Abstract
Capsular polysaccharides are considered as major virulence factors associated with the ability of multidrug-resistant (MDR) Acinetobacter baumannii to cause severe infections. In this study, LysAB1245, a novel bacteriophage-encoded endolysin consisting of a lysozyme-like domain from phage T1245 was successfully expressed, purified, and evaluated for its antibacterial activity against distinct capsular types associated with A. baumannii resistance. The results revealed a broad spectrum activity of LysAB1245 against all clinical MDR A. baumannii isolates belonging to capsular type (KL) 2, 3, 6, 10, 47, 49, and 52 and A. baumannii ATCC 19606. At 2 h following the treatment with 1.7 unit/reaction of LysAB1245, more than 3 log reduction in the numbers of bacterial survival was observed. In addition, LysAB1245 displayed rapid bactericidal activity within 30 min (nearly 3 log CFU/mL of bacterial reduction). Thermostability assay indicated that LysAB1245 was stable over a broad range of temperature from 4 to 70°C, while pH sensitivity assay demonstrated a wide range of pH from 4.5 to 10.5. Furthermore, both minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) of LysAB1245 against all MDR A. baumannii isolates and A. baumannii ATCC 19606 were 4.21 μg/mL (0.1 unit/reaction). Conclusively, these results suggest that LysAB1245 possesses potential application for the treatment of nosocomial MDR A. baumannii infections.
Introduction
Recently, an outbreak of A. baumannii infections in coronavirus disease 2019 (COVID-19) patients has been reported [1–3], resulting in increased morbidity and mortality rates as well as high treatment costs. This emerging pathogen is responsible for several healthcare-associated infections, such as bacteremia, ventilator-associated pneumonia, urinary tract infections, burn and wound infections, and meningitis [4–7]. Nosocomial A. baumannii infections usually correlate with the production of capsular polysaccharide (CPS), which plays an important role in bacterial pathogenesis by protecting from environmental stresses, antimicrobial penetration, and host immune responses [8,9]. In 2019, more than 100 distinct capsular types (KL) of A. baumannii were discovered, with the variations in K unit structures and sugar composition [10]. In a previous study, three capsular genotypes, including KL6, 10, and 47, showed a frequency more than 10% among A. baumannii isolates from three tertiary care hospitals in Thailand [11]. Moreover, carbapenem-resistant A. baumannii isolates belonging to KL2, 10, 22, and 52 showed higher incidence and mortality rates than isolates belonging to other KL groups [12]. Nowadays, isolates of A. baumannii rapidly develop resistance to several currently employed antibiotics, including carbapenems, aminoglycosides and polymyxin [13–18]. Therefore, novel and effective antibacterial agents targeting emerging antibiotic-resistant A. baumannii strains are urgently required.
Bacteriophages (phages) are known as natural enemies of bacteria that have no harmful effects on the human microbiome. Therefore, the use of bacteriophages has been considered as an alternative therapeutic option for drug-resistant A. baumannii infections [19,20]. Moreover, phage-encoded enzymes, such as endolysins, are are effective in eradicating or reducing antibiotic-resistant pathogenic bacteria [21,22]. Endolysins are peptidoglycan lytic enzymes capable of breaking down bacterial cell walls, and these enzymes can be used as recombinant proteins to attack invading bacterial cells. Endolysins are widely used as antibacterial agents due to their major advantages over phages and antibiotics, including rapid killing activity, high efficiency, and a broad spectrum of lytic activity against pathogenic bacteria without showing toxicity on human cells [23–25]. In addition, the C-terminal cell wall binding domain of phage-encoded endolysin is responsible for rapid kinetic, which is highly specific to the peptidoglycan of bacterial cells and poses a low risk of resistance development [26,27].
In our previous study, a novel virulent phage T1245 specifically infecting A. baumannii with KL10 was isolated, characterized, and subjected to biological property tests and whole genome sequencing [28]. However, it is necessary to evaluate the antibacterial activity of its endolysin. In this study, the gene-encoding endolysin of phage T1245, named LysAB1245 was cloned into the expression vector, expressed, and produced as purified proteins. Purified endolysin, LysAB1245 was tested for its catalytic properties against different major capsular types associated with A. baumannii resistance. Overall, this study aimed to produce a protein suitable for further development as an alternative antibacterial agent against MDR A. baumannii isolates belonging to common capsular types.
Materials and methods
Bacteria, bacteriophage, and culture conditions
The bacterial strains, phage, plasmids, and primers used in this study are listed in Table 1. All MDR A. baumannii strains belonged to sequence type 2 with different major capsular types, including KL2, 3, 6, 10, 47, 49, and 52. All bacteria were inoculated in Luria Bertani (LB) broth or LB agar (Difco Laboratories, Detroit, MI, USA) at 37°C and maintained in 20% glycerol (v/v) at -80°C for long-term storage. This study was approved by the Human Research Ethics Committee (HREC) of the Faculty of Medicine at Prince of Songkla University (reference number: 64–284–14–1).
