Abstract
The PheP protein is a high-affinity phenylalanine-specific permease of the bacterium Escherichia coli. A topological model based on genetic analysis involving the construction of protein fusions with alkaline phosphatase has previously been proposed in which PheP has 12 transmembrane segments with both N and C termini located in the cytoplasm (J. Pi and A. J. Pittard, J. Bacteriol. 178:2650–2655, 1996). Site-directed mutagenesis has been used to investigate the functional importance of each of the 16 proline residues of the PheP protein. Replacement of alanine at only three positions, P54, P341, and P442, resulted in the loss of 50% or more activity. Substitutions at P341 had the most dramatic effects. None of these changes in transport activity were, however, associated with any defect of the mutant protein in inserting into the membrane, as indicated by [35S]methionine labelling and immunoprecipitation using anti-PheP serum. A possible role for each of these three prolines is discussed. Inserting a single alanine residue at different sites within span IX and the loop immediately preceding it also had major effects on transport activity, suggesting an important role for a highly organized structure in this region of the protein.
The phenylalanine-specific permease (PheP) is an integral cytoplasmic membrane protein which mediates the active transport of phenylalanine into Escherichia coli (12). The cloned pheP sequence indicates a polypeptide of 458 amino acids with a molecular weight of 50,645 (12). Hydropathicity and genetic analyses involving the construction of pheP′-phoA-′pheP sandwich gene fusions have suggested the presence of 12 transmembrane segments with both N and C termini located in the cytoplasm (11). Studies employing site-directed mutagenesis have identified some charged residues of PheP, including E118, K168, E226, and R252, that are essential for phenylalanine uptake (13).
Sequence analyses have shown that PheP shares extensive sequence homology with a variety of bacterial and eukaryotic proteins that transport various amino acids (11, 15). In particular, PheP shows 60% sequence identity with the general aromatic amino acid permease (AroP) of E. coli (4). The number of proteins identified as members of this superfamily of transporters has increased rapidly in recent years. All of these amino acid transporters are predicted to contain 12 hydrophobic transmembrane segments when analyzed by TopPred IV, a program based on the algorithm of Gunnar von Heijne (16). These proteins are involved exclusively in the transport of various amino acids. These permeases show little local or overall homology with members of the family of sugar transporters (7).
Proline residues in proteins generally, and in integral membrane proteins in particular, are of significant interest because of their unique structural and functional properties. A statistical analysis of Brandle and Deber (2) revealed that proline residues are frequently found in the transmembrane segments of ion channels and transporters but not in the transmembrane segments of proteins that have no transport function. This has led to the suggestion that proline residues located in the transmembrane segments of transport proteins have a key functional role. However, proline residues are not favored in α-helices because the backbone nitrogen is not available for hydrogen bonding and because of steric constraints caused by their ring structure. Consequently, prolines introduce “kinks” into transmembrane α-helices (19). Proline-kinked α-helices may pack to form either funnel- or cage-like structures, which have the potential to form either a channel vestibule or ion binding site(s) (17a). A variety of studies have shown an important role of proline residues within the transmembrane region of transport proteins (2, 17a–19).
In this report, we describe the results of site-directed mutagenesis of proline residues of PheP. Alanine substitutions at three of the proline residues, P54, P341, and P442, cause a marked decrease in phenylalanine uptake. The complete loss of transport activity in mutant PheP permeases with various substitutions at P341 suggests that it plays a critical role in the function of the protein.
MATERIALS AND METHODS
Bacteria, phage, plasmid, and growth media.
Table 1 lists the bacterial strains (all derived from E. coli K-12), phage, and plasmid used in this study. The minimal media used were the half-strength medium 56 described by Monod et al. (10) supplemented with 0.2% glucose and required growth factors. Luria agar and Lennox broth were used as complete media. Kanamycin was used at a final concentration of 25 mg/ml.
TABLE 1.
