In this study, Lee et al. provide structural insight into the domain-specific functions that collaboratively contribute to the role of the RING-type E3 ligase PHF7 in histone ubiquitination. They further describe the molecular interactions that are essential for histone recognition, E2 ubiquitin-conjugating enzyme recruitment, and nucleosome binding by PHF7.
Keywords: E2 ubiquitin-conjugating enzyme, plant homeodomain (PHD), plant homeodomain finger protein 7 (PHF7), RING E3 ubiquitin ligase, histone H3, structure, ubiquitination
Abstract
The RING-type E3 ligase has been known for over two decades, yet its diverse modes of action are still the subject of active research. Plant homeodomain (PHD) finger protein 7 (PHF7) is a RING-type E3 ubiquitin ligase responsible for histone ubiquitination. PHF7 comprises three zinc finger domains: an extended PHD (ePHD), a RING domain, and a PHD. While the function of the RING domain is largely understood, the roles of the other two domains in E3 ligase activity remain elusive. Here, we present the crystal structure of PHF7 in complex with the E2 ubiquitin-conjugating enzyme (E2). Our structure shows that E2 is effectively captured between the RING domain and the C-terminal PHD, facilitating E2 recruitment through direct contact. In addition, through in vitro binding and functional assays, we demonstrate that the N-terminal ePHD recognizes the nucleosome via DNA binding, whereas the C-terminal PHD is involved in histone H3 recognition. Our results provide a molecular basis for the E3 ligase activity of PHF7 and uncover the specific yet collaborative contributions of each domain to the PHF7 ubiquitination activity.
The really interesting new gene (RING) domain is a defining functional domain of RING and RING-between-RING E3 ubiquitin ligases that works in conjunction with E2 ubiquitin-conjugating enzymes (E2s) to ubiquitinate specific substrates (Freemont et al. 1991; Bailly et al. 1997). Despite considerable efforts to unravel the mechanisms of RING-type E3 ligases, the precise mechanism remains an intensely ongoing area of investigation. RING E3 ligases share some common features, including binding to E2s via the RING domain and transferring ubiquitin from E2 to the substrate without self-ubiquitination (Deshaies and Joazeiro 2009; Metzger et al. 2014). However, it is now apparent that the specific aspects of mechanisms vary considerably among different enzymes. Each RING E3 ligase is associated with a distinct set of E2s with which it can couple, and the interaction between them exhibits a wide range of binding affinities, with dissociation constant (KD) values ranging from submicromolar to hundreds of micromolar (Bentley et al. 2011; Buetow and Huang 2016; DiBello et al. 2016; Wright et al. 2016; Behera et al. 2018). Some RING E3 ligases require cofactors for full functionality, whereas others are fully functional as monomers. Additionally, some enzymes homodimerize or heterodimerize to regulate their activities (Metzger et al. 2014; Fiorentini et al. 2020). Although the RING domain is the functionally defining domain, E3 ligase activity is often a collaborative effort of multidomains or even multiproteins that ensues in a diversity of mechanisms within RING E3 ligases (Scott et al. 2016; Dove et al. 2017; Rennie et al. 2020). In light of recent advances in protein engineering and biotechnological tools such as proteolysis targeting chimera (PROTAC), it has become more important to understand the diverse molecular mechanisms by which RING-type E3 ligases operate (Burslem and Crews 2020).
Plant homeodomain finger protein 7 (PHF7) is a recently identified RING-type E3 ligase that specifically ubiquitinates histone H2AK119 and H3K14, making it an epigenetic writer (Wang et al. 2019b; Kim et al. 2020). It is primarily expressed in the male germline of mammals (Yang et al. 2012; Uhlén et al. 2015; Wang et al. 2019b; Karlsson et al. 2021) and plays a crucial role in the normal formation and maturation of male gametes, promoting histone-to-protamine exchange via histone ubiquitination in the chromatin of elongating spermatids (Kimmins and Sassone-Corsi 2005; Liu et al. 2006; Ge et al. 2017; Wang et al. 2019a; Yang 2021; Bhaskar et al. 2022). PHF7 consists of three independent zinc finger domains: the N-terminal extended plant homeodomain (ePHD), RING domain, and C-terminal plant homeodomain (PHD) (Fig. 1A). While the involvement of the RING domain in ubiquitination is clear, the contribution of the remaining two domains has remained uncertain.
Figure 1.
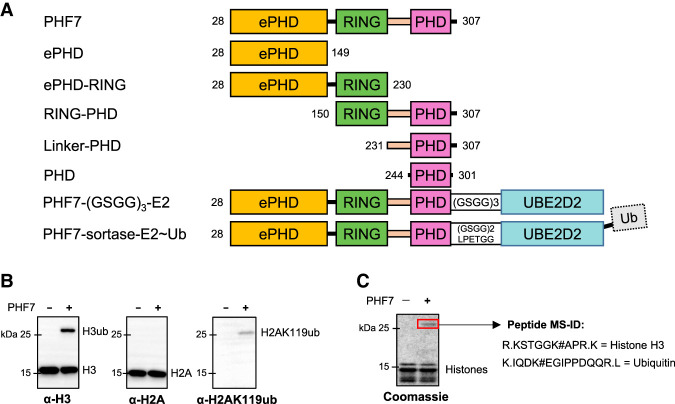
PHF7 preferentially ubiquitinates histone H3. (A) A schematic diagram of PHF7 constructs used in this study. (B) In vitro ubiquitination assay of the NCP by PHF7 shown with Western blots. Each Western blot was stained with antibodies specific against N-terminal histone H3, C-terminal histone H2A, and H2AK119ub as indicated. (C) Coomassie stain of the NCP ubiquitinated by PHF7. The red box indicates histone H3 ubiquitinated at K14 that was identified by mass spectrometry (MS-ID).
The PHD is a zinc finger, similar to the RING domain, with two zinc atoms coordinated by a Cys4–His–Cys3 motif in a cross-brace topology (Sanchez and Zhou 2011). Because of its similarity to the RING domain, the PHD has been suggested to be involved in E3 ligase activity, but no strong evidence exists to support this hypothesis (Bienz 2006). More importantly, the PHD has a well-established role as a “reader” of methylated lysines on histone H3 tails, as evidenced by several crystal structures solved in complex with the H3 tail (Li et al. 2006; Peña et al. 2006; Lan et al. 2007; Ooi et al. 2007; Musselman et al. 2012). However, the function of the N-terminal ePHD is less well known. It is referred to as “extended” PHD because it contains an additional zinc binding motif, coordinating three zinc atoms in total. The ePHD, previously identified in PHF6, has been shown to bind to DNA and H2B acetylated at K12 (Liu et al. 2014; Oh et al. 2020). Therefore, the ePHD and PHD of PHF7 are likely involved in substrate recognition, but the mechanism remains elusive.
Here, to uncover the molecular mechanism of PHF7 as a histone ubiquitin ligase, we determined the crystal structure of PHF7 bound to its E2 partner, UBE2D2. We performed a series of binding affinity studies and in vitro ubiquitination assays to investigate whether all three domains of PHF7 are essential for nucleosome ubiquitination and to determine their distinct roles.
Results
PHF7 recognizes histone H3K4me2/3 via the C-terminal PHD
Previous studies have reported that PHF7 can ubiquitinate both H3K14 and H2AK119 (Wang et al. 2019b; Kim et al. 2020). To determine the substrate preference of PHF7 between H3K14 and H2AK119, we conducted an in vitro ubiquitination assay using the nucleosome core particle (NCP) and analyzed the reaction products by Western blot (Fig. 1B). While ubiquitinated H3 was clearly detected with H3 antibody, H2A ubiquitination at K119 was only detectable with an antibody specific for ubiquitinated H2AK119, likely due to the low efficacy of H2A ubiquitination (Fig. 1B). In addition, mass spectrometry analysis of a single prominent upshifted band in a Coomassie-stained SDS-PAGE gel of the ubiquitinated NCP only identified ubiquitinated H3K14 (Fig. 1C), suggesting that H3K14 is the preferred substrate over H2AK119, at least under our experimental conditions. Hence, we focused on characterizing the interaction between PHF7 and H3, with H3K14 being the primary ubiquitination site in our in vitro system.
To understand how each domain in PHF7 is involved in substrate recognition, we designed several constructs of PHF7. We used Mus musculus PHF7 (mPHF7), as it has a high sequence homology with human PHF7 (91% sequence similarity in amino acids 1–307) but lacks the long unstructured C-terminal tail (308–381) (Supplemental Fig. S1). We truncated the first 27 amino acids from the full-length mPHF7 to improve the protein yield and refer to this construct (mPHF7 28–307) as PHF7 throughout this report. Three constructs containing each domain were designed, but because the RING domain alone expressed poorly, we also made combinatorial constructs ePHD–RING and RING–PHD (Fig. 1A). For the RING–PHD construct, we introduced a single point mutation, C164A, in the linker region connecting the RING domain and PHD to prevent aggregation.
