In this study, Russo et al. describe the functional synergy between the adaptor protein Symplekin, the protein phosphatase 1 (PP1) regulatory subunit (PNUTS), and the Restrictor complex in controlling extragenic transcription termination in yeast. Their findings show that Symplekin is competitively recruited by Restrictor- or CPSF-containing transcription termination machineries and has a role in CPSF- and cleavage-independent transcriptional repression of noncoding RNAs and endogenous retroviral elements.
Keywords: RNA polymerase II, transcription, transcription termination
Abstract
Transcription termination pathways mitigate the detrimental consequences of unscheduled promiscuous initiation occurring at hundreds of thousands of genomic cis-regulatory elements. The Restrictor complex, composed of the Pol II-interacting protein WDR82 and the RNA-binding protein ZC3H4, suppresses processive transcription at thousands of extragenic sites in mammalian genomes. Restrictor-driven termination does not involve nascent RNA cleavage, and its interplay with other termination machineries is unclear. Here we show that efficient termination at Restrictor-controlled extragenic transcription units involves the recruitment of the protein phosphatase 1 (PP1) regulatory subunit PNUTS, a negative regulator of the SPT5 elongation factor, and Symplekin, a protein associated with RNA cleavage complexes but also involved in cleavage-independent and phosphatase-dependent termination of noncoding RNAs in yeast. PNUTS and Symplekin act synergistically with, but independently from, Restrictor to dampen processive extragenic transcription. Moreover, the presence of limiting nuclear levels of Symplekin imposes a competition for its recruitment among multiple transcription termination machineries, resulting in mutual regulatory interactions. Hence, by synergizing with Restrictor, Symplekin and PNUTS enable efficient termination of processive, long-range extragenic transcription.
Mammalian genomes harbor a massive potential for transcription initiation that depends on the hundreds of thousands of cis-regulatory elements (enhancers and promoters) that provide modular platforms for the combinatorial binding of transcription factors (TFs) and the subsequent recruitment of RNA polymerase II (Pol II) (De Santa et al. 2010; Kim et al. 2010; Koch et al. 2011; Djebali et al. 2012; Andersson et al. 2014). Whereas transcription initiation occurs pervasively in the genome, its output is dramatically different inside genes and outside of them (Schlackow et al. 2017), with the abundance of long noncoding RNAs (lncRNAs), such as promoter antisense lncRNAs (pa-lncRNAs) and enhancer-generated lncRNAs (e-lncRNAs), being at least one order of magnitude lower than that of genic transcripts (Castello et al. 2012; Field and Adelman 2020). Although transcript stability also contributes to these differences in abundance, synthesis rates of most lncRNAs are overall much lower than those of pre-mRNAs (Mukherjee et al. 2017). Such a difference is exemplified by the case of bidirectional gene promoters (Andersson et al. 2015), at which divergent noncoding transcription is much less productive than that of the paired sense (coding) transcription unit. A mechanism proposed to account for such directional bias is the cleavage-dependent termination caused by polyA signals (PASs) in the promoter-divergent transcripts, whereas usage of PAS sequences inside genes is suppressed by U1 snRNP recruitment to 5′ splice sites (“telescripting”) (Kaida et al. 2010; Almada et al. 2013; Ntini et al. 2013). However, many such bidirectional transcription units contain promoter-proximal canonical splice sites also on the noncoding side yet show highly skewed transcription, thus clearly indicating additional control mechanisms.
Low processivity of extragenic Pol II may in principle be due to widespread defects in the “maturation” processes that normally equip Pol II with the properties required for efficient elongation and cotranscriptional RNA processing (Zhou et al. 2012; Bentley 2014). Indeed, several lines of evidence support the notion that Pol II maturation is an inefficient process that is subjected to active enforcement early in the transcription cycle in order to license Pol II molecules for entry into productive elongation. First, in up to 80% of cases, transcription initiation is nonproductive and is terminated when Pol II undergoes promoter-proximal pausing (Zimmer et al. 2021), a universal event separating initiation from productive elongation (Core and Adelman 2019). Second, the two mammalian nascent RNA cleavage complexes CPSF (cleavage and polyadenylation specificity factor) and Integrator (INT) act genome-wide in proximity to transcription start sites (TSSs) to cleave nascent RNAs and rapidly terminate transcription by immature (i.e., poorly processive) Pol II (Elrod et al. 2019; Lykke-Andersen et al. 2021; Rouvière et al. 2022; Stein et al. 2022; Wagner et al. 2023). The immaturity of such early-terminated Pol II molecules is inferred on the basis of the observation that upon inactivation of CPSF- and/or INT-mediated cleavage, these Pol II molecules elongate for only a short distance but do not reach the 3′ ends of the transcription units, indicating low processivity (Elrod et al. 2019; Lykke-Andersen et al. 2021; Stein et al. 2022).
In this conceptual framework, we and others recently reported the existence of a protein complex with a nonredundant role in the control of extragenic Pol II termination (Austenaa et al. 2015, 2021; Estell et al. 2021; Park et al. 2022). This complex, subsequently renamed Restrictor (Nojima and Proudfoot 2022), is composed of ZC3H4, a C3H1-type zinc finger RNA-binding protein, and WDR82, which specifically interacts with the initiating Pol II phosphorylated at Ser5 in the C-terminal repeat domain (CTD) (Lee and Skalnik 2008). Notably, the affinity of WDR82 toward p-Ser5 in the CTD is increased when it is bound to ZC3H4 (Park et al. 2022). Restrictor preferentially terminates extragenic transcription while having an overall limited impact on protein-coding genes (Austenaa et al. 2021; Hughes et al. 2023), as exemplified by the differential effects of its depletion on nascent transcripts generated from bidirectional gene promoters that generate a sense mRNA and a divergent lncRNA (Austenaa et al. 2015, 2021). In cells lacking Restrictor, extragenic Pol II complexes that are normally subjected to early termination acquire the ability to efficiently elongate, as shown by the strong increase in CTD phosphorylation at Ser2 and H3K36 trimethylation inside derepressed transcription units (Austenaa et al. 2015), and generate high levels of spliced and polyadenylated lncRNAs that are otherwise almost undetectable. Therefore, while disabling INT- and CPSF-enforced early termination of immature Pol II results in the release of Pol II complexes with a short-range elongation capacity (Elrod et al. 2019; Lykke-Andersen et al. 2021; Stein et al. 2022), Pol II released upon Restrictor inactivation can elongate for extended genomic distances (Austenaa et al. 2015, 2021).
Mechanisms involved in Restrictor-mediated termination are incompletely understood. The core WDR82–ZC3H4 Restrictor complex is very stable and resistant to stringent purification conditions, with no additional stoichiometric components potentially informative of its mechanism of action (our unpublished data). Substoichiometric interactors that have been reported include ARS2, a protein that binds nascent RNAs early in the transcription cycle and contributes to termination and its coordination with transcript degradation (Estell et al. 2023; Rouvière et al. 2023). However, a complete understanding of players and mechanisms involved in the control of transcription termination by Restrictor is still lacking.
Here we report that termination at extragenic transcription units controlled by Restrictor involves the recruitment of PNUTS (PP1 nuclear targeting subunit) and the adaptor protein Symplekin. PNUTS (PPP1R10) is a regulatory subunit of protein phosphatase 1 (PP1) that, together with WDR82 and the adapter protein TOX4, is part of a nuclear phosphatase complex (Lee et al. 2010) involved in termination at the 3′ ends of genes (Austenaa et al. 2015) because of its ability to dephosphorylate the SPT5 elongation factor (Cortazar et al. 2019). Symplekin is a large and flexible protein physically and functionally linked to both the CPSF and the histone cleavage complex, which terminates transcription of replication-dependent histone genes (Marzluff and Koreski 2017). Both mammalian Symplekin and its poorly conserved yeast ortholog, Pta1, make direct and stable interactions with the hinge domain of CSTF2 (Takagaki et al. 1990), an RNA-binding protein specific for GU-rich sequences often located downstream from the PAS and contributing to alternative PAS selection. The Symplekin–CSTF2 core complex is incorporated in both the histone cleavage complex (Marzluff and Koreski 2017) and the CPSF (Takagaki and Manley 2000; Chan et al. 2014). However, while Symplekin is absolutely required for the endonucleolytic activity of CPSF3 in the context of the histone cleavage complex (Sun et al. 2020), it is dispensable for CPSF3 activity within the CPSF (Boreikaite et al. 2022). Similarly, Symplekin mutants unable to contact CSTF2 impair histone 3′ RNA processing but not cleavage and polyadenylation of all other pre-mRNAs (Ruepp et al. 2011). While the precise function of Symplekin within the CPSF complex is unclear, its depletion in Drosophila melanogaster cells results in the destabilization of other complex subunits, including CPSF3, suggesting a structural role in complex integrity (Sullivan et al. 2009).
Notably, the yeast ortholog of Symplekin, Pta1, is also part of the APT (associated with Pta1) complex, which in addition to Swd2 (the ortholog of WDR82) contains the yeast PP1 and PNUTS orthologs Glc7 and Ref2, respectively, and the CSTF2 ortholog Pti1 (Nedea et al. 2003). The APT complex has a distinctive activity consisting of the cleavage-independent termination of noncoding transcription (Casañal et al. 2017; Lidschreiber et al. 2018). While all APT complex subunits but one have mammalian orthologs, a similar complex has not been identified in mammals.
The data reported here show that PNUTS and Symplekin act synergistically with Restrictor to dampen processive extragenic transcription, suggesting that enforcement of termination at extragenic transcription units involves convergent mechanisms.
Results
Stringent proximity labeling indicates contiguity among ZC3H4, Symplekin, and PNUTS
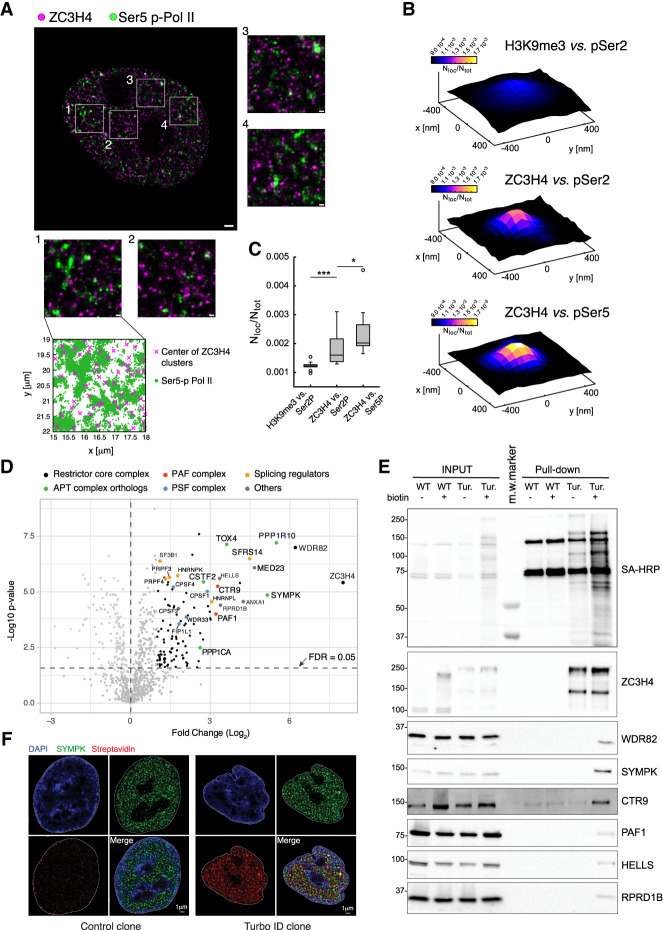
Genomic analyses indicate a frequent overlap in the distribution of ZC3H4 and Pol II, particularly in proximity to transcription start sites (TSSs) (Austenaa et al. 2021; Estell et al. 2023; Hughes et al. 2023). To directly measure proximity between ZC3H4 and Pol II, we used superresolution microscopy. We used two-color dSTORM (direct stochastic optical reconstruction microscopy) to generate images at an average final resolution of 15 nm and measured colocalization of ZC3H4 and either phospho-Ser5 or phospho-Ser2 Pol II CTDs. Antibodies against an abundant heterochromatic mark (H3K9me3) were used as an internal control to measure distances relative to a nuclear feature not associated with active transcription.
ZC3H4 formed well-defined nuclear clusters of 80- to 100-nm average size (Fig. 1A) that in ∼30% of instances colocalized with Pol II. While colocalization was detected with both pSer5 and pSer2 Pol II, ZC3H4 was significantly more associated with pSer5 Pol II (Fig. 1B,C), suggesting that, in keeping with previous data (Austenaa et al. 2015, 2021), initiating and early-elongating Pol II complexes may represent the main target of ZC3H4-driven transcription termination.
Figure 1.
A high-stringency ZC3H4 proximity interactome. (A) dSTORM image of a representative nucleus of HeLa cells stained with anti-ZC3H4 (magenta) and anti-pSer5 CTD Pol II (green) antibodies. Magnified images corresponding to the four boxed regions are also shown. At the bottom, the distribution of the pSer5 Pol II localizations relative to the centers of ZC3H4 clusters in one area is shown. Scale bars indicate 1 µm for the whole nuclei and 200 nm for the boxed regions. (B) Partial colocalization analysis of pSer2/pSer5 Pol II localizations around ZC3H4 clusters in STORM data. pSer2 and pSer5 Pol II localizations were mapped in a 30-nm grid, and the ZC3H4 clusters where the pSer2 or pSer5 Pol II density was higher than its own average density (i.e., ∼30% of clusters) were identified. The cumulative pSer2 and pSer5 Pol II nuclear map around ZC3H4 clusters was computed, the signal was normalized by the total map intensity, and the value of at least 15 nuclei was averaged. Position (0,0) in the plot represents the center of ZC3H4 clusters, and the intensity is the pSer2 or pSer5 Pol II normalized signal, which is proportional to the probability of finding the signal in that spatial position. (C) Box plots represent the colocalization strength; i.e., the average normalized signal of the pSer2 or pSer5 Pol II within 30 nm from the ZC3H4 cluster center. Every point represents the colocalization strength for a single nuclear map. As a control, we performed the same analysis for H3K9me3 localizations around pSer2 Pol II clusters. (D) Volcano plot showing the proteins identified by proximity labeling in HCT116 cells carrying a ZC3H4-Turbo-ID fusion gene. Biotin was added for 10 min to biotin-depleted cells, followed by the preparation of total lysates, streptavidin pull-down, and mass spectrometry. The identified proteins are shown according to their relative abundance (log2 fold change) and statistical significance in ZC3H4-TurboID cells versus biotin-treated wild-type cells. Proteins belonging to different complexes are indicated with different colors. n = 5 independent biological replicates. Significant hits are indicated as black dots. (E) Western blot analysis of selected proteins identified by proximity labeling in ZC3H4-TurboID cells. The top panel shows the detection of the pulled-down material by Western blot with streptavidin-HRP (SA-HRP). Input lysates and pulled-down proteins are shown. Molecular weight markers (in kilodaltons) are indicated at the left. Note the presence of a few biotinylated proteins in WT cells, which represent the three known endogenously biotinylated, long-half-life carboxylases (pyruvate carboxylase, 130 kDa; 3-methylcrotonyl CoA carboxylase, 75 kDa; and propionyl CoA carboxylase, 72 kDa) (Chandler and Ballard 1986; Ahmed et al. 2014). (F) DeepSIM superresolution images of representative nuclei from wild-type cells (control) and a ZC3H4-TurboID knock-in HCT116 clone (Turbo-ID) stained with an anti-Symplekin antibody (green) and streptavidin (red). Nuclear counterstaining with DAPI and a merged image are also shown.
We next sought to identify proteins involved in Restrictor-mediated termination. Immunoprecipitation experiments coupled to mass spectrometry revealed that the only stochiometric core components of Restrictor are WDR82 and ZC3H4 (our unpublished data), a finding in line with quantitative WDR82 IP mass spectrometry data reported by others (van Nuland et al. 2013).
Therefore, we resorted to in vivo proximity labeling, which captures stable and transient interactions as well as contiguity in a 1- to 10-nm radius surrounding the protein of interest fused to a promiscuous biotin ligase (Supplemental Fig. S1A; Qin et al. 2021). To maximize the stringency of this assay, we first knocked in a high-activity biotin ligase, Turbo-ID (Branon et al. 2018), into the ZC3H4 locus to generate a fusion with the ZC3H4 C terminus that is expressed at endogenous levels (Supplemental Fig. S1B). Next, we cultured HCT116 cells in biotin-deficient medium and dialyzed serum for 5 d in order to nearly completely deplete endogenous biotin. Finally, we exposed cells to biotin for a very short time (10 min), thus minimizing nonspecific labeling events caused by low-frequency contacts. DeepSIM (structured illumination microscopy), a superresolution microscopy technique with a resolution of ∼100 nm, showed that ZC3H4-TurboID generated very localized biotinylation halos with a high overlap with ZC3H4, indicating the confined deposition of biotin in the immediate surroundings of ZC3H4 (Supplemental Fig. S1C).