E. coli strains, plasmid, and phage used in this study
| Strain, plasmid, or phage | Relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| JM101 | Δ(lac-pro) supE thi (F′ traD36 proA+B+ lacIqZΔM15) | 9 |
| JP6488 | JP777 Δ(aroP::Tn10) Δ(pheP367::Tn10) | 12 |
| Plasmid pLG339 | Kmr Tcr; low-copy-number cloning vector | 14 |
| Phage mpMU3137 | NdeI (was introduced at Met1) mutant of mpMU2148 | 11 |
The genetic nomenclature is that described by Bachmann (1).
Site-directed mutagenesis of the pheP gene.
Oligonucleotide-directed site-specific mutagenesis (17) was used to introduce amino acid substitutions at each of the 16 prolines within the pheP gene, carried on mpMU3137 (Table 1). The 2.3-kb EcoRI-SalI fragment containing the pheP gene was cloned into the corresponding sites on low-copy-number plasmid pLG339 and transformed into strain JP6488 (Table 1).
[14C]phenylalanine uptake assays.
Active transport was measured in E. coli JP6488 (aroP pheP) transformed with plasmids expressing wild-type or mutant pheP genes as previously described (20). Mid-log-phase cells were washed in half-strength medium 56 buffer containing 0.2% glucose–80-μg/ml chloramphenicol and resuspended in the same buffer to an optical density at 600 nm of approximately 0.5. Cells were preincubated at 30°C for 5 min, and radioactive phenylalanine (final concentration, 10 μM) was added. Aliquots of 150 μl were removed at the appropriate time intervals and filtered through cellulose acetate filters, which were then washed twice with half-strength medium 56 buffer. Intracellular radioactivity was determined by liquid scintillation counting.
Radiolabelling of mutant proteins.
JP6488 cells carrying plasmids expressing various pheP mutants were grown in minimal media to an optical density at 600 nm of 0.5. For pulse-labelling, aliquots of 0.5 ml of cells were incubated for 1 min with 50 μCi of l-[35S]methionine-cysteine (1,175 Ci/mmol; 7.9 mCi/ml; NEN/Du Pont) at 37°C before commencing the immunoprecipitation experiments (see the following paragraph).
Immunoprecipitation of PheP proteins.
Immunoprecipitations were performed by the method specific for integral membrane proteins described by Ito and Akiyama (4) by using antiserum TTP7, which was obtained by immunizing a rabbit with a synthetic peptide of PheP conjugated with tetanus toxoid. Samples were electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel, and the dried gel was exposed to X-ray film for at least 24 h. Broad-range protein molecular weight standards (Bio-Rad) were used to estimate the molecular weights of mutant proteins. The densities of the radioactive bands were measured by scanning the autoradiographs with a Molecular Dynamics scanning densitometer, and these were used to estimate the levels of mutant proteins relative to that of the wild type.
RESULTS
Locations of proline residues within the PheP structure and their conservation within the family.
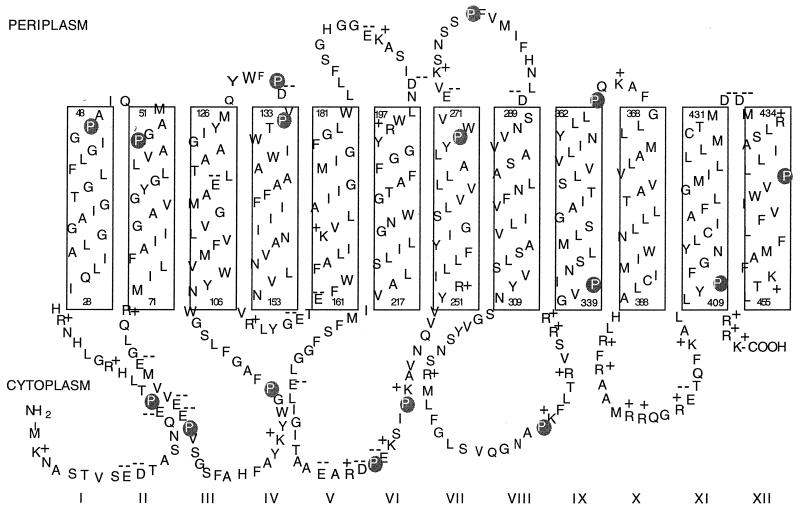
As can be clearly seen in Fig. 1, of the 16 proline residues present in PheP, 6, i.e., P17, P82, P97, P238, P243, and P329, are clearly located in the cytoplasmic loops, 2, i.e., P131 and P279, are clearly within the periplasmic loops, and 8, i.e., P47, P54, P134, P269, P341, P363, P411, and P442, are located either within the interface region between the membrane and the aqueous environment or within the membrane. The precision of the topological map is not sufficient to differentiate between these two possibilities for residues such as P341. Only one of the proline residues, namely, P442, is clearly located toward the middle of a membrane span.
