SM. 2009. Use of microRNA sponges to explore tissue-specific microRNA functions in vivo. Nat Methods 6: 873–874. 10.1038/nmeth1209-873 [DOI] [PubMed] [Google Scholar]
- Couvillion MT, Sachidanandam R, Collins K. 2010. A growth-essential Tetrahymena piwi protein carries tRNA fragment cargo. Genes Dev 24: 2742–2747. 10.1101/gad.1996210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. 2012. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell 48: 509–520. 10.1016/j.molcel.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Lollo B, Freier S, Esau C. 2006. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res 34: 2294–2304. 10.1093/nar/gkl183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou S, Wang Y, Lu J. 2019. Metazoan tsRNAs: biogenesis, evolution and regulatory functions. Noncoding RNA 5: 18. 10.3390/ncrna5010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. 2010a. Emerging roles for natural microRNA sponges. Curr Biol 20: R858–R861. 10.1016/j.cub.2010.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. 2010b. MicroRNA sponges: progress and possibilities. RNA 16: 2043–2050. 10.1261/rna.2414110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726. 10.1038/nmeth1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968. 10.1074/jbc.M109.077560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5: R1. 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau CC. 2008. Inhibition of microRNA with antisense oligonucleotides. Methods 44: 55–60. 10.1016/j.ymeth.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Fulga TA, McNeill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, DeLuca T, et al. 2015. A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nat Commun 6: 7279. 10.1038/ncomms8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Rubin T, Poidevin M, Maroni B, Le Rouzic A, Parvy JP, Montagne J. 2015. Fatty acid synthase cooperates with glyoxalase 1 to protect against sugar toxicity. PLoS Genet 11: e1004995. 10.1371/journal.pgen.1004995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, Zywicki M, Künzi A, Polacek N. 2012. tRNA-derived fragments target the ribosome and function as regulatory noncoding RNA in Haloferax volcanii. Archaea 2012: 260909. 10.1155/2012/260909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N. 2017. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 14: 1364–1373. 10.1080/15476286.2016.1257470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. 2015. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802. 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Agapite J, Attrill H, Calvi BR, Crosby MA, Dos Santos G, Goodman JL, Goutte-Gattat D, Jenkins VK, Kaufman T, et al. 2022. Flybase: a guided tour of highlighted features. Genetics 220: iyac035. 10.1093/genetics/iyac035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimková K, Sommarin MNE, Munita R, Lubas M, et al. 2018. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173: 1204–1216.e26. 10.1016/j.cell.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16: 673–695. 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, Zhang Q, Zhang L, Xue Z, Li Y, et al. 2017. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res 77: 3194–3206. 10.1158/0008-5472.CAN-16-3146 [DOI] [PubMed] [Google Scholar]
- Huang K, Chen W, Zhu F, Li PW, Kapahi P, Bai H. 2019. Ribotag translatomic profiling of Drosophila oenocytes under aging and induced oxidative stress. BMC Genomics 20: 50. 10.1186/s12864-018-5404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. 2013. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261. 10.1016/j.celrep.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakova A, Fehlmann T, Keller A, Quake SR. 2020. A mouse tissue atlas of small noncoding RNA. Proc Natl Acad Sci 117: 25634–25645. 10.1073/pnas.2002277117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. 2015. PIWI-Interacting RNA: its biogenesis and functions. Annu Rev Biochem 84: 405–433. 10.1146/annurev-biochem-060614-034258 [DOI] [PubMed] [Google Scholar]
- Keam SP, Sobala A, Ten Have S, Hutvagner G. 2017. tRNA-Derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J Proteome Res 16: 413–420. 10.1021/acs.jproteome.6b00267 [DOI] [PubMed] [Google Scholar]
- Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, et al. 2017. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552: 57–62. 10.1038/nature25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S, Raghavan S, DasGupta R, Palakodeti D. 2021. tRNA-derived fragments (tRFs): establishing their turf in post-transcriptional gene regulation. Cell Mol Life Sci 78: 2607–2619. 10.1007/s00018-020-03720-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Futschik ME. 2007. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2: 5–7. 10.6026/97320630002005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A. 2014. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12: 78. 10.1186/s12915-014-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kuscu C, Dutta A. 2016. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci 41: 679–689. 10.1016/j.tibs.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. 2018. tRNA fragments (tRFs) guide ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24: 1093–1105. 10.1261/rna.066126.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SM, Zhang C, Wang Z, Ni Z, Zhang S, Yang S, Huang X, Mo L, Li J, Lee B, et al. 2021. A multi-omics investigation of the composition and function of extracellular vesicles along the temporal trajectory of COVID-19. Nat Metab 3: 909–922. 10.1038/s42255-021-00425-4 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749. 10.1074/jbc.M510356200 [DOI] [PubMed] [Google Scholar]