By purifying biotinylated proteins with streptavidin-coated paramagnetic beads followed by mass spectrometry analysis on five biological replicates, we obtained a high-quality data set with only 14 proteins (including ZC3H4) significantly enriched more than eightfold over the control and additional less—but still significantly—enriched interactors (Fig. 1D). As expected, the most enriched proteins were ZC3H4 and WDR82, immediately followed by Symplekin and PNUTS (PPP1R10). The proximity between ZC3H4 and PNUTS was consistent with an independent report (Estell et al. 2023). An organized list of selected proximal proteins divided by the corresponding complex or biological function is shown in Table 1, while the complete list is in Supplemental Table S1. As discussed below, nearly all of the highly enriched proteins are either known Pol II interactors (such as PAF1 and RPRD1B) or proteins binding to nascent RNAs (including several splicing factors).
Table 1.
Partial list of ZC3H4-proximal proteins identified by streptavidin pull-down in ZC3H4-TurboID cells
Validation of selected interactions by Western blot on lysates purified by streptavidin beads revealed that among all proximal proteins tested, only Symplekin was highly enriched relative to the input material (Fig. 1E), which hinted at a close proximity of ZC3H4 with a large share of Symplekin molecules in cells, a finding corroborated by DeepSIM analysis (Fig. 1F). This was not the case for WDR82, which, based on quantitative mass spectrometry data (Andersen et al. 2013), is much more abundantly associated with the PNUTS–PP1–TOX4 complex (Lee et al. 2010) and the SET1 H3K4 methyltransferase complexes (Lee and Skalnik 2008; Wu et al. 2008) than with ZC3H4.
While both Symplekin and its direct interactor, CstF2 (Takagaki and Manley 2000), were among the top ZC3H4-proximal proteins (Fig. 1D; Table 1), no proximity was detected with CPSF3, which is shared between CPSF and the histone cleavage complex (Fig. 1D; Supplemental Table S1; Sullivan et al. 2009). Hence, ZC3H4-proximal Symplekin appears to be not associated with complexes endowed with RNA cleavage activity. Along the same line, no component of the histone cleavage complex was retrieved (Supplemental Table S1).
However, we detected proximity with the entire PSF (polyadenylation specificity factor) subcomplex (Boreikaite and Passmore 2023), which is responsible for PAS recognition and is composed of CPSF1, FIP1L1, CSPF4, and WDR33 (the latter two representing the PAS-binding subunits) (Chan et al. 2014; Schönemann et al. 2014).
Additional proximal proteins of potential relevance included components of the PAF (polymerase-associated factor) complex (Chen et al. 2018), several regulators of splicing, and the CTD-associated protein RPRD1B, which may control CTD phosphorylation by recruiting the RPAP2 phosphatase (Ni et al. 2014). Previous data also reported an association between ZC3H4–WDR82 and casein kinase 2 (CK2), which was shown to phosphorylate the SPT5 N terminus and inhibit its activity (Park et al. 2022). In our proximity data, the α subunit of CK2 was indeed retrieved, but its enrichment was of low magnitude and below statistical significance (Supplemental Table S1).
Overall, stringent proximity labeling identified a close contiguity of ZC3H4 with Symplekin and PNUTS without evidence of proximity to the CPSF core cleavage complex or histone cleavage complex components.
Synergy between Symplekin and Restrictor
To determine whether proximity between ZC3H4 and Symplekin has functional relevance, we generated RNA-seq data sets from control and ZC3H4- and/or Symplekin-depleted cells pulsed for 10 min with 4-thiouridine (4sU) to label nascent transcripts. To this aim, we first inserted an auxin-inducible degron (mini-AID) (Nishimura et al. 2009) at the 3′ end of the ZC3H4 gene in HCT116 cells, which allowed us to obtain the efficient depletion of ZC3H4 after a 4-h incubation with auxin (Supplemental Fig. S2A,B). A partial reduction of Symplekin mRNA levels (∼60%) was instead obtained by siRNA-mediated depletion in untreated or auxin-treated cells (Supplemental Fig. S2C).
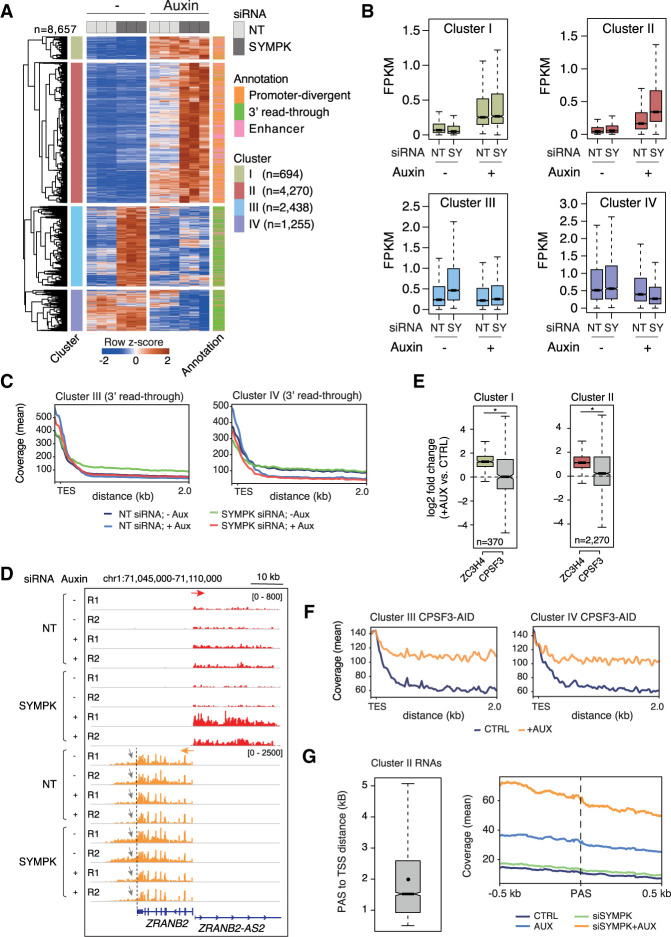
Analysis of the differentially expressed extragenic transcripts (n = 8657) in the 4sU RNA-seq data revealed four well-defined clusters (Fig. 2A, clusters I–IV from top to bottom; Supplemental Table S2).
Figure 2.
Transcriptional effects of combined ZC3H4 depletion and partial siRNA-mediated Symplekin depletion. (A) Heat map showing extragenic transcripts differentially expressed in auxin-treated (ZC3H4-depleted) and/or SYMPK siRNA transduced HCT116 cells (FDR ≤ 0.05, |log2FC| ≥ 0.8, FPKM mean value ≥ 0.1). Row Z-scores are shown. Four clusters (I–IV from top to bottom) were identified that were characterized by different responses to ZC3H4 and/or Symplekin depletion. Each transcript was assigned to the nearest enhancer, promoter, or gene 3′ end (annotation flags at the right). Data are from n = 3 independent biological replicates. (B) Signal intensity of differentially expressed extragenic transcripts in clusters I–IV in the different experimental conditions used. The median value is indicated by a horizontal black line. Boxes show values between the first and third quartiles. The top and bottom whiskers show the smallest and the highest values, respectively. Outliers are not shown. The notches correspond to ∼95% confidence interval for the median. (C) Metaplots of transcripts in clusters III and IV, showing the effects of different perturbations on readthrough transcription at genes’ 3′ ends. (TES) Transcription end site. (D) A representative genomic region showing the effects of auxin-driven ZC3H4 depletion and partial siRNA-mediated Symplekin depletion on the ZRANB2 gene promoter-divergent transcription (ZRANB2-AS2) and on readthrough transcription (indicated by gray arrows) at the 3′ end of the ZRANB2 gene. The red and orange arrows indicate the plus and the minus strand signals, respectively. (E) Effects of auxin-induced CPSF3 depletion on promoter-divergent transcripts assigned to clusters I and II. Log2FC in auxin-treated versus untreated cells is shown. The effects of ZC3H4 depletion are shown for comparison. Statistical significance was assessed by a Wilcoxon paired test (cluster I, P = 2.732294 × 10−19; cluster II, P = 4.253707 × 10−65). (F) Metaplot showing the effects of auxin-induced CPSF3 depletion on readthrough transcripts assigned to clusters III and IV. Data from TES + 2 kb are shown; one bin = 20 bp. (G) Abundance of nascent extragenic transcripts synergistically regulated by ZC3H4 and Symplekin in a window of ±500 nt before and after the first PAS. We considered transcripts belonging to cluster II overlapping with a PRO-seq peak (n = 3090), which was used for the accurate identification of the main TSS. After removing transcripts with the first PAS at <500 nt from the TSS, we retained n = 2335 transcripts and measured the distance between their TSSs and the first PAS. (Left) The distance between the start of the PRO-seq peaks and the PAS is shown in a box plot. The mean (2002 nt) is shown as a dot. Median = 1522 nt. (Right) The coverage around the PAS (±500 nt) is shown in a metaplot.
Both clusters I and II included transcripts that were not affected by the depletion of Symplekin but were up-regulated upon ZC3H4 depletion. While in cluster I (n = 694) the codepletion of Symplekin was devoid of additional effects, transcripts in cluster II (n = 4270) were superinduced upon codepletion of ZC3H4 and Symplekin (Fig. 2A,B).
Cluster III (n = 2438) included transcripts strongly up-regulated by Symplekin depletion and unaffected by the depletion of ZC3H4. Notably, the codepletion of ZC3H4 almost completely reversed the effects of Symplekin depletion (Fig. 2A,B).
Finally, cluster IV (n = 1255) included extragenic transcripts whose abundance was again strongly reduced by the codepletion of ZC3H4.
Remarkably, the genomic distribution of transcripts in clusters I and II was completely different from that of transcripts in clusters III and IV (Fig. 2A, annotation column). Indeed, while transcripts up-regulated upon ZC3H4 depletion (clusters I and II) were associated with enhancers and promoter-divergent transcriptional units, transcripts in clusters III and IV were almost entirely accounted for by transcriptional readthrough beyond the 3′ ends of genes.
These data suggest that the transcriptional readthrough caused by reduced RNA cleavage efficiency in cells with reduced Symplekin levels (cluster III) or by constitutively inefficient transcript processing (cluster IV) was reverted by ZC3H4 depletion (Fig. 2C). At the same time, however, deregulated extragenic transcription caused by ZC3H4 depletion was further enhanced by the depletion of Symplekin (cluster II), suggesting its requirement for efficient transcription termination at these sites.
The locus containing the ZRANB2 gene and its divergent noncoding transcriptional unit (ZRANB2-AS2) typifies these findings (Fig. 2D). The ZRANB2 gene showed constitutive 3′ readthrough transcription (Fig. 2D, gray arrows) that was enhanced upon Symplekin depletion and greatly reduced when ZC3H4 was codepleted. However, transcription of the promoter-divergent noncoding unit ZRANB2-AS2 was mildly increased by ZC3H4 depletion and greatly up-regulated upon codepletion of Symplekin and ZC3H4 (Fig. 2D). Additional representative snapshots are shown in Supplemental Figure S2D.
It is important to stress that whereas ZC3H4 depletion upon auxin treatment was substantial, it was nevertheless incomplete. Hence, the transcripts most affected by ZC3H4 depletion and not showing synergistic regulation by Symplekin codepletion (cluster I) may simply represent those with the highest affinity for (and/or susceptibility to) Restrictor, rather than being subjected to regulatory mechanisms different from those in place at the transcription units that displayed synergistic regulation.
Extragenic transcripts in clusters I and II were not affected by auxin-induced CSPF3 depletion (Fig. 2E). Instead, transcripts in clusters III and IV, which were generated by transcriptional readthrough at genes’ 3′ ends, were very strongly up-regulated by CPSF3 depletion (Fig. 2F), suggesting that, in line with analyses reported by others (Rouvière et al. 2023), ZC3H4-mediated termination may not require nascent transcript cleavage by CPSF. The lack of a role of Integrator-mediated nascent transcript cleavage in termination at transcription units suppressed by Restrictor has already been shown (Austenaa et al. 2021).
A hypothetical model to explain these findings posits that, in the presence of a limited nuclear pool, Symplekin co-option by ZC3H4 to terminate transcription at extragenic sites could reduce its availability for CPSF-mediated termination at the 3′ ends of genes. When ZC3H4 is codepleted, reduced recruitment of Symplekin to extragenic sites would increase its availability for usage by CPSF, thus reducing transcriptional readthrough.
This model implies that the synergy provided by Symplekin in the suppression of extragenic transcription may be independent of its role in the CPSF. However, an alternative possibility is that reduced CPSF activity in Symplekin-depleted cells results in defective cleavage of extragenic transcripts derepressed in ZC3H4-deficient cells, thus impairing termination and enabling further elongation after the PASs. In this scenario, Symplekin codepletion would result in the generation of longer transcripts but not in their increased abundance before the first PAS emerges from the elongating Pol II. We set out to discriminate between these two models by determining the relationship between nascent extragenic transcript abundance in cells depleted of ZC3H4 and Symplekin and the first PAS in the same transcription units. If Symplekin codepletion impacted extragenic transcription because of defective PAS recognition and cleavage by the CPSF, increased transcript abundance would be detectable only after the PAS sequence. When taking into consideration transcripts in cluster II, which were synergistically affected by the codepletion of ZC3H4 and Symplekin (Fig. 2A), the median distance between the TSS (based on PRO-seq data) and the PAS was 1522 nt (Fig. 2G, left). When considering a 1-kb window surrounding the PAS, it became clear that the increased abundance of the nascent transcripts caused by codepletion of Symplekin and ZC3H4 was maximal before the PAS, indicating that Symplekin loss enhanced Pol II processivity before PAS encounter (Fig. 2G, right). This interpretation is in line with the common observation that when Symplekin was depleted in cells lacking ZC3H4, the additive effects primarily involved boosting the levels of extragenic transcripts from their very beginning, with this increase in transcript abundance in some, but not all, cases being associated with an extension in their length (for instance, see snapshots in Fig. 2D; Supplemental Fig. S2D). Possibly because of the efficient incorporation of Symplekin in the histone cleavage complex (Marzluff and Koreski 2017), no strong effects on termination at replication-dependent histone genes were observed in these conditions of partial Symplekin depletion.
Hence, these data indicate that the effects of Symplekin in enforcing extragenic transcription in collaboration with Restrictor include a CPSF-independent component.
Effects of a near-total depletion of Symplekin on mutual regulation with ZC3H4
The Symplekin competition model implies some testable predictions. The first one is that a complete depletion of Symplekin would prevent the rescue of the transcriptional readthrough that we observed when ZC3H4 was codepleted. This is because in the presence of no or minimal residual amounts of Symplekin, the depletion of ZC3H4 cannot increase its availability for usage by the CPSF.
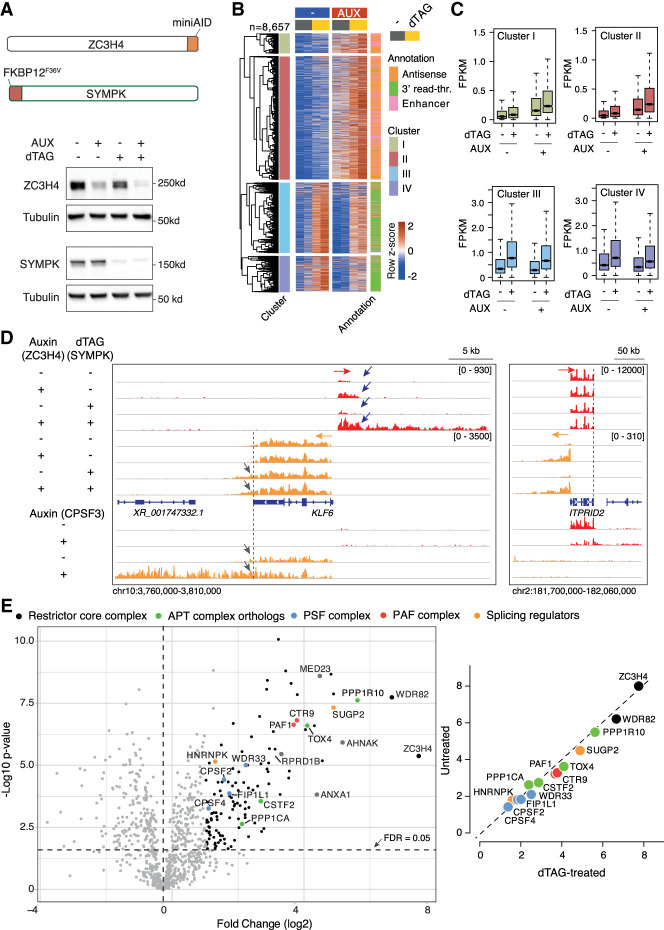
To test this prediction, we generated double-mutant HCT116 cells in which the dTAG-regulated FKBP12F36V degron (Nabet et al. 2018) flanked by an HA tag was inserted in frame at the 5′ end of the SYMPK gene in cells carrying the ZC3H4-AID fusion gene (Supplemental Fig. S3A).