FIG. 1.
Proposed membrane topology of the PheP permease (11) showing the positions of the 16 proline residues.
A database search for proteins with significant sequence similarity to PheP revealed a family of at least 31 proteins from bacteria and yeast, comprising both characterized and putative amino acid transporters. Sixteen of these are bacterial proteins representing 13 different amino acid transporters. Three of them (i.e., PROY, YIFK, and ANSP) have been cloned from both E. coli and Salmonella typhimurium, and only the E. coli transporters were used in the analysis reported in Table 2. The remaining 15 are yeast amino acid transporters. As seen in Table 2, most of the prolines are highly conserved among the bacterial transporters. Eleven of the 16 prolines are conserved in at least 10 of the 13 bacterial proteins. Two of these, P238 and P341, are present in all 13. On the other hand, P17 and P47 are only found in PheP, and P134 and P442 are conserved in 8 and 9 of the 13 proteins, respectively. If the analysis is extended to the entire family (bacteria and yeast), P54, P238, P269, P279, P329, and P341 are all retained in 26 or more of the proteins. On the other hand, P131 and P363, both of which are highly conserved among the bacterial proteins, are represented infrequently among the yeast transporters.
TABLE 2.
Properties of proline mutant PhePs
| Residue | No. of family members in which residue is conserveda | % of wild type
|
Protein leveld | |
|---|---|---|---|---|
| Steady-state level of accumulationb | Initial ratec | |||
| Pro17 | 1 | 97 | 98 | |
| Pro47 | 1 | 90 | 92 | |
| Pro54 | 12 | 50 | 60 | 100 |
| Pro82 | 11 | 102 | 97 | |
| Pro97 | 7 | 98 | 95 | |
| Pro131 | 11 | 98 | 101 | |
| Pro134 | 8 | 80 | 78 | 69 |
| Pro238 | 13 | 100 | 103 | |
| Pro243 | 10 | 99 | 101 | |
| Pro269 | 12 | 72 | 72 | 100 |
| Pro279 | 12 | 141 | 136 | 85 |
| Pro329 | 11 | 80 | 83 | 84 |
| Pro341 | 13 | 5 | 11 | 100 |
| Pro363 | 10 | 135 | 129 | |
| Pro411 | 11 | 80 | 87 | 124 |
| Pro442 | 9 | 46 | 42 | 90 |
Including AROP, PROY, YIFK, CYCA, ANSP, YKFD, LYSP, and GABP from E. coli; ROCE, ROCC, and HUTM from Bacillus subtilis; and AROP from Corynebacterium glutamicum.
Percentage of steady-state level of [14C]phenylalanine uptake compared to the wild type (100% is 10.1 nmol/mg of dry weight).
Percentage of initial rate of [14C]phenylalanine uptake compared to the wild type (100% is 45.4 nmol/mg of dry weight/min).
Values are percentages of the wild-type protein level derived from immunoprecipitation experiments.
Effect of a proline-to-alanine substitution at each position.
Site-directed mutagenesis using synthetic oligonucleotides was used to separately change each of the proline residues of PheP to alanine. The presence of the mutations was verified by sequencing the entire mutant pheP genes. The replicative form of M13mp18 DNA containing each of the specified mutations was isolated, and the EcoRI-SalI fragment containing pheP was cloned into the corresponding sites on pLG339. Uptake of [14C]phenylalanine (10 μM) was measured in transformants of E. coli JP6488 (aroP pheP) with plasmid pLG339 carrying the mutated pheP gene. The results of these assays are shown in Table 2. Of the 16 prolines that were replaced with alanine, changing only 3 of these had a significant effect on transport activity. These were P54, P442, and P341. Of these three, both P54 and P341 are highly conserved within the family, and of these two, only P341 appears to play a critical role in PheP function. Replacement of the other highly conserved residues, such as P238, P269, P279, and P329, caused little or no loss of activity, and in the case of P279, the alanine-substituted mutant showed increased activity.