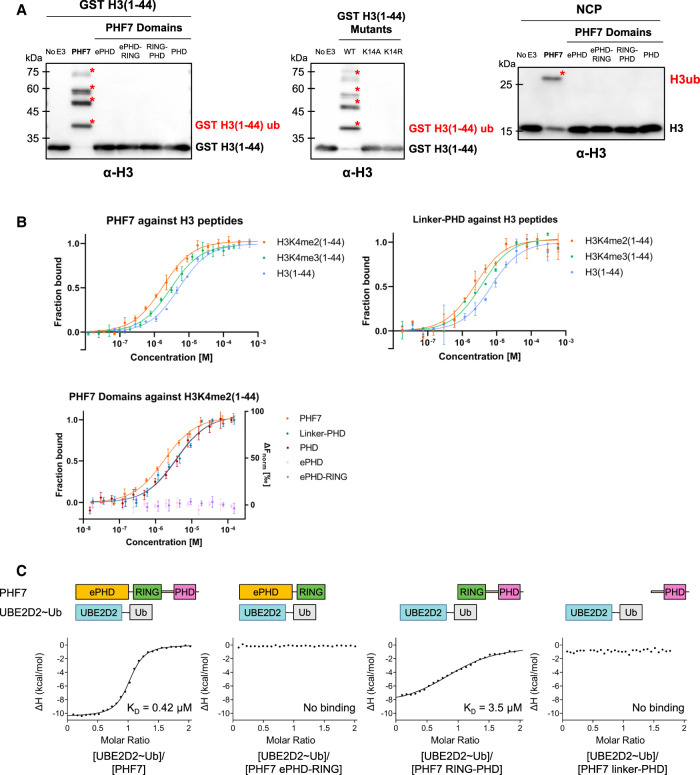
Using purified PHF7, we confirmed that the N-terminal tail of H3(1–44) was sufficient for ubiquitination, at least under in vitro conditions (Fig. 2A). Polyubiquitination of H3(1–44) was observed, whereas only monoubiquitination of H3 was detected with the NCP as the substrate. This suggests that polyubiquitination is an artifact resulting from the use of the peptide form of the substrate. In a previous study, a similar trend of polyubiquitination in the form of the H3(1–44) peptide by PHF7 was reported, and the K14R mutation of full-length H3 resulted in the loss of ubiquitination (Kim et al. 2020). In our study, we further confirmed that the K14A/K14R mutant of H3(1–44) resulted in a complete loss of ubiquitination, demonstrating that PHF7 specifically targets K14 for polyubiquitination in the form of a peptide (Fig. 2A). While physical restraint may be imposed to prevent polyubiquitination for H3K14 in the context of the NCP, it can be reasonably hypothesized that the interaction between PHF7 and H3 remains consistent in both the NCP and peptide forms, given the retained substrate specificity of PHF7 for H3K14.
Figure 2.
PHF7 uses the PHD for histone H3 substrate binding and RING–linker–PHD for E2 interaction. (A) In vitro ubiquitination assay with the indicated PHF7 domains using GST-tagged H3(1–44), H3(1–44) mutants, or the NCP as the substrate. Red asterisks mark ubiquitinated H3. The antibody used is specific against histone H3. (B) MST binding curves showing the fraction of bound H3(1–44) peptides (unmodified, K4me2, and K4me3) against different PHF7 constructs. For the ePHD and ePHD–RING, a baseline level of ΔFnorm (right axis) indicates that binding cannot be detected. Error bars represent the standard deviation of three technical replicates. (C) Isothermal titration calorimetry binding curves of the indicated PHF7 domains with UBE2D2∼Ub. The resulting KD values are indicated.
Although PHF7 has been suggested to bind H3K4 in either the dimethylation or trimethylation state (H3K4me2/3) (Yang et al. 2012; Wang et al. 2019b), whether H3 methylation is required for the ubiquitination activity of PHF7 remains uncertain (Kim et al. 2020). Notably, previous studies observed PHF7 ubiquitinating H2AK119 and H3K14 in the unmethylated NCP (Wang et al. 2019b; Kim et al. 2020). Thus, to investigate the dependency of PHF7 binding on H3 methylation at K4, we prepared H3(1–44) peptides with dimethylation or trimethylation at K4 [H3K4me2/3(1–44)] and measured their binding affinities with PHF7. Methyl-lysine was chemically introduced to H3(1–44) with a K4C mutation using a previously described procedure (Simon et al. 2007). We performed microscale thermophoresis (MST) to measure the affinity of PHF7 with H3(1–44) in the absence or presence of methylation at K4. As a result, PHF7 bound to all three forms of the H3 tail (unmodified, dimethylated, and trimethylated) but showed a slight bias toward methylated H3K4 with a threefold stronger affinity toward H3K4me2(1–44) compared with unmethylated H3(1–44) (Fig. 2B; Table 1). The difference in affinity is small, however, indicating that H3K4 methylation itself is unlikely to have a significant effect on H3(1–44) ubiquitination under the in vitro conditions tested. It appears that PHF7 can ubiquitinate H3 regardless of the H3K4 methylation status in vitro, as suggested by Kim et al. (2020).
Table 1.
Dissociation constant (KD) of PHF7 domains against histone H3 peptides measured using microscale thermophoresis
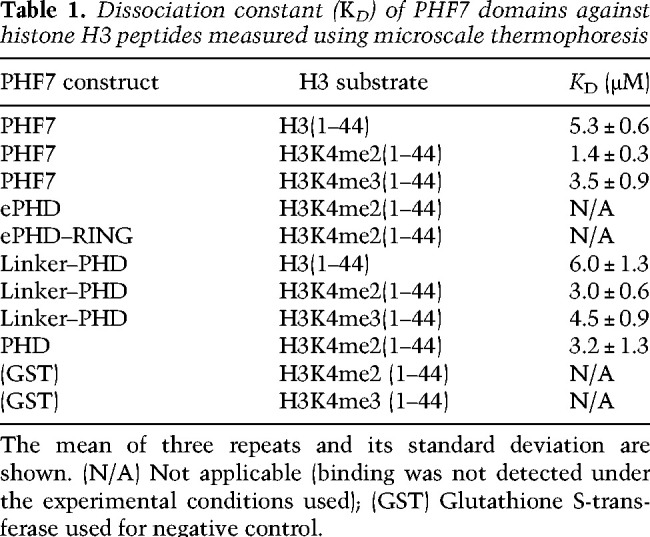
To identify the PHF7 domain responsible for substrate recognition, we measured the binding affinity of the ePHD and PHD with H3K4me2(1–44). Previous studies suggested that the ePHD was the histone binding site, as mutations in this domain were found to impair histone ubiquitination (Wang et al. 2019b; Kim et al. 2020). However, we did not observe any interaction between the ePHD and H3K4me2(1–44) using MST. Instead, we found that the PHD exhibited a binding affinity comparable with that of PHF7, with KD values in the range of 3–6 µM (Fig. 2B; Table 1). Including the linker between RING and the PHD had no effect on H3 binding (Table 1). In addition, the PHD showed slightly better binding to the methylated H3 tail, which is consistent with the binding properties of PHF7. The studies proposing the ePHD as the substrate recognition site were based on experiments using constructs with mutated zinc-coordinating Cys and His residues of the ePHD or with the ePHD deleted in the middle of its domain, which would likely cause destabilization of the protein integrity and result in misfolded and nonfunctional proteins (Low et al. 2002; Wang et al. 2019b; Kim et al. 2020). Our results suggest that the C-terminal PHD is the primary histone H3 recognition domain in PHF7 (Fig. 2B).
All three domains of PHF7 are essential for E2 recruitment
As we identified the PHD as the H3 tail binding site, we hypothesized that RING–PHD would be the necessary and sufficient catalytic core for ubiquitination. However, RING–PHD failed to ubiquitinate H3(1–44) in our in vitro ubiquitination assays (Fig. 2A). We also observed the same phenomenon in the ubiquitination assay with the NCP (Fig. 2A). These unexpected results left us puzzled, as the ePHD did not appear to interact with the histone in our MST experiment (Fig. 2B). Thus, we hypothesized that the ePHD might play a role in recruiting the E2 ubiquitin-conjugating enzyme (E2).
UBE2D2 (UbcH5b) was identified as the cognate E2 of PHF7 (Wang et al. 2019b; Kim et al. 2020). As some E3 ligases exhibit different affinity toward the E2 and ubiquitin-conjugated E2 (Pruneda et al. 2012), we measured the binding affinities of PHF7 toward both UBE2D2 and ubiquitin-conjugated UBE2D2 S22R/C85K (UBE2D2∼Ub) using isothermal titration calorimetry (ITC). The S22R mutation was introduced to prevent the backside binding of neighboring ubiquitin that may complicate the analysis of binding affinity (Brzovic et al. 2006; Buetow et al. 2015), and the C85K mutation allows hydrolysis-resistant ubiquitin conjugation to form through an isopeptide bond instead of a thioester bond (Plechanovová et al. 2012). We observed that PHF7 binds to UBE2D2∼Ub with a KD of 0.4 µM, which is threefold stronger than that of UBE2D2 alone (Table 2; Fig. 2C; Supplemental Fig. S2). Of note, these KD values are much lower than those reported for other RING and E2 interactions, which are typically in the range of 60–200 µM (Das et al. 2013; Metzger et al. 2014).
Table 2.