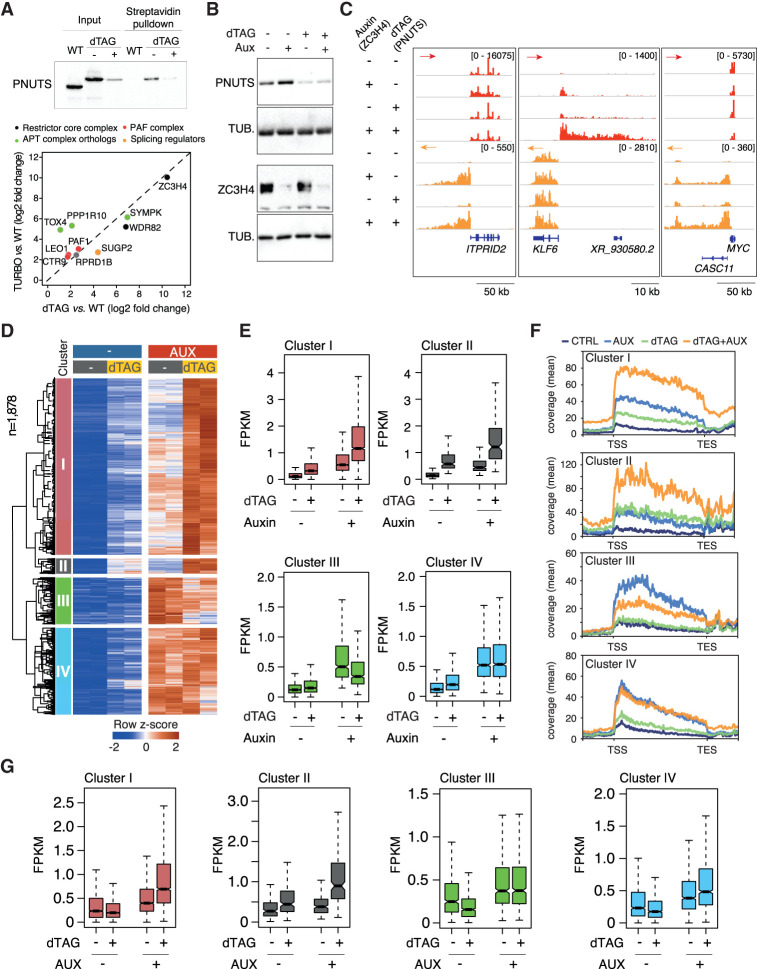
dTAG treatment of these double-knock-in cells generated an extensive depletion of Symplekin, which was associated with the efficient depletion of ZC3H4 when cells were cotreated with auxin (Fig. 3A). Using these cells, we first analyzed the Symplekin interactome by carrying out an IP-MS experiment in untreated versus dTAG-treated cells. Symplekin coprecipitated its direct interactor, CstF2, and all the subunits of the CPSF complex but neither ZC3H4 nor other ZC3H4-proximal proteins, indicating that proximity was not determined by stable interactions (Supplemental Fig. S3B; Supplemental Table S3). Importantly, an MS analysis of the proteome of Symplekin-depleted cells showed no detectable reduction in the abundance of any of its interactors and specifically the core cleavage complex components CPSF3 and CPSF2 (Supplemental Fig. S3C,D; Supplemental Table S4), which were instead shown to require Symplekin for their stability in D. melanogaster cells (Sullivan et al. 2009).
Figure 3.
Transcriptional effects of dTAG-induced depletion of Symplekin in combination with auxin-driven ZC3H4 depletion. (A, top) Schematic representation of the ZC3H4 and SYMPK degron knock-in alleles (not to scale). (Bottom) Depletion of ZC3H4 and Symplekin in double-knock-in HCT116 cells upon treatment with 100 µM auxin and/or 500 nM dTAG for 24 h. Molecular weight markers are shown at the right. (B) Heat map showing extragenic transcripts differentially expressed in auxin-treated (ZC3H4-depleted) and/or dTAG-treated (Symplekin-depleted) HCT116 cells. Row Z-scores are shown. The four clusters (I–IV) indicated in the heat map correspond to those in Figure 2A. Data are from n = 2 independent biological replicates. (C) Signal intensity of differentially expressed extragenic transcripts in clusters I–IV in the indicated experimental conditions. The median value is indicated by a horizontal black line. Boxes show values between the first and third quartiles. The top and bottom whiskers show the smallest and the highest values, respectively. Outliers are not shown. The notches correspond to ∼95% confidence interval for the median. (D) Two representative genomic regions showing the effects of individual or combined degron-driven degradation of Symplekin (dTAG) and ZC3H4 (auxin) on promoter-divergent (plus strand; red) and 3′ readthrough transcription (minus strand; orange). For comparison, the effects of auxin-mediated CPSF3 depletion in the same regions are shown at the bottom. (E, left) Volcano plot showing the proteins identified by proximity labeling upon dTAG-driven Symplekin depletion in HCT116 cells carrying a ZC3H4-Turbo ID fusion gene. The identified proteins are shown according to their relative abundance (log2 fold change) and statistical significance in ZC3H4-Turbo-ID cells versus biotin-treated control cells. n = 5 independent biological replicates. Significant hits are indicated as black dots. Selected statistically significant proteins are indicated in different colors depending on the functional group or complex to which they belong. (Right) Correlation between protein enrichment in mass spectrometry experiments in untreated versus dTAG-treated cells. Data are shown as fold enrichment (log2) in ZC3H4-Turbo-ID cells relative to wild-type cells.
Next, we generated 4sU-seq data sets to determine the impact of individual or combined Symplekin and ZC3H4 depletions (Supplemental Table S2). In keeping with its requirement for the structural and functional integrity of the histone cleavage complex, Symplekin depletion caused strong readthrough transcription at replication-dependent histone genes, with no detectable effects of the codepletion of ZC3H4 (Supplemental Fig. S3D,E).
Similar to what was observed with the partial siRNA-mediated depletion of Symplekin, the more extensive reduction of Symplekin abundance caused by dTAG robustly increased extragenic transcription induced by ZC3H4 depletion at enhancers and promoter-divergent transcription units (Fig. 3B,C, clusters I and II). Instead, differently from what was observed in cells with higher residual Symplekin levels, the more extensive depletion of the same protein obtained by dTAG-mediated degradation greatly attenuated or abrogated the rescue effect of ZC3H4 depletion on readthrough transcription (cf. clusters III and IV in Fig. 2A,B vs. Fig. 3B,C). An exemplary case is provided by the KLF6 gene, in which the depletion of Symplekin caused readthrough transcription at the gene termination site, with this effect being unaffected by the codepletion of ZC3H4 (gray arrows in Fig. 3D, left). Instead, dTAG-induced degradation of Symplekin greatly enhanced the increase in KLF6 gene promoter-divergent transcription caused by ZC3H4 depletion, bringing about both an early increase in the nascent RNA abundance and an extension beyond the 3′ end of the transcript generated in ZC3H4-depleted cells (blue arrows in Fig. 3D, left).
Importantly, although widespread, these synergistic effects were not universal, as exemplified by the case of promoter-divergent transcription at ITPRID2, which was strongly up-regulated by ZC3H4 depletion and not further affected by the codepletion of Symplekin (Fig. 3D, right). The effects of CPSF3 depletion are shown for comparison. Additional representative snapshots are shown in Supplemental Figure S3F.
Given that Symplekin is considered to operate as an adaptor protein, we tested the effects of its depletion on the ZC3H4 proximity interactome. We inserted the FKBP12F36V degron in the SYMPK gene in HCT116 cells carrying the ZC3H4-Turbo-ID fusion gene and generated streptavidin pull-down mass spectrometry data from control or Symplekin-depleted cells exposed to biotin for a 10-min labeling time (Supplemental Fig. S3G,H). Notably, Symplekin depletion did not significantly affect ZC3H4 proximity to any of its main interactors (Fig. 3E; Supplemental Table S5), suggesting that in this context it does not exert the role of an adapter.
Overall, the comparison between the effects of partial and near-complete depletion of Symplekin suggests the existence of a limited nuclear pool of this protein whose co-option by Restrictor limits its availability for CPSF-mediated termination.
Effects of ZC3H4 overexpression on Symplekin-dependent transcription termination
A second prediction of the Symplekin competition model is that by titrating Symplekin away, the overexpression of ZC3H4 should reduce the efficiency of other Symplekin-containing termination complexes.
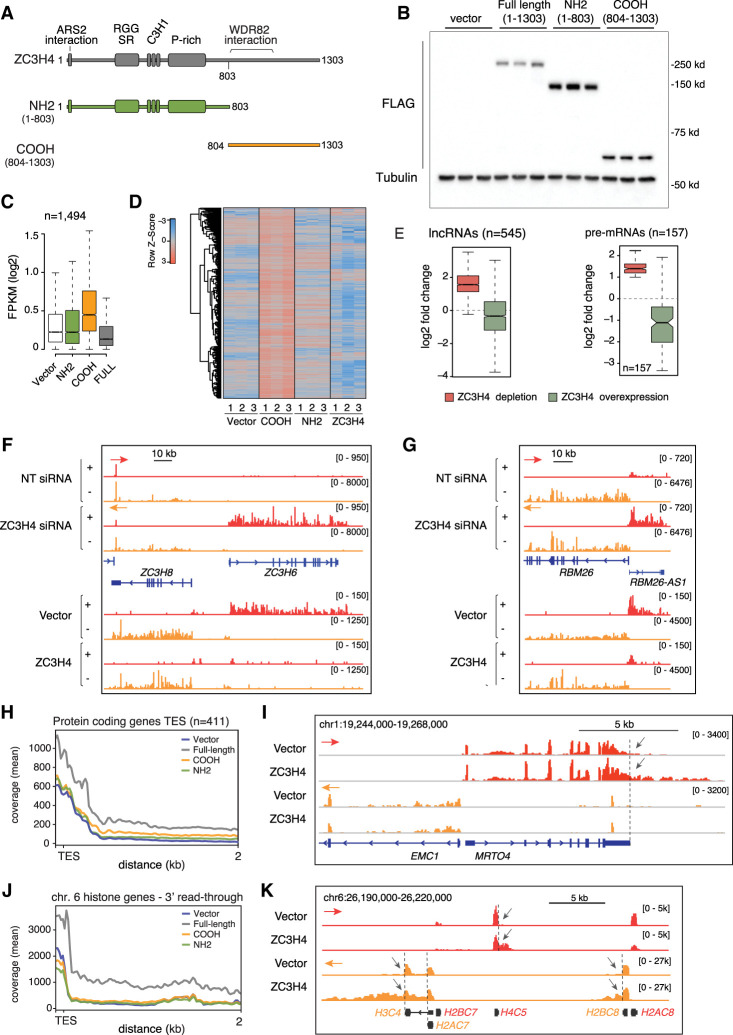
To test this possibility, we generated HeLa clones with a single genomic integration of tetracycline-inducible, FLAG epitope-tagged versions of full-length (amino acids 1–1303), N-terminal (amino acids 1–803), and C-terminal (amino acids 804–1303) ZC3H4 (Fig. 4A,B; Supplemental Fig. S4A). The N-terminal domain included a short ARS2 interaction motif (Rouvière et al. 2023), a highly conserved RGG- and SR-rich region that distantly resembled both the RG repeats and the SR dipeptide repeats present in RNA-binding and splicing proteins (Godin and Varani 2007; Thandapani et al. 2013; Howard and Sanford 2015), and finally, three C3H1-type zinc fingers. The C-terminal fragment contained the WDR82-binding region (amino acids 804–1057) and when overexpressed acted as a dominant-negative mutant (Austenaa et al. 2021), possibly by sequestering WDR82 in the cytoplasm where this mutant accumulates due to the lack of the N-terminal nuclear localization signals (Supplemental Fig. S4A).
Figure 4.
Transcription termination defects caused by the overexpression of ZC3H4. (A) Schematic representation of ZC3H4 and its N-terminal and C-terminal fragments overexpressed in HeLa Flp-In-TREx cells. (B) Anti-FLAG Western blot showing the expression of ZC3H4 and its N-terminal and C-terminal fragments in three independent clones each. Cells were treated with 100 ng/mL doxycycline for 48 h before harvesting. (C) Effects of the overexpression of ZC3H4 and its N-terminal and C-terminal fragments on a set of transcripts previously reported to be up-regulated upon ZC3H4 depletion in HeLa cells (Austenaa et al. 2021). (D) The same data as in C are shown as a heat map. Row Z-scores are shown. (E) Box plot showing the effects of ZC3H4 depletion or overexpression on a set of n = 568 extragenic transcripts (left) and n = 157 pre-mRNAs (right) up-regulated upon ZC3H4 depletion, as well as detectable expression in control, nondepleted cells. The median value is indicated by a horizontal black line. Boxes show values between the first and third quartiles. The top and bottom whiskers show the smallest and the highest values, respectively. Outliers are not shown. The notches correspond to ∼95% confidence interval for the median. (F,G) Representative genomic regions showing the effects of ZC3H4 depletion or overexpression on the ZC3H6 gene transcript (F) and the RBM26-AS1 transcript (G). Note that in the bottom part of the two panels, the tracks were rescaled to show the basal expression of these transcripts. The red and orange arrows indicate the plus and minus strand signals, respectively. (H) Metaplot showing readthrough transcription at genes’ 3′ ends in HeLa cells overexpressing ZC3H4. Data are shown up to 2 kb after the TES; one bin = 20 bp. (I) Representative genomic region showing readthrough transcription (gray arrows) at the MRTO4 gene in cells overexpressing ZC3H4. (J) Metaplot showing readthrough transcription at replication-dependent histone genes in HeLa cells overexpressing ZC3H4. (K) Representative genomic snapshot showing readthrough transcription (gray arrows) at a group of histone genes in the chromosome 6 cluster.
We generated 4sU RNA-seq data sets to measure the effects of ZC3H4 overexpression on a previously defined gold standard set of n = 1494 extragenic RNAs up-regulated upon ZC3H4 depletion in HeLa cells (Austenaa et al. 2021). Consistent with previous findings (Austenaa et al. 2021), the ZC3H4 C-terminal fragment strongly increased the expression of ZC3H4-suppressed transcripts (Fig. 4C,D; Supplemental Table S6), while the N-terminal fragment, in spite of its strong expression (Fig. 4B) and proper nuclear localization (Supplemental Fig. S4A), as well as the presence of the ARS2-interacting motif, was completely devoid of functional effects on these transcripts (Fig. 4C,D; Supplemental Table S6). Interestingly, the overexpression of the full-length ZC3H4 caused a significant down-regulation of a fraction of the same transcripts that were up-regulated when ZC3H4 was depleted, suggesting that the nuclear abundance of ZC3H4 is partially limiting (Fig. 4C,D). An issue with these data is that many RNAs induced when ZC3H4 is depleted have very low, if any, detectable expression in control conditions. Therefore, we focused on a subset (n = 545) of transcripts that was characterized by detectable basal expression. Overexpression of ZC3H4 caused a mild yet significant reduction of their expression (Fig. 4D,E) and similarly reduced the expression of a small number of coding genes that were induced by ZC3H4 depletion (Fig. 4E), as exemplified by the cases of ZC3H6, a ZC3H4 paralog (Fig. 4G), and the RBM26 gene promoter-divergent noncoding transcript RBM26-AS1 (Fig. 4F, note the rescaling of the tracks in the bottom half of the figure, which allowed us to visualize repression of the low basal levels of these transcripts).
These data suggest that, coherently with its autoregulation (Austenaa et al. 2021), the expression levels of ZC3H4 are finely tuned in cells.