Further mutational analysis of P54, P341, and P442.
The possible role of each of these three prolines was examined further by replacing each with glycine or leucine in the case of P54 and P442 and with a number of other amino acids in the case of P341. The results are shown in Table 3. Replacing P54 with glycine restored full transport activity, whereas replacement with leucine reduced transport further, to only 26%. Replacement of P442 with either glycine or leucine resulted in mutant proteins with the same transport activity as the P442A mutant. There was no transport activity when P341 was replaced with glycine, glutamine, lysine, or arginine. Replacement with serine and threonine produced 3 and 17% of the transport activity of the wild-type permease, respectively.
TABLE 3.
Properties of mutant PhePs with substitutions at P54, P341, and P442
| Residue | % of wild-type levela (relative protein level)b
|
||||||
|---|---|---|---|---|---|---|---|
| Gly | Leu | Gln | Ser | Thr | Lys | Arg | |
| P54 | 100 | 26 (101) | |||||
| P341 | 0 (106) | 0 (83) | 3 (110) | 17 (63) | 0 (73) | 0 (84) | |
| P442 | 52 (103) | 43 (121) | |||||
Percentage of the steady-state level of [14C]phenylalanine uptake by the wild type.
Values in parentheses are relative levels of protein detected by immunoprecipitation experiments.
Detection of mutant permeases within the cytoplasmic membrane.
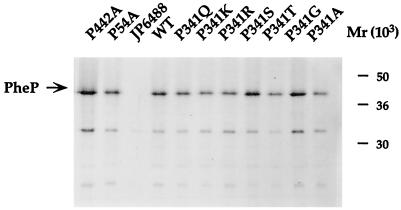
To investigate whether, in any case, loss of function was caused by the inability of the mutant proteins to successfully insert into the membrane, the cytoplasmic membranes of strains showing significant loss of transport activity were probed with PheP-specific antiserum as described in Materials and Methods. The results, which are shown in Fig. 2 and are summarized in Tables 2 and 3, indicate that there were no significant variations between the levels of PheP protein in the membranes of any of the mutant strains and the wild type.
FIG. 2.
Comparison of levels of wild-type (WT) and proline mutant PheP proteins produced by [35S]methionine-cysteine-labelled JP6488 cells by immunoprecipitation. Representative mutant proteins were pulse-labelled for 1 min before being solubilized with NaOH and immunoprecipitated with an antibody prepared against a synthetic PheP peptide. The radioactivity in gels of immunoprecipitated PheP proteins was quantitated as described in Materials and Methods. Similar values were obtained in two separate experiments.
Further studies of span IX and the cytoplasmic loop preceding it.
As can be seen in Fig. 1, P341 is located either at the beginning of span IX or at the end of the loop immediately preceding it. This loop contains a number of highly conserved motifs, i.e., NSG(hydrophobic)Y, RMLF/Y, and R/KR/HGVP (Fig. 3). Furthermore, we have shown that R317, like P341, is essential for PheP function, as even a lysine substitution destroyed transport activity (data not shown). In order to test whether this region of the protein including span IX contains an organized structure that is essential for transport activity, we decided to insert a single alanine residue at different positions in the loop and the span. The consequences of these insertions are shown in Table 4. Inserting an alanine immediately after P341 or even S344 completely destroyed transport activity. As the insertions were made farther up the span, the effect diminished, an insertion after S348 retaining 30% activity and an insertion after L354 yielding a mutant that retained 80% of wild-type activity. In addition, insertions at three different sites within the loop after N309, N327, and V340 also had very dramatic effects on transport activity (Table 4).
FIG. 3.