Dissociation constant (KD) of PHF7 domains against UBE2D2 or UBE2D2∼Ub measured using ITC
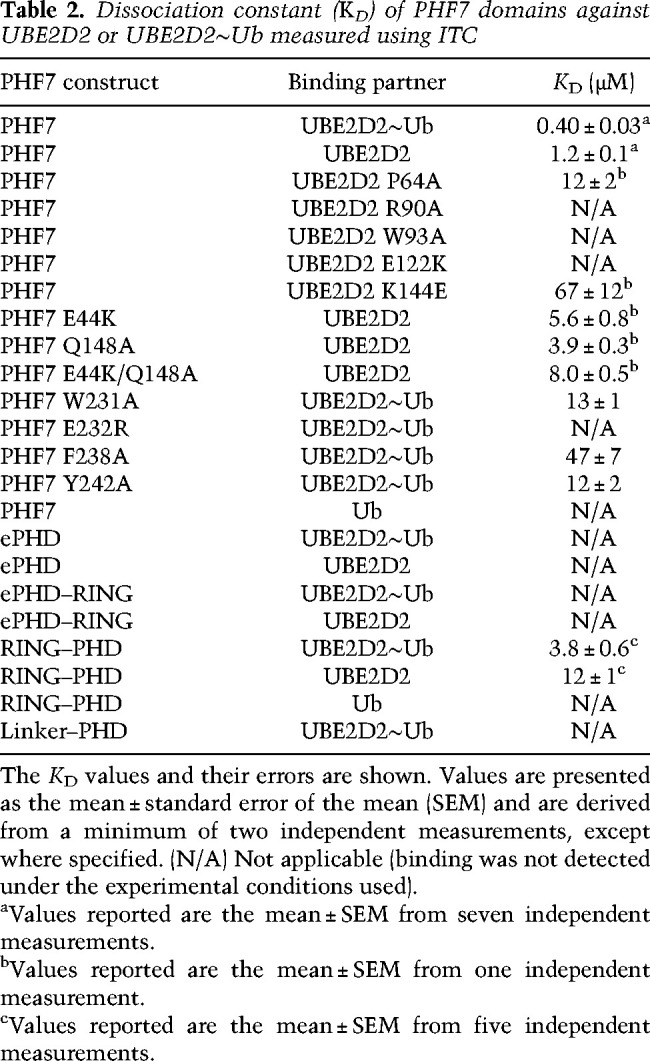
We measured the affinities between various constructs of PHF7 and UBE2D2∼Ub to identify the E2-interacting domains in PHF7. Contrary to our hypothesis, we did not observe any binding between UBE2D2∼Ub and the ePHD by ITC. Furthermore, none of the ePHD–RING, RING–linker, or PHD constructs showed binding to UBE2D2∼Ub. Only RING–PHD exhibited weak binding to UBE2D2∼Ub, with affinity seven times lower than that of PHF7, implying that the ePHD is still functionally necessary for high-affinity binding to UBE2D2∼Ub (Fig. 2C). The weak affinity of RING–PHD to UBE2D2∼Ub mirrors the inability of this construct to ubiquitinate (Fig. 2A). Altogether, we concluded that all three domains need to cooperate for the full activity of PHF7 for UBE2D2∼Ub recruitment and H3K14 ubiquitination. However, our binding and in vitro ubiquitination assay failed to elucidate the molecular mechanism of how the three domains collaborate to enable H3K14 ubiquitination. To answer this question, we determined the structure of PHF7 in complex with UBE2D2∼Ub.
Structure of PHF7 bound to UBE2D2
We initially attempted to crystallize PHF7 in complex with UBE2D2∼Ub, as PHF7 exhibited a higher affinity for UBE2D2∼Ub compared with UBE2D2 alone (Table 2). We maintained the UBE2D2 S22R mutation to prevent additional noncovalent ubiquitin interaction that may interfere with crystallization. Although the two proteins formed a stable complex following purification via size exclusion chromatography (Supplemental Fig. S3), our initial crystallization screening only yielded crystals of dissociated UBE2D2∼Ub, not the desired complex. To further stabilize the complex, we devised two strategies to physically tether UBE2D2 to PHF7.
In the first strategy, we generated a fusion construct by directly tethering PHF7 to UBE2D2 S22R/C85K via a (GSGG)3 linker. After purification, we attempted to conjugate ubiquitin to the fusion protein (Fig. 1A). Although the purification yield was high, the efficiency of ubiquitin conjugation was low, resulting in a heterogeneous protein sample. Thus, we crystallized this fusion complex without ubiquitin conjugation. In the second strategy, PHF7 and UBE2D2∼Ub were purified separately and then fused together via enzymatic linkage using sortase A (Theile et al. 2013). In both strategies, we confirmed that each fusion construct was able to ubiquitinate histone H3(1–44) (Supplemental Fig. S4).
Both strategies produced diffracting crystals, and the crystal structures were determined at a resolution of 3.58 Å for the (GSGG)3 linker complex and 2.96 Å for the sortase A fusion complex (Supplemental Table S1). Alignment of the two structures revealed that despite using different tethering strategies, the two complexes maintained a similar overall structure, confirming that the fusion strategies did not affect the overall structure of the complex (Supplemental Fig. S5). Subsequent descriptions of the PHF7 and UBE2D2 complex structure are based on the higher-resolution structure of the sortase A-fused construct (Fig. 3A).
Figure 3.
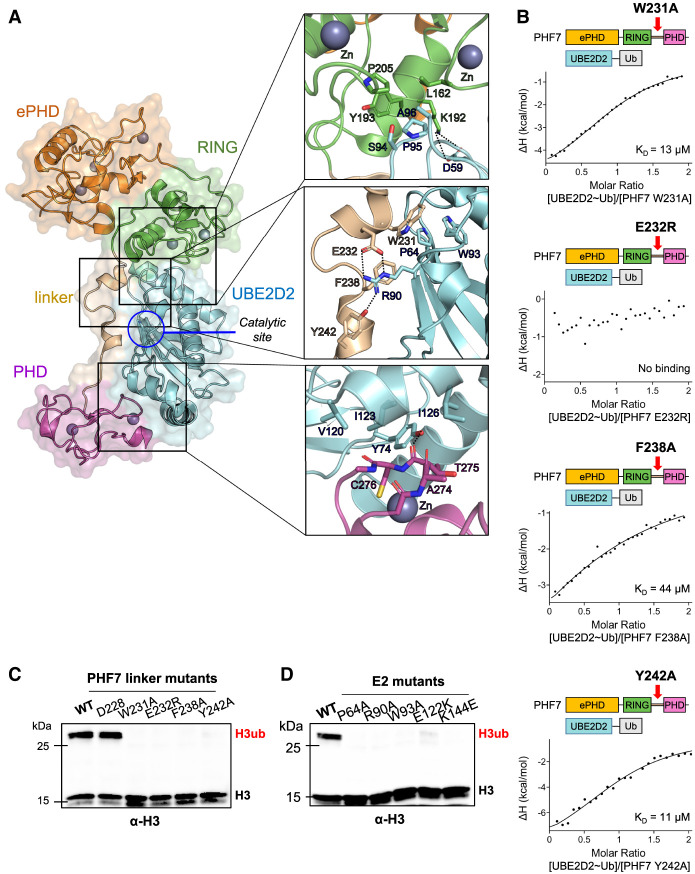
Domain architecture of the PHF7–UBE2D2 ubiquitination module. (A) Crystal structure of the sortase A-fused PHF7–UBE2D2 complex. (Boxes) The binding interface of the RING domain (top), linker (middle), and PHD (bottom) with UBE2D2. Interactions within 4 Å are shown. (B) Isothermal titration calorimetry binding curves of the indicated PHF7 mutants with UBE2D2∼Ub. The KD values are indicated. (C,D) In vitro ubiquitination assay with the indicated PHF7 (C) and UBE2D2 (D) mutants using the NCP as a substrate. PHF7 D228R was used as a negative control. Ubiquitinated histone H3 (H3ub) was detected with an H3-specific antibody.
The final model of the sortase fusion complex included two molecules of the PHF7–UBE2D2 complex in the asymmetric unit. While some RING domains require homodimerization or heterodimerization for their activity (Metzger et al. 2014), size exclusion chromatography analysis of our sample in solution and our complex structure suggest that PHF7 exists in a 1:1 complex with the E2 (Supplemental Fig. S6). When the two complexes of the asymmetric unit are aligned with respect to RING–UBE2D2, the two PHF7 molecules display movement up to 8 Å in the N-terminal ePHD and 10 Å in the C-terminal PHD, suggesting flexibility of PHF7 even in the UBE2D2∼Ub-bound state (Supplemental Fig. S6).
