In this review, Mendez-Dorantes and Burns discuss the molecular mechanisms and genomic consequences of LINE-1 retrotransposition. LINE-1 retrotransposons promote genomic instability and immune activation in cancer, and the authors explore their targetability in diagnostic and therapeutic interventions.
Keywords: LINE-1 retrotransposons, epigenetics, genome instability
Abstract
Long interspersed element 1 (LINE-1) is the only protein-coding transposon that is active in humans. LINE-1 propagates in the genome using RNA intermediates via retrotransposition. This activity has resulted in LINE-1 sequences occupying approximately one-fifth of our genome. Although most copies of LINE-1 are immobile, ∼100 copies are retrotransposition-competent. Retrotransposition is normally limited via epigenetic silencing, DNA repair, and other host defense mechanisms. In contrast, LINE-1 overexpression and retrotransposition are hallmarks of cancers. Here, we review mechanisms of LINE-1 regulation and how LINE-1 may promote genetic heterogeneity in tumors. Finally, we discuss therapeutic strategies to exploit LINE-1 biology in cancers.
Barbara McClintock's discovery of transposable elements (TEs; DNA sequences that can mobilize from one genomic location to another) in maize (McClintock 1950) provided a foundation for understanding genome composition and the dynamic nature of DNA across taxa.
Since this discovery, we have recognized that much of the content of eukaryotic genomes, including the human genome, is composed of interspersed repeats derived from TE activity (Britten and Kohne 1968; Smit 1996; Lander et al. 2001). Retrotransposons, which mobilize via RNA intermediates, are the major class of interspersed repeats in humans. Although most retrotransposons are incapable of mobilization in humans, subfamilies of LINE-1 (L1PA1 and L1PA2) are still active for retrotransposition and hence are a potential source of heritable genetic variation, somatic mosaicism, and genome instability via germline and somatic activity (Boissinot et al. 2000; Beck et al. 2010; Huang et al. 2010; Iskow et al. 2010).
In 1988, physician scientists discovered LINE-1 insertional mutations in the blood-clotting gene factor VIII in two unrelated individuals with hemophilia, providing the foundation that LINE-1 is an active mobile element in humans (Kazazian et al. 1988). In the 35 yr since this landmark discovery, >100 LINE-1-mediated insertional mutations resulting from germline retrotransposition have been shown to cause human genetic diseases (Hancks and Kazazian 2016). In each example, LINE-1 activity inserted a retrotransposon sequence at a critical gene sequence and caused a loss-of-function allele.
Shortly after LINE-1 insertional mutagenesis was recognized as a cause of constitutional genetic disease, somatic retrotransposition was discovered as a driver of tumorigenesis. Investigators evaluating the adenomatous polyposis coli (APC) locus in 150 individuals with colon cancer found a somatic LINE-1 insertion that disrupted the tumor suppressor gene in the malignant cells of an individual (Miki et al. 1992). Since this report, others have corroborated that LINE-1 retrotransposition is an uncommon though recurrent mechanism of APC loss in colon cancer (Scott et al. 2016; Cajuso et al. 2019).
Recently, other reports and studies from large cancer genome consortia have highlighted how pervasive somatic retrotransposition is across many distinct types of cancers (Lee et al. 2012; Helman et al. 2014; Tubio et al. 2014; Rodriguez-Martin et al. 2020), indicating that mutagenesis by retrotransposition is a hallmark of malignancies. While initially the field anticipated that insertions might more commonly “drive” cancers as insertional mutagens, this seems not to be the rule. Interest now is shifting toward understanding the influence of LINE-1 and retrotransposons on cancer biology, including whether their dysregulation or associated DNA damage may contribute to cancer initiation and evolution and whether this biology could be exploited for therapeutic opportunities. Here, we provide an orientation to LINE-1 biology in cancers with an emphasis on these emerging topics.
Overview of non-LTR retrotransposons in humans
TEs are classified as DNA transposons or retrotransposons (Wicker et al. 2007; Bourque et al. 2018). DNA transposons (3% of the human genome) mobilize by excising as DNA fragments and relocating in the genome (Fig. 1A; Hoyt et al. 2022). These transposons are no longer active in humans; however, some of their sequences have been “domesticated” as genes for host function (Joly-Lopez and Bureau 2018). For example, recombination-activating genes (RAGs) involved in V(D)J recombination were domesticated from the Transib DNA transposon (Agrawal et al. 1998; Roth and Craig 1998; Huang et al. 2016). Other domesticated genes in humans include CENPB, THAP9, and PGBD5 (Smit and Riggs 1996; Majumdar et al. 2013; Henssen et al. 2015; Jangam et al. 2017).
Figure 1.
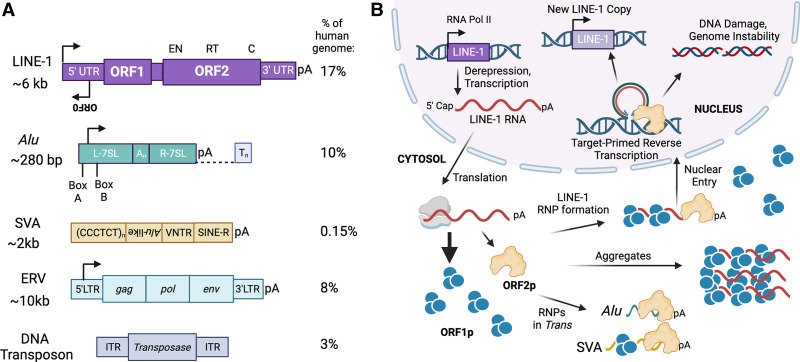
Retrotransposons in the human genome. (A) Shown are the sequence structures of transposable elements and their composition in the human genome. The LINE-1 retrotransposon encodes two proteins required for mobilization: ORF1p contains RNA binding properties, and ORF2p contains endonuclease (EN) and reverse transcriptase (RT) activities. Alu elements and SVAs are non-protein-coding retrotransposons and rely on LINE-1-encoded proteins for retrotransposition. Endogenous retroviruses (ERVs) and DNA transposons are no longer mobile in humans. However, our genome contains ERV copies that are transcriptionally active with partially intact open reading frames. (B) Shown is a diagram of the life cycle of LINE-1. In somatic cells, a full-length copy of LINE-1 escapes epigenetic silencing and is transcribed by RNA polymerase II. The LINE-1 RNA is translated into ORF1p and ORF2p, which together form ribonucleoproteins (RNPs) in cis. LINE-1-encoded proteins can also form RNPs in trans with other transcripts, including Alu elements and SVAs. LINE-1 RNPs are commonly found in cytosolic aggregates, which are poorly characterized. Once RNPs access the nucleus, LINE-1 retrotransposition is initiated by ORF2p via target-primed reverse transcription to generate a new copy of the element in the genome. Alternatively, LINE-1 retrotransposition can be a source of DNA damage and genome instability.
In contrast, retrotransposons mobilize by making cDNA copies of their transcribed RNAs and inserting these into the genome, which is a process called retrotransposition (Fig. 1; Burns 2017). Retrotransposons can be further classified in several ways. One type includes long terminal repeat (LTR) retrotransposons (8%) that are derived from past retrovirus infections in the germline and hence are also called endogenous retroviruses (ERVs) (Dewannieux et al. 2006; Hoyt et al. 2022). Like DNA transposons, there are no known propagating ERVs active in humans, although there are ERV copies in the genome that are transcriptionally active with partially intact protein-coding open reading frames (ORFs; gag, pol, or env) (Bannert and Kurth 2006). Another type is non-LTR retrotransposons (Fig. 1A), which include elements that encode their own proteins for retrotransposition, such as LINE-1 (17%), or that exploit LINE-1 proteins for retrotransposition, such as short interspersed elements (SINEs); e.g., Alu (10%) and SINE–VNTR–Alu composite elements (SVAs; 0.15%) (Payer and Burns 2019; Hoyt et al. 2022). Hence, all ongoing retrotransposition in humans is attributable to LINE-1 and its proteins.
An active LINE-1 unit and its retrotransposition life cycle
Our current estimate is that we each inherit a complement of 100 copies of LINE-1 (L1PA1 and L1PA2) that are retrotransposition-competent (Brouha et al. 2003; Beck et al. 2010). The rest of the LINE-1 copies in our genome are inactive commonly due to incomplete insertions resulting from 5′ truncations or acquired mutations (Beck et al. 2011). An intact functional LINE-1 copy in humans is 6 kb, and its sequence structure contains a 5′ untranslated region (UTR), two open reading frames (ORF1 and ORF2) separated by a short 63-bp sequence, and a 3′ UTR with a polyadenylation (polyA) signal (Fig. 1A; Dombroski et al. 1991).
The retrotransposition cycle of LINE-1 begins with its transcription by RNA polymerase II, which is regulated by the internal promoter in the 5′ UTR (Fig. 1B). The bicistronic LINE-1 transcript is 5′-capped, 3′-polyadenylated, and exported to the cytosol for translation of ORF1p and ORF2p, both of which are essential for propagation of the element. ORF1p is a 40-kDa RNA binding protein that forms homotrimers that chaperone LINE-1 RNA (Martin et al. 2003, 2005; Khazina et al. 2011). ORF2p is a 150-kDa protein with endonuclease (EN) and reverse transcriptase (RT) activities and a strong cis preference for the polyA of the LINE-1 transcript that encodes it (Mathias et al. 1991; Feng et al. 1996; Wei et al. 2001; Doucet et al. 2015). Together, ORF1p trimers and ORF2p with the LINE-1 RNA form a functional ribonucleoprotein (RNP) that need to access the nucleus for retrotransposition (Kulpa and Moran 2006; Taylor et al. 2013). In addition, the LINE-1 5′ UTR contains antisense promoter (ASP) activity and a primate-specific third ORF (ORF0) encoding a small protein, ORF0p (Fig. 1A; Speek 2001; Cruickshanks and Tufarelli 2009; Denli et al. 2015), that are dispensable for LINE-1 retrotransposition. Interestingly, ORF0 contains two splice donor sites, which could generate ORF0 chimeric transcripts with neighboring genes (Denli et al. 2015).
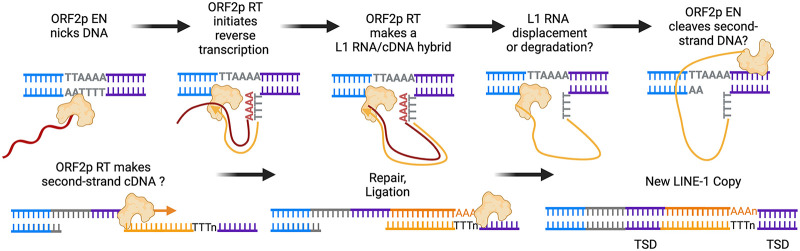
Mechanistically, canonical LINE-1 retrotransposition occurs via target-primed reverse transcription (TPRT) (Fig. 2; Luan et al. 1993; Cost et al. 2002). The working model involves ORF2p EN generating a DNA nick at a flexible target sequence (3′-AA/TTTT-5′) to liberate a 3′ OH (Feng et al. 1996). This reveals a short stretch of polyT single-stranded DNA (ssDNA) that can complementarily bind the polyA of the LINE-1 transcript to form a primer–template structure (Fig. 2). ORF2p RT can extend from the 3′ OH using the LINE-1 RNA as a template to synthesize the first stand of LINE-1 cDNA (Fig. 2; Cost et al. 2002). The cDNA intermediate is then processed into a de novo LINE-1 copy in the genome flanked by <20-bp target site duplications (TSDs) (Symer et al. 2002; Beck et al. 2011), completing the life cycle of a LINE-1 unit. Hence, the genomic “scars” of ORF2p activity include inserted LINE-1 sequences that terminate with polyA tails, are found at EN target sequences, and are flanked by short TSDs (Fig. 2).
Figure 2.
Mechanism of LINE-1 target-primed reverse transcription (TPRT). Shown is a working model for canonical LINE-1 retrotransposition via TPRT. ORF2p EN initiates TPRT by nicking DNA at a relaxed target site (3′-AA/TTTT-5′) to liberate a 3′ OH at a short stretch of polyT, which binds the polyA tail of LINE-1 RNA to form a primer–template structure. ORF2p RT can then extend from the 3′ OH to generate LINE-1 cDNA using LINE-1 RNA as a template, resulting in a presumed LINE-1 RNA/cDNA hybrid intermediate. The subsequent steps of LINE-1 retrotransposition are poorly understood but may include the displacement or degradation of the LINE-1 RNA template, the cleavage of the second strand DNA, the synthesis of the second strand DNA, and the joining of the 5′ end of a double-stranded cDNA intermediate to genomic DNA. New copies of LINE-1 in the genome are characterized by signature features: They are enriched at EN target sequences, contain polyA tails in their 3′ end, and are flanked by short target site duplications (TSDs).