In addition to the down-regulation of ZC3H4-suppressed transcription units, ZC3H4 overexpression caused three distinct groups of termination defects, resulting in the increased transcription of extragenic sequences.
First, it caused moderate-level transcription readthrough at the 3′ end of hundreds of protein-coding genes (Fig. 4H,I; Supplemental Table S7), a phenotype consistent with the partially impaired CPSF function caused by Symplekin depletion (Fig. 2A–D).
Second, it induced transcription readthrough beyond the 3′ end of a subset of replication-dependent histone genes (Fig. 4J) that are terminated by the histone cleavage complex, as shown by a representative region of the main histone gene cluster on chromosome 6 (Fig. 4K). Replication-independent histone genes, which are terminated via canonical CPSF-dependent mechanisms, were in general unaffected, as exemplified by the H2AJ gene that is in close proximity to the replication-dependent H4-16 gene on chromosome 12, as well as by the replication-independent H3-3A gene (Supplemental Fig. S4B).
ZC3H4 overexpression unveils a role for Symplekin in the repression of endogenous retroviral elements
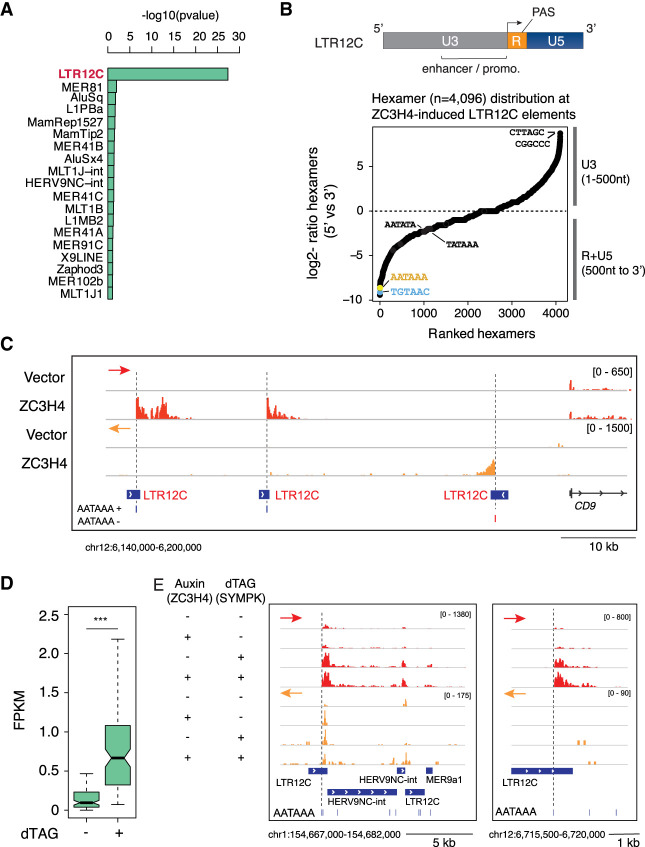
The third group of termination defects caused by ZC3H4 overexpression consisted of the increased transcription of a defined group of transposable elements (TEs). Because of the action of purifying selection inside genes, noncoding transcription units are relatively enriched for TE-derived sequences compared with genic sequences (Kelley and Rinn 2012; Kapusta et al. 2013). When considering the entire set of noncoding transcripts expressed in HeLa cells, we found that extragenic transcripts expressed at higher levels because of the termination defects caused by ZC3H4 overexpression showed a significant enrichment of sequences derived from endogenous retroviruses (ERVs), in particular the LTR12C elements, which belong to the HERV9 group (Fig. 5A; Kapusta et al. 2013).
Figure 5.
Derepression of solo LTR-driven transcription caused by Symplekin titration or depletion. (A) Overrepresented subfamilies of transposable elements enriched in extragenic transcripts up-regulated in response to ZC3H4 overexpression relative to unaffected extragenic transcripts of similar length. The statistical significance was assessed by a Fisher test (“alternative = greater,” significance for P < 0.01). Subfamilies were ranked based on the most significant −log10 transformed P-value. (B, top) Schematic structure of solo LTR12C elements containing the U3 enhancer–promoter element, the R region with the TSS (arrow) and the PAS, and the U5 element. (Bottom) Relative distribution of all possible hexamers in the 5′ versus 3′ fragments of LTR12C elements. (C) A representative genomic region containing three LTR12C elements that, upon ZC3H4 overexpression in HeLa Flp-In TREx cells, generated transcripts extending into the adjacent genomic regions. The position of the canonical PAS in the R region of each LTR is shown. Arrowheads indicate transcription start sites. The red and orange arrows indicate the plus and minus strand signals, respectively. (D) RNA-seq read counts at LTR12C elements induced upon dTAG-driven Symplekin depletion in HCT116 cells. (***) P = 9.630827 × 10−20 by Wilcoxon paired test. (E) Representative genomic regions showing LTR12C elements induced upon Symplekin depletion in HCT116 cells. The red and orange arrows indicate the plus and minus strand signals, respectively.
Most ERV-derived sequences in the human genome are solitary (or solo) LTRs; namely, ERVs that, due to homologous recombination between the identical 5′ and 3′ LTRs of the integrated proviruses, lost the retroviral genes normally comprised between them (Stoye 2012; Babaian and Mager 2016). Since LTRs are endowed with cis-regulatory activity, they were frequently exapted as promoters or enhancers, with the LTR12 elements and in particular the LTR12C representing a prominent subgroup in terms of frequency of exaptation (Kelley and Rinn 2012; Kapusta et al. 2013) and strength of promoter activity (van Arensbergen et al. 2017).
LTRs can be divided into two unique regions (U3 and U5) and a repeated (R) region in between them (Fig. 5B). U3 contains transcription factor binding sites and has strong promoter activity (van Arensbergen et al. 2017), with transcription initiating at the beginning of the R region. Notably, LTRs contain a PAS very close to the transcription start site, followed by downstream GU sequences (15–60 nt from the PAS) (Guntaka 1993), with an organization resembling the pattern of PAS and GU-rich CstF2-bound DSEs (downstream sequence elements) in canonical genes (MacDonald et al. 1994; Yao et al. 2012). The combination of PAS and downstream GU sequences results in efficient termination of LTR-initiated transcription (Guntaka 1993). The analysis of the relative distribution of all possible hexamers (n = 4096) at the 5′ portion (including the U3) versus the 3′ part (R and U5) of the LTR12C elements induced upon ZC3H4 overexpression showed a strong skew of the canonical PAS and TGT elements in the 3′ end, a finding consistent with the typical organization of these repeats (Fig. 5B).
Strikingly, increased LTR12C activity in HeLa cells overexpressing ZC3H4 precisely started downstream from the PAS, as shown by a representative genomic region containing three induced LTR12C elements (Fig. 5C), thus indicating relief from PAS-mediated termination.
Another consequence of defective termination at LTRs is the activation of otherwise silent promoter elements. For instance, an LTR12C element inside the DHRS2 gene acquired promoter activity and generated spliced DHRS2 transcripts in ZC3H4-overexpressing cells (Supplemental Fig. S5). In the case of the GBP2 and RPL3L genes, reactivation of LTR12 elements upstream of the conventional gene promoter resulted in the generation of spliced transcripts containing extra sequences at their 5′ ends (Supplemental Fig. S5).
If this group of termination defects was caused by Symplekin titration, then Symplekin depletion is expected to cause a similar phenotype. Indeed, analysis of the dTAG-Symplekin data generated in HCT116 cells confirmed the existence of termination defects at a subset of LTR12C elements (Fig. 5D,E), although the strength of the effect was not as strong as the one caused by ZC3H4 overexpression.
PNUTS synergizes with ZC3H4 at a subset of Restrictor-suppressed transcription units
We next analyzed whether depletion of PNUTS affected ZC3H4-repressed extragenic transcription in a manner similar to that caused by the depletion of Symplekin. We surmised that proximity of both proteins to ZC3H4 may be indicative of functional partnership even if—differently from their yeast orthologs, Pta1 and Ref2—Symplekin and PNUTS are not physically associated into a stable multimolecular complex analogous to the APT.
Importantly, PNUTS has recently been proposed to be required for ZC3H4-driven transcription termination (Estell et al. 2023), implying the existence of a linear pathway linking PNUTS and ZC3H4. In this model, the codepletion of ZC3H4 and PNUTS would not enhance the effects caused by the individual depletion of the two proteins. However, this model was based on the lack of additive effects of the codepletion of ZC3H4 (by siRNA) and PNUTS (by degron-mediated degradation) on the expression level of a few representative RNAs selected among those maximally induced upon Restrictor depletion (Estell et al. 2023).
In order to obtain a comprehensive view of the relationships between PNUTS and ZC3H4, we carried out two groups of experiments. First, we inserted the dTAG-regulated FKBP12F36V degron into both alleles of the PNUTS gene in a HCT116 clone carrying the ZC3H4-Turbo-ID fusion gene. Then, we generated streptavidin pull-down MS data in control and PNUTS-depleted cells exposed to biotin for a short (10-min) labeling time. PNUTS depletion brought about a reduction in the retrieval of TOX4 (Fig. 6A; Supplemental Table S8), indicating that proximity between ZC3H4 and PNUTS involved the whole WDR82–PNUTS–TOX4 complex. Reciprocally, we detected a moderate increase in the retrieval of WDR82, possibly because its release from PNUTS/TOX4, which represents the most abundant WDR82-containing complex in cells (van Nuland et al. 2013), increased its availability for binding to ZC3H4. Other ZC3H4-proximal proteins, such as the components of the PAF complex, were unaffected (Fig. 6A).
Figure 6.
Effects of individual and combined depletions of ZC3H4 and PNUTS on extragenic transcription termination. (A) Effects of PNUTS depletion on the proximity interactome of ZC3H4. A dTAG-regulated degron was inserted into both PPP1R10 alleles in ZC3H4-Turbo-ID cells, and the proximity interactome of ZC3H4 was determined before and after PNUTS depletion. (Top) Western blot showing the depletion of degron-containing PNUTS upon dTAG treatment. (Bottom) Volcano plot showing selected proteins identified by proximity labeling upon dTAG-driven PNUTS depletion. The complete list of proteins is in Supplemental Table S8. (B) Levels of PNUTS and ZC3H4 after individual or combined degron-mediated depletion were analyzed by Western blot. Tubulin was used as a loading control. (C) Representative 4sU RNA-seq snapshots showing extragenic transcription changes induced by individual and combined depletion of ZC3H4 and PNUTS. The red and orange arrows indicate the plus and minus strand signals, respectively. (D) Clustered extragenic transcripts differentially expressed in the indicated depletions. The heat map includes promoter-divergent and enhancer-associated RNAs. Row Z-scores are shown. All transcripts in the heat map are significant in both clones in the ZC3H4-depleted condition versus control (FDR ≤ 0.01 and log2 transformed fold change of ≥2). The complete list of differentially expressed transcripts is in Supplemental Table S9. Cluster I: n = 1009, cluster II: n = 89, cluster III: n = 271, cluster IV: n = 509. Data are from n = 2 biological replicates of a single clone. (E) Levels (FPKM) of differentially expressed transcripts in the four clusters. The median value is indicated by a horizontal black line. Boxes show values between the first and third quartiles. The bottom and top whiskers show the smallest and highest values, respectively. Outliers are not shown. The notches correspond to ∼95% confidence interval for the median. (F) The metaplot shows the replicate average of the 4sU-seq signal in the four clusters of extragenic transcription units in C. (TSS) Transcription start site, (TES) transcription end site. (G) The levels of transcripts in clusters I–IV were measured in data sets obtained from cells depleted of Symplekin (by dTAG-mediated degradation) and/or ZC3H4 (by auxin-mediated degradation).
Second, to obtain a transcriptome-wide view of the effects of the codepletion of ZC3H4 and PNUTS, we generated double-degron PNUTS-dTAG and ZC3H4-AID clones (Fig. 6B) and used 4sU RNA-seq to measure nascent transcription upon individual or combined depletion of these two proteins.
Initial exploration of the data suggested the existence of two broad classes of responses. Some transcription units (exemplified by the promoter-divergent transcription at the ITPRID2 gene) showed very strong induction upon ZC3H4 depletion and marginal, if any, effects of PNUTS depletion both alone and in combination with ZC3H4 (Fig. 6C). However, for other transcription units, individual depletion of both ZC3H4 and PNUTS caused a detectable increase in transcription that was of much lower magnitude than that caused by their codepletion, as shown in the case of the MYC and even more so the KLF6 gene promoter-divergent transcription units (Fig. 6C). These data hinted at the possibility that at some extragenic transcription units efficiently terminated by ZC3H4, PNUTS may not be involved at all, while at others ZC3H4 and PNUTS may work in a collaborative manner, thus explaining the superinduction observed upon their codepletion.
This model was consistent with the identification of two clusters of differentially expressed extragenic transcripts (clusters I and II) (Fig. 6D–F; Supplemental Table S9) showing strongly enhanced induction upon ZC3H4 and PNUTS codepletion compared with individual depletions. The two clusters differed mainly because of the relative magnitude of the effects of individual depletions: Cluster I (n = 1009) was composed of transcripts affected more by ZC3H4 depletion than by PNUTS depletion, while the much smaller cluster II (n = 89) included transcripts more sensitive to the depletion of PNUTS than that of ZC3H4. Cluster III and IV contained transcripts not (or only marginally) affected by PNUTS depletion, strongly up-regulated by the depletion of ZC3H4, and not further induced in codepleted cells (Fig. 6D–F). Although some variability among the double-knock-in cell clones was observed, qualitatively similar effects were consistently observed in all clones analyzed (Supplemental Fig. S6).
Finally, if PNUTS and Symplekin impinge on the same extragenic termination pathway, it is expected that the effects of the depletion of Symplekin on extragenic transcription should be similar to those caused by the depletion of PNUTS. Therefore, we analyzed the levels of expression of the transcripts affected by PNUTS depletion in cells depleted of Symplekin and/or ZC3H4. The two main sets of effects observed upon codepletion of ZC3H4 and PNUTS were recapitulated by the codepletion of Symplekin. Specifically, in clusters I and II, codepletion of Symplekin and ZC3H4 was strongly synergistic, while no synergy at all or only a small increase was observed in clusters III and IV, respectively, when Symplekin was codepleted with ZC3H4 (Fig. 6G). Overall, these data indicate that extragenic transcripts jointly repressed by Restrictor and PNUTS are also convergently regulated by Restrictor and Symplekin.
Discussion
This study aimed to identify players and mechanisms involved in Restrictor-mediated extragenic transcription termination. The ZC3H4 proximity interactome reported here includes (1) proteins associated with Pol II, such as PAF1 complex components (interacting with the body of Pol II), RPRD1B, and PNUTS complex components (all interacting with the CTD), and (2) proteins interacting with the nascent RNA such as the PAS-binding PSF complex, CstF2 (which recognizes the GU-rich DSE), and several splicing factors. In addition to data showing the extensive overlap of the genomic distribution of ZC3H4 and the initiating Pol II (Austenaa et al. 2021; Hughes et al. 2023), the results shown here point to the recruitment of Restrictor to initiating or early-elongating Pol II complexes, which may occur by multiple concurring mechanisms, including WDR82-mediated interactions with the Ser5-p CTD of Pol II, ARS2-mediated recognition of the 5′CAP on the nascent RNA (Rouvière et al. 2023), and possibly direct recognition of extragenic transcripts by ZC3H4. The likely relevance of WDR82-mediated tethering to the Ser5-p CTD (Lee and Skalnik 2008; Park et al. 2022) is suggested by the efficient pull-down of ZC3H4 by Ser5-phoshorylated CTD peptide baits (Ebmeier et al. 2017) and indirectly by the lack of effects of the overexpression of the N-terminal ZC3H4 fragment.
Aside from WDR82, the only stable and stoichiometric ZC3H4 interactor, the two most enriched proteins in our data set were Symplekin and PNUTS. However, we could not coprecipitate PNUTS with ZC3H4 in standard immunoprecipitation experiments (Austenaa et al. 2021), and a Symplekin IP-MS experiment showed no detectable interactions with either PNUTS or ZC3H4. Along the same line, a PNUTS–PP1 complex purified from cells did not include ZC3H4 as a stoichiometric component (Lee et al. 2010). Overall, proximity and functional interactions of PNUTS and Symplekin with Restrictor in the absence of robust binding are compatible with different scenarios, including (1) direct low-affinity interactions disrupted in immunoprecipitation experiments; (2) the requirement of multimolecular assemblies that, similarly to splicing complexes, can only occur on RNA templates and thus are disrupted upon cell lysis (Wahl et al. 2009); and (3) WDR82-mediated interactions of distinct Restrictor and PNUTS–PP1 complexes with adjacent Ser5 phosphorylated repeats on the same Pol II CTD tail.