Sequence alignment of 13 amino acid transporters of bacteria showing some highly conserved motifs (in boldface) in the region near P341 of PheP.
TABLE 4.
Properties of mutant PhePs with alanine insertions
| Residue | % of steady-state wild-type activitya | Protein levelb |
|---|---|---|
| N309 | 8 | 121 |
| N327 | 14 | 105 |
| V340 | 11 | 103 |
| P341 | 0 | 103 |
| S344 | 0 | 108 |
| S348 | 30 | 118 |
| L354 | 80 |
Percentage of the steady-state level of [14C]phenylalanine uptake by the wild type.
Values are percentages of wild-type protein level derived from immunoprecipitation experiments.
Residue P411, like P341, is found at the beginning of an outward-facing span and is highly conserved within the family. However, unlike P341, it is not essential for function, and inserting an alanine residue immediately after P411 only reduces transport activity marginally (to 70% of the wild-type level).
DISCUSSION
The three proline residues of PheP whose individual replacement with alanine results in reduced transport activity appear to play markedly different roles in PheP function. Replacement of P54 with alanine or leucine reduced transport to 50 and 26% of wild-type activity, respectively. Replacement with glycine, however, had no effect on activity. Proline 54 is part of a very highly conserved AGPA motif and may either participate in a turn between spans I and II or, as suggested by modelling studies, occupy position 2 of the N cap sequence for the helix of span II (3).
Replacement of P442 with either alanine, glycine, or leucine resulted in a protein with about 50% of wild-type activity, and these results, coupled with the position of P442 in the middle of span XII, may indicate that although it only exerts a twofold influence on activity, it does so by the introduction of a kink in the middle of a helix.
Proline 341 appears to play a critical role in the function of the PheP protein, since its replacement with four other amino acids resulted in total loss of transport activity. Replacement with alanine or serine resulted in 5 and 3% of wild-type activity, respectively, and replacement with threonine produced a mutant permease with 17% of wild-type activity. Prolines have been proposed to play an important role in many protein-protein interactions (6). Proline residues in the regions flanking interacting sites are proposed to act as “brackets” to position and preserve the interaction site structure. The results obtained by P341 substitutions and alanine insertions within span IX may indicate that P341 acts as such a bracket to present an interaction site which may be present along one face of the helix of span IX toward the cytoplasmic side of the span. In this case, the proposed interaction could be with another span of the same protein rather than with another protein.
Transport activity is completely destroyed by alanine insertion either directly before or after P341. Furthermore, when either N343 or S344 was deleted from the mutant with alanine inserted directly after P341, transport activity was not restored (data not shown), even though such deletions should have restored the face of the helix to its original position. This result may imply that the bulky side chains of valine and isoleucine which flank P341 may sterically restrict the proline side chain in a manner necessary for its function.
Single amino acid insertions within the loop preceding P341 also had dramatic effects on PheP function, indicating that an organized structure of the protein in this region is essential for transport activity.
Unlike with the Lac permease, the majority of the proline residues are not located within the transmembrane spans of the protein. However, as with the Lac permease, only one proline residue (P341) appears to be critical for transport activity. Although a number of proline residues are highly conserved among the members of the extensive family of amino acid transporters, they could be readily changed to alanine without adversely affecting transport activity. This is further confirmation that conservation alone does not necessarily indicate the functional significance of any residue, as detected in our assays.
In an extensive study of the hydrophilic loops of the Lac permease using insertional mutagenesis, McKenna et al. (8) showed that insertions into 10 of the 13 hydrophilic loops of the Lac permease had no effect on transport activity. Insertion of six histidine residues into hydrophilic domain 3, 9, or 10 of the Lac permease did, however, cause a marked decrease in transport activity. Hydrophilic domain 9 corresponds to the loop between spans VIII and IX in PheP, which similarly exhibits a low tolerance for insertions. This loop in the Lac permease contains a highly conserved NRIGGK motif. Although this particular motif is not present in the corresponding loop of PheP, it is perhaps worth noting that this loop nevertheless contains three motifs that are highly conserved in the family of amino acid transporters.