The mechanisms for how LINE-1 cDNA intermediates are resolved into complete, double-stranded insertions remain elusive and may require host DNA repair factors. For example, the LINE-1 RNA template needs to be displaced or degraded from a presumed LINE-1 RNA:cDNA intermediate likely by host factors, since ORF2p lacks RNase H activity (Fig. 2; Mathias et al. 1991; Malik et al. 1999). In addition, the formation of TSDs suggests that a second DNA nick is generated by ORF2p EN or host factors in the top strand DNA downstream proximal to the initial DNA nick at the EN target sequence when it is located at the bottom strand DNA to form staggered DNA nicks, which is likely a mechanism that can lead to short TSDs flanking the inserted sequence (Fig. 2). The mechanism of second strand LINE-1 cDNA synthesis is unknown, although ORF2p RT may mediate this step (Fig. 2). The remaining steps are unclear but may involve the joining of the 5′ end of a double-stranded LINE-1 cDNA to genomic DNA to resolve the insertion (Fig. 2). The 5′ junctions of resolved LINE-1 insertions sometimes contain nontemplated nucleotides (Symer et al. 2002; Kojima 2010), but how these are generated or used in the resolution of the insertion is unknown. Finally, the role of ORF1p during TPRT remains unknown, although its function in the LINE-1 life cycle may be limited to the cytosol. Biochemical reconstitution of LINE-1 retrotransposition has the potential to elucidate these processes, including addressing the contribution of ORF1p, ORF2p, or host DNA repair factors in each step of TPRT.
Noncoding parasites of the LINE-1 retrotransposition machinery
Although LINE-1-encoded ORF2p has cis preference for reverse-transcribing the RNA encoding it (Wei et al. 2001), non-protein-coding retrotransposons can hijack LINE-1 proteins for amplification in the human genome (Fig. 1). These include Alu elements, which are the most successful retrotransposons in humans, having generated >1 million germline copies (Batzer and Deininger 2002; Dewannieux et al. 2003). Alu elements are ∼280 bp in length and are composed of two related monomer sequences derived from the 7SL RNA of the signal recognition particle (SRP) (Fig. 1A). Alu elements contain an internal promoter (A box and B box) in the left monomer for transcription initiation mediated by RNA polymerase III but lack a terminator sequence and instead use downstream T-rich genomic DNA for transcription termination. The 3′ end of an Alu sequence also contains a long A-rich region, which is required for Alu RNAs to associate with ORF2p in trans (Doucet et al. 2015). RNP formation is likely mediated via the interaction of Alu transcripts with SRP9-14 proteins at the ribosome that together stall ORF2p translation in close proximity to Alu RNAs (Weichenrieder et al. 2000). In contrast to ORF2p, ORF1p is dispensable for Alu retrotransposition (Dewannieux et al. 2003).
SVAs are the youngest family of retrotransposons in humans, with ∼2700 copies in the genome (Ono et al. 1987; Hancks and Kazazian 2010). SVAs are a composite element of five repeats: a hexamer repeat [(CCCTCT)n], two antisense Alu-like fragments, a variable number tandem repeat (VNTR), a SINE derived from the LTR of an ERV (HERV-K10), and a polyA signal (Fig. 1A; Hancks and Kazazian 2010). The full-length size of SVAs can vary drastically due to their VNTRs, ranging from 50 bp to 2 kb, although most SVAs are 2 kb long (Wang et al. 2005; Chu et al. 2023). Transcription of SVAs is poorly understood but appears to be RNA polymerase II-dependent. In contrast to Alu elements, retrotransposition of SVAs requires both ORF1p and ORF2p (Hancks et al. 2011).
Gene transcripts (mRNAs) can also be reverse-transcribed by ORF2p, a process that has generated ∼8000 processed pseudogenes (retrocopies) in our genome (Esnault et al. 2000). Importantly, these retrocopies lack promoters, and hence most are not transcriptionally active. Finally, U6 ribosomal RNA sequences are infrequently reverse-transcribed by LINE-1 proteins in the form of U6-3′L1 chimeras (Buzdin et al. 2002; Moldovan et al. 2019). Like LINE-1 insertions, inserted sequences derived from canonical ORF2p activity terminate with polyA tails, are found at EN cleavage sequences in the genome, and are flanked by short TSDs (Fig. 2).
Regulation of LINE-1 retrotransposition in somatic cells
Epigenetic repression of LINE-1
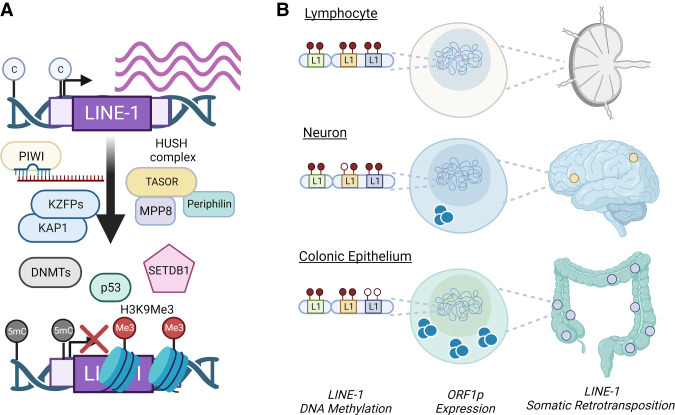
Human cells have evolved several defenses to restrain LINE-1 retrotransposition, including multiple related epigenetic silencing mechanisms (Fig. 3A). These include DNA cytosine methylation and repressive histone modifications, which are established in the germline and in early development and are maintained in somatic cells via PIWI-interacting RNAs (piRNAs), DNA methyltransferases (DNMTs), and multiple protein complexes, including Kruppel-associated box (KRAB) domain-containing zinc finger proteins (KZFPs)/KAP1 and the human silencing hub (HUSH) complex (Fig. 3A; Castro-Diaz et al. 2014; Jacobs et al. 2014; Newkirk et al. 2017; Robbez-Masson et al. 2018). In somatic cells, DNA methylation is found at CpG islands of the 5′ UTR of LINE-1 copies (Woodcock et al. 1997). However, such LINE-1 methylation is found compromised in human cancers with global DNA hypomethylation and is associated with ORF1p expression and somatic retrotransposition (Chalitchagorn et al. 2004; Estécio et al. 2007; Rodić et al. 2014; Ardeljan et al. 2017), supporting that DNA methylation suppresses LINE-1. Indeed, DNA-hypomethylating agents in cultured cells induce LINE-1 expression (Yang et al. 2004; de Cubas et al. 2020; Sato et al. 2023). In some cases, loss of methylation at a specific LINE-1 locus helped to identify it as the “source element” responsible for a somatic retrotransposition event (Scott et al. 2016; Nguyen et al. 2018). However, this type of locus-specific analysis is not commonplace due to technical challenges that make it difficult to relate a de novo insertion found with short read sequencing to a specific source element. As a result, we do not have a comprehensive census of active LINE-1 loci in cancers.
Figure 3.
Epigenetic silencing of LINE-1. (A) Multiple protein complexes, including KAP1 and HUSH, function in concert to deposit repressive marks at LINE-1 sequences, such as DNA methylation via DNMTs or H3K9 trimethylation via SETDB1. (B) Shown is a working model for how epigenetic variation (e.g., alterations in DNA methylation patterns) of individual LINE-1 sequences across somatic cell types influences ORF1p expression and somatic retrotransposition in human tissues.
Our understanding of epigenetic variation of individual LINE-1 loci across human tissues is limited: the precise promoter sequence and CpG content of each inherited L1 allele, its DNA methylation state in each cell type, and the associated pattern of expression of the element (Fig. 3B). Genome-wide analysis of LINE-1 methylation not only would identify source elements responsible for somatic retrotransposition but would also indicate influences of LINE-1 methylation on genome function, such as neighboring gene expression, regional chromatin states, and three-dimensional genome organization. Testing these relationships might then be possible using CRISPR activation and interference systems to manipulate methylation at individual LINE-1 loci to directly probe its consequences. Future studies addressing the epigenetic variation of LINE-1 loci may be particularly significant in the context of human malignancies with epigenetic dysregulation and in Mendelian diseases caused by mutations in epigenetic “writer,” “eraser,” and “reader” genes and chromatin remodelers (Baylin and Jones 2016).
In addition to DNA methylation, recent studies identified that the HUSH complex, which is a heterotrimer composed of MPP8, TASOR, and periphilin, silences LINE-1 elements via recruitment of the histone methyltransferase SETDB1 for deposition of repressive histone H3K9 trimethylation (Fig. 3A; Liu et al. 2018; Robbez-Masson et al. 2018; Tunbak et al. 2020). In normal cells, the HUSH complex is proposed to selectively silence LINE-1 copies via the detection of long, intronless transcription units in the genome, which are distinct characteristics of evolutionarily young LINE-1 elements (Seczynska et al. 2022; Seczynska and Lehner 2023). In human cancers, compromised HUSH-mediated heterochromatinization, potentially due to down-regulation of MPP8, may contribute to the activation of LINE-1 expression (Tunbak et al. 2020). Importantly, the HUSH complex also targets transgenes for silencing. Thus, it is critical that investigations into HUSH functions in different cell type and disease contexts include analyses of endogenous LINE-1 loci in addition to relying on LINE-1 transgene reporters.
Finally, the tumor suppressor p53, which is mutated in over half of human cancers, has been implicated as a transcriptional repressor of LINE-1 (Fig. 3A; Harris et al. 2009; Tiwari et al. 2020). Namely, loss of p53 causes ORF1p expression in cells (Tiwari et al. 2020); however, this result is cell type-specific, suggesting that there are likely multiple layers of LINE-1 suppression. Thus, investigating the interactions between DNA methylation, histone methylation, and p53 at individual LINE-1 loci will provide insights into the multiple layers of LINE-1 epigenetic regulation.
Retrotransposition-competent LINE-1 RNAs
Transcription of LINE-1 elements initiates at the start of the 5′ UTR and can terminate at the polyA signal within the 3′ UTR or continue past the 3′ UTR until a polyA signal is encountered in downstream genomic DNA (Skowronski et al. 1988; Philippe et al. 2016; Deininger et al. 2017). Hence, retrotransposition-competent LINE-1 transcripts contain intact ORFs and are ≥6 kb in length.
It is important to note that the majority of LINE-1 RNAs in cells are nonfunctional for retrotransposition and stem from “readthrough” of LINE-1 embedded in intronic intervals or encompassed by long noncoding RNA (lncRNAs) (Deininger et al. 2017). Thus, measurements of LINE-1 RNA by RT-PCR or in situ hybridizations can be misleading and should not be used as a surrogate for retrotransposition potential. However, several bioinformatics tools have been developed to assay LINE-1 subfamily expression using consensus sequences (e.g., L1PA1 contains a diagnostic “5′-ACA-3′” trinucleotide in its 3′ UTR) or locus-specific expression using the 3′ unique readthrough sequences of LINE-1 chimeric RNAs (Jin et al. 2015; Jung et al. 2018; Yang et al. 2019; McKerrow and Fenyö 2020). These analyses can be further supported with detection of chromatin marks at upstream sequences of the elements, including histone H3K4 trimethylation (Philippe et al. 2016).
Long read sequencing will allow us to accurately distinguish retrotransposition-competent “unit transcripts” versus other LINE-1 RNAs in cells (Berrens et al. 2022). These tools are expected to open avenues of investigation into the functional roles of LINE-1-containing transcripts. Indeed, there are ∼5000 LINE-1 loci that can be transcriptionally active in our genome (Deininger et al. 2017). The ASP activity of LINE-1 5′ UTR can generate spliced chimeric transcripts extending to exons of nearby genes (Nigumann et al. 2002; Denli et al. 2015; Attig et al. 2018), including a spliced variant of the MET oncogene in bladder cancers (Weber et al. 2010; Wolff et al. 2010). The extent of LINE-1 ASP causing aberrant proteins such as oncogenes in cancers requires additional investigation.
Regulation of LINE-1 RNA, encoded proteins, and RNPs
In the cytosol, bicistronic LINE-1 transcripts can be translated into ORF1p and ORF2p. ORF1p is abundantly translated and can be readily detected in cells using standard protein detection techniques (e.g., immunoblotting and immunohistochemistry) (Rodić et al. 2014). Hence, ORF1p detection can be a metric for LINE-1 expression and retrotransposition potential. ORF1p contains a coiled-coil domain (CCD) that mediates homotrimer formation (Martin et al. 2003; Khazina et al. 2011) and an RNA recognition motif (RRM) and a C-terminal domain (CTD) that together mediate LINE-1 RNA binding in a non-sequence-specific manner (Januszyk et al. 2007; Khazina and Weichenrieder 2009), promoting the protein's role as a LINE-1 RNA chaperone.
ORF2p performs the essential enzymatic activities for retrotransposition and is an endogenous source of DNA damage in cells (Cost et al. 2002; Gasior et al. 2006). ORF2p contains an apurinic/apyrimidinic endonuclease (APE)-like EN domain and a telomerase-like RT, as well as a cystine-rich domain with a poorly understood function required for retrotransposition (Mathias et al. 1991; Feng et al. 1996; Kopera et al. 2011; Adney et al. 2019). In contrast to ORF1p, ORF2p is difficult to detect in cells (Ardeljan et al. 2020b), suggesting that LINE-1 and its host cells may limit ORF2p levels, likely to restrain its genotoxicity. Nonetheless, the pervasive genetic signature of germline and somatic retrotransposition provides unequivocal evidence that ORF2p is produced and functional in human cells.
The regulation of ORF2p production from the LINE-1 RNA remains elusive but may involve multiple mechanisms. For one, ORF2p is translated via an unconventional ribosomal termination/reinitiation mechanism (Alisch et al. 2006), which may be a mechanism that limits its production; however, this process is only partly understood. Moreover, the clearance of ORF2p from cells remains unclear, although a role of autophagy in the clearance of LINE-1 RNPs has been implicated (Guo et al. 2014). Other recent studies highlighted N6-methyladenosine (m6A) modification of LINE-1 RNA or let-7 microRNAs binding to LINE-1-RNA as mechanisms that regulate ORF2p levels in cells (Tristán-Ramos et al. 2020; Hwang et al. 2021).