While a previous RT-PCR analysis of the abundance of a few extragenic transcripts in cells depleted of PNUTS and ZC3H4 showed a lack of synergy (Estell et al. 2023), our nascent transcriptome data indicate a more complex scenario. Schematically, we identified a group of ZC3H4-suppressed transcripts (exemplified by the ITPRID2 promoter-divergent RNA) that, although very sensitive to ZC3H4 depletion, were not affected by PNUTS depletion and showed no superinduction upon ZC3H4–PNUTS codepletion. At these extragenic regions, termination appears to be executed by Restrictor without any involvement whatsoever of PNUTS, which in turn implies the essential concept that ZC3H4 does not absolutely require PNUTS–PP1 to execute termination.
Instead, at a second group of extragenic regions, the codepletion of PNUTS and ZC3H4 was moderately to strongly additive or synergistic. In this context, synergy may be interpreted according to two alternative mechanistic frameworks that cannot be distinguished based on current data. First, the two pathways may be completely distinct and control the elongating Pol II complexes via different mechanisms. Alternatively, at some transcription units, the activity of PNUTS–PP1 may increase the ability of Restrictor to execute termination, possibly by reducing Pol II speed (Cortazar et al. 2019) and making it more termination-prone. This scenario would reconcile the observed synergy with the notion that at some transcription units, PNUTS–PP1 and Restrictor may work in a linear pathway. The WDR82–PNUTS complex has also been reported to dephosphorylate Pol II at CTD Ser5 and to promote its proteasomal degradation, eventually reducing its residence time on chromatin (Landsverk et al. 2020). However, in the absence of a clear understanding of how Restrictor induces Pol II termination and what the critical PNUTS–PP1 targets in this context are, the mechanistic bases of the observed synergy remain speculative.
In this context, it is remarkable that in addition to being in close spatial proximity to ZC3H4, Symplekin and PNUTS similarly synergized with ZC3H4 to enforce extragenic transcription termination, although the participation of Symplekin in multiple complexes complicates the interpretation of these data. Indeed, a critical issue is whether the additive or synergistic effects of Symplekin depletion in ZC3H4-deficient cells could simply be explained by the reduced activity of the CPSF complex on the extragenic transcripts. If this was the case, codepletion of Symplekin would exclusively lead to the production of elongated transcripts without increasing their abundance before the first PAS emerges from the transcribing Pol II. We posit that our data are in keeping with the notion that a large share of the effects of Symplekin in this context are not accounted for by its inclusion in the CPSF. First, the additive or synergistic effects of Symplekin depletion on transcript abundance were clearly detectable between the transcription start site and the first PAS. This finding indicates that in ZC3H4-depleted cells, the codepletion of Symplekin increased processivity of early-elongating extragenic Pol II independently of the PAS sequence. Second, ZC3H4 proximity labeling data robustly identified Symplekin; its direct interactor, CstF2; and all components of the PAS-recognizing PSF complex (CPSF1, WDR33, CPSF4, and FIP1L1) but not CPSF3, suggesting that whereas Symplekin coimmunoprecipitated the whole CPSF complex, it may be recruited in proximity to ZC3H4 independently of it. Third, differently from what was observed in Drosophila cells (Sullivan et al. 2009), the depletion of Symplekin did not reduce the abundance of any of the CPSF subunits, thus ruling out any impact on CPSF abundance. Finally, while Symplekin depletion caused very strong and pervasive termination defects at histone genes, it had a comparatively smaller effect on 3′ readthrough at protein-coding genes, indicating a considerably higher Symplekin requirement for the activity of the histone cleavage complex compared with that of the CPSF. Consistent with this interpretation, while the Symplekin N-terminal domain is absolutely required for the endonucleolytic activity of the histone cleavage complex (Sun et al. 2020), it is dispensable for that of the CPSF (Boreikaite et al. 2022). Hence, the precise functional role of Symplekin in the mammalian CPSF complex is still unclear. Nonetheless, it is indisputable that at least at a subset of extragenic transcription units, Symplekin depletion could also enable Pol II elongation beyond PAS sites due to a decrease in CPSF-dependent cleavage.
Based on the data reported here, it is tempting to speculate that Symplekin and PNUTS–PP1, while not stably interacting within a multiprotein complex, may act as a functionally integrated unit with biological roles in part analogous to those of the yeast APT complex. APT, which includes Pta1, the WDR82 ortholog Swd2, the PNUTS ortholog Ref2, and the phosphatases Glc7 (PP1) and Ssu72 (Nedea et al. 2003), exists as both a distinct module of the CPSF that is endowed with phosphatase activity and contributes to PAS-dependent termination and a stand-alone complex (which includes Syc1, an additional subunit with no clear mammalian ortholog) with a specific role in the control of cleavage-independent extragenic transcription termination (Casañal et al. 2017; Lidschreiber et al. 2018). Hence, similarities may exist in the different roles of Symplekin/Pta1 in PAS-dependent and -independent termination in mammals and yeast. Another enzymatic component of APT is the Ser5 Pol II phosphatase Ssu72 (Lidschreiber et al. 2018). In mammals, SSU72 is in close proximity to Symplekin (St-Denis et al. 2016) and can be cocrystallized with it (Xiang et al. 2010). However, based on current data sets (St-Denis et al. 2016), there is no evidence supporting Ssu72 proximity to PNUTS–PP1 in mammalian cells, suggesting once again that during evolution, the APT complex may have been dispersed into multiple separate complexes and isolated subunits. Consistent with this interpretation, we did not find any enrichment of SSU72 in our data set. However, the role of Ssu72 in extragenic transcription termination cannot be ruled out and warrants further investigation.
Our depletion and overexpression data also point to the existence of a limited nuclear pool of Symplekin, whose recruitment by Restrictor restrains its availability to the CPSF and the histone cleavage complex. Indeed, ZC3H4 depletion rescued the transcription readthrough occurring at the 3′ end of protein-coding genes as a consequence of a partial Symplekin depletion. Notably, siRNA-mediated, partial Symplekin depletion did not impact termination at histone genes either because the high efficiency of Symplekin incorporation into the histone cleavage complex neutralized the consequences of a partial reduction of its abundance or because the residual Symplekin sufficed to activate its cleavage activity.
Along the same line, in the ZC3H4 overexpression experiments, we found a comparatively higher impact of the increased ZC3H4 abundance on termination at histone genes and LTR12C repeats relative to the 3′ ends of all other genes. Hypothesizing that overexpressed ZC3H4 may titrate Symplekin away from the CPSF and the histone cleavage complex, these data may be interpreted as the consequence of the different Symplekin requirements for their endonucleolytic activity. Moreover, it has been reported that direct interaction of Symplekin with CstF2 is essential for histone RNA 3′ end processing but not similarly important for pre-mRNA cleavage and processing (Ruepp et al. 2011). Finally, UG-rich sequences (and in particular UGU) recognized by CstF2 (Martin et al. 2012) are located 15–60 nt downstream from the PAS in ERV LTRs and are relevant for termination (Guntaka 1993). Therefore, these data are consistent with the hypothesis that ZC3H4 overexpression preferentially titrates away a Symplekin–CstF2 complex particularly relevant for histone RNA 3′ end processing and LTR12C termination.
Overall, the notion of a nuclear pool of Symplekin–CstF2 core complexes that can be dynamically recruited to different termination machineries may be more correct than that of highly stable pre-existing multimolecular complexes.
ZC3H4 orthologs are highly conserved in metazoan evolution, from worms to humans. Indeed, the discovery of Restrictor was preceded by the identification of the role of Suppressor of sable, the D. melanogaster ZC3H4 ortholog, in termination of transcription of transposable elements inserted into coding transcripts (Fridell et al. 1990), suggesting that this mechanism may have originally evolved to counteract detrimental effects of active transposons and then adapted to attenuate extragenic transcription. In addition, in keeping with the functional conservation of this pathway in metazoans, several screenings for factors involved in transcription termination in Caenorhabditis elegans highlighted the role of Zfp3, the worm ortholog of ZC3H4, in transcription termination in different contexts and experimental designs (Cui et al. 2008; LaBella et al. 2020). Strikingly, in these screens, Zfp3 scored positive together with other proteins that extensively overlap our proximity interactome, including PAF and PSF complex components (but not the cleavage subunit CPSF3) (Cui et al. 2008; LaBella et al. 2020) as well as Pta1/Symplekin and Cstf2 (Cui et al. 2008). Moreover, Zfp3 scored as a possible binding partner of Cstf2 in a two-hybrid screen in C. elegans (Li et al. 2004), again pointing to regulatory networks involved in termination that are conserved from worms and flies to humans.
At this stage, two major issues remain unaddressed: notably, the mechanism linking Restrictor to the execution of transcription termination and a complete understanding of the bases for its preferential action at extragenic transcription units. Data reported in this and other studies (Austenaa et al. 2021; Estell et al. 2023; Rouvière et al. 2023) indicate that Restrictor drives termination in a transcript cleavage-independent manner, implying fundamentally different mechanisms compared with the other known pathways controlling transcription termination in mammals.
Materials and methods
Cell culture and reagents
HCT-116 (from ATCC), ZC3H4-mAID, ZC3H4-Turbo-ID, ZC3H4-mAID:SYMPK-dTAG, ZC3H4-mAID:PNUTS-dTAG, ZC3H4-Turbo-ID:SYMPK-dTAG, and ZC3H4-Turbo-ID:PNUTS-dTAG HCT-116 cells were cultured in McCoy's 5A medium modified with 10% South American serum, 1% penicillin/streptomycin (Sigma P4333), and 1% L-Glutamax (Gibco 35050061). HeLa TREx Flp-In cell lines were cultured in DMEM with 10% tetracycline-free serum (Euroclone ECS0182L), 1% penicillin/streptomycin, 250 µg/mL hygromycin B (Invitrogen 10687010), and 1 µg/mL blasticidin (Gibco R210-01). Cell lines were authenticated by the Tissue Culture Facility of the European Institute of Oncology using the GenePrint10 system (Promega) and routinely screened for mycoplasma contamination. For the streptavidin pull-down experiment, ZC3H4-Turbo-ID and wild-type HCT-116 cells were maintained in DMEM with 10% dialyzed fetal calf serum (Thermo Fisher 26400044) and 1% penicillin/streptomycin for 5 d. At day 5, 50 µM biotin (Sigma B4639) was added to the cells for 10 min. Auxin (Sigma I5148) was added at a final concentration of 100 µM for 1, 2, 4, or 24 h as indicated. dTAG-13 (Tocris Bioscience 6605) was added at a final concentration of 500 nM for 4 or 24 h. Tetracycline hydrochloride (Sigma T7660) was added for 48 h at a final concentration of 100 ng/mL. 4sU (Santa Cruz Biotechnologies sc-204628A) was added to the medium at a final concentration of 500 μM for 10 min for HCT-116 cells or 300 µM for 45 min for HeLa TREx Flp-In cells before harvesting.
Direct stochastic optical reconstruction microscopy (dSTORM) imaging
HeLa cells were seeded on 35-mm glass-bottom dishes (MatTek P35G-1.5-10-C) 24 h before being processed for indirect immunofluorescence. Briefly, HeLa cells were fixed with PFA for 10 min, permeabilized with 0.1% Triton X-100, blocked, and stained with 2 µg/mL anti- ZC3H4 (Sigma HPA040934), 4 µg/mL Pol II-Ser2p (Diagenode C15200005), 4 µg/mL Pol II-Ser5p (Diagenode C15200007), and 10 µg/mL H3K9me3 (Abcam ab8898). Secondary antibodies used were AF 647 antirabbit (Thermo Fisher A31573) and AF 568 antimouse (Thermo Fisher A10037). Samples were then postfixed in 2% PFA for 5 min before being storing in PBS at 4°C until imaging. Streptavidin-conjugated fluorescent nanodiamonds (FNDs) measuring 40 nm in size (Adamas Nanotechnologies) (Pelicci et al. 2022) were added to the cells at a final concentration of 20 mg/mL in PBS, and the cells were incubated for 1 h at room temperature before imaging. dSTORM imaging was carried out using a Nikon N-Storm microscope that was equipped with a 1.49 NA CFI Apochromat TIRF objective. The Alexa fluor 647 (AF647) and Alexa fluor 568 (AF568) dyes were excited with 647-nm and 561-nm lasers, respectively, in HILO (highly inclined and laminated optical sheet) mode (Tokunaga et al. 2008). Whenever necessary, the 405-nm laser (activation laser) was used for reactivating the fluorophores into a fluorescent state.
Starting from the AF647, 15,000 images per channel were acquired using an Orca-Flash4.0 sCMOS camera (Hamamatsu Photonics K.K.) with an exposure time of 20 msec, a pixel size of 161.5 nm, and a field of view of 256 × 256 pixels. Throughout dSTORM imaging, cells were immersed in imaging buffer (100 mM MEA, 1% glucose, 560 mg/mL glucose oxidase, 34 mg/mL catalase in PBS). The localization of single molecules was performed in three steps in Fiji/ImageJ using the ThunderSTORM plug-in (Ovesný et al. 2014) and custom-made macros.
Fluorophore localization: The AF647 and AF568 stacks were preprocessed with a Wavelet filter (B-Spline), the approximate positions of molecules were identified with the local maximum method (connectivity: eight neighborhoods, threshold: two times the STD [Wave.F1]), and the subpixel localization of molecules was performed with a Gaussian fit and the maximum likelihood method to determine the Gaussian parameters.
Drift correction: The AF647 and AF568 localization tables obtained in step 1 were concatenated, the image was reconstructed and visualized in Fiji, and a small region of interest (ROI) with the FND signal was cropped. The drift was estimated tracking the FNDs (fiducial markers), and the trajectory of the relative sample drift was saved and then applied to the whole concatenated data set.
Localization filtering: Localizations with an uncertainty >30 nm were discarded, and localizations <20 nm between each other were merged.
For each nucleus, four regions of 9 mm2 were chosen and processed for the partial colocalization analysis.
Partial colocalization analysis of ZC3H4 and Pol II in two-color STORM
Partial colocalization analysis was performed in sequential steps as follows: (1) ZC3H4 cluster analysis to extract cluster centers, (2) calculation of the percentage of ZC3H4 clusters colocalizing with Pol II, and (3) measurement of the colocalization strength between ZC3H4 clusters and Pol II.
ZC3H4 cluster analysis was performed by combining the SuperStructure method and DBSCAN (Ester et al. 1996; Marenda et al. 2021). By visually investigating SuperStructure cluster connectivity curves, we identified the clustering length scale and therefore the value of the spatial parameter R to fix for a cluster analysis with DBSCAN. We then performed the cluster analysis for that R by fixing the second DBSCAN parameter; i.e., the minimum number of localizations to define a cluster (Nmin = 0). Finally, the investigation of the cluster size distribution and the visual inspection of the data allowed us to define a cutoff to remove clusters below a certain size. In the case of ZC3H4, we identified R = 16 nm and a minimum cluster size equal to 60 localizations.
In order to perform a partial colocalization analysis, we initially classified each ZC3H4 cluster as either colocalizing or not colocalizing with Pol II. In particular, a cluster was considered as colocalizing if the density of the Pol II signal within 100 nm from the ZC3H4 cluster center was over the Pol II average density. From this result, we extracted the percentage of colocalizing and not colocalizing clusters.
In the final step, we only considered colocalizing clusters to measure the colocalization strength of ZC3H4 clusters with the Pol II signal. In particular, we calculated an intensity map of Pol II localizations in the nucleus by coarse-graining the localizations in pixels 30 nm in size. By doing so, we obtained a 2D matrix in which each 30-nm bin contained the sum of the Pol II localizations. We then associated to each ZC3H4 cluster center the respective Pol II intensity map by considering the 1-µm × 1-µm 2D map around the cluster center. A cumulative intensity of Pol II around ZC3H4 clusters was calculated for each nucleus and then normalized by the total intensity of Pol II. Such a value is assumed to be proportional to the probability of finding Pol II at each pixel. We therefore created a 3D map with dimensions 1 µm × 1 µm divided into 30-nm bins for the probability of finding Pol II at a certain distance in the X and Y directions from ZC3H4 clusters. We then averaged over these maps across at least n = 15 nuclei over three replicates. Finally, we quantified the colocalization strength by averaging for each nucleus the Pol II intensity value in the ranges −30 nm < X < 30 nm and −30 nm < Y < 30 nm (i.e., in the neighborhood of the cluster center).