Table 1. Bacterial strains, bacteriophage, plasmids, and oligonucleotide primers.
| Strain, plasmid, phage, or primer | Relevant characteristic(s), description, or sequence | Source or reference |
|---|---|---|
| Strains | ||
| A. baumannii ABMYH-1245 | Multidrug-resistant, clinical isolate with KL10; primary host bacteria of phage T1245 | [11] |
| A. baumannii ABAPSP-55 | Multidrug-resistant, clinical isolate with KL10 | [11] |
|
A. baumannii ABAPSP-64 A. baumannii ABMYSP-109 |
Multidrug-resistant, clinical isolate with KL10 Multidrug-resistant, clinical isolate with KL10 | [11] |
| A. baumannii ABMYSP-101 | Multidrug-resistant, clinical isolate with KL10 | [11] |
| A. baumannii ABMYSP-182 | Multidrug-resistant, clinical isolate with KL10 | [11] |
| A. baumannii AB1039 | Multidrug-resistant, clinical isolate with KL2 | [11] |
| A. baumannii AB3396 | Multidrug-resistant, clinical isolate with KL2 | [11] |
| A. baumannii ABJNH-403 | Multidrug-resistant, clinical isolate with KL3 | [11] |
| A. baumannii ABMYSP-185 | Multidrug-resistant, clinical isolate with KL6 | [11] |
| A. baumannii ABMYSP-210 | Multidrug-resistant, clinical isolate with KL6 | [11] |
| A. baumannii ABMYSP-216 | Multidrug-resistant, clinical isolate with KL6 | [11] |
| A. baumannii ABMYSP-419 | Multidrug-resistant, clinical isolate with KL6 | [11] |
| A. baumannii ABMYH-1033 | Multidrug-resistant, clinical isolate with KL6 | [11] |
| A. baumannii AB15 | Multidrug-resistant, clinical isolate with KL47 | [11] |
| A. baumannii ABAPP-61 | Multidrug-resistant, clinical isolate with KL47 | [11] |
| A. baumannii AB724 | Multidrug-resistant, clinical isolate with KL49 | [11] |
| A. baumannii AB2792 | Multidrug-resistant, clinical isolate with KL49 | [11] |
| A. baumannii ABMYSP-21 | Multidrug-resistant, clinical isolate with KL52 | [11] |
| A. baumannii ABMYSP-444 | Multidrug-resistant, clinical isolate with KL52 | [11] |
| E. coli Top10 | Laboratory strain for TA cloning use | Invitrogen, San Diego, USA |
| E. coli BL21 (DE3 | Laboratory strain for protein expression | Invitrogen, San Diego, USA |
| Plasmids | ||
| pGEM-T-easy | 3,015-bp E. coli vector, Ampr, Plac, lacZ | Promega, San Diego, USA |
| pET30b(+) | Expression vector; 5,421-bp E. coli vector, Kmr, PT7, His-Tag | Novagen, Wisconsin, USA |
| Phages | ||
| A. baumannii phage T1245 | Accession No. ERS3583556 | [28] |
| Primers | ||
| Forward primer: FP-EcoRIEndo | GAATTCGATGATTCTGACTAAAGACGG | This study |
| Reverse primer: RP-XholEndo | CTCGAGTAAGCTCCGTAGAG | This study |
Bioinformatics analysis
The whole genome of phage T1245 was deposited in GenBank under accession number ERS3583556. The gene-encoding endolysin (LysAB1245) in phage T1245 was blasted in the NCBI protein database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequences of LysAB1245 and other reported Acinetobacter phage endolysins were aligned using ClustalOmega multiple sequence alignment (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Construction of LysAB1245 expression vector
LysAB1245-encoding gene in phage T1245 was amplified by polymerase chain reaction (PCR). Gene information and the PCR primers are listed in Table 1. LysAB1245, 569-bp PCR products were purified using the GenepHlowTM Gel/PCR Cleanup Kit (Geneaid, Taiwan). Purified LysAB1245 gene was cloned into the pGEM-T cloning vector according to standard procedures [29]. Ligation reaction was performed and transformed into Escherichia coli Top 10 by heat shock method. The transformed cells were plated onto LB agar plates containing ampicillin (100 μg/mL), isopropyl b-D-1 thiogalactopyranoside (IPTG), and X-Gal. Plates were incubated at 37°C for 18 h. White-positive colonies were selected and verified using PCR and sequencing. Purified LysAB1245 genes were digested with EcoRI and Xhol, purified from the agarose gel using a QIAquick gel extraction kit (Qiagen, Hilden, Germany), and assembled into EcoRI/Xhol digested pET30b(+). The resulting plasmids (pET30b(+)-LysAB1245) were transformed into an E. coli BL21 (DE3) strain for over-expression. The transformed cells were plated onto agar plates containing kanamycin (50 μg/mL) and incubated overnight at 37°C for 16–18 h. Glycerol stocks were prepared from positive clones and sequencing was performed to confirm LysAB1245 expression.
Expression and purification of endolysin LysAB1245
An ExiProgen automated protein synthesis system (ExiProgenTM, Bioneer, Korea) with cell-free protein synthesis and magnetic bead-based His-Tag affinity purification was used to express and purify endolysin LysAB1245. Briefly, 6 μg of plasmid DNA (pET30b(+)-LysAB1245) was prepared for LysAB1245 synthesis. Ten microliters of DNA was added to the reaction well of the ExiProgenTM EC1 protein synthesis kit’s protein expression cartridge. The reaction was performed using E. coli cellular lysate and Bioneer’s master mix for the transcription and translation of coding sequence to protein and purification of the target protein. After 6 h, 250 μL of purified protein samples were collected from each elution tube. Bradford assay using bovine serum albumin was performed to determine the concentration of LysAB1245. Endolysin protein synthesis has an efficiency of over 30% yield, according to the manufacturer’s maximum efficacy.