ORF1p and ORF2p associate with LINE-1 RNA to form dynamic, heterogenous LINE-1 ribonucleoproteins (RNPs), as revealed through interactomics (Goodier et al. 2013; Taylor et al. 2013, 2018; Luqman-Fatah et al. 2023). These studies show that LINE-1-encoded proteins are associated with diverse cellular host factors that can either promote or restrict retrotransposition. For example, the RNA helicase MOV10 and SAMHD1 limit retrotransposition by sequestering LINE-1 RNPs in stress granules (Goodier et al. 2012; Hu et al. 2015), whereas polyA binding proteins PABPN1/4 promote retrotransposition by aiding in LINE-1 RNP formation (Dai et al. 2012). Interestingly, phase separation of ORF1p in the cytosol appears to be a prerequisite for retrotransposition (Sil et al. 2023), highlighting that intrinsic properties of ORF1p mediating RNP formation are critical. Together, these studies indicate that the spatial organization and composition of LINE-1 RNPs may be important for translation and stability of LINE-1-encoded proteins and their assembly into functional intermediates of retrotransposition.
After assembly, retrotransposition-competent LINE-1 RNPs are proposed to enter the nucleus during nuclear envelope breakdown (Mita et al. 2018). However, LINE-1 retrotransposition is detected in postmitotic cells (Macia et al. 2017), highlighting that LINE-1 RNPs may access the nucleus via several mechanisms. Future single-molecule live-cell imaging analysis of LINE-1 RNA and encoded proteins might provide insights into the formation, localization, and regulation of LINE-1 RNPs.
Regulation of target-primed reverse transcription: resolving vs. removing insertion intermediates
In the nucleus, a LINE-1 RNP can initiate retrotransposition via TPRT (Luan and Eickbush 1995; Cost et al. 2002; Wilkinson et al. 2023). However, LINE-1 retrotransposition in somatic cells rarely generates full-length insertions and instead results in a variety of genomic outcomes and alterations, including most commonly 5′ truncations (Fig. 4; Gilbert et al. 2002; Symer et al. 2002; Rodriguez-Martin et al. 2020; Nam et al. 2023). Host DNA repair mechanisms likely limit LINE-1 retrotransposition through several mechanisms (Fig. 4A). Here, we discuss the roles of host DNA repair factors in removing or resolving LINE-1 insertion intermediates.
Figure 4.
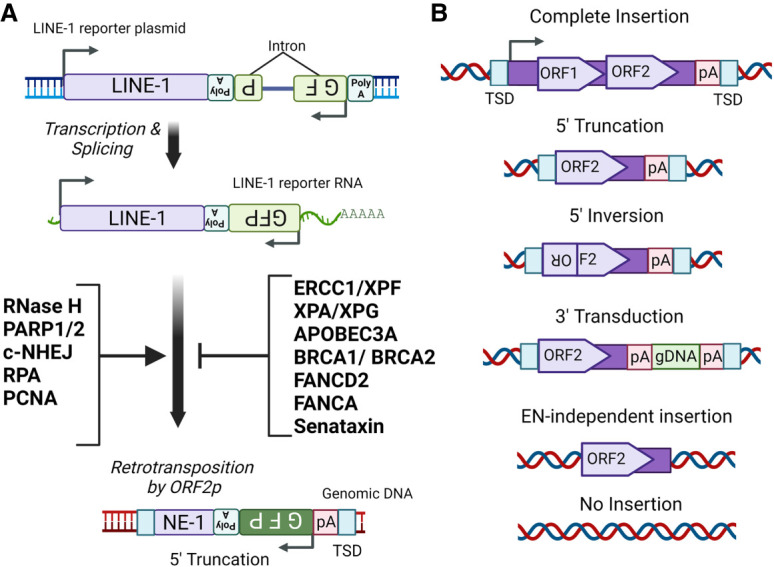
Host DNA repair mechanisms regulate LINE-1 retrotransposition. (A) Shown is a diagram of the cell-based LINE-1 retrotransposition functional assay. A LINE-1 retrotransposition reporter sequence can be expressed in cells using an ectopic expression plasmid. The LINE-1 sequence contains an antisense reporter gene (e.g., eGFP) interrupted by a sense intron in its 3′ UTR, which ensures the reporter gene is only expressed in cells after splicing and retrotransposition of the LINE-1 transcript. Also shown is a list of host DNA repair factors that mediate or inhibit LINE-1 retrotransposition based on functional studies using the cell-based LINE-1 assay. (B) Shown are the sequence structures of genomic outcomes generated by LINE-1 retrotransposition, including full-length insertions, 5′ truncations, 5′ inversions, 3′ transductions, and EN-independent insertions.
Much of our understanding of the roles of host DNA repair factors on TPRT is based on a cell-based functional assay for LINE-1 retrotransposition (Fig. 4A; Moran et al. 1996; Kopera et al. 2016). This elegant assay includes a LINE-1 sequence with its 3′ UTR containing an antisense reporter gene cassette disrupted by a sense-oriented intron, which ensures the reporter gene will be expressed in cells only after splicing and retrotransposition of the LINE-1 transcript (Fig. 4A; Kopera et al. 2016). Importantly, this assay primarily indicates 5′ truncated LINE-1 insertions with a minimal length of ∼2 kb (Fig. 4A; Gilbert et al. 2002). Using this assay, several DNA repair factors have been identified as regulators of LINE-1 retrotransposition (Fig. 4A; Liu et al. 2018; Mita et al. 2020). An interpretation of these studies is that those factors that inhibit retrotransposition may remove LINE-1 insertion intermediates, whereas those factors that promote LINE-1 retrotransposition may resolve intermediates into insertions. For example, the DNA sliding clamp PCNA is required for efficient LINE-1 retrotransposition; it is suggested to regulate reverse transcription via the PCNA-interacting protein (PIP) box of ORF2p (Taylor et al. 2013). RNase H2 enzymes also promote LINE-1 retrotransposition (Benitez-Guijarro et al. 2018) likely by the degradation of the LINE-1 RNA template from the presumed hybrid intermediate to allow for second stand cDNA synthesis. In contrast to RNase H2, the RNA/DNA helicase senataxin inhibits LINE-1 retrotransposition likely via unwinding LINE-1 RNA:cDNA intermediates to limit reverse transcription (Liu et al. 2018). Additionally, PARP1/PARP2 and the singe-stranded DNA binding complex RPA promote LINE-1 retrotransposition (Fig. 4A) and are proposed to function in concert to protect the newly synthesized cDNA from degradation or mutations after removal of the LINE-1 RNA template (Miyoshi et al. 2019). In support of this model, the APOBEC cytidine deaminases—including APOBEC3A—that target single-strand DNA substrates inhibit LINE-1 retrotransposition (Muckenfuss et al. 2006; Richardson et al. 2014). Moreover, the 3′ DNA flap endonuclease XPF inhibits retrotransposition and was proposed to cleave LINE-1 3′ cDNA intermediates (Gasior et al. 2008).
In addition, landmark genetic screens identified multiple suppressors of LINE-1 retrotransposition that are involved in DNA repair pathways, including factors of the intracross-link (ICL) repair pathway Fanconi anemia (FA; e.g, FANCD2 and FANCA) and the chromosomal break repair pathway homologous recombination (e.g., BRCA1 and BRCA2) (Liu et al. 2018; Mita et al. 2020). Consistent with these results, a recent study highlighted that core factors in the FA pathway (e.g., FANCD2, FANCA, and SLX4) are epistatic with XPF for suppression of LINE-1 retrotransposition (Bona and Crossan 2023), proposing a model in which factors of the FA pathway function in concert to recruit XPF to cleave LINE-1 3′ cDNA intermediates. In addition to recruitment via the FA pathway, XPF was previously proposed to be recruited to LINE-1 cDNA intermediates by factors (e.g, XPA, XPD, and XPC) in the nucleotide excision repair pathway (Servant et al. 2017). These results suggest that there are likely redundant DNA repair mechanisms for suppression of LINE-1 retrotransposition. Future studies are required to address the mechanisms of how factors in homologous recombination individually or in concert limit LINE-1 retrotransposition. Together, these results highlight the intricate roles of host DNA repair factors in regulating LINE-1 retrotransposition.
Regulation of LINE-1 retrotransposition by DNA replication
Several biochemical and genetics studies have linked LINE-1 retrotransposition to DNA replication. For example, proteomics analyses found ORF2p to be associated with factors detected at DNA replication forks (Mita et al. 2018; Taylor et al. 2018), including PCNA, MCM proteins, RPA, and PARP1. In addition, high-throughput mapping of 3′ junctions of thousands of de novo LINE-1 insertions in cultured cells revealed a correlation between retrotransposition site preference and DNA replication timing but not transcription or chromatin state (Flasch et al. 2019; Sultana et al. 2019). Namely, LINE-1 EN-dependent insertions are enriched in early replicating genomic regions. Consistent with these results, somatic LINE-1 insertions in cancer genomes also correlate with replication timing (Rodriguez-Martin et al. 2020); however, these are enriched at late replicating genomic regions. These discrepancies in replication timing could be explained by differing dosages of LINE-1 exposure and selective processes on cells with a high occurrence of retrotransposition during cancer development.
Moreover, although LINE-1 retrotransposition can be detected in nondividing cells (Macia et al. 2017), measurements of retrotransposition in replicating cells have found LINE-1 insertions to be enriched during S phase (Shi et al. 2007; Mita et al. 2018), suggesting that DNA replication may provide an ideal context for efficient retrotransposition. In support of this model, many factors involved in replication-coupled DNA repair, including the FA pathway and ATR signaling (e.g., FANCD2, FANCM, BRCA1, and ATRIP), were found to be required for growth of p53-deficient cells expressing LINE-1 (Ardeljan et al. 2020a). These results support a working model in which LINE-1 may efficiently exploit DNA replication forks for integration in cells deficient for replication-coupled DNA repair. Future studies are required to address the mechanisms of how LINE-1 RNPs exploit ongoing DNA replication forks for integration versus how LINE-1 RNPs integrate into the genome independent of DNA replication.
Genomic outcomes of LINE-1 retrotransposition
5′ truncations
Somatic LINE-1 insertions are commonly 5′-truncated (Fig. 4B). In addition to the 5′ truncations, these differ from full-length insertions in that the 5′ junctions of insertions contain microhomology (Symer et al. 2002; Zingler et al. 2005), which is a signature (i.e., a genomic “scar”) of chromosomal break repair via end-joining pathways (Cisneros-Aguirre et al. 2022). Consistent with this observation, factors involved in end-joining repair pathways (e.g., KU70, LIG4, and PARP1) are required for LINE-1 retrotransposition (Fig. 4A; Suzuki et al. 2009; Miyoshi et al. 2019), suggesting that these repair pathways complete the ligation of the truncated LINE-1 cDNA to genomic DNA. The mechanisms that cause 5′ truncations are unclear but may involve the degradation or cleavage of LINE-1 RNA or insertion intermediates during TPRT by host DNA repair factors. Interestingly, the lengths of somatic LINE-1 insertions in cancer genomes show a bimodal distribution, with most being 5′-truncated shorter than 2 kb and the rest being nearly full length (Tubio et al. 2014; Nam et al. 2023). We speculate that 5′ truncations might occur early during reverse transcription and that the generation of a long cDNA likely bypasses the mechanisms that cause 5′ truncations. In contrast to LINE-1, somatic insertions of Alu and SVAs are commonly complete (Rodriguez-Martin et al. 2020).
5′ inversions
Other common outcomes of LINE-1 retrotransposition include 5′ inversions, which are composed of inverted 5′ LINE-1 sequences joined to 3′ LINE-1 sequences (Fig. 4B). These events may be mediated by a proposed twin priming model in which a second priming event occurs at the target site of TPRT, and the resulting cDNAs from both priming events are joined using microhomology to resolve the insertion (Ostertag and Kazazian 2001). However, this proposed model of twin priming requires experimental validation.
3′ transductions
Another common outcome includes LINE-1 3′ transductions (Fig. 4B), which are generated by retrotransposition of LINE-1 transcripts containing 3′ genomic readthrough sequences. These events result in the duplication of genomic sequences downstream from LINE-1 source elements, which can include coding and regulatory sequences with the potential to influence genome function (Moran et al. 1999; Tubio et al. 2014). Resolved LINE-1 3′ transductions contain the genomic “scars” of ORF2p activity, and most are commonly 5′-truncated with some only containing 3′ genomic readthrough sequences, called orphan transductions (Solyom et al. 2012; Tubio et al. 2014). Importantly, these events are technically valuable for the identification of source elements for somatic retrotransposition (Tubio et al. 2014).