In both copies of the complex, we did not obtain well-resolved density for ubiquitin despite confirming the presence of ubiquitin linked to UBE2D2 using SDS-PAGE analysis of the crystals (Supplemental Fig. S7). To visualize ubiquitin, we modeled ubiquitin with the PHF7–UBE2D2 complex via AlphaFold2 (Supplemental Fig. S8). The model showed ubiquitin juxtaposed between the RING domain of PHF7 and UBE2D2, as observed in other catalytically primed RING–E2∼Ub structures (Supplemental Fig. S8; Dou et al. 2012; Plechanovová et al. 2012; Buetow et al. 2015; Middleton et al. 2020). To verify this model, we performed ubiquitination assays using PHF7 mutants (mutations located in the binding interface between PHF7 and ubiquitin: S181A, Q206A, and N208A). All of them resulted in a loss of ubiquitination activity (Supplemental Fig. S8). This binding interface explains how PHF7 bound to ubiquitin-charged UBE2D2 with higher affinity than to the E2 alone (Supplemental Fig. S2). However, the lack of density for ubiquitin in the crystal structure denotes flexibility in the complex, indicating that PHF7 could not lock ubiquitin in place. In the available structures of the RING-E2∼Ub complexes, a linchpin arginine of the RING domain inserts between the E2 and ubiquitin and stabilizes the “closed” conformation vital for ubiquitin transfer (Supplemental Fig. S8; Buetow and Huang 2016; Horn et al. 2019). In PHF7, this residue was substituted with Asn, which does not make extensive interactions with the E2 and ubiquitin compared with a linchpin Arg (Supplemental Fig. S8).
PHF7 wraps around UBE2D2 via RING–linker–PHD
Consistent with our binding assay results, our structure demonstrates that the RING domain is not the sole domain interacting with UBE2D2. UBE2D2 is sandwiched between the RING domain and PHD of PHF7. The binding interface between the RING domain and UBE2D2 exhibits similarities to that of other RING domain and UBE2D2 complexes (Deshaies and Joazeiro 2009). The RING domain of PHF7 adopts a typical cross-brace fold that coordinates two zinc ions. Residues L162 and P205, located in finger-like projections of two bipartite zinc-coordinating loops, and Y193 form contacts with UBE2D2 through the SPA motif: S94E2, P95E2, and A96E2 (superscript E2 represents UBE2D2) (Fig. 3A; Zhang et al. 2005; Mace et al. 2008; Stewart et al. 2016; Gundogdu and Walden 2019). Assisting this interaction, PHF7 K192 forms a salt bridge with D59E2.
In addition to the canonical interaction with the RING domain, UBE2D2 also makes contacts with the PHD and the linker region connecting the RING domain to the PHD of PHF7. The residues within the linker region of PHF7 are well coordinated by UBE2D2. W231 interacts with W93E2 and P64E2. E232, F238, and Y242 form electrostatic, π-cation, and hydrogen bond interactions with R90E2, respectively (Fig. 3A). Point mutations of these residues that interact with UBE2D2 (W231A, E232R, F238A, and Y242) all exhibited a significant decrease in binding affinity to UBE2D2 (Fig. 3B). In particular, the electrostatic interaction of E232 appeared to be critical, as the E232R mutation completely abolished the binding. These mutations accordingly failed to ubiquitinate H3K14, as demonstrated by the in vitro ubiquitination assay with the NCP (Fig. 3C). It is interesting to note that the PHF7 linker interacts with the “front side” of the E2, close to the ubiquitin-charged catalytic site C85E2 (Fig. 3A). This unique binding pattern differs from the “backside” interaction observed in the structures of some RING E3 ligases that use extended regions for their interactions with partner E2s (Das et al. 2013; Li et al. 2015). To confirm this novel binding mode, we designed a series of UBE2D2 mutants in the binding interface with the PHF7 linker region: P64AE2, R90AE2, and W93AE2 (Fig. 3A). All of these E2 mutants showed greatly reduced binding affinity to PHF7 and exhibited reduced efficiency in NCP ubiquitination by PHF7, providing compelling support for our structural model (Fig. 3D; Table 2; Supplemental Fig. S9).
Following the linker, the PHD, which is also a zinc-coordinating domain, grabs UBE2D2 from the opposite end of the RING domain interaction site. The zinc finger loop of the PHD (A274, T275, and C276) inserts into a hydrophobic groove formed by Y74E2, V120E2, I123E2, and I126E2 (Fig. 3A). While the PHD exhibits structural flexibility within PHF7, this interaction network is maintained in the (GSGG)3 linker complex, highlighting the crucial role of these interactions in ensuring the stability of the complex (Supplemental Fig. S5). In each complex structure, additional nonconserved polar and van der Waals interactions were observed to stabilize the complex. Notably, while the PHD shows a domain movement in the two copies of the crystal structure, R244 and E299 within the PHD form ionic interactions with E122E2 and K144E2, respectively, in each conformation (Supplemental Fig. S9). Reversing the charges of these amino acids in the E2 led to reduced binding affinity to PHF7 and loss of NCP ubiquitination, suggesting the contribution of these interactions to the PHF7's activity (Fig. 3D; Table 2; Supplemental Fig. S9). The flexibility of the PHD is likely to play a role in recruiting the H3 tail to the catalytic site for efficient ubiquitin transfer. To obtain the information about the putative substrate binding site within the PHD, we designed loss-of-function mutations within the PHD based on structural alignment of the PHD of PHF7 with the PHD bound to the H3 peptide (PDB ID 5WXH) (Supplemental Fig. S10). It is evident that several hydrophobic residues (Y245, F262, L270, and L272) that are located at the potential H3 tail binding site played an important role in substrate binding, as demonstrated by the reduction in NCP ubiquitination by the mutations of these residues (Fig. 4A).
Figure 4.
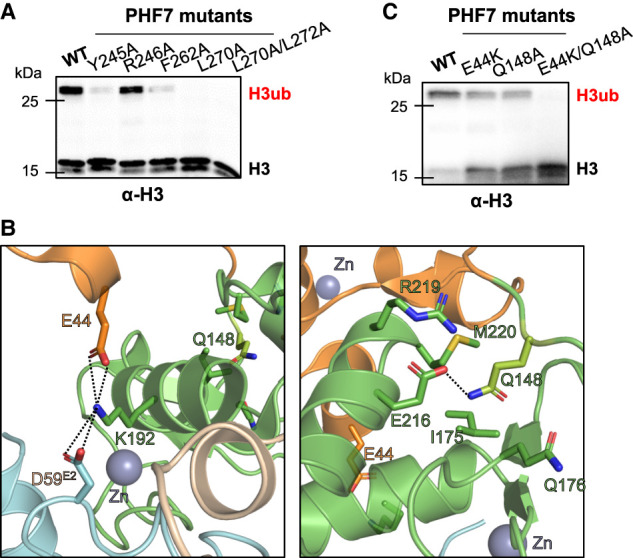
Structural features of PHF7 important for H3 ubiquitination. (A) In vitro ubiquitination assay with the indicated PHF7 mutants targeting the potential H3 binding site of PHF7 using the NCP as a substrate. For details, see Supplemental Figure S10. (B) An amino acid network that stabilizes the ePHD and RING domain of PHF7. (C) In vitro ubiquitination assay of the NCP using PHF7 mutants targeting ePHD residues in B. Ubiquitinated histone H3 (H3ub) was detected with an H3-specific antibody.
Our crystal structure did not reveal any direct and critical interactions between the ePHD and UBE2D2, which was unexpected considering the reduced affinity of PHF7 toward UBE2D2 in the absence of the ePHD (Fig. 2C). However, further investigation uncovered an intriguing indirect interaction between the ePHD and UBE2D2 mediated by the intrachain RING domain. At one end of the ePHD, electrostatic interactions encompassing the ePHD, RING, and E2s are observed, involving E44 of the ePHD, K192 of the RING domain, and D59E2 (Fig. 4B). On the other side of the ePHD, Q148, located in the solvent-exposed loop connecting the ePHD and the RING domain, inserts into the pocket within the RING domain composed of I175, Q176, E216, R219, and M220. This loop immediately extends into the zinc-coordinating region (159–165) of the RING domain that contacts UBE2D2 (Fig. 4B). Although the ePHD does not make extensive contact with RING domain, it plays a role in orienting the connecting loop in a manner that facilitates binding of E2s. (Fig. 3A). It is worth noting that individual point mutations E44K and Q148A exhibited relatively minimal impact on ubiquitination activity and E2 binding, but the E44K/Q148A double mutation resulted in a defect in the ubiquitination activity of PHF7 and a sixfold reduction in binding affinity for E2s (Fig. 4C; Table 2; Supplemental Fig. S11). Interestingly, we observed that while purified PHF7 by itself was prone to cleavage between N149 and I150 when left at ambient temperature, the presence of UBE2D2 prevented this cleavage (Supplemental Fig. S12). This phenomenon is consistent with our structural findings where the cleaved region is in extensive contact with the RING domain. The stabilization of this loop between the ePHD and the RING domain completes the tripartite interaction of PHF7 domains with UBE2D2 and explains the indirect influence of the ePHD on E2 binding.