SDS-PAGE and western blot analysis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analyses were performed to assess the purity of endolysin LysAB1245. Samples collected from ExiProgen including purified protein in elution tubes, unbound, expression, and washing were mixed with sample buffer (62.5 mM Tris–HCl, pH 6.8, containing 5% 2-mercaptoethanol, 2% sodium dodecyl sulfate, 10% glycerol, and 0.01% bromophenol blue) and heated for 5 min in boiling water. All samples were separated using 12% SDS-PAGE and blotted onto a 0.45-μm nitrocellulose membrane (Bio-Rad).
For the immunodetection of a 6×His-tagged protein, the membrane was blocked with 3% bovine serum albumin in phosphate-buffered saline (10 mM PBS; pH 7.4) for 18 h, followed by incubation with a mouse anti-His-tag antibody (1:3000) (Bio-Rad, Hercules, CA, USA) for 1 h. After three washes with PBS containing Tween 20 (PBST), the membranes were incubated with alkaline phosphatase-labeled goat anti-mouse IgG antibody (1:3000) (KPL, Gaithersburg, MD, USA) for 1 h. After four washes with PBST, the blot was developed using 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and nitro blue tetrazolium (NBT) (Sigma, Deisen- hofen, Germany).
Effects of LysAB1245 on MDR A. baumannii ABMYH1245
The effects of purified LysAB1245 were examined on a primary host of phage T1245 according to the protocol of Lai et al. (2011) [30], with slight modifications. Briefly, the log phase of A. baumannii AMMYSP-1245 was grown in tryptic soy broth (TSB; Difco) and adjusted to 105 colony-forming units (CFU/mL). The bacterial cells were centrifuged at 9,000 rpm for 5 min and the supernatant was discarded. Thereafter, bacterial pellets were treated with 50 μL of LysAB1245 (134.71 μg/mL) or 10 mM of PBS (as a control) followed by incubation at 37°C under constant shaking at 150 rpm. Samples were collected at 0, 2, and 24 h, and the log CFU/mL was calculated. Data obtained from two independent experiments performed in triplicate are presented as mean ± standard deviation (SD). Significant differences between groups were determined using one-way analysis of variance (ANOVA), followed by Tukey’s multiple comparison test. Statistical significance was set at 99% confidence interval (p < 0.05). One unit of enzyme activity was defined as the amount of enzyme required to kill bacterial cells at 2 logs per 2 h.
Determination of the lytic range of LysAB1245
The antibacterial activity of LysAB1245 was determined against 19 isolates of MDR A. baumannii belonging to seven different major capsular types (KL2; 2 isolates, KL3; 1, KL6; 5, KL10; 5, KL47; 2, KL49; 2, and KL52; 2) and ATCC 19606. Briefly, the bacterial pellets at 105 CFU/mL were resuspended with 50 μL of LysAB1245 (1.7 unit/reaction) or PBS and incubated at 37°C with shaking at 150 rpm. Samples were collected at 0, 2, and 24 h and calculated the number of log CFU/mL. Data obtained from two independent experiments performed in triplicate are presented as mean ± SD.
Sensitivity of LysAB1245 to temperature and pH
Thermal and pH stability test for endolysin LysAB1245 were examined. Briefly, LysAB1245 (0.4 unit/reaction) was incubated at six different temperatures (4, 25, 37, 50, 60, and 70°C) for 30 min. Subsequently, the cell pellets of A. baumannii AMMYSP-1245 (105 CFU/mL) were resuspended with LysAB1245 from various temperatures and incubated at 37°C with shaking (150 rpm) for 24 h.
For pH sensitivity assay, the pH of LysAB1245 was adjusted using PBS with different pH values (4.5, 5.5, 7.4, 8.5, and 10.5), followed by incubation at 37°C. After 30 min, the pH of LysAB1245 was adjusted to 7.4 and incubated at 37°C with shaking for 24 h. For both experiments, a mixture of bacteria and PBS (pH 7.4) at 37°C was served as the control group. The number of log CFU/mL was determined and calculated the percentage of bacterial reduction. Each experiment was performed in triplicate with two independent replicates.
Kinetic analysis of LysAB1245
A mixture of MDR A. baumannii ABMYH-1245 (at 105 CFU/mL) and LysAB1245 (at 1.7 unit/reaction) or PBS (as a control) was incubated at 37°C with shaking at 150 rpm. The samples were withdrawn at 0, 15 min, and 30-min intervals for 0.5–2.5 h and the number of log CFU/mL was determined. Data obtained from two independent experiments performed in triplicate are presented as mean ± SD.
Antimicrobial activity of LysAB1245
The minimum inhibitory concentration (MIC) of LysAB1245 was determined using the broth microdilution method, according to the Clinical and Laboratory Standard Institute (CLSI) guidelines [31]. Briefly, a single colony of 20 clinical MDR A. baumannii isolates belonging to KL 2, 3, 6, 10, 47, 49, and 52 and A. baumannii ATCC 19606 was grown in Mueller-Hinton broth (MHB; Difco) and incubated at 37°C with shaking until the cells reached the logarithmic phase. Endolysin LysAB1245 (134.71 μg/mL) was serially diluted (1:2) in microtiter plate (50 μL/well). Subsequently, 50 μL of A. baumannii culture with approximately 106 colony-forming units (CFU/mL) was inoculated to the microplate, containing different diluted LysAB1245 and further incubated at 37°C for 18 h. MIC was defined as the of the antibacterial agent that inhibited the visible growth of bacteria, while minimal bactericidal concentration (MBC) was defined as the lowest concentration of the antibacterial agent required to kill bacteria. The experiment was performed in triplicate in two independent experiments.