EN-independent insertions
ORF2p can also generate LINE-1 insertions independent of its EN activity (Feng et al. 1996; Morrish et al. 2002), although EN-independent (ENi) insertions are inefficient compared with EN-dependent insertions. ENi events commonly lack TSDs, do not have a preference to insert at canonical EN cleavage motifs, and contain 5′ and 3′ truncations (Fig. 4B). The working model for these insertions is that ORF2p RT generates cDNA at a pre-existing DNA lesion containing a free 3′ OH in the genome such as a chromosomal break, where de novo cDNA bridges the broken chromosome. In support of this model, EN mutated ORF2p can integrate LINE-1 at Cas9-mediated chromosomal breaks and at deprotected telomere ends (Morrish et al. 2002; Tao et al. 2022). In addition, deficiencies of DNA repair factors (e.g., FANCD2 and BRCA1) that cause increased DNA breaks also increase ENi insertions (Flasch et al. 2019; Mita et al. 2020; Bona and Crossan 2023). The mechanisms for how a LINE-1 RNP is recruited to an endogenous site of DNA damage in the genome remain unknown. However, these ENi insertions might represent a rare mechanism of RNA-templated DNA repair by ORF2p RT in cells, which is implicated for other RTs such as a group II intron-like RT in bacteria (Park et al. 2022) and telomerase and polymerase θ in humans (Kopera et al. 2011; Chandramouly et al. 2021). In summary, LINE-1 retrotransposition results in a variety of genomic outcomes, the mechanisms for which are only partly understood.
LINE-1 overexpression and retrotransposition are hallmarks of human cancers
ORF1p overexpression in human malignancies
LINE-1 overexpression as evaluated by ORF1p expression is a hallmark of human epithelial cancers, particularly those with p53 mutations (Rodić et al. 2014; McKerrow et al. 2022). These include ovarian, esophageal, colon, lung, breast, and pancreatic cancers, although ORF1p accumulates at varying levels and in varying proportions of cases depending on the type of malignancy. In high-grade serous ovarian cancers (HGSOCs), ORF1p levels are quite consistently high, and the marker has been detected in precursor lesions of the disease (serous tubal intraepithelial carcinoma [STIC]) (Pisanic et al. 2019; Xia et al. 2019; Sato et al. 2023), indicating that induction of LINE-1 expression is an early event in tumor development, concomitant with histologic transformation and the fixation of mutations in TP53. Timing and precise molecular causes of induction of ORF1p and its duration of expression during cancer evolution mostly remain unknown, including whether cancer cells might transiently express LINE-1. Interestingly, in contrast to many epithelial cancers, blood cancers and glioblastomas lack ORF1p expression, and these cancer genomes are not characterized by frequent insertion events (Rodić et al. 2014; Achanta et al. 2016; Carreira et al. 2016). Furthermore, although ORF1p expression appears to be an indicator of retrotransposition potential, ORF1p expression levels and the number of somatic LINE-1 insertions do not always correlate in cancers (Rodić et al. 2014; Rodriguez-Martin et al. 2020; McKerrow et al. 2022). These observations suggest that there are likely additional determinants of somatic LINE-1 retrotransposition in cancers, which might include host factors that regulate the LINE-1 life cycle.
Strikingly, ORF1p was recently detected in peripheral blood draws from cancer patients, including women with ovarian cancers, using ultrasensitive detection assays in the attomolar range (Sato et al. 2023; Taylor et al. 2023). This finding suggests that ORF1p has the potential to be used as a blood-based biomarker of malignancy. Additionally, this finding motivates future studies to determine whether circulating ORF1p can be measured in early-stage malignancies for applications in early detection and to address its potential as a biomarker to monitor cancers over time or to evaluate for minimal residual disease after therapy.
Mechanistically, the contribution of LINE-1 to cellular transformation is yet unclear, and while ORF2p can function as an endogenous mutagen, functional consequences of the readily detected overexpressed ORF1p are more opaque. In addition to binding to LINE-1 RNA, ORF1p can bind other cellular RNAs in trans, including mRNAs and circular RNAs (Martin et al. 2005; Briggs et al. 2021), and the consequences of these interactions in cancer cells are unknown. It is plausible that ORF1p can sequester not only LINE-1 RNA but also other endogenous mRNAs or other RNA species in cancers. It is intriguing to consider addressing whether genetic vulnerabilities can be identified in cells overexpressing ORF1p, although overexpression of ORF1p alone does not appear to be toxic in cultured cells (Ardeljan et al. 2020a).
LINE-1 retrotransposition in cancer genomes
A recent pan-cancer analysis of whole genomes confirmed that somatically acquired LINE-1 copies are genetic hallmarks of human cancers (Rodriguez-Martin et al. 2020), which was previously revealed by several landmark studies (Iskow et al. 2010; Lee et al. 2012; Helman et al. 2014; Tubio et al. 2014; Rodić et al. 2015). However, the burden of somatic LINE-1 insertions vastly varies across cancer types. For example, esophageal cancers can contain up to hundreds of insertions, colon cancers contain dozens of insertions, and ovarian cancers contain few insertions (Cajuso et al. 2019; Rodriguez-Martin et al. 2020), as ascertained using whole-genome sequencing by paired-end short reads of bulk tumor samples. Genomic outcomes of somatic LINE-1 retrotransposition detected in cancer genomes include full-length insertions (rare), 5′ truncations (most common), 5′ inversions, and 3′ transductions (∼20%) (Rodriguez-Martin et al. 2020). In addition to LINE-1, somatically-acquired copies of Alu, SVAs, and processed pseudogenes are also found in cancer genomes, although at a much lower frequency (Cooke et al. 2014; Rodriguez-Martin et al. 2020). Regarding 3′ transductions, over half of these events detected in cancer genomes can be attributable to five conserved source LINE-1 elements, highlighting that there are “hot” LINE-1 source elements in cancer genomes (Tubio et al. 2014; Pradhan et al. 2017; Rodriguez-Martin et al. 2020).
A major finding of mapping somatic LINE-1 insertions in cancer genomes is that these most commonly spare protein-coding regions and that no genomic sites are recurrent “hot spots” for insertions, which indicate that most insertions likely represent passenger mutations (Burns 2017; Rodriguez-Martin et al. 2020). On rare occasions, however, LINE-1 insertions can cripple tumor suppressor genes, such as the adenomatous polyposis coli (APC) gene in colorectal cancers (Miki et al. 1992; Scott et al. 2016; Cajuso et al. 2019). Importantly, our current measurements of LINE-1 retrotransposition in cancer genomes using short read sequencing are likely an underestimate. Indeed, precise retrotransposition rates and selection pressures exerted by this mutagenesis as tumor subclones evolve are not yet well understood. Long read sequencing and single-cell analyses of cancer genomes are both expected to reveal more comprehensive pictures of LINE-1 retrotransposition in human malignancies. Somatic LINE-1 insertions in cancer genomes may prove an important genomic signature for future therapeutic strategies exploiting LINE-1 biology (Burns 2022).
p53 mutations and LINE-1 expression and retrotransposition
LINE-1 ORF1p expression and retrotransposition both correlate with p53 mutations in cancers (Rodić et al. 2014; Rodriguez-Martin et al. 2020; McKerrow et al. 2022); however, the sequence of events in cellular transformation and the precise mechanistic relationships between LINE-1 expression and p53 compromise are unclear. One working hypothesis is that p53 is a direct transcriptional repressor of LINE-1 by binding to its 5′ UTR (Tiwari et al. 2020). However, so-called p53 signature lesions in the fallopian tube, which are thought to precede cellular transformation to the STIC precursors of HGSOC, are characterized by TP53 mutations but lack the ORF1p expression seen in STIC and HGSOC (Pisanic et al. 2019; Xia et al. 2019). This progression suggests that disruption of p53 alone is not sufficient to induce ORF1p expression during cancerous transformation. An alternative hypothesis is that p53 functions as a “guardian of the genome” downstream from LINE-1 expression, limiting the proliferation of cells with DNA damage from retrotransposition. In support of this hypothesis, ectopic expression of LINE-1 in p53-proficient cells induces cell cycle arrest and significantly limits cell fitness and clonogenic potential (Ardeljan et al. 2020a). These findings suggest that p53-mediated responses may represent a selective mechanism to eliminate or curtail the growth of cells with deregulated LINE-1 expression. A corollary of this is that cancer cells need to acquire p53 mutations to survive with the genotoxic pressure of LINE-1 expression. We expect unraveling the relationship between p53 and LINE-1 will provide insights into the contribution of LINE-1 to cellular transformation.
LINE-1 is a source of endogenous DNA damage and genome instability
LINE-1 expression has been known to cause DNA damage and genome instability in cultured cells for two decades (Gilbert et al. 2002; Symer et al. 2002; Gasior et al. 2006). However, this area was relatively understudied while the field focused on LINE-1 as an insertional mutagen in cancers; we have now only a partial understanding of the nature of DNA damage induced by LINE-1 activity and the consequences of LINE-1-mediated DNA damage on genome integrity (Fig. 5). The recent pan-cancer analysis revealed that LINE-1 insertion intermediates are sources of chromosomal rearrangements (Rodriguez-Martin et al. 2020), supporting a hypothesis that DNA lesions generated by LINE-1 activity may be a major source of genome instability in ORF1p (+) cancers.
Figure 5.
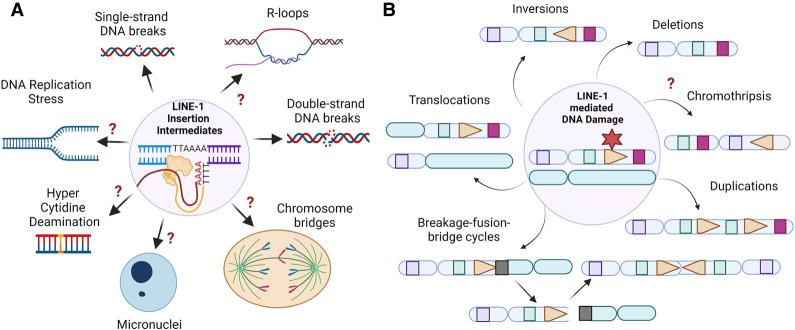
LINE-1 retrotransposition is a source of DNA damage and genome instability. (A) LINE-1 retrotransposition causes DNA damage in cells. Shown is a diagram highlighting the multiple types of DNA damage that might be induced by LINE-1 retrotransposition, including chromosomal breaks, single-strand DNA breaks, DNA replication stress, DNA–RNA hybrids, abnormal nuclear structures such as chromosome bridges and micronuclei, and indirect activation of aberrant cytidine deamination by APOBEC proteins. (B) LINE-1-mediated DNA damage can be a source of chromosomal structural variants (SVs): deletions, duplications, translocations, inversions, breakage–fusion–bridge cycles, and chromothripsis.
DNA damage
LINE-1 has the potential to generate diverse types of DNA lesions via multiple mechanisms (Fig. 5A). For one, LINE-1 retrotransposition causes the most toxic type of DNA damage—chromosomal breaks—in cells via the enzymatic activities of ORF2p (Fig. 5A; Gasior et al. 2006). Consistent with this, cells induced with LINE-1 expression contain elevated levels of several targets of ATM, including γH2AX (S139) and phosphorylated RAD50 (S635) (Gasior et al. 2006; Miyoshi et al. 2019; McKerrow et al. 2022). Nonetheless, a comprehensive analysis of the status of the DNA damage response in cells induced with LINE-1 expression is required to characterize the DNA-damaging effects of LINE-1. In addition, the canonical target sequence of ORF2p EN cutting in the genome has been indirectly determined based on genome-wide mapping of resolved LINE-1 insertions in cells. Future experiments directly mapping ORF2p binding sites and ORF2p-mediated chromosomal breaks using sequencing-based approaches (Canela et al. 2016; Yan et al. 2017) can comprehensively profile the sites of DNA breakage in the genome caused by LINE-1 activity, which are likely sources of chromosomal instability in cancers.
In addition to chromosomal breaks, the TPRT model suggests that LINE-1 retrotransposition may generate other types of toxic DNA lesions in the genome, such as DNA nicks, cDNA–RNA hybrids, and LINE-1 3′ cDNA flaps (Figs. 2, 5A). These structures have been detected in LINE-1 biochemical assays (Feng et al. 1996; Cost et al. 2002; Miyoshi et al. 2019) but have yet to be characterized in cells undergoing retrotransposition. Moreover, not only may LINE-1 cause genotoxicity locally at sites targeted by ORF2p, but LINE-1 retrotransposition and cellular host responses may indirectly cause global genotoxic effects. For example, LINE-1 insertion intermediates may cause DNA replication stress (Fig. 5A) by posing as barriers to DNA replication forks and by activating checkpoint responses that broadly influence DNA replication progression. Additional studies are needed to address the influence of LINE-1 expression on the dynamics of DNA replication. Last, cytidine deaminases such as APOBEC proteins are activated in response to LINE-1 retrotransposition (Muckenfuss et al. 2006; Richardson et al. 2014) and hence have the potential to be sources of aberrant APOBEC-related cytidine deamination and associated mutagenesis in the genome (Fig. 5A). It will be important to address whether LINE-1 directly or indirectly results in specific mutational signatures beyond retrotransposition that can be appreciated in cancer genomes.