This procedure was performed to evaluate the partial colocalization of ZC3H4 clusters with Pol II phosphorylated at Ser2 (Pol II Ser2p) and at Ser5 (Pol II Ser5p).
As a negative control, we evaluated the partial colocalization of Pol II Ser2p clusters with the H3K9me3 signal. In this case, we identified R = 12 nm and a minimum cluster size of 60 localizations as clustering parameters.
dSTORM image reconstruction
dSTORM images were reconstructed by a Gaussian rendering with the standard deviation set to 20 nm. Only the nuclear signal is shown. In the figures, the scale bars indicate 1 µm for the whole nuclei and 200 nm for the boxed regions.
DeepSIM imaging
WT HCT-116 and ZC3H4-Turbo-ID cells were grown on glass coverslips, and 50 µM biotin was added for 10 min before PFA fixation. Cells were then permeabilized with 0.1% Triton X-100, blocked, and stained with DAPI, 2 µg/mL anti-ZC3H4 (Sigma HPA040934) or 10 µg/mL anti-SYMPK (Fortis Life Sciences A301-465A) antibodies, and antistreptavidin (1:200; Invitrogen S11226). Cells were imaged by a DeepSIM superresolution module (CrestOptics S.p.A.) mounted on an Eclipse Ti2 fluorescence microscope (Nikon Europe B.V.) equipped with solid-state lasers (Celesta light engine, Lumencor), a sCMOS camera (Kinetix, Teledyne Photometrics), and a 100×/1.49 NA oil immersion objective lens. The standard DeepSIM imaging mask was used to produce the multispot lattice pattern to excite the fluorophores, requiring the acquisition of 37 images per channel to obtain the superresolution image with a pixel size of 35 nm.
Confocal imaging
Dual-color immunofluorescence and confocal analysis were performed on HeLa TREx Flp-In cells grown on glass coverslips. Briefly, PFA-fixed cells were permeabilized with 0.1% Triton X-100, blocked, and stained overnight with an anti-FLAG antibody at 10 µg/mL. Nuclei were counterstained with DAPI, and samples were mounted with glycerol. Images were acquired by a LeicaSP8 AOBS confocal microscope with an HC PL APO CS2 63×/1.40 oil immersion objective lens (Leica Microsystems GmbH) and a pixel size of 80 nm. A single confocal section is shown.
Plasmid cloning and engineered cell lines
ZC3H4-mAID, ZC3H4-Turbo-ID, ZC3H4-mAID:SYMPK-dTAG, ZC3H4-mAID:PNUTS-dTAG, ZC3H4-Turbo-ID:SYMPK-dTAG, and ZC3H4-Turbo-ID:PNUTS-dTAG HCT-116 cells were generated using CRISPR/Cas9-mediated homology-directed repair (HDR). The following sgRNAs were designed using Benchling and cloned into px330A-1x2 (Addgene 58766) and px330 (Addgene 42230): ZC3H4 For_sgHD2 (5′-CACCGTAGTGTCCAGCCAGAGCTG-3′), ZC3H4 Rev_sgHD2 (5′-AAACCAGCTCTGGCTGGACACTAC-3′), SYMPK For_sg (5′-CACCGTGGAGACAGCGTCACCCGT-3′), SYMPK Rev_sg (5′-AAACACGGGTGACGCTGTCTCCAC-3′), PNUTS For_sg (5′-CACCGATGGTGGTTTCTATGGTAAG-3′), and PNUTS Rev_sg (5′-AAACCTTACCATAGAAACCACCATC-3′). To generate ZC3H4-mAID cells, we designed a pUC57-based donor vector (Biomatik) containing the ZC3H4 left homology arm (chromosome 19: 47,066,359–47,066,658), a GGGS spacer, a 3xFlag mini-AID insert, a P2A sequence, the sequence for hygromycin resistance, and the ZC3H4 right homology arm (chromosome 19: 47,066,056–47,066,355).
HCT-116 cells were cotransfected with the ZC3H4 sgRNA-expressing px300A-1x2 plasmid and the donor plasmid. After transfection and hygromycin selection at 250 µg/mL for 5 d, single cells were seeded in 96-well plates by limiting dilution and expanded. Clones containing the 3xFlag mini-AID-P2A-hygromycin cassette were screened by PCR using the following primers: For_HDc2 (5′-ACCTTCCCAGACACCAACTG-3′), Rev_HDc2 (5′-ATTTCGGCTCCAACAATGTC-3′), For_WT check (5′-AGGGTGAGGAGCGTTCAATA-3′), and Rev_WT check (5′-AACAGCCAGAGACAGGGAAG-3′). Positive clones were validated by Western blot.
Selected homozygous clones were then infected with the pRRL-SFFV-OsTir1_3xMyc-tag-T2A-eBFP2 plasmid (Muhar et al. 2018) in order to stably express the Oryza sativa TIR protein OsTir1. After BFP sorting, cells were seeded in 96-well plates by limiting dilution and expanded. A single homozygous clone was selected for the subsequent experiments.
The Turbo-ID pUC57-based donor vector was generated by substituting the 3xFlag mini-AID sequence from pUC57-ZC3H4 AID plasmid with a Turbo-ID insert obtained from the C1(1–29)-TurboID-V5_pCDNA3 plasmid (Addgene 107173) using the following primers: TurboID_FOR (5′-TCGGGAGGTGGATCGAAAGACAATACTGTGCCTCTGAAGCTGATCGC-3′) and TurboID_REV (5′-AGTAGCTCCGCTTCCCTTTTCGGCAGACCGCAGAC-3′).
As before, HCT-116 WT cells were cotransfected with the ZC3H4 sgRNA-expressing px300A-1x2 plasmid and the Turbo-ID donor plasmid. After transfection and hygromycin selection at 250 µg/mL for 5 d, single cells were seeded in 96-well plates by limiting dilution and expanded. Clones containing the Turbo-ID-P2A-hygromycin cassette were screened using PCR (same primers as above) and validated by Western blot. A single homozygous clone was selected for the subsequent experiments.
Finally, to generate the double-degron ZC3H4-mAID:SYMPK-dTAG cell line and the ZC3H4-Turbo-ID:SYMPK-dTAG HCT-116 cells, a dTAG-SYMPK pUC57-based donor vector containing the SYMPK left homology arm (chromosome 19: 45,854,495–45,854,795), the puromycin resistance cassette, the 2xHA dTAG insert, and the SYMPK right homology arm (chromosome 19: 45,854,192–45,854,492) was assembled using Gibson assembly (New England Biolabs). The dTAG insert (FKBP_F36V) was derived from pCRIS-PITChv2-Puro-dTAG (BRD4; Addgene 91793). In this case, ZC3H4-mAID and ZC3H4-Turbo-ID cells were cotransfected with the SYMPK sgRNA-expressing px330 plasmid and the dTAG-SYMPK donor plasmid. After transfection and puromycin selection at 1 µg/mL for 3 d, single cells were seeded in 96-well plates by limiting dilution and expanded. The same strategy was used to generate the double-degron ZC3H4-mAID:PNUTS-dTAG and the ZC3H4-Turbo-ID:PNUTS-dTAG HCT-116 cells with the subsequent PNUTS left (chromosome 6 :30,609,944–30,610,244) and right (chromosome 6: 30,609,641–30,609,941) homology arms. Homozygous clones containing the puromycin-P2A-2xHA-dTAG cassette were screened by PCR using the following primers: F_HALSy (5′-GTTCATGTGGCCCATCGTTCAGC-3′), R_SYPCR1 (5′-CCCTCACCTGTTTGAGCACT-3′), F_HA_5′_PNUTS (5′-TAGGAAGGATGCTGCTGGGA-3′), and R_3′_HA_PNUTS 5′-CAGTTTCCATTATGGTCAGAA-3′). Positive clones were then validated by Western blot.
To generate the HeLa TREx Flp-In cells expressing full-length ZC3H4 (1–1303 amino acids), the N-terminal fragment (1–803 amino acids), or the C-terminal portion (804–1303 amino acids), different pcDNA5/FRT/TO expression plasmids (Thermo Fisher Scientific) were generated and used together with the pOG44 vector to cotransfect HeLa Flp-In TREx cells generated by the Tissue Culture Facility of the European Institute of Oncology. After transfection and 250 µg/mL hygromycin and 1 µg/mL blasticidin selections, single cells were seeded in 96-well plates by limiting dilution and expanded. Clones were screened by Western blot, and three different clones for each construct were selected for subsequent experiments.
Mass spectrometry
WT and ZC3H4-Turbo-ID HCT-116 cells were harvested, washed twice with ice-cold PBS, and centrifuged at 1500 rpm for 5 min. Cell pellets were lysed with RIPA buffer (150 mM NaCl, 1% NP40, 0.1% Na deoxycholate, 0.1% SDS, 50 mM Tris HCl at pH 8.0) containing a mixture of protease inhibitors (Complete EDTA-free, Roche 5056489001) and 1 mM PMSF, incubated by rotation for 30 min at 4°C, and centrifuged in a microfuge at the maximum speed for 10 min at 4°C. The supernatant was collected and quantified, and 1 mg was used for streptavidin pull-down. Fifty microliters of Dynabeads MyOne streptavidin C1 beads (Invitrogen 65002) was then added to each sample and incubated overnight at 4°C with rotation. Beads were then washed using RIPA buffer (seven 5-min washes followed by three 15-min washes) and then 1× PBS (two 5-min washes). For the HA-IP, control and SYMPK-depleted cells were harvested, washed twice with ice-cold PBS, and centrifuged at 1500 rpm for 5 min. Cell pellets were lysed with buffer B (250 mM NaCl, 50 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 0.5 mM EGTA, 0.2% NP40), incubated for 30 min at 4°C with rotation, and centrifuged in a microfuge at the maximum speed for 10 min at 4°C. One milligram of total lysate was incubated for 3 h at 4°C with Dynabeads M-280 sheep antimouse IgG (Thermo Fisher 11202D) coupled with 5 µg of HA antibody (Invitrogen 26183). Immunoprecipitates were washed extensively using buffer B and eluted in 0.1 M glycine (pH 3.0), followed by an overnight acetone precipitation step. For the MS proteome experiment, 150 µg of whole-cell extracts from control and SYMPK-depleted cells was precipitated overnight using acetone. The whole-cell extract pellets, the HA-IP-derived pellets, and the beads from the streptavidin pull-downs were then prepared for MS using the iST sample preparation kit (Preomics 00027) following the manufacturer's specifications.
In all cases, peptides derived from on-bead digestion were then eluted in 200 µL of buffer B (80% ACN, 0.1% formic acid [FA]). Samples were dried using a speed-vac concentrator (Eppendorf), and the volume of the eluates was adjusted to 5 µL with 1% TFA and then analyzed by LC-MS/MS using an Easy-nLC 1200 (Thermo Fisher Scientific LC140) connected to a Q-Exactive HF (Thermo Fisher Scientific) through a nanoelectrospray ion source (EasySpray, Thermo Fisher Scientific). The nano-LC system was operated in one-column setup with an EasySpray Pepmap RSLC C18 column (Thermo Fisher Scientific) kept at a constant temperature of 45°C. Solvent A was 0.1% formic acid (FA), and solvent B was 0.1% FA in 80% ACN. Samples were injected in aqueous 1% (TFA) at a constant pressure of 980 bars. Peptides were separated with a gradient of 5%–20% solvent B over 47 min followed by a gradient of 20%–30% for 10 min and 30%–65% over 5 min at a flow rate of 300 nL/min. The MS instrument was operated in the data-dependent acquisition (DDA) mode. The 15 most intense peptide ions with charge states ≥2 were sequentially isolated to a target value of 3 × 106 and fragmented in the high-collision dissociation (HCD) cell using a normalized collision energy setting of 27%. MS spectra were detected in the Orbitrap using a resolution R = 60,000 at m/z 200 within an m/z range corresponding to 375–1650. The maximum allowed ion accumulation times were 20 msec for full scans and 90 msec for MSMS. The dynamic exclusion time was set to 15 sec.
IP mass spectrometry data processing and statistics
Acquired raw data were analyzed using MaxQuant version 1.6.2.3 integrated with the Andromeda search engine (Cox et al. 2011). False discovery rate (FDR) was set to a maximum of 1% at both the peptide and protein levels. Carbamidomethylcysteine and methionine oxidation were selected as fixed and variable modifications, respectively. The UniProt human Fasta database UP000005640 (85,678 entries) was specified for the search. The LFQ intensity calculation and the “match between run” (MBR) function were enabled (Cox et al. 2014). The “protein groups” output file from MaxQuant was first inspected using Perseus software to filter out common contaminant proteins (keratin, desmoplakin, plectin, and actin) and false positive hits (reverse hits from the decoy database), and four out five valid values of data completeness in at least one group (WT or ZC3H4) were required. After data filtering, the missing values were then replaced by random numbers drawn from a normal distribution under the assumption that these values would belong to the low-intensity spectrum of the distribution (downshift = 1.8, width = 0.3) (Tyanova et al. 2016). Normalized intensities (LFQ) were log2 transformed. To determine significantly changing proteins between the two groups, a two-sample Student's t-test was used. The original P-value was than corrected for an FDR of 0.05 by the Benjamini–Hochberg method. A list of all proteins confidently identified in each experiment is in Supplemental Table S1.
Cell lysates and Western blots
For whole-cell extracts, cells were harvested, washed twice with ice-cold PBS, and centrifuged at 1500 rpm for 5 min. Cell pellets were resuspended with NP-40 lysis buffer (250 mM NaCl, 50 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 0.5 mM EGTA, 0.2% NP40) or RIPA buffer (150 mM NaCl, 1% NP40, 0.1% Na deoxycholate, 0.1% SDS, 50 mM Tris HCl at pH 8.0) and incubated for 30 min on ice. Lysates were then centrifuged in microfuge tubes at 13,000 rpm for 10 min at 4°C. A cocktail of protease inhibitors and 1 mM PMSF was added to all lysis buffers used. Protein extracts were resolved on SDS–polyacrylamide gel, blotted onto nitrocellulose membranes, and probed with the following antibodies: ZC3H4 (Sigma HPA040934), FLAG (Sigma F1804), Vinculin (Santa Cruz Biotechnologies sc-73614), and Tubulin (Santa Cruz Biotechnologies sc-32293). For the validation of MS results, streptavidin pull-down was performed as described above, and beads at the final step were eluted in Laemmli buffer (Bio-Rad 1610747). Protein inputs and pulled-down extracts were resolved on SDS–polyacrylamide gel, blotted onto nitrocellulose membranes, and probed with the following antibodies: ZC3H4, SYMPK (Fortis Life Sciences A301-463A and A301-465A, and Cell Signaling Technology 13071), CTR9 (Fortis Life Sciences A301-395A), PAF1 (Cell Signaling Technology 12883S), RPRD1B (Fortis Life Sciences A303-782A), HELLS (Cell Signaling Technology 7998), WDR82 (Cell Signaling Technology 99715S), streptavidin-HRP (Abcam ab7403), PNUTS (Cell Signaling Technology 14171), CPSF3 (Fortis Life Science A301-091A), CPSF2 (Sigma HPA024238), CPSF1 (Cell Signaling Technology 73993), CPSF4 (Fortis Life Science A301-585A), CPSF6 (Fortis Life Science A301-356A), and CSTF2 (Fortis Life Science A301-092A).
RT-qPCR
Total RNA was extracted using the Zymo Quick-RNA kit (Zymo Research R1055), and 1 µg was reverse-transcribed with the ImProm-II reverse transcription system (Promega A3800). RT-qPCR was assembled with the Fast SYBR Green master mix (Applied Biosystems 4385614) and run on a QuantStudio 6 real-time PCR machine (Applied Biosystems). Analysis was done on the Thermo Fisher Cloud platform.
Primers for SYMPK (SYMPK F2: 5′-CATCGCATTCCAAGCAGACA-3′ and SYMPK R2: 5′-CACCTTGTAGAGCTGGGTCA-3′) and GAPDH (GAPDH_F: 5′-GTGGAAGGGCTCATGACCA-3′ and GAPDH_R: 5′-GGATGCAGGGATGATGTTCT-3′) were designed using Primer3.
siRNA-mediated protein depletion in HCT-116 ZC3H4-AID cells
siRNAs were purchased from Santa Cruz Biotechnologies (siRNA Sympk: sc-97297 and control siRNA: sc-37007) and transfected using Lipofectamine RNAiMAX reagent (Thermo Fisher 13778150) according to the manufacturer's protocol.