Results and discussion
Characterization of endolysin LysAB1245
The endolysin gene of phage T1245, named LysAB1245 contains 558 base pairs and 185 amino acids. BLAST analysis showed that the LysAB1245 gene had 98.92% sequence similarity to putative chitinase-like endolysin from Acinetobacter phage phiAB6 (accession no. YP_009288673.1). The results of conserved domain analysis using the Pfam database revealed that amino acids of LysAB1245 contain lysozyme-like (N-acetyl-β-D-muramidase) domains between residues 79 and 136, which are the catalytic domains of LysAB1245. The catalytic activities of purified endolysin LysAB1245 are attributable to glycosidases that cleave β-1,4-N-acetyl-D-glucosamine bonds between N-acetylmuramic acid and N-acetylglucosamine in glycan chains [32]. Previously, phage endolysins from different host genera and species such as PlyE146 from E. coli phage [33], LysSS from Salmonella enterica phage [20] and LysAB2, PlyAB1, LysABP-01, and Ply6A3 from A. baumannii phages [24,30,34,35] were studied their catalytic activities, which belong to glycosidase hydrolase family.
Multiple sequence alignments of LysAB1245 with two other reported A. baumannii phage endolysins showed high similarity in the domain region with 12 amino acid polymorphisms identified (Fig 1). However, in silico sequence analysis indicated that six mutations (amino acids 96, 97, 100, 103, 108, and 111) were founded in the conserved domain, which play a vital role in enhancing the catalytic function of the enzyme
Fig 1. Amino acid sequence alignment using Clustal Omega multiple sequence alignment.
The multiple sequence alignments of three phage endolysins revealed similar and dissimilar amino acids of LysAB1245, LysAB2 (accession no. ADX62345), and PlyAB1 (accession no. YP_008058242). Amino acid polymorphisms are indicated by asterisks (*).
Cloning, expression, and purification of endolysin LysAB1245
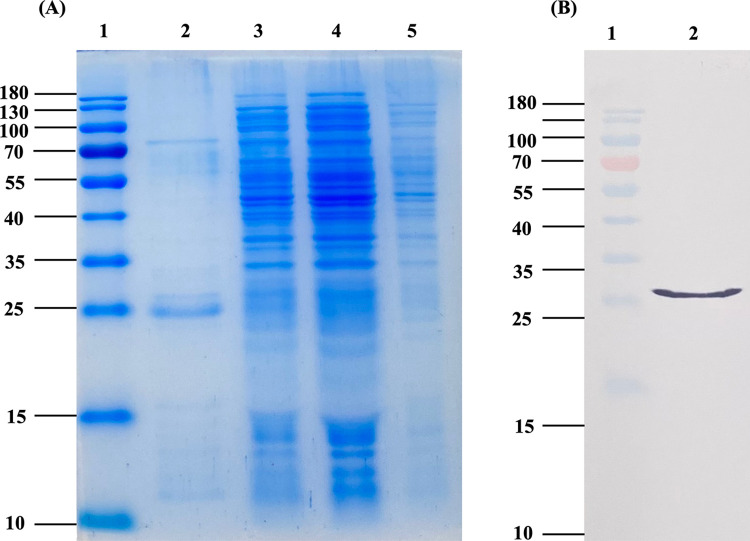
SDS-PAGE results indicated that LysAB1245 was effectively expressed and purified using the ExiProgen protein synthesis system (Fig 2A). Western blot analysis using specific His-tagged antibodies revealed an expected size of approximately 26 kDa (Fig 2B). Moreover, the concentration of purified LysAB1245 was approximately 134.71 μg/mL (S1 Fig).
Fig 2.
SDS-PAGE analysis of LysAB1245 (A). Lane 1, molecular size marker; lane 2, purified LysAB1245; lane 3, expression sample; lanes 4, washed sample; lanes 5, unbound sample. Western blot analysis of LysAB1245 (B). Lane 1, molecular size marker; lane 2, purified LysAB1245.