Chromosomal instability
LINE-1 retrotransposition and its associated DNA damage can result in a variety of genomic alterations, from small target site alterations to large-scale chromosomal structural variants (SVs) (Fig. 5B). For example, LINE-1 retrotransposition has been known to cause genomic deletions at target sites (target site deletions) (Gilbert et al. 2002). These events are commonly found at EN target sequences, contain 5′-truncated LINE-1 sequences with polyA tails, lack TSDs, and contain genomic deletions of the target site sequences ranging from a few to thousands of base pairs (Gilbert et al. 2002; Symer et al. 2002; Rodriguez-Martin et al. 2020). Strikingly, the pan-cancer analysis uncovered that these retrotransposition-mediated deletions can cause the loss of up to tens of megabase pairs in cancer genomes, resulting in large interstitial deletion rearrangements (Fig. 5B) that can encompass regulatory regions and genes, including tumor suppressor genes (Rodriguez-Martin et al. 2020). In some cases, these deletions also encompass centromeres, which implicates LINE-1 retrotransposition in mitotic errors and the formation of chromosome aneuploidies.
Mechanistically, one model for the formation of retrotransposition-mediated deletions involves the pairing of a LINE-1 insertion intermediate with a distant endogenous chromosomal break upstream of the EN target site that causes the loss of the intervening sequence. Similarly, LINE-1 insertion intermediates on one chromosome can pair with endogenous DNA lesions on another distinct chromosome, causing retrotransposition-mediated translocations (Fig. 5B; Rodriguez-Martin et al. 2020). Nonetheless, the mechanisms of these retrotransposition-mediated deletions and translocations remain to be elucidated. LINE-1 can also cause other types of retrotransposition-mediated SVs, such as chromosomal inversions or duplications that can amplify oncogenes (Fig. 5B), but their etiologies are unknown.
The pan-cancer analysis also revealed that LINE-1 retrotransposition induces complex rearrangements via chromosome breakage–fusion–bridge (BFB) cycles (Fig. 5B; Rodriguez-Martin et al. 2020), which is a mechanism of chromosomal instability originally described by McClintock (1939). One mechanism that LINE-1 is proposed to trigger is a BFB cycle by bridging two broken chromosomes in inverted orientation, generating a dicentric chromosome that forms an anaphase bridge (Fig. 5A), which is an initiating event for rounds of BFB cycles. Another possible outcome of abnormally segregated chromosomes is the formation of micronuclei (Fig. 5A), which are structures with compromised nuclear envelopes containing lagging acentric chromosomes or chromosome fragments (Zhang et al. 2015). However, the frequency of anaphase bridges or micronucleation in cells undergoing retrotransposition has not been experimentally addressed. Both anaphase bridges and micronuclei can trigger chromothripsis (Zhang et al. 2015; Umbreit et al. 2020), which is a rapid mechanism of chromosomal instability characterized by massive, clustered rearrangements of chromosomes often generated “all at once” (Fig. 5B; Stephens et al. 2011). Nonetheless, the contribution of LINE-1 retrotransposition to chromothripsis remains to be addressed.
In summary, these observations suggest that LINE-1-mediated DNA lesions can have profound impact on cancer genome evolution. We expect that long read sequencing of cancer genomes and development of bioinformatic detection tools specially focused on LINE-1-mediated SVs will indicate the frequencies of retrotransposition-mediated SVs in human malignancies. In addition, it is plausible LINE-1 retrotransposition and its associated DNA damage could generate SVs with junctions that lack genomic “scars” of LINE-1 activity, although experiments are needed to fully characterize the chromosomal instability signature induced by LINE-1 retrotransposition. Characterizing the DNA-damaging effects and mutational impact of LINE-1 will require modeling the representative levels and the duration of LINE-1 expression observed in cancers, which might become possible using CRISPR activation systems to induce endogenous LINE-1 expression in experimental settings.
Repeat-mediated genome instability independent of retrotransposition
The abundant and homologous nature of Alu and LINE-1 sequences poses a threat to genome integrity via the formation of chromosomal structural variants (SVs) between nonallelic repeats (repeat-mediated SVs) (Batzer and Deininger 2002; Carvalho and Lupski 2016). For example, germline deletion rearrangements between Alu sequences, which are TEs enriched in introns of genes, can disrupt tumor suppressor genes such as BRCA1, MSH2, and VHL and cause cancer predisposition (Song et al. 2018). In addition to deletions, other repeat-mediated SVs can include translocations, duplications, inversions, and complex rearrangements (Elliott et al. 2005; Gu et al. 2015; Carvalho and Lupski 2016).
Mechanistically, these repeat-mediated SVs are likely generated during the repair of broken chromosomes via the annealing of two repeat sequences from nonallelic loci (Bhargava et al. 2016; Morales et al. 2018; Balachandran et al. 2022). Interestingly, the most common TE substrates involved in repeat-mediated SVs are young Alu sequences, which share the highest degree of sequence homology (Batzer and Deininger 2002; Song et al. 2018; Balachandran et al. 2022). These observations are consistent with cell-based assays demonstrating that sequence divergence between repeat substrates is a major barrier to the formation of repeat-mediated SVs via heteroduplex rejection mechanisms (Morales et al. 2015; Mendez-Dorantes et al. 2018, 2020).
The prevalence of repeat-mediated SVs in cancers (somatic) and in normal tissues (germline) has been challenging to address due to the limitation of resolving rearrangement junctions containing TE sequences. Two recent studies that leveraged long read sequencing approaches revealed that repeat-mediated SVs may be more common than previously reported (Balachandran et al. 2022; Pascarella et al. 2022). For example, a group identified nearly 500 germline repeat-mediated SVs across only three human genomes (Balachandran et al. 2022), suggesting that these events contribute to human variation. Another group estimated one to four somatic repeat-mediated SVs per cell from normal human tissues (Pascarella et al. 2022), indicating that these events may be a source of somatic mosaicism, although sequencing analyses of single cells or single-cell clones are required to corroborate these findings. Repeat-mediated SVs in normal cells are likely double-edged swords, such that they may restore broken chromosomes to prevent whole-chromosome losses at the risk of generating potential harmful mutations. However, in human cancers, repeat-mediated SVs are likely sources of genetic heterogeneity for cancer genome evolution. Long read sequencing of cancer genomes will reveal the prevalence and impact of these events in human malignancies.
TE expression including LINE-1 activates immune responses
Recent impactful studies established that induction of TE expression by exposing cells to epigenetic small molecule inhibitors activates cellular interferon responses (Chiappinelli et al. 2015; Roulois et al. 2015). One mechanism is the generation of cytosolic nucleic acids by TEs that are recognized by sensors that in turn induce a type I interferon (IFN-I) response (Fig. 6; Ishak et al. 2018). This activation of pattern recognition receptor (PRR) pathways by TEs has been termed “viral mimicry,” since these sensors are usually activated by invading viruses (Chan and Gack 2016). These sensors include RIG-I and MDA5, which detect dsRNAs, and cGAS–STING, which detects dsDNAs or DNA:RNA hybrids (Chan and Gack 2016). Understanding how TEs generate immunogenic nucleic acids and how these affect cells in the context of chronic TE activation is important to develop strategies to modulate the immunologic properties of cancer cells.
Figure 6.
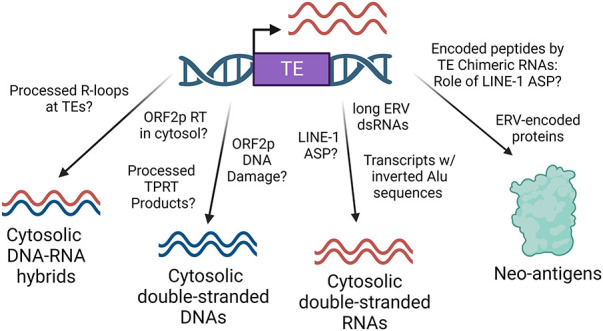
Activation of retrotransposons induces immune responses. TE expression can result in the generation of cytosolic nucleic acids that can induce a type I interferon response via multiple mechanisms—processes termed viral mimicry. For example, TE RNA transcripts, including Alu, ERVs, and LINE-1, have the potential to generate double-stranded RNAs via intramolecular or intermolecular base pairing, which could be sensed by MDA5 and RIG-I to elicit an innate immune response. In addition, DNA-damaging effects of ORF2p EN or cDNA products of ORF2p RT activity could be sources for cytosolic double-stranded DNAs or DNA–RNA hybrids, which could be sensed by cGAS–STING to elicit an innate immune response. Moreover, TE-derived neoantigens could elicit an adaptive immune response, which could be generated via translation of peptides encoded by ERV transcripts containing partially intact ORFs or ORF0 chimeric transcripts generated via aberrant splicing of LINE-1 copies with nearby genes.
Regarding cytosolic dsRNAs, sequencing analysis of cytosolic RNAs protected by MDA5 in cells treated with 5-aza-CdR, an DNMT inhibitor, revealed that most immunogenic dsRNAs are derived from transcripts containing inverted Alu sequences, which are structures relaxed by ADAR1-mediated adenosine deamination (Mehdipour et al. 2020). In addition, expression of ERVs can generate complementary RNAs that form long dsRNAs and induce an IFN-I response via MDA5 and RIG-I (Chiappinelli et al. 2015). Induction of LINE-1 expression using an inducible transgene system in hTERT RPE-1 cells, which lack cGAS–STING, can similarly induce an IFN-I response, but the mechanism for this is less clear (Ardeljan et al. 2020a). Although not modeled by this transgene experimental system, induction of endogenous LINE-1 loci may generate dsRNAs by intramolecular or intermolecular secondary structures involving LINE-1 RNA, including via antisense transcripts induced by the LINE-1 ASP that may complementarily bind the 5′ UTR of sense LINE-1 transcripts (Fig. 6).
Moreover, LINE-1 is proposed to be a source of cytosolic dsDNA and DNA:RNA hybrids that elicit an IFN-I response through cGAS activation (De Cecco et al. 2019; Simon et al. 2019). However, the exact mechanisms and the genetic determinants are poorly understood. Here we speculate on possible mechanisms of how LINE-1 could generate cytosolic dsDNA and DNA:RNA hybrids (Fig. 6). One possible source of cytosolic DNA:RNA hybrids is from LINE-1 retrotransposition in the nucleus, where intermediates of TPRT may be cleaved and exported into the cytosol. Alternatively, ORF2p may synthesize cDNA from LINE-1 RNA in the cytosol independent of retrotransposition in the nucleus. In support of this hypothesis, ORF2p is proposed to make Alu cDNA in the cytosol via an Alu self-priming mechanism (Fukuda et al. 2021a,b). Another likely source of cytosolic LINE-1 DNA may be related to the genotoxic effects of ORF2p that can result in cytosolic dsDNA fragments, including those contained in micronuclei (Fig. 5A). Last, immunogenic DNA–RNA hybrids were recently found to be derived and cleaved from R-loops, which are three-stranded structures harboring an RNA–DNA hybrid and a displaced strand of DNA generated during transcription (Crossley et al. 2023). Nonetheless, it remains to be elucidated whether dysregulated TE loci in cancers are sources of R-loops that can be processed into immunogenic DNA–RNA hybrids.
In addition to immunogenic nucleic acids, another mechanism by which TEs can induce immunologic responses in cancer cells is via generation of neoantigens (Fig. 6). For example, proteins expressed from a subfamily of ERV (HERV-K) are sources of antibodies detected in cancer patients (Boller et al. 1997; Cegolon et al. 2013), suggesting that encoded proteins from TEs can be immunogenic. A recent pan-cancer transcriptome study identified that TEs can generate chimeric transcripts with neighboring genes that result in tumor-specific neoantigens (Shah et al. 2023). The role of the LINE-1 ASP in the formation of ORF0 chimeric transcripts that could in turn produce fusion proteins that serve as neoantigens remains to be addressed (Fig. 6). In summary, there are many potential mechanisms by which TE-derived transcripts and proteins may contribute to cancer cell immunogenicity. Identifying key loci and the underlying mechanism of action may allow us to recognize key biomarkers and develop strategies to enhance immunologic responses to cancers.
Translating LINE-1 and TE deregulation to the clinic
It is now becoming clear that LINE-1 is both a marker and a mutator in human cancers. Previous clinical work investigating global LINE-1 promoter hypomethylation with cancer prognosis laid the foundation for translating LINE-1 biology to the clinic, although these studies resulted in mixed success (Ogino et al. 2008; Saito et al. 2010; Wu et al. 2012; Kupcinskas et al. 2017; Lavasanifar et al. 2019). Recent insights into LINE-1 biology provide relatively untapped directions for translational cancer research, offering a novel cancer protein biomarker. Indeed, recent evidence that ORF1p can be detected inexpensively and noninvasively in blood draws of cancer patients motivates future studies to address whether it may have utility for early diagnosis, disease monitoring, minimal residual disease detection, and patient stratification (Taylor et al. 2023). Significant work remains to be done to validate the clinical utility of plasma ORF1p measurements in particular patient cohorts, including whether it may be most useful as one of the several pan-cancer biomarkers in multianalyte panels. Detection of circulating ORF1p may also prove to be useful as a companion diagnostic if exploiting the genotoxicity and immunogenicity of LINE-1 expression represents new therapeutic strategies for targeting human malignancies.