4sU RNA-seq
4sU (Santa Cruz Biotechnologies sc-204628A) was added to the medium at 500 μM for 10 min for HCT-116 cells or at 300 μM for 45 min for HeLa TREx Flp-In cells before collection. The 4sU-labeled RNA was extracted from 30–50 μg of total Trizol-isolated RNA. We used 80–100 ng of the 4sU-labeled RNA for cDNA library synthesis using the TruSeq stranded total RNA sample preparation kit (Illumina RS-122-9007) with the ribosomal depletion step. Libraries were quantified with the Quantifluor reagent (Promega E2670) and analyzed using TapeStation (Agilent) with the high-sensitivity assay HD5000 (Agilent 5067-5592). cDNA libraries were sequenced on an Illumina NovaSeq platform with 51-bp paired-end reads.
Analysis of 4sU RNA-seq data sets
Strand-specific paired-end reads (51 bp) were trimmed and clipped for quality control with Trimmomatic v0.38. The quality of the reads was then checked using FastQC v0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were aligned to the hg38 reference genomes (GENCODE, https://www.gencodegenes.org/human/release_33.html) using TopHat v2.1.1 (Trapnell et al. 2012; Kim et al. 2013), allowing up to two mismatches and using the option ‐‐b2-very-sensitive ‐‐library-type fr-firststrand. Indels due to sequencing errors were identified using Bowtie2 v2.6 (Langmead and Salzberg 2012). Only uniquely mapping reads were retained (–g 1). For the dTAG-induced depletion experiments, multimapping reads were also retained using STAR v2.7.7a (Robinson and Oshlack 2010) with the options ‐‐sjdbOverhang 100 ‐‐winAnchorMultimapNmax 200 ‐‐outFilterMultimapNmax 100.
Analysis of combined ZC3H4 depletion and siRNA-mediated Symplekin depletion
First, all mapped reads that overlapped by >10 nt with annotated protein-coding genes according to the GENCODE annotation (https://www.gencodegenes.org/human/release_33.html) were excluded. Then, SICER v.2 was used to detect all genomic extragenic transcripts sensitive or not to Symplekin depletion (Zang et al. 2009). All possible combinations among the experimental conditions were taken into account with respect to the control (single and double depletions vs. control and vice versa). For each comparison, the entire genome was partitioned into blocks of nonoverlapping 500-bp windows with a gap size of <1000 nt (effective genome fraction = 1, fragment size = 0, FDR = 1). After sorting and merging all the resulting transcripts, we obtained 35,709 extragenic transcripts detected in HCT116. In order to determine how many of these transcripts increased after Symplekin depletion or codepletion of ZC3H4 and Symplekin, a differential expression analysis was carried out using edgeR v3.10.5 with the limma v3.24.15 Bioconductor package (Robinson and Smyth 2008; Robinson et al. 2010; McCarthy et al. 2012). Read counts for edgeR analysis were obtained with BamScale v1.0 (Pongor et al. 2020), admitting the option cov ‐‐libtype paired ‐‐frag ‐‐stran. To eliminate genes with very low expression levels, we retained only those genes with at least 10 reads in all conditions. The standard counts per million (CPM) for all conditions were adjusted with the trimmed mean of M-values (TMM) using the calcNormFactors() function from the edgeR Bioconductor package. With respect to standard normalization, the TMM normalization has the advantage of reducing the false positive rate. We modeled the data variability by estimating the dispersion of the negative binomial model using the quantile-adjusted conditional maximum likelihood (qCML) method. We first used the (qCML) estimateCommonDisp() function and then the (qCML) estimateTagwiseDisp() function from the edgeR Bioconductor package. Finally, the exact P-values for the negative binomial distribution were computed using the function exactTest(), making a pairwise comparisons between the groups. A total of n = 8657 extragenic transcripts was detected based on an FDR of ≤0.05 (using the Benjamini–Hochberg correction), a fold change of ≥0.8 (log2 transformed), and FPKM ≥ 0.1.
We assigned each transcript to the nearest annotated enhancer or the nearest gene TSS or TES of the coding genes, as described before (Austenaa et al. 2021). The n = 8657 extragenic transcripts were hierarchically clustered using the Ward's method as the algorithm and the Pearson correlation as the distance metric. A Z-score scaling on the log2 transformed FPKM values was performed by subtracting the mean and then dividing it by the standard deviation. This calculation was carried out on a “transcript-by-transcript” (i.e., row-by-row) basis.
Analysis of CPSF3 depletion
We reanalyzed previously published RNA-seq data sets from CPSF3-depleted HCT116 cells (GSE137727) (Eaton et al. 2020). Strand-specific single-end reads were trimmed and clipped for quality control with Trimmomatic v0.38 and then aligned to the hg38 reference genome (GENCODE, https://www.gencodegenes.org/human/release_33.html) using TopHat v2.1.1 (Trapnell et al. 2012; Kim et al. 2013), allowing up to two mismatches and using the option ‐‐b2-very-sensitive ‐‐library-type fr-firststrand. Only uniquely mapping reads were retained (–g 1). Indels due to sequencing errors were identified using Bowtie2 v2.6 (Langmead and Salzberg 2012). Tracks were generated using bamCoverage (-bs 1 ‐‐normalizeUsing RPKM –outFileFormat bigwig) from deepTools v.3.5.0 (Ramírez et al. 2016). To create strand-specific bigWig files, the option ‐‐filterRNAstrand forward or ‐‐filterRNAstrand reverse was used. The response to CPSF3 depletion in clusters I and II was evaluated. All antisense extragenic transcripts belonging to cluster I (n = 370) and to cluster II (n = 2270) were selected. Counts and FPKM (fragments per kilobase per million mapped fragments) were evaluated in each cluster with BamScale v1.0 (Pongor et al. 2020) with the option cov ‐‐frag –stran. The enrichment (fold change, log2 transformed) between the CPSF3 depletion and control was calculated with edgeR v3.10.5 with the limma v3.24.15 Bioconductor package and is shown in the box plot in comparison with the enrichment measured in the comparison between ZC3H4 depletion and control. For clusters III and IV, metaplots showing the average coverage of the distance from the TES + 2 kb were generated with deepTools v3.5.0 (Ramírez et al. 2016).
Analysis of combined ZC3H4 depletion and dTAG-driven Symplekin depletion
A heat map was generated using as anchors the 8657 transcripts identified as differentially expressed upon ZC3H4 depletion and/or siRNA-mediated Symplekin depletion with respect to the control. For each of the four previously identified clusters, the expression in all conditions was quantified with FPKM (BamScale v1.0 [Pongor et al. 2020] with the option cov ‐‐libtype paired ‐‐frag –stran.). In addition, all extragenic transcripts up-regulated after dTAG-induced Symplekin depletion were collected. We used SICER v2 (Zang et al. 2009) to detect the differentially expressed extragenic transcripts (-rt 100000 -w 500 -f 0 -egf 1 -g 1000 -fdr 1). Only clustered transcripts up-regulated in both replicates were retained (fold change greater than twofold and FDR < 0.05, with a minimum acceptable overlap of 50% between the two replicates). For the overlap, we used the intersecBed function from the BEDTools v2.29.2 suite as follows: –sorted –e –f 0.5 –F 0.5 (Quinlan and Hall 2010). In order to measure the effects of dTAG-mediated Symplekin depletion on 3′ readthrough at replication-dependent histone genes, all histone genes on chromosome 6 were considered (n = 55), and their TESs were identified based on the GENCODE database annotations (https://www.gencodegenes.org/human/release_33.html). The TESs were then used as reference points for the computeMatrix function of deepTools v3.5.0. In order to highlight the effect of the depletion of Symplekin in relation to 3′ readthrough, a 1-kb region after the TES is shown in a metaplot (one bin = 20 bp).
Analysis of the combined depletion of ZC3H4 and PNUTS
We used SICER v2 (Zang et al. 2009) to detect extragenic transcripts up-regulated after ZC3H4 depletion with respect to the control condition (-rt 100000 -w 500 -f 0 -egf 1 -g 1000 -fdr 0.01). Only transcripts up-regulated in both replicates were retained (fold change of twofold or more and FDR ≤ 0.01, with a minimum acceptable overlap of 50% between the two replicates). For the overlap, we used the intersecBed function from the BEDTools v2.29.2 suite as follows: –sorted –e –f 0.5 –F 0.5 (Quinlan and Hall 2010). We obtained n = 2321 transcripts that were categorized based on their genomic location. A total of n = 997 transcripts was assigned to the nearest annotated enhancer based on the FANTOM database (http://fantom.gsc.riken.jp/5); n = 881 transcripts were in divergent orientation relative to the TSSs of protein-coding genes, and 443 transcripts corresponded to the transcription end sites of protein-coding genes, according to the GENCODE annotations (https://www.gencodegenes.org/human/release_33.html).
All promoter-divergent and enhancer-associated RNAs (n = 1878) were collected, and their expression changes are shown in a heat map using the Ward's method as the algorithm and the Pearson correlation as the distance metric. The FPKM values were calculated with BamScale v1.0 (Pongor et al. 2020), admitting the option cov ‐‐libtype paired ‐‐frag ‐‐stran. The extragenic transcripts were clustered in four groups (cluster1: n = 1009, cluster2: n = 89, cluster3: n = 271, and cluster4: n = 509), and for each cluster the expression is shown as a box plot. In the metaplot, scores calculated with the computeMatrix function of deepTools v3.5.0 (‐‐regionBodyLength 6000 ‐‐missingDataAsZero -bs 20 -b 600 -a 600) are shown (Ramírez et al. 2016). Next, the expression of transcripts associated with the four clusters upon Symplekin depletion and codepletion was measured with deepTools v3.5.0 and is shown in a box plot.
Differential gene expression in HeLa TREx cells overexpressing ZC3H4
The response to ZC3H4 overexpression of 1494 transcripts previously shown to be up-regulated after ZC3H4 depletion in HeLa cells (Austenaa et al. 2021) was evaluated. The log2 transformed FPKMs were calculated with BamScale v1.0 (Pongor et al. 2020) using the option cov ‐‐libtype paired ‐‐frag –stran. Differences in the expression between transcripts are shown with a heat map that integrates the complete linkage method as the algorithm and the Pearson correlation as the distance metric. Differentially expressed coding and noncoding transcripts with basal expression in the previously published siRNA experiment (Austenaa et al. 2021) were collected. We selected n = 545 extragenic transcripts (RPKM > 0.06 in at least two replicates in the control condition) and n = 157 coding genes (RPKM > 0.1 in at least two replicates in the control condition) with basal expression that was up-regulated upon ZC3H4 depletion. For the read count quantification related to the experiment in which ZC3H4 was overexpressed, we used featureCounts v1.6.4 (Liao et al. 2014) normalizing based on the FPKM. For each replicate, we calculated the fold change in the comparison between overexpression condition and empty vector. For the coding genes, the fold change was detected using edgeR v3.10.5 with the limma v3.24.15 Bioconductor package (Robinson and Smyth 2008; Robinson and Oshlack 2010; McCarthy et al. 2012) in the pairwise comparisons between ZC3H4 and empty vector. For the identification of the extragenic transcripts up-regulated after ZC3H4 overexpression, we applied the same strategy described before (Austenaa et al. 2021). We first excluded all mapped reads that overlapped by >10 nt with annotated protein-coding genes according to the GENCODE annotation (https://www.gencodegenes.org/human/release_33.html). We then used SICER v2 (Zang et al. 2009) to detect the extragenic transcripts regulated in response to ZC3H4 overexpression relative to cells transfected with empty vector (-rt 100000 -w 500 -f 0 -egf 1 -g 1000 -fdr 0.01). The FDR was calculated using P-value adjusted for multiple testing, following the approach developed by Benjamini and Hochberg. In the SICER analysis, only clustered transcripts with more than twofold enrichment with respect to the empty vector and at least 100 reads were retained. For each experiment, only transcripts up-regulated in at least two replicates were retained, with a minimum acceptable overlap of 50% between different replicates using the intersecBed function from the BEDTools v2.29.2 suite: –sorted –e –f 0.5 –F 0.5 (Quinlan and Hall 2010). We obtained n = 1407 transcripts that were assigned to the nearest annotated enhancer based on the FANTOM database (http://fantom.gsc.riken.jp/5) or to the nearest TSS/TES-proximal regions using the ClosestBed tool from the BEDTools suite with the parameter -t first (Quinlan and Hall 2010). All transcripts corresponding to the TESs of the coding genes were collected (n = 411), and the coverage around the TESs is shown in a metaplot. Using the TES as an anchor, we extended 2 kb (one bin = 20 nt) and calculated the scores with the computeMatrix function of deepTools v3.5.0 (Ramírez et al. 2016).
Analysis of transcription termination at histone genes
We considered replication-dependent histone genes in the chromosome 6 cluster. For each cell type, using the TES as an anchor, a metaplot (+2 kb from the TES, bin = 20) was created with the computeMatrix and plotProfile functions of deepTools v3.5.0 (Ramírez et al. 2016).
LTR12C analysis in HeLa TREx
In order to have two sets of extragenic transcripts comparable in number and nucleotide length, a golden set of transcripts up-regulated upon ZC3H4 overexpression (n = 918, length average = 13,520, FC > 1, and FDR < 0.05 in at least two replicates) and another golden set of transcripts not affected by ZC3H4 (n = 1074, length average = 18,365, FC < 1, FDR > 0.05 in all replicates) were selected. For each subfamily of repeats (https://hgdownload.cse.ucsc.edu/goldenpath/hg38/database), a contingency table with the number of overlaps and the number of nonoverlaps versus both groups of transcripts (up-regulated by ZC3H4 and not affected after ZC3H4 depletion) was evaluated with the Fisher test (“alternative = greater,” significance for P < 0.01). Subfamilies were ranked according to the most significant −log10 transformed P-value. A total of n = 263 LTR12C elements overlapped with n = 243 extragenic transcripts up-regulated after ZC3H4 overexpression. Those >1 kb (n = 246) were collected for the distribution of all 4096 possible hexamers. For each hexamer, the total number of occurrences was counted and the log2 ratio of the occurrence on 5′ (+500 nt) and 3′ (−500 nt) was calculated. Hexamers were ranked by enrichment.
Analysis of PRO-seq data sets
We analyzed two previously published PRO-seq samples, GSM3714462 and GSM3714463, which are part of the GSE129501 series (Steinparzer et al. 2019). To ensure data quality, we performed several preprocessing steps on strand-specific single-end reads. Initially, we used Trimmomatic v0.38 to trim and clip the reads. Then, we mapped these reads to the human genome in two stages. In the first mapping step, we aligned the reads with respect to human annotated repeats obtained from Repbase (https://www.girinst.org) using STAR v2.7.7a. We applied specific parameters for this alignment, including “‐‐outFilterMultimapNmax 30,” “‐‐outFilterMismatchNmax 10,” “‐‐outSAMattributes All,” “‐‐outFilterMultimapScoreRange 1,” “‐‐outFilterScoreMin 10,” and “‐‐alignEndsType EndToEnd.” In the second mapping step, any unmapped reads from the previous step were remapped to the hg38 reference genome downloaded from GENCODE (https://www.gencodegenes.org/human/release_33.html) using STAR v2.7.7a with parameters “‐‐outFilterMultimapNmax 10,” “‐‐outFilterMismatchNmax 10,” “‐‐outFilterMismatchNoverLmax 0.3,” and “‐‐alignIntronMin 21,” among others. We used SAMtools v1.3.1 to filter the mapped reads based on the MAPQ score with a threshold of “-q 10.” Subsequently, we divided the alignment files into reads that mapped to the forward and reverse strands, which allowed us to perform strand-specific peak calling using MACS2 v2.2.7.1 (Zhang et al. 2008). The MACS2 parameters applied were “-g hs,” “-s 34,” “‐‐keep-dup all,” “‐‐nomodel,” “‐‐shift 76,” “‐‐extsize 1,” “‐‐mfold 3 500,” “‐‐pvalue 1e-10,” “‐‐slocal 100,” “‐‐llocal 5000,” “‐‐max-gap 100,” and “‐‐min-length 10.” In the end, our analysis identified a total of n = 125,603 PRO-seq peaks.
PolyA site (AATAAA) analysis
From the 8657 transcripts identified as differentially expressed upon ZC3H4 depletion and/or siRNA-mediated Symplekin depletion, those belonging to cluster II (n = 4270) were selected. Transcripts that overlapped with a PRO-seq peak (n = 3090) and those that contained the PAS site at a distance >500 nt from the start of the PRO-seq (n = 2,35) were collected. In the presence of multiple PRO-seq peaks overlapping the same transcript, the PRO-seq peak with the highest Q-value cutoff was selected. The coverage around the PAS (±500 bp, one bin = 1bp) is shown in a metaplot for SYMPK depletion (siRNA and dTAG). The distance between the start of the PRO-seq peaks and the PAS is shown in a box plot.