Lytic effects of purified endolysin LysAB1245 on A. baumannii ABMYH-1245 cells
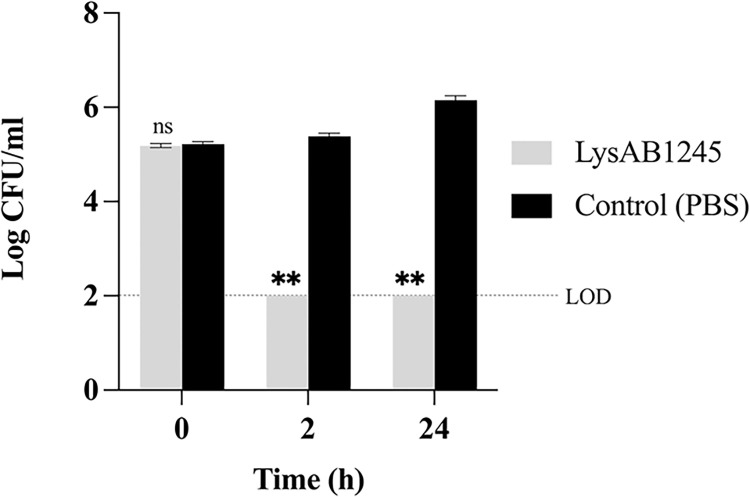
Treatment with LysAB1245 at 134.71 μg/mL significant decreased (P <0.01) the growth of A. baumannii ABMYSP-1245 when compared with control at 2 h and 24 h (Fig 3). At 2 h and 24 h, LysAB1245 reduced the viability of A. baumannii cells up to 3.39 log CFU/mL (>99.9% reduction) and 4.16 log CFU/mL (>99.99% reduction), respectively, compared with control (S2 Fig). Compared with other phage endolysins such as LysSP1 (Salmonella phage) and LysPN09 (Pseudomonas phage), LysAB1245 exhibited efficient bactericidal activity, even in the absence of outer membrane permeabilisers [36,37]. Generally, endolysins exert their effects against Gram-positive bacteria by binding directly to the cell walls. In contrast, the presence of an outer membrane can prevent the entry of several antimicrobials into Gram-negative bacterial cells. The mechanism of LysAB1245 on Gram-negative bacteria could be attributed to the highly positively charged region at its C-terminus, which has the potential to destabilize the outer membrane of Gram-negative bacteria. Consequently, the N-terminal enzymatic domain can penetrate the peptidoglycan layer and induce bacterial lysis [38]. In addition, Düring et al. reported that helix-forming amphipathic peptides at the C-terminus of T4 lysozyme can interact with negatively charged lipopolysaccharides in Gram-negative bacteria, resulting in antimicrobial activities [39].
Fig 3. The antibacterial activity of purified endolysin LysAB1245 on A.
baumannii ABMYSP-1245 cells. Bacterial pellets (105 CFU/mL) were treated with LysAB1245 (134.71 μg/mL). A significant reduction in bacterial growth was compared with control, **P < 0.01, and ns means non-significant. Two independent experiments were performed in triplicate and error bars represent the standard deviation. Limit of detection (LOD) for surviving bacterial cells was 2 log CFU/mL.
Lytic spectrum of purified endolysin LysAB1245 on MDR A. baumannii isolates with major different capsular types
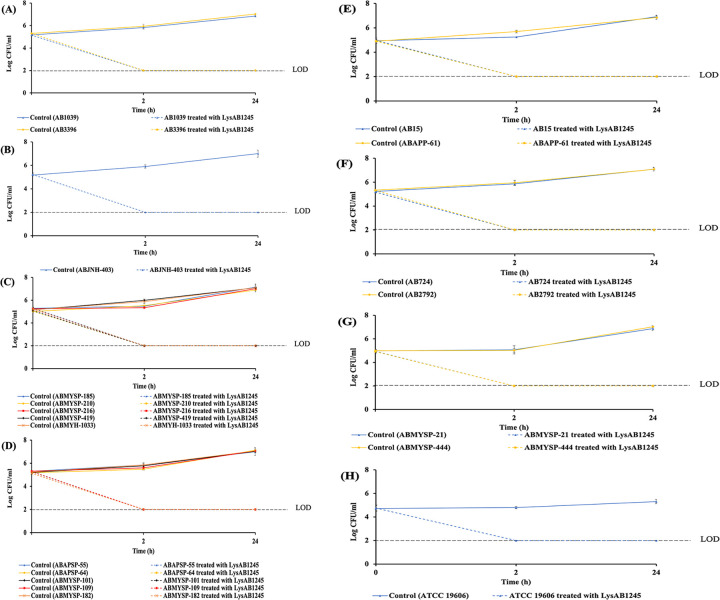
Recently, a newly isolated lytic phage targeting the MDR A. baumannii isolates, phage T1245 was isolated and characterized. However, phage T1245 specifically infects only A. baumannii with KL10 and KL3 isolates. Capsular structure has been recognized as an important virulence factor among A. baumannii strains [40]. LysAB1245 was further investigated whether it could lyse MDR A. baumannii isolates belonging to major capsular types identified in the Thai collection. The results of lytic spectrum revealed that LysAB1245 not only killed A. baumannii with KL10 but also lysed all the tested clinical MDR A. baumannii isolates with KL2, 3, 6, 47, 49, and 52 and A. baumannii ATCC 19606. A more than 3-log reduction (>99.9% reduction) in the viability of all tested MDR A. baumannii isolates belonging to KL2, 3, 6, 10, 47, 49 and 52 and A. baumannii ATCC 19606 was observed when treated with LysAB1245 at 1.7 unit/reaction, compared with control at 2 h (Fig 4). Moreover, no re-growth of any A. baumannii isolates was observed after 24 h of treatment with LysAB1245. Notably, MDR A. baumannii with KL2 and KL49 which were not lysed by any isolated phages from previous study, were killed by endolysin LysAB1245.