Inducing genotoxicity in cells using exogenous agents (ionizing radiation, DNA-damaging agents, or small molecule inhibitors targeting DNA repair inhibitors) is a cornerstone of cancer therapy (Cleary et al. 2020). It is conceivable then that LINE-1 may be exploited as an endogenous source of DNA damage in cancer cells via several strategies. For example, epigenetic therapies that selectively induce LINE-1 expression in p53-proficient cancers may induce genotoxic cell death. In support of this hypothesis, a recent study indicated that disruption of the HUSH complex causes genotoxicity via LINE-1 expression in myeloid leukemia cells (Gu et al. 2021). This strategy could also be exploited for ORF1p (+), p53 mutant cancer cells to amplify the genotoxic burden of LINE-1. While levels of the genotoxic ORF2p are low in ORF1p (+) cancers (Ardeljan et al. 2020b), mechanisms that limit this are unknown and may be targetable in a manner that then uniquely exposes malignant cells to ORF2p as compared with normal cells that lack expression of retrotransposition-competent LINE-1 RNAs. Thus, it is intriguing to pursue strategies that modulate the levels of ORF2p by increasing its translation and/or promoting its stability to selectively target ORF1p (+) cancer cells.
Mechanisms that mediate the repair of LINE-1-associated DNA damage will be important to understand and may be targetable in combination with strategies that enhance LINE-1 expression. Recent findings from our group showed that LINE-1 (+) cells are dependent on DNA repair factors for cell growth, including those involved in homologous recombination, the Fanconi anemia pathway, and ATR signaling (e.g., BRCA1, FANCD2, FANCJ, FANCM, BLM, WRN, and ATRIP) (Ardeljan et al. 2020a). These results suggest that LINE-1-expressing cancers may be sensitized to small molecule inhibitors targeting factors in the DNA damage response or DNA repair pathways or to DNA-damaging agents (Cleary et al. 2020; Hopkins et al. 2022). Similarly, strategies that induce LINE-1 expression may be selectively damaging to BRCA-deficient cancers. Finally, nucleoside reverse transcriptase inhibitors (NRTIs) are potent inhibitors of retrotransposition and may alleviate or compound LINE-1-associated DNA damage. NRTIs were recently found to sensitize colorectal cancer cell models with deregulated TE expression via increased DNA damage (Rajurkar et al. 2022), suggesting that possible inhibition of ORF2p via chain terminators may generate toxic DNA lesions. However, additional work is required to determine the exact mechanisms by which NRTIs sensitize cancer cells with deregulated TEs, including addressing whether ORF2p RT or other cellular RTs or error-prone polymerases are the direct target. Development of specific small molecule inhibitors to LINE-1 ORF2p EN and RT are needed to address the consequences of disrupting the enzymatic activities of LINE-1 in human cancers.
Finally, cytosolic nucleic acids and neoantigens in cancers stemming from the expression of TEs appear to be stimulants of antitumor immunity (Shah et al. 2023), although much remains to be understood about which TE loci are most critical in which biologic contexts. The recognition that TEs are sources of immunogenic nucleic acids and neoantigens suggests that antitumor immunity could be regulated via the modulation of TE expression and activity, including via epigenetic therapies such as DNA-hypomethylating agents, HDAC inhibitors, and EZH2 inhibitors (Chiappinelli et al. 2015; Simon et al. 2019; Deblois et al. 2020). Additionally, the immunogenic properties of TE-derived nucleic acids may be directly targeted, such as with ADAR1 inhibition, which promotes dsRNA-mediated immunogenicity (Mehdipour et al. 2020). Neoantigens encoded by TEs may be leveraged as targets for cancer vaccines or cellular therapies. Indeed, TE methylation, TE-derived RNAs, and TE-encoded neoantigens may serve as important indicators of response to checkpoint blockade and other immunotherapeutic strategies (Morel et al. 2021; Ng et al. 2023).
Conclusion
LINE-1 retrotransposons are widely activated in human malignancies, yet contributions of the retroelements to cancer cell biology require elucidation. Research efforts using biochemical, genetic, and bioinformatic approaches have provided insights into the life cycle of LINE-1 and its regulation and established foundations to address the influence of LINE-1 and its associated DNA damage on cancer initiation and evolution. Bridging these connections between LINE-1 biology and cancer biology has the potential to address whether LINE-1 is a cause or a consequence of cellular transformation, as well as shed light on innovative cancer therapeutic strategies.
Acknowledgments
K.H.B. is supported by the National Institutes of Health (R01CA240816, R01CA276112, and UG3NS132127) and by the Dana-Farber Cancer Institute (Innovations Research Fund). C.M.-D. is supported by the Jane Coffin Childs Memorial Fund for Medical Research. Figures were generated with BioRender.com.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.351051.123.
Freely available online through the Genes & Development Open Access Option.
Competing interest statement
The authors declare no competing interests.
References
- Achanta P, Steranka JP, Tang Z, Rodić N, Sharma R, Yang WR, Ma S, Grivainis M, Huang CRL, Schneider AM, et al. 2016. Somatic retrotransposition is infrequent in glioblastomas. Mob DNA 7: 22. 10.1186/s13100-016-0077-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adney EM, Ochmann MT, Sil S, Truong DM, Mita P, Wang X, Kahler DJ, Fenyö D, Holt LJ, Boeke JD. 2019. Comprehensive scanning mutagenesis of human retrotransposon LINE-1 identifies motifs essential for function. Genetics 213: 1401–1414. 10.1534/genetics.119.302601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Eastman QM, Schatz DG. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394: 744–751. 10.1038/29457 [DOI] [PubMed] [Google Scholar]
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. 2006. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev 20: 210–224. 10.1101/gad.1380406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeljan D, Taylor MS, Ting DT, Burns KH. 2017. The human long interspersed element-1 retrotransposon: an emerging biomarker of neoplasia. Clin Chem 63: 816–822. 10.1373/clinchem.2016.257444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeljan D, Steranka JP, Liu C, Li Z, Taylor MS, Payer LM, Gorbounov M, Sarnecki JS, Deshpande V, Hruban RH, et al. 2020a. Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat Struct Mol Biol 27: 168–178. 10.1038/s41594-020-0372-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeljan D, Wang X, Oghbaie M, Taylor MS, Husband D, Deshpande V, Steranka JP, Gorbounov M, Yang WR, Sie B, et al. 2020b. LINE-1 ORF2p expression is nearly imperceptible in human cancers. Mob DNA 11: 1. 10.1186/s13100-019-0191-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attig J, Agostini F, Gooding C, Chakrabarti AM, Singh A, Haberman N, Zagalak JA, Emmett W, Smith CWJ, Luscombe NM, et al. 2018. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell 174: 1067–1081.e17. 10.1016/j.cell.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran P, Walawalkar IA, Flores JI, Dayton JN, Audano PA, Beck CR. 2022. Transposable element-mediated rearrangements are prevalent in human genomes. Nat Commun 13: 7115. 10.1038/s41467-022-34810-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet 7: 149–173. 10.1146/annurev.genom.7.080505.115700 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379. 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2016. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol 8: a019505. 10.1101/cshperspect.a019505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170. 10.1016/j.cell.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. 2011. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12: 187–215. 10.1146/annurev-genom-082509-141802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Guijarro M, Lopez-Ruiz C, Tarnauskaitė Ž, Murina O, Mian Mohammad M, Williams TC, Fluteau A, Sanchez L, Vilar-Astasio R, Garcia-Canadas M, et al. 2018. RNase H2, mutated in Aicardi-Goutières syndrome, promotes LINE-1 retrotransposition. EMBO J 37: e98506. 10.15252/embj.201798506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrens RV, Yang A, Laumer CE, Lun ATL, Bieberich F, Law CT, Lan G, Imaz M, Bowness JS, Brockdorff N, et al. 2022. Locus-specific expression of transposable elements in single cells with CELLO-seq. Nat Biotechnol 40: 546–554. 10.1038/s41587-021-01093-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Onyango DO, Stark JM. 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet 32: 566–575. 10.1016/j.tig.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17: 915–928. 10.1093/oxfordjournals.molbev.a026372 [DOI] [PubMed] [Google Scholar]
- Boller K, Janssen O, Schuldes H, Tönjes RR, Kurth R. 1997. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J Virol 71: 4581–4588. 10.1128/jvi.71.6.4581-4588.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bona N, Crossan GP. 2023. Fanconi anemia DNA crosslink repair factors protect against LINE-1 retrotransposition during mouse development. Nat Struct Mol Biol 30: 1434–1445. 10.1038/s41594-023-01067-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, et al. 2018. Ten things you should know about transposable elements. Genome Biol 19: 199. 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs EM, McKerrow W, Mita P, Boeke JD, Logan SK, Fenyö D. 2021. RIP-seq reveals LINE-1 ORF1p association with p-body enriched mRNAs. Mob DNA 12: 5. 10.1186/s13100-021-00233-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE. 1968. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161: 529–540. 10.1126/science.161.3841.529 [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285. 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH. 2017. Transposable elements in cancer. Nat Rev Cancer 17: 415–424. 10.1038/nrc.2017.35 [DOI] [PubMed] [Google Scholar]
- Burns KH. 2022. Repetitive DNA in disease. Science 376: 353–354. 10.1126/science.abl7399 [DOI] [PubMed] [Google Scholar]
- Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics 80: 402–406. 10.1006/geno.2002.6843 [DOI] [PubMed] [Google Scholar]
- Cajuso T, Sulo P, Tanskanen T, Katainen R, Taira A, Hänninen UA, Kondelin J, Forsström L, Välimäki N, Aavikko M, et al. 2019. Retrotransposon insertions can initiate colorectal cancer and are associated with poor survival. Nat Commun 10: 4022. 10.1038/s41467-019-11770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, Nussenzweig A. 2016. DNA breaks and end resection measured genome-wide by end sequencing. Mol Cell 63: 898–911. 10.1016/j.molcel.2016.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira PE, Ewing AD, Li G, Schauer SN, Upton KR, Fagg AC, Morell S, Kindlova M, Gerdes P, Richardson SR, et al. 2016. Evidence for L1-associated DNA rearrangements and negligible L1 retrotransposition in glioblastoma multiforme. Mob DNA 7: 21. 10.1186/s13100-016-0076-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Lupski JR. 2016. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet 17: 224–238. 10.1038/nrg.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. 2014. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409. 10.1101/gad.241661.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegolon L, Salata C, Weiderpass E, Vineis P, Palù G, Mastrangelo G. 2013. Human endogenous retroviruses and cancer prevention: evidence and prospects. BMC Cancer 13: 4. 10.1186/1471-2407-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. 2004. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 23: 8841–8846. 10.1038/sj.onc.1208137 [DOI] [PubMed] [Google Scholar]
- Chan YK, Gack MU. 2016. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol 14: 360–373. 10.1038/nrmicro.2016.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouly G, Zhao J, McDevitt S, Rusanov T, Hoang T, Borisonnik N, Treddinick T, Lopezcolorado FW, Kent T, Siddique LA, et al. 2021. Polθ reverse transcribes RNA and promotes RNA-templated DNA repair. Sci Adv 7: eabf1771. 10.1126/sciadv.abf1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Lin EW, Tran A, Jin H, Ho NI, Veit A, Cortes-Ciriano I, Burns KH, Ting DT, Park PJ. 2023. The landscape of human SVA retrotransposons. Nucleic Acids Res 51: 11453–11465. 10.1093/nar/gkad821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros-Aguirre M, Ping X, Stark JM. 2022. To indel or not to indel: factors influencing mutagenesis during chromosomal break end joining. DNA Repair 118: 103380. 10.1016/j.dnarep.2022.103380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JM, Aguirre AJ, Shapiro GI, D'Andrea AD. 2020. Biomarker-guided development of DNA repair inhibitors. Mol Cell 78: 1070–1085. 10.1016/j.molcel.2020.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke SL, Shlien A, Marshall J, Pipinikas CP, Martincorena I, Tubio JM, Li Y, Menzies A, Mudie L, Ramakrishna M, et al. 2014. Processed pseudogenes acquired somatically during cancer development. Nat Commun 5: 3644. 10.1038/ncomms4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J 21: 5899–5910. 10.1093/emboj/cdf592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley MP, Song C, Bocek MJ, Choi JH, Kousorous J, Sathirachinda A, Lin C, Brickner JR, Bai G, Lans H, et al. 2023. R-loop-derived cytoplasmic RNA–DNA hybrids activate an immune response. Nature 613: 187–194. 10.1038/s41586-022-05545-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks HA, Tufarelli C. 2009. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 94: 397–406. 10.1016/j.ygeno.2009.08.013 [DOI] [PubMed] [Google Scholar]
- Dai L, Taylor MS, O'Donnell KA, Boeke JD. 2012. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol Cell Biol 32: 4323–4336. 10.1128/MCB.06785-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblois G, Tonekaboni SAM, Grillo G, Martinez C, Kao YI, Tai F, Ettayebi I, Fortier AM, Savage P, Fedor AN, et al. 2020. Epigenetic switch-induced viral mimicry evasion in chemotherapy-resistant breast cancer. Cancer Discov 10: 1312–1329. 10.1158/2159-8290.CD-19-1493 [DOI] [PubMed] [Google Scholar]