Functional role of the PHF7 ePHD in nucleosome binding
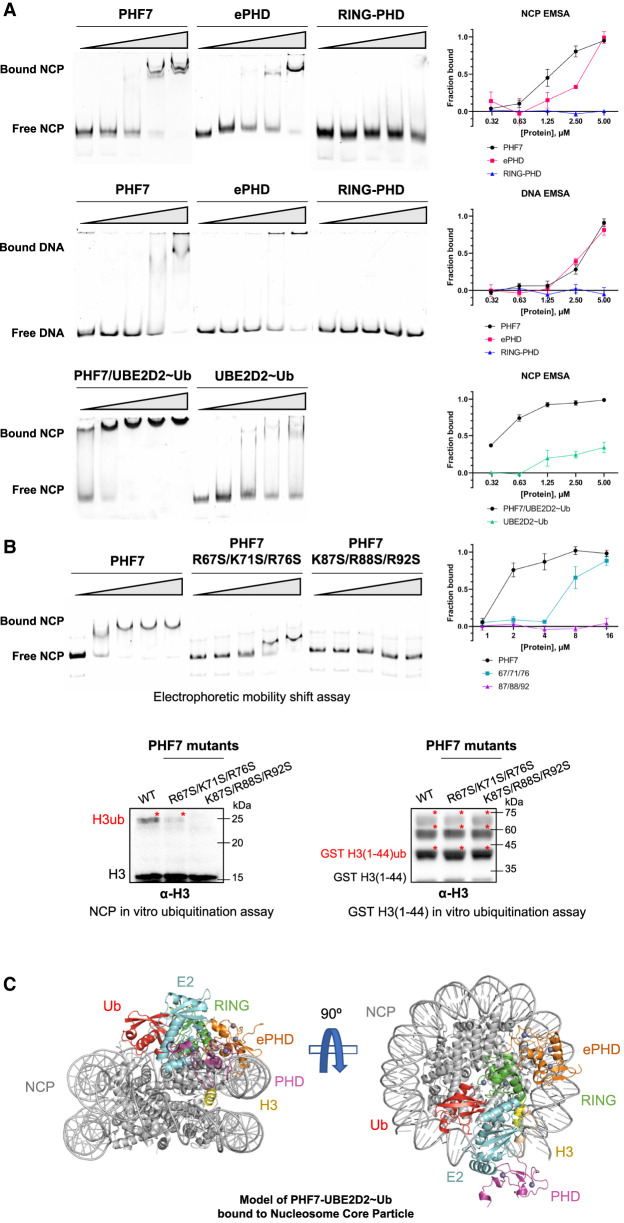
We have clearly shown the structural basis of requirement of all three domains of PHF7 for E2 recruitment and ubiquitination activity. As an epigenetic reader and writer, PHF7 would recognize the N-terminal H3 tail as part of the nucleosome in physiological conditions. Because the ePHD of PHF6 has been shown to interact with DNA (Liu et al. 2014), we hypothesized that the ePHD of PHF7 may play an additional role in substrate recruitment by participating in interaction with the NCP via nucleosomal DNA. The surfaces of ePHDs in both PHF6 and PHF7 are covered with positively charged amino acids, supporting this hypothesis (Supplemental Fig. S13). To confirm the formation of PHF7 and the nucleosome complex, we performed an electrophoretic mobility shift assay (EMSA), which showed a clear band shift (Fig. 5A). We then investigated which PHF7 domains are responsible for NCP binding. As we hypothesized, we found that the ePHD is the main domain interacting with the NCP, while RING–PHD failed to bind the NCP by itself (Fig. 5A).
Figure 5.
PHF7/E2∼Ub binds to the NCP via multivalent interactions. (A) Electrophoretic mobility shift assay (EMSA) of the NCP and Widom 601 free DNA with increasing concentrations of PHF7 or UBE2D2∼Ub. A fraction of the bound NCP is plotted against the indicated protein concentrations. (B) EMSA and in vitro ubiquitination assay of PHF7 with mutations on the surface basic patches. Red asterisks denote ubiquitinated histone H3. (C) Binding model of PHF7–UBE2D2∼Ub to the NCP. An AlphaFold2 model of PHF7–UBE2D2∼Ub was aligned to the E2 of 7JZV. For details, see Supplemental Figure S14.
We next confirmed that DNA is the primary interaction site between the ePHD and nucleosome by performing EMSA with free Widom 601 DNA that we used to reconstitute the nucleosome. We observed a clear upshift of bands, indicating the formation of the DNA–ePHD complex (Fig. 5A). To determine whether the nucleosomal DNA is the main interaction site for the ePHD, we conducted an EMSA with the NCP using the PHF7 mutants R67S/K71S/R76S and K87S/R88S/R92S, both of which were designed to disrupt the positively charged patch on the surface of the ePHD (Supplemental Fig. S13). Intriguingly, R67S/K71S/R76S and K87S/R88S/R92S triple mutants exhibited reduced interaction with the NCP and a loss of interaction with the NCP, respectively (Fig. 5B). These results were consistently reflected in NCP ubiquitination activity, the former mutant showing the reduced level of NCP ubiquitination, while the latter failed to ubiquitinate the NCP (Fig. 5B). Notably, these mutants exhibit the wild-type-like activity for H3 peptide ubiquitination (Fig. 5B). These mutational data provide support for our model that PHF7 binds to the NCP primarily through the interaction between the highly basic ePHD and DNA of the NCP.
While the ePHD exhibited a capacity to bind free DNA similar to that of the intact nucleosome, PHF7 bound to the NCP with a higher affinity than free DNA. This suggests that the ePHD and DNA interaction is the main driving force behind the binding between PHF7 and the nucleosome and that RING–PHD provides additional interactions to strengthen the binding (Fig. 5A). The H3 tail is known to be sheathed by intranucleosomal DNA via electrostatic interactions to prevent nonspecific targeting by enzymes (Ikebe et al. 2016; Li and Kono 2016; Morrison et al. 2018). It would first need to be unwrapped by a specific event before becoming available as a substrate (Gatchalian et al. 2017; Gibson et al. 2017). We propose that the interaction between the ePHD and DNA could initiate such a conformational change.
Recently, structural studies have shown that UBE2D2 and UBE2D3 possess an innate ability to bind to the nucleosome through DNA backbone interaction (McGinty et al. 2014; Ai et al. 2023). We observed a direct interaction of UBE2D2∼Ub with the NCP via EMSA, but smearing of the migration complex indicated a heterogeneous or weak binding (Fig. 5A; Hellman and Fried 2007). In contrast, the PHF7/UBE2D2∼Ub complex bound to the nucleosome with much higher affinity compared with equimolar amounts of PHF7 or UBE2D2∼Ub (Fig. 5A). This suggests that PHF7 and UBE2D2∼Ub cooperate allosterically to position UBE2D2∼Ub for ubiquitination of the NCP, which is further improved when PHF7 contains the RING–PHD that can directly recruit UBE2D2.
Binding model of the PHF7–UBE2D2–ubiquitin complex with the NCP
Our findings propose a model in which PHF7 recruits ubiquitin-charged UBE2D2 to the nucleosome through multivalent interactions, and it is likely that the ePHD initiates this binding by interacting with nucleosomal DNA. Given that ubiquitin is partially stabilized between PHF7 and UBE2D2, we compared the ubiquitination module of PHF7 with the available structures of RING–E2–Ub complexes bound to the NCP. Notably, the structures of RING ligases such as PRC1 and BRCA1–BARD1 bound to the NCP demonstrate the binding of the core histones to position E2 for site-specific ubiquitination (McGinty et al. 2014; Witus et al. 2021; Ai et al. 2023). However, aligning the PHF7–UBE2D2–ubiquitin complex to the available structures with respect to the E2 resulted in a severe clash of the PHF7 ePHD and ubiquitin with the nucleosome body, except for the BRCA1–BARD1–UBE2D3 ubiquitination module bound to the NCP (Hu et al. 2021). This model presents PHF7 bound to the nucleosome body through interactions between the positively charged residues of the ePHD and DNA, as well as between the RING domain and histone, stabilizing the conformation of the E2 and ubiquitin (Fig. 5C; Supplemental Fig. S14). The PHD is oriented toward the site of nucleosome opening, located near the flexible histone H3 tail. As such, upon binding of the PHD to H3, particularly around the K4 region, the H3 tail becomes stretched, positioning K14 in proximity to the catalytic C85E2 and triggering ubiquitin transfer. To validate this structural model, we attempted to solve the cryo-EM structure of the PHF7–UBE2D2–NCP complex. However, we encountered challenges due to the flexibility of the positioning of PHF7 onto the NCP. While further investigations are required to elucidate the precise conformation of the PHF7–UBE2D2–NCP complex, our study provides valuable insights into the structural basis of PHF7-mediated histone H3 ubiquitination.
Discussion
Most RING E3 ligases are multidomain proteins; however, the importance of domains beyond the RING domain in ubiquitination activity has often been overlooked. Our study emphasizes the importance of considering the entire protein structure of RING E3 ligases, as domains outside the RING domain play significant roles by cooperating in substrate recognition and catalytic activity. Binding and functional assays as well as the determination of the crystal structure of the PHF7–E2 complex provide valuable insights into the mechanism of PHF7 function.