Fig 4. The lytic spectrum of LysAB1245 on multi-drug resistant (MDR) A.
baumannii isolates. Bacterial pellets (105 CFU/mL) were treated with LysAB1245 (1.7 unit/reaction). Acinetobacter baumannii with KL2 (A), KL3 (B), KL6 (C), KL10 (D), KL47 (E), KL49 (F), and KL52 (G) and ATCC 19606 (H). Two independent experiments were performed in triplicate and error bars represent the standard deviation. Limit of detection (LOD) for surviving bacterial cells was 2 log CFU/mL.
Stability of LysAB1245 under various temperature and pH conditions
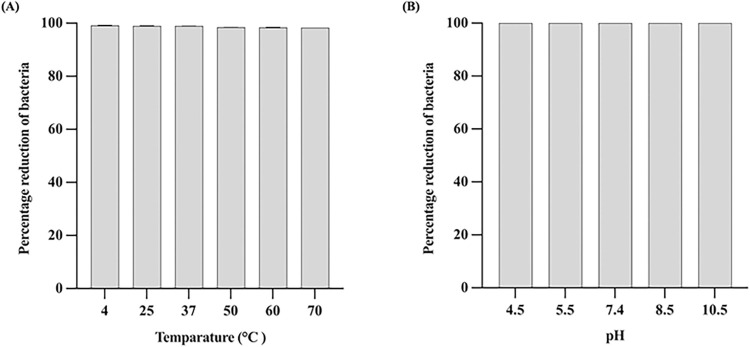
Thermal and pH stability are desirable properties of antibacterial agents during storage. Therefore, the stability of LysAB1245 under different temperatures and pH conditions was examined. LysAB1245 remained stable and highly bactericidal at temperatures ranging from 4 to 70°C (>99% reduction in bacterial cells when compared with the control) (Fig 5A). Additionally, the activity of LysAB1245 was relatively stable to pH changes over a range from 4.5 to 10.5 (Fig 5B) (S2 Fig). The results indicated that LysAB1245 could be used as a potential alternative antibacterial agent due to its stable activity across a broad range of thermal and pH conditions.
Fig 5. The stability of purified endolysin LysAB1245 at various temperatures and pH conditions.
Bacterial pellets (105 CFU/mL) were mixed with LysAB1245 at 4, 25, 37, 50, 60, and 70°C (A) and pH 4.5, 5.5, 7.4, 8.5, and 10.5 (B). The percentage of bacterial reduction was compared with the control. The experiment was performed in triplicate and error bars represent the standard deviation.
Kinetic analysis of LysAB1245
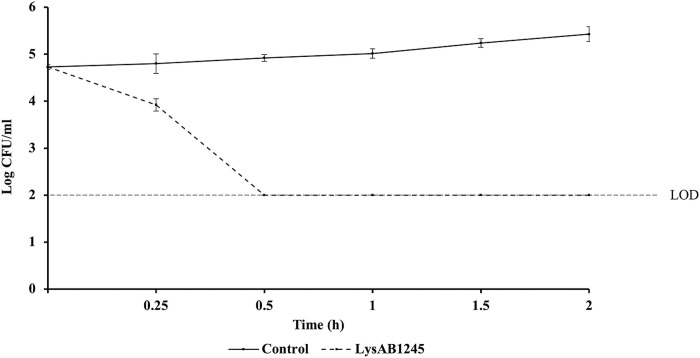
Killing curve analysis revealed that LysAB1245 at 1.7 unit/reaction displayed rapid bactericidal activity against A. baumannii ABMYH-1245 (Fig 6). A 2.92 log reduction (>99% reduction) in viable bacteria was observed within 30 min, compared with control (S2 Fig). In this study, the phage T1245-produced endolysin LysAB1245 was successfully expressed and purified using an automated high-throughput protein synthesis system for producing highly pure, stable, and soluble active proteins.
Fig 6. Time-kill curve of A. baumannii ABMYH-1245 incubated with LysAB1245.
Two independent experiments were performed in triplicate and error bars represent the standard deviation. Limit of detection (LOD) for surviving bacterial cells was 2 log CFU/mL.
Antimicrobial activity of LysAB1245
Furthermore, we examined the antimicrobial activity of LysAB1245 against 20 MDR A. baumannii isolates and ATCC 19606. The MIC value of LysAB1245 was 4.21 μg/mL (0.1 unit/reaction) for all MDR A. baumannii isolates and A. baumannii ATCC 19606. The lowest concentration of LysAB1245 with bactericidal activity was 4.21 μg/mL, which was identical to the MIC value (Table 2). In general, the peptidoglycan structure of Gram-negative bacteria is highly conserved. Therefore, the conservation of the peptidoglycan structure among the tested isolates, which serves as the target site of endolysin, might have resulted in the same MIC values of LysAB1245.
Table 2. Minimal inhibitory concentrations (MICs) and minimal bactericidal concentrations (MBCs) of LysAB1245 against Acinetobacter baumannii.
| Acinetobacter baumannii | Antibacterial activities (μg/mL) | |
|---|---|---|
| MIC | MBC | |
| 20 clinical isolates | 4.21 | 4.21 |
| ATCC 19606 | 4.21 | 4.21 |
According to previous report, no cytotoxic effect of endolysin LysSS was observed on human lung cell line A549 at concentrations less than 250 μg/mL [21]. Additionally, the therapeutic effects of phage endolysins in mouse models of infection have been extensively reported [38,41,42]. For example, the direct administration of endolysin by inhalation improved survival rate by 80% in a mouse model of pneumococcal pneumonia [43]. Moreover, treatment with endolysin SAL200 by inhalation did not induce abnormal inflammatory response in mice with pneumonia [41]. Furthermore, the toxicity and safety of phage endolysin SAL200 administered via intravenous injection has been assessed in tested animals for the drug development process [44]. In 2017, endolysin SL200 was successful administered to target drug-resistant staphylococcal infections in humans [45].