- Li S, Xu Z, Sheng J. 2018. tRNA-derived small RNA: a novel regulatory small noncoding RNA. Genes (Basel) 9: 246. 10.3390/genes9050246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dong P, Yang Y, Guo T, Zhao Q, Miao D, Li H, Lu T, Xia F, Lyu J, et al. 2022. Metabolic control of progenitor cell propagation during Drosophila tracheal remodeling. Nat Commun 13: 2817. 10.1038/s41467-022-30492-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- López-Maury L, Marguerat S, Bähler J. 2008. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 9: 583–593. 10.1038/nrg2398 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, He F, Luo J, Dou S, Wang Y, Guo A, Lu J. 2018. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res 46: 5250–5268. 10.1093/nar/gky189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. 2013. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci 110: 1404–1409. 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkey AB, Wong CK, Hoshizaki DK, Gibbs AG. 2011. Energetics of metamorphosis in Drosophila melanogaster. J Insect Physiol 57: 1437–1445. 10.1016/j.jinsphys.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Mosher J, Zhang W, Blumhagen RZ, D'Alessandro A, Nemkov T, Hansen KC, Hesselberth JR, Reis T. 2015. Coordination between Drosophila Arc1 and a specific population of brain neurons regulates organismal fat. Dev Biol 405: 280–290. 10.1016/j.ydbio.2015.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666. 10.1101/gad.1210204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121. 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolff J, Johnston PR, Reynolds S. 2019. Complete metamorphosis of insects. Philos Trans R Soc Lond B Biol Sci 374: 20190063. 10.1098/rstb.2019.0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595. 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351: 391–396. 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, Franklin R, Shahbazi M, Mackinlay K, Liu S, et al. 2021. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol 23: 424–436. 10.1038/s41556-021-00652-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobala A, Hutvagner G. 2013. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 10: 553–563. 10.4161/rna.24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Lam SM, Fan X, Cao WJ, Wang SY, Tian H, Chua GH, Zhang C, Meng FP, Xu Z, et al. 2020. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab 32: 188–202.e5. 10.1016/j.cmet.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Wilson B, Kumar P, Dutta A. 2020. Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu Rev Genet 54: 47–69. 10.1146/annurev-genet-022620-101840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308. 10.1016/j.cell.2007.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu M, Zuo Z, Chen C, Zhang X, Wang S, Chen C, Sun Y. 2023. Transfer RNA-derived small RNAs (tsRNAs) sequencing revealed a differential expression landscape of tsRNAs between glioblastoma and low-grade glioma. Gene 855: 147114. 10.1016/j.gene.2022.147114 [DOI] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905. 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- Varghese J, Cohen SM. 2007. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev 21: 2277–2282. 10.1101/gad.439807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt CM, Hahn O, Arlt K, Gruhn M, Jahn AJ, Esser J, Werner JA, Klein C, Buschges A, Gronke S, et al. 2019. Loss of miR-210 leads to progressive retinal degeneration in Drosophila melanogaster. Life Sci Alliance 2: e201800149. 10.26508/lsa.201800149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Hironaka KI, Habara O, Morishita Y, Nishimura T. 2020. A developmental checkpoint directs metabolic remodelling as a strategy against starvation in Drosophila. Nat Metab 2: 1096–1112. 10.1038/s42255-020-00293-4 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42. 10.1083/jcb.200811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dou S, He F, Luo J, Wei L, Lu J. 2018. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol 16: e2003903. 10.1371/journal.pbio.2003903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yang J, Li H, You J, Chatterjee N, Zhang X. 2020. Development of the transcriptome for a sediment ecotoxicological model species, Chironomus dilutus. Chemosphere 244: 125541. 10.1016/j.chemosphere.2019.125541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.