Our structure reveals a novel binding mode of the RING E3 ligase for the E2 partner. PHF7 exhibited high affinity to the E2 through additional interaction sites outside the RING domain, including the linker region and PHD. In contrast to certain other RING E3 ligases containing extended regions that interact with the “backside” of the E2 (Das et al. 2013; Li et al. 2015), PHF7 establishes interactions with the “front side” of the E2 through its linker. Excessively high-affinity binding to the E2 through the backside surface, such as in the case of RNF25 with KD in the nanomolar range, decreases the rate of ubiquitination (Li et al. 2015). Although PHF7 wraps around the E2 via RING–linker–PHD, it still leaves the “backside” surface of the E2 exposed, potentially allowing free ubiquitin to bind and allosterically activate the E2 for processive polyubiquitination (Buetow et al. 2015). However, during our study and also in previous studies, only monoubiquitinated histone bands were observed in the ubiquitination assays of PHF7 with the NCP (Wang et al. 2019b; Kim et al. 2020). This may be explained by the model of the PHF7/E2∼Ub complex bound to the NCP, in which the catalytic C85 of the E2 is attached to ubiquitin on one side and shielded by the linker region of PHF7 on the other side. Steric hindrance by the linker would modulate PHF7 activity, which may be critical to prevent inadvertent ubiquitination in histones.
Furthermore, our study reveals the crucial role of the PHF7 PHD in UBE2D2 recruitment, shedding light onto the involvement of PHD-containing proteins in ubiquitination beyond their known function as histone epigenetic readers. PHDs have varying sequence and structure and play diverse roles in eukaryotic nuclear proteins (Sanchez and Zhou 2011). In the case of PHF7, the N-terminal ePHD does not bind to either the E2 or histone H3 but instead interacts with the NCP via nucleosomal DNA and allosterically regulates PHF7 ubiquitination activity. We demonstrate the relay of interactions between the ePHD and RING domain, which indirectly stabilize the RING–E2 conformation.
While the methylation state of the H3 peptide and NCP did not significantly affect the binding of PHF7 in vitro, in vivo studies have shown colocalization of PHF7 with H3K4me2/3 marks (Wang et al. 2019b; Kim et al. 2020; Garry et al. 2021). The substrate selection of PHF7 in vivo is likely influenced by various factors, such as DNA enhancers and other available epigenetic markers. These factors may regulate the choice of histone ubiquitination site between H3K14 and H2A K119 depending on the developmental context, especially during extensive epigenetic remodeling such as spermatogenesis. The model of the PHF7 ubiquitination module bound to the NCP highlights the importance of multivalent binding interactions for histone H3K14 ubiquitination that may allow ubiquitination in vitro without methylation but prevent nonspecific ubiquitination in vivo with further restrictions.
Further research is required to elucidate the precise molecular mechanism underlying the substrate specificity of PHF7 in vivo. Understanding the functional mechanism of PHF7 is essential for its therapeutic applications, as in addition to its natural role in spermatogenesis, it has demonstrated potential in cardiac reprogramming in conjunction with the SWI/SNF complex (Garry et al. 2021). In summary, our results advance the understanding of the molecular mechanism of PHF7 action and uncover a new potential role for the PHD in general.
Materials and methods
Cloning, protein expression, and purification
Standard cloning and site-directed mutagenesis were performed by PCR with the Q5 high-fidelity DNA polymerase (New England Biolabs). Genes encoding various constructs of PHF7 (PHF7 [28–307], ePHD [28–149], ePHD–RING [28–230], RING–PHD [150–307], linker–PHD [231–307], and PHD [244–301]) (Fig. 1A) were cloned into the modified pGEX-4T-1 vector containing a HRV 3C protease cut site between the GST tag and PHF7. RING–PHD required a C164A mutation for successful purification. The PHF7–UBE2D2 (S22R/C85K) fusion construct was designed by adding a (GSGG)3 linker between PHF7 and UBE2D2 [PHF7–(GSGG)3–E2]. The sequences of all plasmids were verified via DNA sequencing.
For PHF7 expression, the plasmids were transformed into competent Escherichia coli Rosetta cells. The cells were grown to an OD600 of ∼0.6, and protein expression was induced by adding 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 0.1 mM ZnSO4. Expression was performed for 16 h at 20°C. The cells were harvested by centrifugation, resuspended in phosphate-buffered saline (PBS) supplemented with 500 mM NaCl, and frozen at −80°C.
Typical purification of recombinant PHF7 was performed as described below, with the choice of buffer (HEPES at pH 7.0 or Tris at pH 8.0) and ion exchange chromatography depending on the isoelectric point of each construct. Cells were thawed and incubated with DNase I and phenylmethylsulfonyl fluoride (PMSF). Lysis was performed using Avestin EmulsiFlex C3 (Avestin, Inc.). After centrifugation, the supernatant was loaded onto glutathione agarose resin (Pierce) in equilibration buffer (20 mM HEPES at pH 7.0, 300 mM NaCl). Bound protein was eluted with equilibration buffer containing 20 mM reduced glutathione after column washing. To remove the GST tag, the eluted sample was incubated with HRV 3C protease overnight at 4°C. The reaction mixture was then loaded onto the HiTrap SP ion exchange column (Cytiva), and PHF7 was eluted with a linear NaCl gradient. PHF7 was further purified by size exclusion chromatography with Superdex 200 Increase 10/300 GL (Cytiva) pre-equilibrated with a buffer containing 20 mM HEPES (pH 7.0) and 150 mM NaCl.
Wild-type UBE2D2 and double-mutant UBE2D2 (S22R/C85K) were cloned into the pET-28a vector with an N-terminal Hisx6-SUMO tag to improve protein expression and a tobacco etch virus protease cut site in between. Hisx6-tagged ubiquitin was cloned into the pET-21d vector. Wild-type and K4C histone H3 (1–44) constructs were cloned into a modified pGEX-4T-1 vector with a PreScission protease cut site. All proteins were expressed in E. coli Rosetta cells with IPTG induction. The purification of wild-type and double-mutant UBE2D2 and ubiquitin was performed as described previously (Buetow et al. 2015). Histone peptides were cleaved off the GST tag, purified by ion exchange chromatography, desalted, and lyophilized. Site-specific installation of methyl-lysine chemical analogs for K4me2 and K4me3 was performed as described previously (Simon et al. 2007).
PHF7/UBE2D2∼Ub protein preparation for crystallization
For sortase-linked PHF7 purification, the PHF7 (28–307) construct was cloned as above but with a C-terminal extension composed of the (GSGG)2 linker, sortase A recognition sequence (LPETG), and Hisx6 tag. Protein expression and purification were performed as described above but without cleavage of the GST tag. UBE2D2 with covalently linked ubiquitin (UBE2D2∼Ub) was prepared as previously described (Plechanovová et al. 2012). Briefly, 100 μM UBE2D2 (S22R/C85K) was incubated with 300 μM ubiquitin and 1 μM homemade UBE1 for 24 h at 37°C in a buffer containing 5 mM ATP, 5 mM MgCl2, 50 mM Tris (pH 8.5), 150 mM NaCl, 5 mM creatine phosphate, and 0.6 U mL−1 creatine kinase. This resulted in a complete depletion of unreacted UBE2D2. UBE2D2∼Ub was further purified by loading it onto a HiPrep 26/10 desalting column (Cytiva) equilibrated with 20 mM HEPES (pH 7.0) and 100 mM NaCl, followed by a HiTrap SP ion exchange column (Cytiva) equilibrated in the same buffer. UBE2D2∼Ub was eluted with a linear NaCl gradient.
Purified GST-PHF7-(GSGG)2-LPETG-Hisx6 and UBE2D2∼Ub were fused by sortase A as described previously (Theile et al. 2013). Briefly, GST-PHF7 and UBE2D2∼Ub were mixed in a 1:1.2 molar ratio in a sortase reaction buffer (50 mM HEPES at pH 7.5, 150 mM NaCl, 10 mM CaCl2). Sortase A was added to the mixture and incubated for 15 min at 37°C. Next, the reaction mixture was loaded onto glutathione agarose resin, and the resulting GST-PHF7/UBE2D2∼Ub fusion complex was eluted. After cleavage of GST by HRV3C, the PHF7/UBE2D2∼Ub fusion protein was purified by a HiTrap SP ion exchange column and size exclusion chromatography (20 mM HEPES at pH 7.5, 150 mM NaCl).
Crystallization
The purified PHF7/UBE2D2∼Ub sortase A fusion complex was concentrated to 7.6 mg mL−1. Crystals were obtained by mixing 2 µL of the protein complex with 1 µL of reservoir solution containing 0.1 M Tris (pH 8.3) and 6% 2-methyl-2,4-pentanediol (MPD) using sitting drop vapor diffusion at 4°C. Initial crystals were harvested and crushed to produce a crystal seed stock, which was diluted and mixed to the original conditions in a 1.5:1.0:0.5 volume ratio of reservoir:protein:seed stock. Crystals were harvested and flash-frozen in 0.1 M Tris (pH 8.3), 6% MPD, and 25% glycerol.
The purified PHF7–(GSGG)3–UBE2D2 fusion complex was concentrated to 7.2 mg mL−1. Crystals were obtained by mixing 1 µL of the protein complex with 1 µL of reservoir solution containing 0.1 M MOPS (pH 7.0), 6% PEG8000, and 3% DMSO. Crystals were harvested and flash-frozen in a reservoir solution supplemented with 25% ethylene glycol.
The N-terminal ePHD was concentrated to 10 mg mL−1. Crystals were obtained by mixing 1 µL of protein with 1 µL of reservoir solution containing 0.1 M bicine (pH 8.7), 0.175 M LiCl, and 26% (w/v) PEG3350. Crystals were flash-frozen in a reservoir solution supplemented with 25% ethylene glycol.