- De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD, et al. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566: 73–78. 10.1038/s41586-018-0784-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cubas AA, Dunker W, Zaninovich A, Hongo RA, Bhatia A, Panda A, Beckermann KE, Bhanot G, Ganesan S, Karijolich J, et al. 2020. DNA hypomethylation promotes transposable element expression and activation of immune signaling in renal cell cancer. JCI Insight 5: e137569. 10.1172/jci.insight.137569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P, Morales ME, White TB, Baddoo M, Hedges DJ, Servant G, Srivastav S, Smither ME, Concha M, DeHaro DL, et al. 2017. A comprehensive approach to expression of L1 loci. Nucleic Acids Res 45: e31. 10.1093/nar/gkw1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, Diedrich JK, Aslanian A, Ma J, Moresco JJ, et al. 2015. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163: 583–593. 10.1016/j.cell.2015.09.025 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48. 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res 16: 1548–1556. 10.1101/gr.5565706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr. 1991. Isolation of an active human transposable element. Science 254: 1805–1808. 10.1126/science.1662412 [DOI] [PubMed] [Google Scholar]
- Doucet AJ, Wilusz JE, Miyoshi T, Liu Y, Moran JV. 2015. A 3′ poly(A) tract is required for LINE-1 retrotransposition. Mol Cell 60: 728–741. 10.1016/j.molcel.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Jasin M. 2005. Chromosomal translocation mechanisms at intronic Alu elements in mammalian cells. Mol Cell 17: 885–894. 10.1016/j.molcel.2005.02.028 [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat Genet 24: 363–367. 10.1038/74184 [DOI] [PubMed] [Google Scholar]
- Estécio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. 2007. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One 2: e399. 10.1371/journal.pone.0000399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916. 10.1016/S0092-8674(00)81997-2 [DOI] [PubMed] [Google Scholar]
- Flasch DA, Macia A, Sánchez L, Ljungman M, Heras SR, García-Pérez JL, Wilson TE, Moran JV. 2019. Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication. Cell 177: 837–851.e28. 10.1016/j.cell.2019.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Narendran S, Varshney A, Nagasaka Y, Wang SB, Ambati K, Apicella I, Pereira F, Fowler BJ, Yasuma T, et al. 2021a. Alu complementary DNA is enriched in atrophic macular degeneration and triggers retinal pigmented epithelium toxicity via cytosolic innate immunity. Sci Adv 7: eabj3658. 10.1126/sciadv.abj3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Varshney A, Fowler BJ, Wang SB, Narendran S, Ambati K, Yasuma T, Magagnoli J, Leung H, Hirahara S, et al. 2021b. Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc Natl Acad Sci 118: e2022751118. 10.1073/pnas.2022751118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL. 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol 357: 1383–1393. 10.1016/j.jmb.2006.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Roy-Engel AM, Deininger PL. 2008. ERCC1/XPF limits L1 retrotransposition. DNA Repair 7: 983–989. 10.1016/j.dnarep.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315–325. 10.1016/S0092-8674(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH Jr. 2012. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 8: e1002941. 10.1371/journal.pgen.1002941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH Jr. 2013. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res 41: 7401–7419. 10.1093/nar/gkt512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Yuan B, Campbell IM, Beck CR, Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, et al. 2015. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet 24: 4061– 4077. 10.1093/hmg/ddv146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Liu Y, Zhang Y, Cao H, Lyu J, Wang X, Wylie A, Newkirk SJ, Jones AE, Lee M, et al. 2021. Silencing of LINE-1 retrotransposons is a selective dependency of myeloid leukemia. Nat Genet 53: 672–682. 10.1038/s41588-021-00829-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Gagnon D, Meng L, Perez-Iratxeta C, Lagace D, Gibbings D. 2014. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun 5: 5276. 10.1038/ncomms6276 [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr. 2010. SVA retrotransposons: evolution and genetic instability. Semin Cancer Biol 20: 234–245. 10.1016/j.semcancer.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7: 9. 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH Jr. 2011. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet 20: 3386–3400. 10.1093/hmg/ddr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, Levine AJ, Hoh J. 2009. p53 responsive elements in human retrotransposons. Oncogene 28: 3857–3865. 10.1038/onc.2009.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. 2014. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res 24: 1053–1063. 10.1101/gr.163659.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssen AG, Henaff E, Jiang E, Eisenberg AR, Carson JR, Villasante CM, Ray M, Still E, Burns M, Gandara J, et al. 2015. Genomic DNA transposition induced by human PGBD5. Elife 4: e10565. 10.7554/eLife.10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins JL, Lan L, Zou L. 2022. DNA repair defects in cancer and therapeutic opportunities. Genes Dev 36: 278–293. 10.1101/gad.349431.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt SJ, Storer JM, Hartley GA, Grady PGS, Gershman A, de Lima LG, Limouse C, Halabian R, Wojenski L, Rodriguez M, et al. 2022. From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. Science (New York, NY) 376: eabk3112. 10.1126/science.abk3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L, Pang X, Cen S, Jin Q, Liang C, et al. 2015. SAMHD1 Inhibits LINE-1 retrotransposition by promoting stress granule formation. PLoS Genet 11: e1005367. 10.1371/journal.pgen.1005367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, et al. 2010. Mobile interspersed repeats are major structural variants in the human genome. Cell 141: 1171–1182. 10.1016/j.cell.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, et al. 2016. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell 166: 102–114. 10.1016/j.cell.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SY, Jung H, Mun S, Lee S, Park K, Baek SC, Moon HC, Kim H, Kim B, Choi Y, et al. 2021. L1 retrotransposons exploit RNA m6A modification as an evolutionary driving force. Nat Commun 12: 880. 10.1038/s41467-021-21197-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishak CA, Classon M, De Carvalho DD. 2018. Deregulation of retroelements as an emerging therapeutic opportunity in cancer. Trends Cancer 4: 583–597. 10.1016/j.trecan.2018.05.008 [DOI] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141: 1253–1261. 10.1016/j.cell.2010.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangam D, Feschotte C, Betrán E. 2017. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet 33: 817–831. 10.1016/j.tig.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszyk K, Li PW, Villareal V, Branciforte D, Wu H, Xie Y, Feigon J, Loo JA, Martin SL, Clubb RT. 2007. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J Biol Chem 282: 24893–24904. 10.1074/jbc.M702023200 [DOI] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M. 2015. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31: 3593–3599. 10.1093/bioinformatics/btv422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly-Lopez Z, Bureau TE. 2018. Exaptation of transposable element coding sequences. Curr Opin Genet Dev 49: 34–42. 10.1016/j.gde.2018.02.011 [DOI] [PubMed] [Google Scholar]
- Jung H, Choi JK, Lee EA. 2018. Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers. Genome Res 28: 1136–1146. 10.1101/gr.231837.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332: 164–166. 10.1038/332164a0 [DOI] [PubMed] [Google Scholar]
- Khazina E, Weichenrieder O. 2009. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci 106: 731–736. 10.1073/pnas.0809964106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E, Truffault V, Büttner R, Schmidt S, Coles M, Weichenrieder O. 2011. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat Struct Mol Biol 18: 1006–1014. 10.1038/nsmb.2097 [DOI] [PubMed] [Google Scholar]
- Kojima KK. 2010. Different integration site structures between L1 protein-mediated retrotransposition in cis and retrotransposition in trans. Mob DNA 1: 17. 10.1186/1759-8753-1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopera HC, Moldovan JB, Morrish TA, Garcia-Perez JL, Moran JV. 2011. Similarities between long interspersed element-1 (LINE-1) reverse transcriptase and telomerase. Proc Natl Acad Sci 108: 20345–20350. 10.1073/pnas.1100275108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopera HC, Larson PA, Moldovan JB, Richardson SR, Liu Y, Moran JV. 2016. LINE-1 Cultured cell retrotransposition assay. Methods Mol Biol 1400: 139–156. 10.1007/978-1-4939-3372-3_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. 2006. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol 13: 655–660. 10.1038/nsmb1107 [DOI] [PubMed] [Google Scholar]
- Kupcinskas J, Steponaitiene R, Langner C, Smailyte G, Skieceviciene J, Kupcinskas L, Malfertheiner P, Link A. 2017. LINE-1 hypomethylation is not a common event in preneoplastic stages of gastric carcinogenesis. Sci Rep 7: 4828. 10.1038/s41598-017-05143-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lavasanifar A, Sharp CN, Korte EA, Yin T, Hosseinnejad K, Jortani SA. 2019. Long interspersed nuclear element-1 mobilization as a target in cancer diagnostics, prognostics and therapeutics. Clin Chim Acta 493: 52–62. 10.1016/j.cca.2019.02.015 [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, Lohr JG, Harris CC, Ding L, Wilson RK, et al. 2012. Landscape of somatic retrotransposition in human cancers. Science 337: 967–971. 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Lee CH, Swigut T, Grow E, Gu B, Bassik MC, Wysocka J. 2018. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553: 228– 232. 10.1038/nature25179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH. 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol 15: 3882–3891. 10.1128/MCB.15.7.3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. 10.1016/0092-8674(93)90078-5 [DOI] [PubMed] [Google Scholar]
- Luqman-Fatah A, Watanabe Y, Uno K, Ishikawa F, Moran JV, Miyoshi T. 2023. The interferon stimulated gene-encoded protein HELZ2 inhibits human LINE-1 retrotransposition and LINE-1 RNA-mediated type I interferon induction. Nat Commun 14: 203. 10.1038/s41467-022-35757-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A, Widmann TJ, Heras SR, Ayllon V, Sanchez L, Benkaddour-Boumzaouad M, Muñoz-Lopez M, Rubio A, Amador-Cubero S, Blanco-Jimenez E, et al. 2017. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res 27: 335–348. 10.1101/gr.206805.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Singh A, Rio DC. 2013. The human THAP9 gene encodes an active P-element DNA transposase. Science (New York, NY) 339: 446–448. 10.1126/science.1231789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. 1999. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 16: 793–805. 10.1093/oxfordjournals.molbev.a026164 [DOI] [PubMed] [Google Scholar]
- Martin SL, Branciforte D, Keller D, Bain DL. 2003. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci 100: 13815–13820. 10.1073/pnas.2336221100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC. 2005. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol 348: 549–561. 10.1016/j.jmb.2005.03.003 [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. 1991. Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810. 10.1126/science.1722352 [DOI] [PubMed] [Google Scholar]
- McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci 36: 344–355. 10.1073/pnas.36.6.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci 25: 405–416. 10.1073/pnas.25.8.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrow W, Fenyö D. 2020. L1EM: a tool for accurate locus specific LINE-1 RNA quantification. Bioinformatics 36: 1167–1173. 10.1093/bioinformatics/btz724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrow W, Wang X, Mendez-Dorantes C, Mita P, Cao S, Grivainis M, Ding L, LaCava J, Burns KH, Boeke JD, et al. 2022. LINE-1 expression in cancer correlates with p53 mutation, copy number alteration, and S phase checkpoint. Proc Natl Acad Sci 119: e2115999119. 10.1073/pnas.2115999119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdipour P, Marhon SA, Ettayebi I, Chakravarthy A, Hosseini A, Wang Y, de Castro FA, Loo Yau H, Ishak C, Abelson S, et al. 2020. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588: 169–173. 10.1038/s41586-020-2844-1 [DOI] [PubMed] [Google Scholar]
- Mendez-Dorantes C, Bhargava R, Stark JM. 2018. Repeat-mediated deletions can be induced by a chromosomal break far from a repeat, but multiple pathways suppress such rearrangements. Genes Dev 32: 524–536. 10.1101/gad.311084.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Dorantes C, Tsai LJ, Jahanshir E, Lopezcolorado FW, Stark JM. 2020. BLM has Contrary effects on repeat-mediated deletions, based on the distance of DNA DSBs to a repeat and repeat divergence. Cell Rep 30: 1342–1357.e4. 10.1016/j.celrep.2020.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. 1992. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 52: 643–645. [PubMed] [Google Scholar]
- Mita P, Wudzinska A, Sun X, Andrade J, Nayak S, Kahler DJ, Badri S, LaCava J, Ueberheide B, Yun CY, et al. 2018. LINE-1 protein localization and functional dynamics during the cell cycle. Elife 7: e30058. 10.7554/eLife.30058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P, Sun X, Fenyo D, Kahler DJ, Li D, Agmon N, Wudzinska A, Keegan S, Bader JS, Yun C, et al. 2020. BRCA1 and S phase DNA repair pathways restrict LINE-1 retrotransposition in human cells. Nat Struct Mol Biol 27: 179–191. 10.1038/s41594-020-0374-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Makino T, Moran JV. 2019. Poly(ADP-Ribose) polymerase 2 recruits replication protein a to sites of LINE-1 integration to facilitate retrotransposition. Mol Cell 75: 1286–1298.e12. 10.1016/j.molcel.2019.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan JB, Wang Y, Shuman S, Mills RE, Moran JV. 2019. RNA ligation precedes the retrotransposition of U6/LINE-1 chimeric RNA. Proc Natl Acad Sci 116: 20612–20622. 10.1073/pnas.1805404116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales ME, White TB, Streva VA, DeFreece CB, Hedges DJ, Deininger PL. 2015. The contribution of alu elements to mutagenic DNA double-strand break repair. PLoS Genet 11: e1005016. 10.1371/journal.pgen.1005016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales ME, Kaul T, Deininger P. 2018. Long-distance relationships: suppression of repeat-mediated deletions. Trends Genet 34: 572–574. 10.1016/j.tig.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. 10.1016/S0092-8674(00)81998-4 [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH Jr. 1999. Exon shuffling by L1 retrotransposition. Science (New York, NY) 283: 1530–1534. 10.1126/science.283.5407.1530 [DOI] [PubMed] [Google Scholar]