The findings of this study suggest that LysAB1245 would be valuable to further development as a new potential therapeutic alternative for controlling of nosocomial MDR A. baumannii infections. However, studies are necessary to elucidate the optimal dosage and bactericidal mechanism of LysAB1245 using mammalian infection models before reaching the phase of clinical trials in humans.
Conclusions
In the present study, the endolysin LysAB1245 from Acinetobacter phage T1245 was successfully expressed and purified using an automated protein synthesis system with high-purity target proteins. A novel purified LysAB1245 exhibited a broad lytic spectrum activity against MDR A. baumannii isolates, which belong to various major capsular types. Additionally, LysAB1245 displayed rapid bactericidal activity and stability under various pH and temperature conditions. This work elucidated a potential of LysAB1245 as a new potential therapeutic agent towards the management of MDR A. baumannii infections.
Supporting information
SDS-PAGE and western blot images for Fig 2. Lane 1, molecular size marker; lane 2, purified LysAB1245; lane 3, expression sample; lane 4, washed sample; lane 5, unbound sample.
(PDF)
The values used to build the graphs included the means and standard deviations.
(PDF)
(ZIP)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by the National Research Council of Thailand (Grant No. N41A640071) and the Postdoctoral Fellowship from Prince of Songkla University, Thailand. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perez S, Innes GK, Walters MS, Mehr J, Arias J, Greeley R, et al. Increase in hospital-acquired carbapenem-resistant Acinetobacter baumannii infection and colonization in an acute care hospital during a surge in COVID-19 admissions—New Jersey, February–July 2020. MMWR Morb Mortal Wkly Rep. 2020;69: 1827–1831. doi: 10.15585/mmwr.mm6948e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boral J, Genç Z, Pınarlık F, Ekinci G, Kuskucu MA, İrkören P, et al. The association between Acinetobacter baumannii infections and the COVID-19 pandemic in an intensive care unit. Sci Rep. 2022;12: 20808. doi: 10.1038/s41598-022-25493-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo A, Gavaruzzi F, Ceccarelli G, Borrazzo C, Oliva A, Alessandri F, et al. Multidrug-resistant Acinetobacter baumannii infections in COVID-19 patients hospitalized in intensive care unit. Infection. 2022;50: 83–92. doi: 10.1007/s15010-021-01643-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, Yao Y, Zhu B, Ren D, Yang Q, Fu Y, et al. Risk factors for acquisition and mortality of multidrug-resistant Acinetobacter baumannii bacteremia: A retrospective study from a Chinese hospital. Medicine. 2019;98: e14937. doi: 10.1097/MD.0000000000014937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohd Sazlly Lim S, Zainal Abidin A, Liew SM, Roberts JA, Sime FB. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J Infect. 2019;79: 593–600. doi: 10.1016/j.jinf.2019.09.012 [DOI] [PubMed] [Google Scholar]
- 6.Motbainor H, Bereded F, Mulu W. Multi-drug resistance of blood stream, urinary tract and surgical site nosocomial infections of Acinetobacter baumannii and Pseudomonas aeruginosa among patients hospitalized at Felegehiwot referral hospital, Northwest Ethiopia: A cross-sectional study. BMC Infect Dis. 2020;20: 92. doi: 10.1186/s12879-020-4811-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez J, Razo-Gutierrez C, Le C, Courville R, Pimentel C, Liu C, et al. Cerebrospinal fluid (CSF) augments metabolism and virulence expression factors in Acinetobacter baumannii. Sci Rep. 2021;11: 4737. doi: 10.1038/s41598-021-81714-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talyansky Y, Nielsen TB, Yan J, Carlino-Macdonald U, Di Venanzio G, Chakravorty S, et al. Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. PLOS Pathog. 2021;17: e1009291. doi: 10.1371/journal.ppat.1009291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akoolo L, Pires S, Kim J, Parker D. The Capsule of Acinetobacter baumannii protects against the innate immune response. J Innate Immun. 2022;14: 543–554. doi: 10.1159/000522232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh JK, Adams FG, Brown MH. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front Microbiol. 2018;9: 3301. doi: 10.3389/fmicb.2018.03301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loraine J, Heinz E, Soontarach R, Blackwell GA, Stabler RA, Voravuthikunchai SP, et al. Genomic and phenotypic analyses of Acinetobacter baumannii isolates from three tertiary care hospitals in Thailand. Front Microbiol. 2020;11: 548. doi: 10.3389/fmicb.2020.00548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh YC, Wang SH, Chen YY, Lin TL, Shie SS, Huang CT, et al. Association of capsular types with carbapenem resistance, disease severity, and mortality in Acinetobacter baumannii. Emerg Microbes Infect. 2020;9: 2094–2104. doi: 10.1080/22221751.2020.1822757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishk R, Soliman N, Nemr N, Eldesouki R, Mahrous N, Gobouri A, et al. Prevalence of aminoglycoside resistance and aminoglycoside modifying enzymes in Acinetobacter baumannii among intensive care unit patients, Ismailia, Egypt. Infect Drug Resist. 