Data collection and processing
For the N-terminal ePHD, diffraction data were collected at Beamline BL-7A of the Pohang Accelerator Laboratory. The native data set was collected to a resolution of 2.0 Å and processed using HKL2000. Initial phases were obtained by molecular replacement with Phaser (McCoy 2007) using the PHF6 ePHD (PDB ID: 4NN2). Iterative model building and refinement were performed with Coot (Emsley and Cowtan 2004) and Phenix (Adams et al. 2010) to obtain a final model containing three copies of the PHF7 ePHD (residues 30–146).
For the sortase A and (GSGG)3 fusion PHF7–UBE2D2 crystals, diffraction data were collected at Beamline BL-5C of the Pohang Accelerator Laboratory, processed, and integrated via XDS-based data processing pipeline AutoProcess (Kabsch 2010). Initial phases were obtained by molecular replacement with Phaser using the PHF7 ePHD (this study), PHF7 PHD (PDB 1WEQ), and UBE2D2 (PDB 6HPR) as search models. All models were built in Coot and refined using Phenix. The PHF7–UBE2D2 complex was refined to a resolution of 2.96 Å and 3.58 Å for sortase A and (GSGG)3 fusion constructs, respectively. The final model contained two copies of PHF7 (residues 30–301) and two copies of UBE2D2 (residues 1–147) for the sortase A fusion construct. Ubiquitin could not be modeled probably due to its flexible nature. All figure models were generated using PyMOL v2.4.0 (https://pymol.org/2).
Nucleosome reconstitution
Recombinant nucleosomes with Widom 601 147-bp DNA and Xenopus laevis histones were reconstituted as described previously (Dyer et al. 2004). Briefly, individual histones were purified under denaturing conditions and refolded into histone octamers in the presence of 2 M NaCl. DNA was added and the salt was dialyzed out to yield folded nucleosome core particles. Reconstituted nucleosomes were stored at 4oC.
Electrophoretic mobility shift assay (EMSA)
Nucleosome and DNA binding assays were performed by incubating 50 nM recombinant nucleosomes or DNA in binding buffer [10 mM Tris-HCl at pH 7.5, 50 mM NaCl, 5 mM MgCl2, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), 5% glycerol] with varying concentrations of PHF7 in 10-μL reactions. The reactions were performed for 30 min on ice before being resolved on a 5% 19:1 acrylamide:bis-acrylamide gel running with 0.5× TBE at 130 V for 60 min in a cold chamber. The gel was stained with SYBR Gold nucleic acid stain (Invitrogen) and imaged using a ChemiDoc MP (Bio-Rad). Each assay was performed at least three times.
Quantification was performed according to the method described by Liu et al. (2020). Free (bottom) and bound (shifted) nucleosome/DNA bands were quantified with ImageJ (v.1.54d). The fraction bound was determined by calculating the integrated densitometry values of the free and bound nucleosome/DNA bands according to Y = (IDVBound − IDVBoundBackground) ÷ (IDVBound − IDVBoundBackground + IDVFree − IDVFreeBackground), where Y is fraction bound.
Isothermal titration calorimetry (ITC)
Each purified protein was buffer-changed to ITC buffer (20 mM HEPES at pH 7.5, 150 mM NaCl) immediately before the experiment. For a typical assay, MicroCal PEAQ-ITC automated (Malvern Panalytical Ltd.) was used to inject 1.2 μL of 300 μM titrant 30 times into 30 μM titrate in the sample cell, with 150-sec spacing. The samples were stirred at 500 rpm at 25°C throughout the assay. MicroCal PEAQ-ITC analysis software (Malvern Panalytical Ltd.) was used to construct a binding curve and determine the binding affinity.
Microscale thermophoresis (MST)
Purified PHF7 proteins were labeled with protein labeling kit RED-NHS second-generation (NanoTemper Technologies GmbH) according to the manufacturer's instructions. Histone peptides were serially diluted in MST buffer (20 mM HEPES at pH 7.5, 150 mM NaCl, 0.05% Tween-20) immediately before use. A typical MST assay was performed by mixing 20 nM labeled PHF7 with serially diluted peptides and incubating for 15 min at room temperature. The samples were loaded into Monolith Premium capillaries (NanoTemper Technologies GmbH) and measured with medium (40%) MST power and 20% excitation power at 22°C using Monolith NT.115 pico (NanoTemper Technologies GmbH). MO.Control was used throughout the process, and MO.Affinity analysis was used to analyze the data and calculate the binding affinity curves. Figures were produced with GraphPad Prism.
In vitro ubiquitination assay
Histone in vitro ubiquitination assays were performed using recombinant nucleosome and H3 peptides as substrates. Typical reaction conditions contained 0.1 μM UBE1, 1 μM UBE2D2, 1 μM PHF7, 10 μM ubiquitin, and 1 μM histone/nucleosome substrates in assay buffer (50 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 5 mM ATP) and were incubated for 30 min at 30°C.
Reaction samples were next run on SDS-PAGE gels and visualized via Western blot. Ubiquitinated histones were detected with antibodies specific for histone H3 (Thermo Fisher 17H2L9 and Abcam ab1791). To verify H3K14 ubiquitination, the upshifted band visualized by Coomassie staining was dissected and sent for analysis. The mass spectrometry analysis was performed at the Taplin Mass Spectrometry Facility of Harvard Medical School (https://taplin.hms.harvard.edu).
The ubiquitination activity of the fusion constructs was confirmed by using PHF7 fused to wild-type UBE2D2 via (GSGG)3 linker and PHF7 fused to wild-type UBE2D2 via sortase A. Purification and enzymatic reactions were performed in the same way as their mutant counterparts.
Data availability
Atomic coordinates and structure factors for PHF7 fused to UBE2D2 via sortase A, PHF7 fused to UBE2D2 via (GSGG)3 linker, and the ePHD of PHF7 have been deposited in the Protein Data Bank under accession codes 8JWJ, 8JWS, and 8JWU, respectively.
Supplementary Material
Acknowledgments
We thank the staff members at Pohang Accelerator Laboratory Beamlines 5C and 7A for their help with data collection. This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government, the Ministry of Education (Creative Research Initiatives Programs; Research Center for Epigenetic Code and Diseases; 2017R1A3B1023387 to S.H.B.), and the Ministry of Science and ICT (Information and Communication Technologies; NRF-2022M3A9I2017587 and NRF-2019M3E5D6063903 to H.-J.C., and NRF-2020R1A2B5B03001517 to J.-J.S.).