- Morel KL, Sheahan AV, Burkhart DL, Baca SC, Boufaied N, Liu Y, Qiu X, Cañadas I, Roehle K, Heckler M, et al. 2021. EZH2 inhibition activates a dsRNA-STING-interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat Cancer 2: 444–456. 10.1038/s43018-021-00185-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 31: 159–165. 10.1038/ng898 [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perković M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 281: 22161–22172. 10.1074/jbc.M601716200 [DOI] [PubMed] [Google Scholar]
- Nam CH, Youk J, Kim JY, Lim J, Park JW, Oh SA, Lee HJ, Park JW, Won H, Lee Y, et al. 2023. Widespread somatic L1 retrotransposition in normal colorectal epithelium. Nature 617: 540–547. 10.1038/s41586-023-06046-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden Berg N, Hogarth CA, Marchetto MCN, Muotri AR, Griswold MD, et al. 2017. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci 114: E5635–E5644. 10.1073/pnas.1701069114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KW, Boumelha J, Enfield KSS, Almagro J, Cha H, Pich O, Karasaki T, Moore DA, Salgado R, Sivakumar M, et al. 2023. Antibodies against endogenous retroviruses promote lung cancer immunotherapy. Nature 616: 563–573. 10.1038/s41586-023-05771-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THM, Carreira PE, Sanchez-Luque FJ, Schauer SN, Fagg AC, Richardson SR, Davies CM, Jesuadian JS, Kempen MHC, Troskie RL, et al. 2018. L1 retrotransposon heterogeneity in ovarian tumor cell evolution. Cell Rep 23: 3730–3740. 10.1016/j.celrep.2018.05.090 [DOI] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Mätlik K, Speek M. 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634. 10.1006/geno.2002.6758 [DOI] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. 2008. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 100: 1734–1738. 10.1093/jnci/djn359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Kawakami M, Takezawa T. 1987. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res 15: 8725–8737. 10.1093/nar/15.21.8725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr. 2001. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res 11: 2059–2065. 10.1101/gr.205701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Mohr G, Yao J, Russell R, Lambowitz AM. 2022. Group II intron-like reverse transcriptases function in double-strand break repair. Cell 185: 3671–3688.e23. 10.1016/j.cell.2022.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascarella G, Hon CC, Hashimoto K, Busch A, Luginbühl J, Parr C, Hin Yip W, Abe K, Kratz A, Bonetti A, et al. 2022. Recombination of repeat elements generates somatic complexity in human genomes. Cell 185: 3025–3040.e6. 10.1016/j.cell.2022.06.032 [DOI] [PubMed] [Google Scholar]
- Payer LM, Burns KH. 2019. Transposable elements in human genetic disease. Nat Rev Genet 20: 760–772. 10.1038/s41576-019-0165-8 [DOI] [PubMed] [Google Scholar]
- Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, Corbin A, Nigumann P, Cristofari G. 2016. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife 5: e13926. 10.7554/eLife.13926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanic TR 2nd, Asaka S, Lin SF, Yen TT, Sun H, Bahadirli-Talbott A, Wang TH, Burns KH, Wang TL, Shih IM. 2019. Long interspersed nuclear element 1 retrotransposons become deregulated during the development of ovarian cancer precursor lesions. Am J Pathol 189: 513–520. 10.1016/j.ajpath.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan B, Cajuso T, Katainen R, Sulo P, Tanskanen T, Kilpivaara O, Pitkänen E, Aaltonen LA, Kauppi L, Palin K. 2017. Detection of subclonal L1 transductions in colorectal cancer by long-distance inverse-PCR and nanopore sequencing. Sci Rep 7: 14521. 10.1038/s41598-017-15076-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajurkar M, Parikh AR, Solovyov A, You E, Kulkarni AS, Chu C, Xu KH, Jaicks C, Taylor MS, Wu C, et al. 2022. Reverse transcriptase inhibition disrupts repeat element life cycle in colorectal cancer. Cancer Discov 12: 1462–1481. 10.1158/2159-8290.CD-21-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. 2014. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife 3: e02008. 10.7554/eLife.02008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbez-Masson L, Tie CHC, Conde L, Tunbak H, Husovsky C, Tchasovnikarova IA, Timms RT, Herrero J, Lehner PJ, Rowe HM. 2018. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res 28: 836–845. 10.1101/gr.228171.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, Hruban RH, Iacobuzio-Donahue CA, Maitra A, Torbenson MS, et al. 2014. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol 184: 1280–1286. 10.1016/j.ajpath.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodic N, Steranka JP, Makohon-Moore A, Moyer A, Shen P, Sharma R, Kohutek ZA, Huang CR, Ahn D, Mita P, et al. 2015. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med 21: 1060–1064. 10.1038/nm.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, Santamarina M, Ju YS, Temes J, Garcia-Souto D, et al. 2020. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet 52: 306–319. 10.1038/s41588-019-0562-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Craig NL. 1998. VDJ recombination: a transposase goes to work. Cell 94: 411–414. 10.1016/S0092-8674(00)81580-9 [DOI] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162: 961–973. 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kawakami K, Matsumoto I, Oda M, Watanabe G, Minamoto T. 2010. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer. Clin Cancer Res 16: 2418–2426. 10.1158/1078-0432.CCR-09-2819 [DOI] [PubMed] [Google Scholar]
- Sato S, Gillette M, de Santiago PR, Kuhn E, Burgess M, Doucette K, Feng Y, Mendez-Dorantes C, Ippoliti PJ, Hobday S, et al. 2023. LINE-1 ORF1p as a candidate biomarker in high grade serous ovarian carcinoma. Sci Rep 13: 1537. 10.1038/s41598-023-28840-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. 2016. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res 26: 745–755. 10.1101/gr.201814.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seczynska M, Lehner PJ. 2023. The sound of silence: mechanisms and implications of HUSH complex function. Trends Genet 39: 251–267. 10.1016/j.tig.2022.12.005 [DOI] [PubMed] [Google Scholar]
- Seczynska M, Bloor S, Cuesta SM, Lehner PJ. 2022. Genome surveillance by HUSH-mediated silencing of intronless mobile elements. Nature 601: 440–445. 10.1038/s41586-021-04228-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Streva VA, Derbes RS, Wijetunge MI, Neeland M, White TB, Belancio VP, Roy-Engel AM, Deininger PL. 2017. The nucleotide excision repair pathway limits L1 retrotransposition. Genetics 205: 139–153. 10.1534/genetics.116.188680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NM, Jang HJ, Liang Y, Maeng JH, Tzeng SC, Wu A, Basri NL, Qu X, Fan C, Li A, et al. 2023. Pan-cancer analysis identifies tumor-specific antigens derived from transposable elements. Nat Genet 55: 631–639. 10.1038/s41588-023-01349-3 [DOI] [PubMed] [Google Scholar]
- Shi X, Seluanov A, Gorbunova V. 2007. Cell divisions are required for L1 retrotransposition. Mol Cell Biol 27: 1264–1270. 10.1128/MCB.01888-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil S, Keegan S, Ettefa F, Denes LT, Boeke JD, Holt LJ. 2023. Condensation of LINE-1 is critical for retrotransposition. Elife 12: e82991. 10.7554/eLife.82991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Van Meter M, Ablaeva J, Ke Z, Gonzalez RS, Taguchi T, De Cecco M, Leonova KI, Kogan V, Helfand SL, et al. 2019. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab 29: 871–885.e5. 10.1016/j.cmet.2019.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. 1988. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 8: 1385–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. 1996. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6: 743–748. 10.1016/S0959-437X(96)80030-X [DOI] [PubMed] [Google Scholar]
- Smit AF, Riggs AD. 1996. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci 93: 1443–1448. 10.1073/pnas.93.4.1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyom S, Ewing AD, Hancks DC, Takeshima Y, Awano H, Matsuo M, Kazazian HH Jr. 2012. Pathogenic orphan transduction created by a nonreference LINE-1 retrotransposon. Hum Mutat 33: 369–371. 10.1002/humu.21663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Beck CR, Du R, Campbell IM, Coban-Akdemir Z, Gu S, Breman AM, Stankiewicz P, Ira G, Shaw CA, et al. 2018. Predicting human genes susceptible to genomic instability associated with Alu/Alu-mediated rearrangements. Genome Res 28: 1228–1242. 10.1101/gr.229401.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21: 1973–1985. 10.1128/MCB.21.6.1973-1985.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana T, van Essen D, Siol O, Bailly-Bechet M, Philippe C, Zine El Aabidine A, Pioger L, Nigumann P, Saccani S, Andrau JC, et al. 2019. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol Cell 74: 555–570.e7. 10.1016/j.molcel.2019.02.036 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Yamaguchi K, Kajikawa M, Ichiyanagi K, Adachi N, Koyama H, Takeda S, Okada N. 2009. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet 5: e1000461. 10.1371/journal.pgen.1000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110: 327–338. 10.1016/S0092-8674(02)00839-5 [DOI] [PubMed] [Google Scholar]
- Tao J, Wang Q, Mendez-Dorantes C, Burns KH, Chiarle R. 2022. Frequency and mechanisms of LINE-1 retrotransposon insertions at CRISPR/Cas9 sites. Nat Commun 13: 3685. 10.1038/s41467-022-31322-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, LaCava J, Mita P, Molloy KR, Huang CR, Li D, Adney EM, Jiang H, Burns KH, Chait BT, et al. 2013. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155: 1034–1048. 10.1016/j.cell.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, Altukhov I, Molloy KR, Mita P, Jiang H, Adney EM, Wudzinska A, Badri S, Ischenko D, Eng G, et al. 2018. Dissection of affinity captured LINE-1 macromolecular complexes. Elife 7: e30094. 10.7554/eLife.30094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, Wu C, Fridy PC, Zhang SJ, Senussi Y, Wolters JC, Cajuso T, Cheng WC, Heaps JD, Miller BD, et al. 2023. Ultrasensitive detection of circulating LINE-1 ORF1p as a specific multi-cancer biomarker. Cancer Discov 10.1158/2159-8290.CD-23-0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari B, Jones AE, Caillet CJ, Das S, Royer SK, Abrams JM. 2020. p53 directly represses human LINE1 transposons. Genes Dev 34: 1439–1451. 10.1101/gad.343186.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristán-Ramos P, Rubio-Roldan A, Peris G, Sánchez L, Amador-Cubero S, Viollet S, Cristofari G, Heras SR. 2020. The tumor suppressor microRNA let-7 inhibits human LINE-1 retrotransposition. Nat Commun 11: 5712. 10.1038/s41467-020-19430-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al. 2014. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345: 1251343. 10.1126/science.1251343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbak H, Enriquez-Gasca R, Tie CHC, Gould PA, Mlcochova P, Gupta RK, Fernandes L, Holt J, van der Veen AG, Giampazolias E, et al. 2020. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat Commun 11: 5387. 10.1038/s41467-020-19170-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Zhang CZ, Lynch LD, Blaine LJ, Cheng AM, Tourdot R, Sun L, Almubarak HF, Judge K, Mitchell TJ, et al. 2020. Mechanisms generating cancer genome complexity from a single cell division error. Science 368: eaba0712. 10.1126/science.aba0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. 2005. SVA elements: a hominid-specific retroposon family. J Mol Biol 354: 994–1007. 10.1016/j.jmb.2005.09.085 [DOI] [PubMed] [Google Scholar]
- Weber B, Kimhi S, Howard G, Eden A, Lyko F. 2010. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29: 5775–5784. 10.1038/onc.2010.227 [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 21: 1429–1439. 10.1128/MCB.21.4.1429-1439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S. 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408: 167–173. 10.1038/35041507 [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. 2007. A unified classification system for eukaryotic transposable elements. Nature reviews Genetics 8: 973–982. 10.1038/nrg2165 [DOI] [PubMed] [Google Scholar]
- Wilkinson ME, Frangieh CJ, Macrae RK, Zhang F. 2023. Structure of the R2 non-LTR retrotransposon initiating target-primed reverse transcription. Science 380: 301–308. 10.1126/science.adg7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. 2010. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet 6: e1000917. 10.1371/journal.pgen.1000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. 1997. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem 272: 7810–7816. 10.1074/jbc.272.12.7810 [DOI] [PubMed] [Google Scholar]
- Wu HC, Delgado-Cruzata L, Flom JD, Perrin M, Liao Y, Ferris JS, Santella RM, Terry MB. 2012. Repetitive element DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Carcinogenesis 33: 1946–1952. 10.1093/carcin/bgs201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Cochrane DR, Tessier-Cloutier B, Leung S, Karnezis AN, Cheng AS, Farnell DA, Magrill J, Schmidt D, Kommoss S, et al. 2019. Expression of L1 retrotransposon open reading frame protein 1 in gynecologic cancers. Hum Pathol 92: 39–47. 10.1016/j.humpath.2019.06.001 [DOI] [PubMed] [Google Scholar]
- Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, Custodio J, Wernersson E, Li Y, Gao L, et al. 2017. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8: 15058. 10.1038/ncomms15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. 2004. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32: 38e. 10.1093/nar/gnh032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WR, Ardeljan D, Pacyna CN, Payer LM, Burns KH. 2019. SQuIRE reveals locus-specific regulation of interspersed repeat expression. Nucleic Acids Res 47: e27. 10.1093/nar/gky1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. 2015. Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, Hanschmann KM, Morrish TA, Löwer J, Schumann GG. 2005. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res 15: 780–789. 10.1101/gr.3421505 [DOI] [PMC free article] [PubMed] [Google Scholar]