ACKNOWLEDGMENTS
We thank Graeme B. Cox and Frank Gibson for reading the manuscript. We also thank J.-H. An, Y. Jiang, and T. Betteridge for technical assistance.
This work was supported by the Australia Research Council Large Grants Scheme.
REFERENCES
- 1.Bachmann B J. Linkage map of Escherichia coli K-12. Microbiol Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandle C J, Deber C M. Hypothesis about the function of membrane-buried proline residues in transport proteins. Proc Natl Acad Sci USA. 1986;83:917–921. doi: 10.1073/pnas.83.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson, F. Personal communication.
- 4.Honore N, Cole S T. Nucleotide sequence of the aroP gene encoding the general aromatic amino acid transport protein of E. coli K-12: homology with yeast transport proteins. Nucleic Acids Res. 1990;18:653. doi: 10.1093/nar/18.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito K, Akiyama Y. In vivo analysis of integration of membrane proteins in Escherichia coli. Mol Microbiol. 1991;5:2243–2253. doi: 10.1111/j.1365-2958.1991.tb02154.x. [DOI] [PubMed] [Google Scholar]
- 6.Kini R M, Evans H J. A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem Biophys Res Commun. 1995;212:1115–1124. doi: 10.1006/bbrc.1995.2084. [DOI] [PubMed] [Google Scholar]
- 7.Maiden M C, Davis E O, Baldwin S A, Moore D C, Henderson P J. Mammalian and bacterial sugar transport proteins are homologous. Nature. 1987;325:641–643. doi: 10.1038/325641a0. [DOI] [PubMed] [Google Scholar]
- 8.McKenna E, Hardy D, Kaback H R. Insertional mutagenesis of hydrophilic domains in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:11954–11958. doi: 10.1073/pnas.89.24.11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 10.Monod J, Cohen-Bazire G, Cohn M. Sur la biosynthese de la b-galactosidase (lactase) chez Escherichia coli. La specificite de l’induction. Biochim Biopys Acta. 1951;7:585–599. doi: 10.1016/0006-3002(51)90072-8. [DOI] [PubMed] [Google Scholar]
- 11.Pi J, Pittard A J. Topology of the phenylalanine-specific permease of Escherichia coli. J Bacteriol. 1996;178:2650–2655. doi: 10.1128/jb.178.9.2650-2655.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pi J, Wookey P J, Pittard A J. Cloning and sequencing of the pheP gene, which encodes the phenylalanine-specific transport system of Escherichia coli. J Bacteriol. 1991;173:3622–3629. doi: 10.1128/jb.173.12.3622-3629.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pi J, Wookey P J, Pittard A J. Site-directed mutagenesis reveals the importance of conserved charged residues for the transport activity of the PheP permease of Escherichia coli. J Bacteriol. 1993;175:7500–7504. doi: 10.1128/jb.175.22.7500-7504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pouwels P H, Enger-Valk B E, Brammar W J. Cloning vectors. Amsterdam, The Netherlands: Elsevier Science Publishers, B.V.; 1985. pp. I-A–ii-1. [Google Scholar]
- 15.Reizer J, Finley K, Kakuda D, Macleod C L, Reizer A, Saier J M H. Mammalian integral membrane receptors are homologous to facilitators and antiporters of yeast, fungi, and eubacteria. Protein Sci. 1993;2:20–30. doi: 10.1002/pro.5560020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sipos L, von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- 17.Vandeyar M A, Weiner M P, Hutton C J, Batt C A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 17a.von Heijne G. Proline kinks in transmembrane a-helices. J Mol Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 18.Williams K A, Deber C M. Proline residues in transmembrane helices: structural or dynamic role? Biochemistry. 1991;30:8919–8923. doi: 10.1021/bi00101a001. [DOI] [PubMed] [Google Scholar]
- 19.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wookey P J, Pittard J, Forrest S M, Davidson B E. Cloning of the tyrP gene and further characterization of the tyrosine-specific transport system in Escherichia coli K-12. J Bacteriol. 1984;160:169–174. doi: 10.1128/jb.160.1.169-174.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]