Author contributions: H.S.L., I.B., and H.-J.C. conceived the study. H.S.L. and I.B. determined the crystal structures and performed the biochemical studies. J.Y., C.R.K., M.H., J.-S.K., and S.H.B. assisted with biochemical research. T.-K.J. and J.-J.S. prepared the nucleosome core particles. H.S.L., I.B., and H.-J.C. wrote the manuscript. H.-J.C. supervised the research.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350989.123.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai HS, Tong ZB, Deng ZH, Tian JK, Zhang LY, Sun MS, Du YX, Xu ZY, Shi Q, Liang LJ, et al. 2023. Synthetic E2–Ub–nucleosome conjugates for studying nucleosome ubiquitination. Chem 9: 1221–1240. 10.1016/j.chempr.2023.01.012 [DOI] [Google Scholar]
- Bailly V, Prakash S, Prakash L. 1997. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol Cell Biol 17: 4536–4543. 10.1128/MCB.17.8.4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera AP, Naskar P, Agarwal S, Banka PA, Poddar A, Datta AB. 2018. Structural insights into the nanomolar affinity of RING E3 ligase ZNRF1 for Ube2N and its functional implications. Biochem J 475: 1569–1582. 10.1042/BCJ20170909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley ML, Corn JE, Dong KC, Phung Q, Cheung TK, Cochran AG. 2011. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J 30: 3285–3297. 10.1038/emboj.2011.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PK, Southard S, Baxter K, Van Doren M. 2022. Germline sex determination regulates sex-specific signaling between germline stem cells and their niche. Cell Rep 39: 110620. 10.1016/j.celrep.2022.110620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. 2006. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31: 35–40. 10.1016/j.tibs.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. 2006. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell 21: 873–880. 10.1016/j.molcel.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Buetow L, Huang DT. 2016. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol 17: 626–642. 10.1038/nrm.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetow L, Gabrielsen M, Anthony NG, Dou H, Patel A, Aitkenhead H, Sibbet GJ, Smith BO, Huang DT. 2015. Activation of a primed RING–E3–E2–ubiquitin complex by non-covalent ubiquitin. Mol Cell 58: 297–310. 10.1016/j.molcel.2015.02.017 [DOI] [PubMed] [Google Scholar]
- Burslem GM, Crews CM. 2020. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181: 102–114. 10.1016/j.cell.2019.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Liang YH, Mariano J, Li J, Huang T, King A, Tarasov SG, Weissman AM, Ji X, Byrd RA. 2013. Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J 32: 2504–2516. 10.1038/emboj.2013.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- DiBello A, Datta AB, Zhang X, Wolberger C. 2016. Role of E2–RING interactions in governing RNF4-mediated substrate ubiquitination. J Mol Biol 428: 4639–4650. 10.1016/j.jmb.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. 2012. BIRC7–E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol 19: 876–883. 10.1038/nsmb.2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove KK, Kemp HA, Di Bona KR, Reiter KH, Milburn LJ, Camacho D, Fay DS, Miller DL, Klevit RE. 2017. Two functionally distinct E2/E3 pairs coordinate sequential ubiquitination of a common substrate in Caenorhabditis elegans development. Proc Natl Acad Sci 114: E6576–E6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. 2004. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375: 23–44. 10.1016/S0076-6879(03)75002-2 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fiorentini F, Esposito D, Rittinger K. 2020. Does it take two to tango? RING domain self-association and activity in TRIM E3 ubiquitin ligases. Biochem Soc Trans 48: 2615–2624. 10.1042/BST20200383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont PS, Hanson IM, Trowsdale J. 1991. A novel cysteine-rich sequence motif. Cell 64: 483–484. 10.1016/0092-8674(91)90229-R [DOI] [PubMed] [Google Scholar]
- Garry GA, Bezprozvannaya S, Chen K, Zhou H, Hashimoto H, Morales MG, Liu N, Bassel-Duby R, Olson EN. 2021. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat Cell Biol 23: 467–475. 10.1038/s41556-021-00668-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J, Wang X, Ikebe J, Cox KL, Tencer AH, Zhang Y, Burge NL, Di L, Gibson MD, Musselman CA, et al. 2017. Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun 8: 1489. 10.1038/s41467-017-01598-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge LQ, Xia T, Huang B, Gu HT, Song QS, Yang GQ, Liu F, Wu JC. 2017. PHF7, a novel male gene influences female fecundity and population growth in Nilaparvata lugens Stål (Hemiptera: Delphacidae). Sci Rep 7: 11611. 10.1038/s41598-017-11524-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MD, Gatchalian J, Slater A, Kutateladze TG, Poirier MG. 2017. PHF1 Tudor and N-terminal domains synergistically target partially unwrapped nucleosomes to increase DNA accessibility. Nucleic Acids Res 45: 3767–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundogdu M, Walden H. 2019. Structural basis of generic versus specific E2–RING E3 interactions in protein ubiquitination. Protein Sci 28: 1758–1770. 10.1002/pro.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman LM, Fried MG. 2007. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc 2: 1849–1861. 10.1038/nprot.2007.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn V, Uckelmann M, Zhang H, Eerland J, Aarsman I, le Paige UB, Davidovich C, Sixma TK, van Ingen H. 2019. Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nat Commun 10: 1751. 10.1038/s41467-019-09756-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Botuyan MV, Zhao D, Cui G, Mer E, Mer G. 2021. Mechanisms of BRCA1–BARD1 nucleosome recognition and ubiquitylation. Nature 596: 438–443. 10.1038/s41586-021-03716-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikebe J, Sakuraba S, Kono H. 2016. H3 histone tail conformation within the nucleosome and the impact of K14 acetylation studied using enhanced sampling simulation. PLoS Comput Biol 12: e1004788. 10.1371/journal.pcbi.1004788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, et al. 2021. A single-cell type transcriptomics map of human tissues. Sci Adv 7: eabh2169. 10.1126/sciadv.abh2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CR, Noda T, Kim H, Kim G, Park S, Na Y, Oura S, Shimada K, Bang I, Ahn JY, et al. 2020. PHF7 modulates BRDT stability and histone-to-protamine exchange during spermiogenesis. Cell Rep 32: 107950. 10.1016/j.celrep.2020.107950 [DOI] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589. 10.1038/nature03368 [DOI] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. 2007. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448: 718–722. 10.1038/nature06034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kono H. 2016. Distinct roles of histone H3 and H2A tails in nucleosome stability. Sci Rep 6: 31437. 10.1038/srep31437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95. 10.1038/nature04802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liang YH, Mariano J, Metzger MB, Stringer DK, Hristova VA, Li J, Randazzo PA, Tsai YC, Ji X, et al. 2015. Insights into ubiquitination from the unique clamp-like binding of the RING E3 AO7 to the E2 UbcH5B. J Biol Chem 290: 30225–30239. 10.1074/jbc.M115.685867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu J, Cao Q, Sha J, Zhou Z, Wang H, Li J. 2006. NYD-SP15: a novel gene potentially involved in regulating testicular development and spermatogenesis. Biochem Genet 44: 409–423. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li F, Ruan K, Zhang J, Mei Y, Wu J, Shi Y. 2014. Structural and functional insights into the human Börjeson-Forssman-Lehmann syndrome-associated protein PHF6. J Biol Chem 289: 10069–10083. 10.1074/jbc.M113.535351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WH, Zheng J, Feldman JL, Klein MA, Kuznetsov VI, Peterson CL, Griffin PR, Denu JM. 2020. Multivalent interactions drive nucleosome binding and efficient chromatin deacetylation by SIRT6. Nat Commun 11: 5244. 10.1038/s41467-020-19018-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low LY, Hernández H, Robinson CV, O'Brien R, Grossmann J, Ladbury J, Luisi B. 2002. Metal dependent folding and stability of nuclear hormone receptor DNA-binding domains. J Mol Biol 319: 87–106. 10.1016/S0022-2836(02)00236-X [DOI] [PubMed] [Google Scholar]
- Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL. 2008. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem 283: 31633–31640. 10.1074/jbc.M804753200 [DOI] [PubMed] [Google Scholar]
- McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41. 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Henrici RC, Tan S. 2014. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514: 591–596. 10.1038/nature13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM. 2014. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta 1843: 47–60. 10.1016/j.bbamcr.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton AJ, Zhu J, Day CL. 2020. The RING domain of RING finger 12 efficiently builds degradative ubiquitin chains. J Mol Biol 432: 3790–3801. 10.1016/j.jmb.2020.05.001 [DOI] [PubMed] [Google Scholar]
- Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J, Musselman CA. 2018. The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife 7: e31481. 10.7554/eLife.31481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nuñez JK, Nickoloff J, et al. 2012. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19: 1266–1272. 10.1038/nsmb.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Boo K, Kim J, Baek SA, Jeon Y, You J, Lee H, Choi HJ, Park D, Lee JM, et al. 2020. The chromatin-binding protein PHF6 functions as an E3 ubiquitin ligase of H2BK120 via H2BK12Ac recognition for activation of trophectodermal genes. Nucleic Acids Res 48: 9037–9052. 10.1093/nar/gkaa626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717. 10.1038/nature05987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103. 10.1038/nature04814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. 2012. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489: 115–120. 10.1038/nature11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. 2012. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell 47: 933–942. 10.1016/j.molcel.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie ML, Chaugule VK, Walden H. 2020. Modes of allosteric regulation of the ubiquitination machinery. Curr Opin Struct Biol 62: 189–196. 10.1016/j.sbi.2020.02.003 [DOI] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. 2011. The PHD finger: a versatile epigenome reader. Trends Biochem Sci 36: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, de Jong A, Ovaa H, Alpi AF, Harper JW, et al. 2016. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166: 1198–1214.e24. 10.1016/j.cell.2016.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. 2007. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012. 10.1016/j.cell.2006.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. 2016. E2 enzymes: more than just middle men. Cell Res 26: 423–440. 10.1038/cr.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theile CS, Witte MD, Blom AE, Kundrat L, Ploegh HL, Guimaraes CP. 2013. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat Protoc 8: 1800–1807. 10.1038/nprot.2013.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347: 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Wang T, Gao H, Li W, Liu C. 2019a. Essential role of histone replacement and modifications in male fertility. Front Genet 10: 962. 10.3389/fgene.2019.00962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kang JY, Wei L, Yang X, Sun H, Yang S, Lu L, Yan M, Bai M, Chen Y, et al. 2019b. PHF7 is a novel histone H2A E3 ligase prior to histone-to-protamine exchange during spermiogenesis. Development 146: dev175547. 10.1242/dev.175547 [DOI] [PubMed] [Google Scholar]
- Witus SR, Burrell AL, Farrell DP, Kang J, Wang M, Hansen JM, Pravat A, Tuttle LM, Stewart MD, Brzovic PS, et al. 2021. BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1. Nat Struct Mol Biol 28: 268–277. 10.1038/s41594-020-00556-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JD, Mace PD, Day CL. 2016. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat Struct Mol Biol 23: 45–52. 10.1038/nsmb.3142 [DOI] [PubMed] [Google Scholar]
- Yang SY. 2021. Germline masculinization by Phf7 in D. melanogaster requires its evolutionarily novel C-terminus and the HP1-family protein HP1D3csd. Sci Rep 11: 6308. 10.1038/s41598-021-85560-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Baxter EM, Van Doren M. 2012. Phf7 controls male sex determination in the Drosophila germline. Dev Cell 22: 1041–1051. 10.1016/j.devcel.2012.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. 2005. Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP–Ubc13–Uev1a complex. Mol Cell 20: 525–538. 10.1016/j.molcel.2005.09.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for PHF7 fused to UBE2D2 via sortase A, PHF7 fused to UBE2D2 via (GSGG)3 linker, and the ePHD of PHF7 have been deposited in the Protein Data Bank under accession codes 8JWJ, 8JWS, and 8JWU, respectively.