Track generation and visualization
Tracks were generated using bamCoverage (-bs 1 ‐‐normalizeUsing RPKM –outFileFormat bigwig) from deepTools v.3.1.373. To create strand-specific bigWig files, the option ‐‐filterRNAstrand forward or ‐‐filterRNAstrand reverse was used.
Statistics and plots
R v3.6.1 was used to compute statistics and generate plots (https://www.r-project.org). Each exact P-value of statistical tests is reported in the figure legends.
Data availability
Raw and processed genomic sequencing data were deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE237460. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (Perez-Riverol et al. 2022) via the Proteomics Identification Database (PRIDE) partner repository with the data set identifier PXD043638.
Supplementary Material
Acknowledgments
We thank David Bentley for discussions and constructive comments. This work was supported by the European Research Council (Advanced ERC grant 692789 to G.N.). M.R. is supported by a long-term fellowship from the Italian Association for Research on Cancer (AIRC). D.P. and G.M.M. are PhD students at the European School of Molecular Medicine (SEMM). G.M.M. is supported by a Marie Sklodowska-Curie Action (Horizon 2020 European Training Network [ETN] Consortium ENHPATHY, grant agreement no. 860002).
Author contributions: M.R., V.P., D.P., and G.N. conceptualized the study. M.R., D.P., E.P., C.B., and S.P. generated the data. V.P., F.B., G.M.M., M.M., D.M., and A.C. analyzed the data. G.N., M.R., and V.P. wrote the manuscript with contributions from all the authors. G.N., S.R., and A.C. supervised the study. G.N. acquired the funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.351057.123.
Competing interest statement
The authors declare no competing interests.
References
- Ahmed R, Spikings E, Zhou S, Thompsett A, Zhang T. 2014. Pre-hybridisation: an efficient way of suppressing endogenous biotin-binding activity inherent to biotin-streptavidin detection system. J Immunol Methods 406: 143–147. 10.1016/j.jim.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. 10.1038/nature12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20: 1367–1376. 10.1038/nsmb.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Chen Y, Core L, Lis JT, Sandelin A, Jensen TH. 2015. Human gene promoters are intrinsically bidirectional. Mol Cell 60: 346–347. 10.1016/j.molcel.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austenaa LM, Barozzi I, Simonatto M, Masella S, Della Chiara G, Ghisletti S, Curina A, de Wit E, Bouwman BA, de Pretis S, et al. 2015. Transcription of mammalian cis-regulatory elements is restrained by actively enforced early termination. Mol Cell 60: 460–474. 10.1016/j.molcel.2015.09.018 [DOI] [PubMed] [Google Scholar]
- Austenaa LMI, Piccolo V, Russo M, Prosperini E, Polletti S, Polizzese D, Ghisletti S, Barozzi I, Diaferia GR, Natoli G. 2021. A first exon termination checkpoint preferentially suppresses extragenic transcription. Nat Struct Mol Biol 28: 337–346. 10.1038/s41594-021-00572-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaian A, Mager DL. 2016. Endogenous retroviral promoter exaptation in human cancer. Mob DNA 7: 24. 10.1186/s13100-016-0080-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreikaite V, Passmore LA. 2023. 3′-end processing of eukaryotic mRNA: machinery, regulation, and impact on gene expression. Annu Rev Biochem 92: 199–225. 10.1146/annurev-biochem-052521-012445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreikaite V, Elliott TS, Chin JW, Passmore LA. 2022. RBBP6 activates the pre-mRNA 3′ end processing machinery in humans. Genes Dev 36: 210–224. 10.1101/gad.349223.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36: 880–887. 10.1038/nbt.4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casañal A, Kumar A, Hill CH, Easter AD, Emsley P, Degliesposti G, Gordiyenko Y, Santhanam B, Wolf J, Wiederhold K, et al. 2017. Architecture of eukaryotic mRNA 3′-end processing machinery. Science 358: 1056–1059. 10.1126/science.aao6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR III, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. 10.1101/gad.250993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler CS, Ballard FJ. 1986. Multiple biotin-containing proteins in 3T3-L1 cells. Biochem J 237: 123–130. 10.1042/bj2370123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Smith ER, Shilatifard A. 2018. Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 19: 464–478. 10.1038/s41580-018-0010-5 [DOI] [PubMed] [Google Scholar]
- Core L, Adelman K. 2019. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33: 960–982. 10.1101/gad.325142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar MA, Sheridan RM, Erickson B, Fong N, Glover-Cutter K, Brannan K, Bentley DL. 2019. Control of RNA Pol II speed by PNUTS–PP1 and Spt5 dephosphorylation facilitates termination by a ‘sitting duck torpedo’ mechanism. Mol Cell 76: 896–908.e4. 10.1016/j.molcel.2019.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Allen MA, Larsen A, Macmorris M, Han M, Blumenthal T. 2008. Genes involved in pre-mRNA 3′-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc Natl Acad Sci 105: 16665–16670. 10.1073/pnas.0807104105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JD, Francis L, Davidson L, West S. 2020. A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev 34: 132–145. 10.1101/gad.332833.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeier CC, Erickson B, Allen BL, Allen MA, Kim H, Fong N, Jacobsen JR, Liang K, Shilatifard A, Dowell RD, et al. 2017. Human TFIIH kinase CDK7 regulates transcription-associated chromatin modifications. Cell Rep 20: 1173–1186. 10.1016/j.celrep.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod ND, Henriques T, Huang KL, Tatomer DC, Wilusz JE, Wagner EJ, Adelman K. 2019. The integrator complex attenuates promoter-proximal transcription at protein-coding genes. Mol Cell 76: 738–752.e7. 10.1016/j.molcel.2019.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estell C, Davidson L, Steketee PC, Monier A, West S. 2021. ZC3H4 restricts non-coding transcription in human cells. Elife 10: e67305. 10.7554/eLife.67305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estell C, Davidson L, Eaton JD, Kimura H, Gold VAM, West S. 2023. A restrictor complex of ZC3H4, WDR82, and ARS2 integrates with PNUTS to control unproductive transcription. Mol Cell 683: 2222–2239.e5. 10.1016/j.molcel.2023.05.029 [DOI] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, Xu X. 1996. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the second international conference on knowledge discovery and data mining (ed. Simoudis E et al. ), pp. 226–231. AAAI Press, Portland, OR. [Google Scholar]
- Field A, Adelman K. 2020. Evaluating enhancer function and transcription. Annu Rev Biochem 89: 213–234. 10.1146/annurev-biochem-011420-095916 [DOI] [PubMed] [Google Scholar]
- Fridell RA, Pret AM, Searles LL. 1990. A retrotransposon 412 insertion within an exon of the Drosophila melanogaster vermilion gene is spliced from the precursor RNA. Genes Dev 4: 559–566. 10.1101/gad.4.4.559 [DOI] [PubMed] [Google Scholar]
- Godin KS, Varani G. 2007. How arginine-rich domains coordinate mRNA maturation events. RNA Biol 4: 69–75. 10.4161/rna.4.2.4869 [DOI] [PubMed] [Google Scholar]
- Guntaka RV. 1993. Transcription termination and polyadenylation in retroviruses. Microbiol Rev 57: 511–521. 10.1128/mr.57.3.511-521.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JM, Sanford JR. 2015. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA 6: 93–110. 10.1002/wrna.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Szczurek AT, Kelley JR, Lastuvkova A, Turberfield AH, Dimitrova E, Blackledge NP, Klose RJ. 2023. A CpG island-encoded mechanism protects genes from premature transcription termination. Nat Commun 14: 726. 10.1038/s41467-023-36236-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. 10.1038/nature09479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D, Rinn J. 2012. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13: R107. 10.1186/gb-2012-13-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, et al. 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18: 956–963. 10.1038/nsmb.2085 [DOI] [PubMed] [Google Scholar]
- LaBella ML, Hujber EJ, Moore KA, Rawson RL, Merrill SA, Allaire PD, Ailion M, Hollien J, Bastiani MJ, Jorgensen EM. 2020. Casein kinase 1δ stabilizes mature axons by inhibiting transcription termination of ankyrin. Dev Cell 52: 88–103.e18. 10.1016/j.devcel.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk HB, Sandquist LE, Bay LTE, Steurer B, Campsteijn C, Landsverk OJB, Marteijn JA, Petermann E, Trinkle-Mulcahy L, Syljuåsen RG. 2020. WDR82/PNUTS–PP1 prevents transcription-replication conflicts by promoting RNA polymerase II degradation on chromatin. Cell Rep 33: 108469. 10.1016/j.celrep.2020.108469 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. 2008. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28: 609–618. 10.1128/MCB.01356-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, You J, Dobrota E, Skalnik DG. 2010. Identification and characterization of a novel human PP1 phosphatase complex. J Biol Chem 285: 24466–24476. 10.1074/jbc.M110.109801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540–543. 10.1126/science.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lidschreiber M, Easter AD, Battaglia S, Rodriguez-Molina JB, Casanal A, Carminati M, Baejen C, Grzechnik P, Maier KC, Cramer P, et al. 2018. The APT complex is involved in non-coding RNA transcription and is distinct from CPF. Nucleic Acids Res 46: 11528–11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Žumer K, Molska ES, Rouvière JO, Wu G, Demel C, Schwalb B, Schmid M, Cramer P, Jensen TH. 2021. Integrator is a genome-wide attenuator of non-productive transcription. Mol Cell 81: 514–529.e6. 10.1016/j.molcel.2020.12.014 [DOI] [PubMed] [Google Scholar]
- MacDonald CC, Wilusz J, Shenk T. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol 14: 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenda M, Lazarova E, van de Linde S, Gilbert N, Michieletto D. 2021. Parameter-free molecular super-structures quantification in single-molecule localization microscopy. J Cell Biol 220: e202010003. 10.1083/jcb.202010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep 1: 753–763. 10.1016/j.celrep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Koreski KP. 2017. Birth and death of histone mRNAs. Trends Genet 33: 745–759. 10.1016/j.tig.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhar M, Ebert A, Neumann T, Umkehrer C, Jude J, Wieshofer C, Rescheneder P, Lipp JJ, Herzog VA, Reichholf B, et al. 2018. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 360: 800–805. 10.1126/science.aao2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Calviello L, Hirsekorn A, de Pretis S, Pelizzola M, Ohler U. 2017. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol 24: 86–96. 10.1038/nsmb.3325 [DOI] [PubMed] [Google Scholar]
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al. 2018. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14: 431–441. 10.1038/s41589-018-0021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem 278: 33000–33010. 10.1074/jbc.M304454200 [DOI] [PubMed] [Google Scholar]
- Ni Z, Xu C, Guo X, Hunter GO, Kuznetsova OV, Tempel W, Marcon E, Zhong G, Guo H, Kuo WW, et al. 2014. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat Struct Mol Biol 21: 686–695. 10.1038/nsmb.2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nojima T, Proudfoot NJ. 2022. Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol 23: 389–406. 10.1038/s41580-021-00447-6 [DOI] [PubMed] [Google Scholar]
- Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20: 923–928. 10.1038/nsmb.2640 [DOI] [PubMed] [Google Scholar]
- Ovesný M, Křížek P, Borkovec J, Švindrych Z, Hagen GM. 2014. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30: 2389–2390. 10.1093/bioinformatics/btu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Zhong J, Jang JS, Kim J, Kim HJ, Lee JH, Kim J. 2022. ZWC complex-mediated SPT5 phosphorylation suppresses divergent antisense RNA transcription at active gene promoters. Nucleic Acids Res 50: 3835–3851. 10.1093/nar/gkac193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci S, Furia L, Scanarini M, Pelicci PG, Lanzanò L, Faretta M. 2022. Novel tools to measure single molecules colocalization in fluorescence nanoscopy by image cross correlation spectroscopy. Nanomaterials (Basel) 12: 686. 10.3390/nano12040686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, et al. 2022. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50: D543–D552. 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongor LS, Gross JM, Vera Alvarez R, Murai J, Jang SM, Zhang H, Redon C, Fu H, Huang SY, Thakur B, et al. 2020. BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks. Epigenetics Chromatin 13: 21. 10.1186/s13072-020-00343-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Cho KF, Cavanagh PE, Ting AY. 2021. Deciphering molecular interactions by proximity labeling. Nat Methods 18: 133–143. 10.1038/s41592-020-01010-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9: 321–332. 10.1093/biostatistics/kxm030 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière JO, Lykke-Andersen S, Jensen TH. 2022. Control of non-productive RNA polymerase II transcription via its early termination in metazoans. Biochem Soc Trans 50: 283–295. 10.1042/BST20201140 [DOI] [PubMed] [Google Scholar]
- Rouviere JO, Salerno-Kochan A, Lykke-Andersen S, Garland W, Dou Y, Rathore O, Molska ES, Wu G, Schmid M, Bugai A, et al. 2023. ARS2 instructs early transcription termination-coupled RNA decay by recruiting ZC3H4 to nascent transcripts. Mol Cell 83: 2240–2257.e6. 10.1016/j.molcel.2023.05.028 [DOI] [PubMed] [Google Scholar]
- Ruepp MD, Schweingruber C, Kleinschmidt N, Schümperli D. 2011. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol Biol Cell 22: 91–104. 10.1091/mbc.e10-06-0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell 65: 25–38. 10.1016/j.molcel.2016.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönemann L, Kühn U, Martin G, Schäer P, Gruber AR, Keller W, Zavolan M, Wahle E. 2014. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 28: 2381–2393. 10.1101/gad.250985.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis N, Gupta GD, Lin ZY, Gonzalez-Badillo B, Veri AO, Knight JDR, Rajendran D, Couzens AL, Currie KW, Tkach JM, et al. 2016. Phenotypic and interaction profiling of the human phosphatases identifies diverse mitotic regulators. Cell Rep 17: 2488–2501. 10.1016/j.celrep.2016.10.078 [DOI] [PubMed] [Google Scholar]
- Stein CB, Field AR, Mimoso CA, Zhao C, Huang KL, Wagner EJ, Adelman K. 2022. Integrator endonuclease drives promoter-proximal termination at all RNA polymerase II-transcribed loci. Mol Cell 82: 4232–4245.e11. 10.1016/j.molcel.2022.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinparzer I, Sedlyarov V, Rubin JD, Eislmayr K, Galbraith MD, Levandowski CB, Vcelkova T, Sneezum L, Wascher F, Amman F, et al. 2019. Transcriptional responses to IFN-γ require mediator kinase-dependent pause release and mechanistically distinct CDK8 and CDK19 functions. Mol Cell 76: 485–499.e8. 10.1016/j.molcel.2019.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10: 395–406. 10.1038/nrmicro2783 [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332. 10.1016/j.molcel.2009.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhang Y, Aik WS, Yang XC, Marzluff WF, Walz T, Dominski Z, Tong L. 2020. Structure of an active human histone pre-mRNA 3′-end processing machinery. Science 367: 700–703. 10.1126/science.aaz7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525. 10.1128/MCB.20.5.1515-1525.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev 4: 2112–2120. 10.1101/gad.4.12a.2112 [DOI] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S. 2013. Defining the RGG/RG motif. Mol Cell 50: 613–623. 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods 5: 159–161. 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- van Arensbergen J, FitzPatrick VD, de Haas M, Pagie L, Sluimer J, Bussemaker HJ, van Steensel B. 2017. Genome-wide mapping of autonomous promoter activity in human cells. Nat Biotechnol 35: 145–153. 10.1038/nbt.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT. 2013. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol 33: 2067–2077. 10.1128/MCB.01742-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Tong L, Adelman K. 2023. Integrator is a global promoter-proximal termination complex. Mol Cell 83: 416–427. 10.1016/j.molcel.2022.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28: 7337–7344. 10.1128/MCB.00976-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. 2010. Crystal structure of the human symplekin–Ssu72–CTD phosphopeptide complex. Nature 467: 729–733. 10.1038/nature09391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. 2012. Transcriptome-wide analyses of CstF64–RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci 109: 18773–18778. 10.1073/pnas.1211101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics 25: 1952–1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143. 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer JT, Rosa-Mercado NA, Canzio D, Steitz JA, Simon MD. 2021. STL-seq reveals pause-release and termination kinetics for promoter-proximal paused RNA polymerase II transcripts. Mol Cell 81: 4398–4412.e7. 10.1016/j.molcel.2021.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed genomic sequencing data were deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE237460. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (Perez-Riverol et al. 2022) via the Proteomics Identification Database (PRIDE) partner repository with the data set identifier PXD043638.