2021;14: 143–150. doi: 10.2147/IDR.S290584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Chen K, Wu Y, Huang L, Fang Y, Lu J, et al. Epidemiological and genetic characteristics of clinical carbapenem-resistant Acinetobacter baumannii strains collected countrywide from hospital intensive care units (ICUs) in China. Emerg Microbes Infect. 2022;11: 1730–1741. doi: 10.1080/22221751.2022.2093134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seleim SM, Mostafa MS, Ouda NH, Shash RY. The role of pmrCAB genes in colistin-resistant Acinetobacter baumannii. Sci Rep. 2022;12: 20951. doi: 10.1038/s41598-022-25226-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabic J, Novovic K, Kekic D, Trudic A, Opavski N, Dimkic I, et al. Comparative genomics and molecular epidemiology of colistin-resistant Acinetobacter baumannii. Comput Struct Biotechnol J. 2023;21: 574–585. doi: 10.1016/j.csbj.2022.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pormohammad A, Mehdinejadiani K, Gholizadeh P, Nasiri MJ, Mohtavinejad N, Dadashi M, et al. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: A systematic review and meta-analysis. Microb Pathog. 2020;139: 103887. doi: 10.1016/j.micpath.2019.103887 [DOI] [PubMed] [Google Scholar]
- 18.Sun B, Liu H, Jiang Y, Shao L, Yang S, Chen D. New mutations involved in colistin resistance in Acinetobacter baumannii. mSphere. 2020;5: e00895–19. doi: 10.1128/mSphere.00895-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan X, Chen H, Zhang M, Zhao Y, Jiang Y, Liu X, et al. Clinical experience of personalized phage therapy against carbapenem-resistant Acinetobacter baumannii lung infection in a patient with chronic obstructive pulmonary disease. Front Cell Infect Microbiol. 2021;11: 631585. doi: 10.3389/fcimb.2021.631585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu N, Dai J, Guo M, Li J, Zhou X, Li F, et al. Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg Microbes Infect. 2021;10: 612–618. doi: 10.1080/22221751.2021.1902754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Lee DW, Jin JS, Kim J. Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J Glob Antimicrob Resist. 2020;22: 32–39. doi: 10.1016/j.jgar.2020.01.005 [DOI] [PubMed] [Google Scholar]
- 22.Gouveia A, Pinto D, Veiga H, Antunes W, Pinho MG, são-José C. Synthetic antimicrobial peptides as enhancers of the bacteriolytic action of staphylococcal phage endolysins. Sci Rep. 2022;12: 1245. doi: 10.1038/s41598-022-05361-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briers Y, Schmelcher M, Loessner MJ, Hendrix J, Engelborghs Y, Volckaert G, et al. The high-affinity peptidoglycan binding domain of Pseudomonas phage endolysin KZ144. Biochem Biophys Res Commun. 2009;383: 187–191. doi: 10.1016/j.bbrc.2009.03.161 [DOI] [PubMed] [Google Scholar]
- 24.Wu M, Hu K, Xie Y, Liu Y, Mu D, Guo H, et al. A novel phage PD-6A3, and its endolysin Ply6A3, with extended lytic activity against Acinetobacter baumannii. Front Microbiol. 2018;9: 3302. doi: 10.3389/fmicb.2018.03302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman MU, Wang W, Sun Q, Shah JA, Li C, Sun Y, et al. Endolysin, a promising solution against antimicrobial resistance. Antibiotics (Basel). 2021;10: 1277. doi: 10.3390/antibiotics10111277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerova M, Halgasova N, Ugorcakova J, Bukovska G. Endolysin of bacteriophage BFK20: Evidence of a catalytic and a cell wall binding domain. FEMS Microbiol Lett. 2011;321: 83–91. doi: 10.1111/j.1574-6968.2011.02312.x [DOI] [PubMed] [Google Scholar]
- 27.Chang Y, Ryu S. Characterization of a novel cell wall binding domain-containing Staphylococcus aureus endolysin LysSA97. Appl Microbiol Biotechnol. 2017;101: 147–158. doi: 10.1007/s00253-016-7747-6 [DOI] [PubMed] [Google Scholar]
- 28.Soontarach R, Srimanote P, Enright MC, Blundell-Hunter G, Dorman MJ, Thomson NR, et al. Isolation and characterisation of bacteriophage selective for key Acinetobacter baumannii capsule chemotypes. Pharmaceuticals (Basel). 2022;15: 443. doi: 10.3390/ph15040443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis. Molecular Cloning: A Laboratory Manual, 2nd (ed.). New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Lai MJ, Lin NT, Hu A, Soo PC, Chen LK, Chen LH, et al. Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both Gram-positive and gram-negative bacteria. Appl Microbiol Biotechnol. 2011;90: 529–539. doi: 10.1007/s00253-011-3104-y [DOI] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI Document m100. Wayne (Pennsylvania): CLSI; 2020. [Google Scholar]
- 32.Simpson DJ, Sacher JC, Szymanski CM. Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr Opin Struct Biol. 2015;34: 69–77. doi: 10.1016/j.sbi.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 33.Larpin Y, Oechslin F, Moreillon P, Resch G, Entenza JM, Mancini S. In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLOS ONE. 2018;13: e0192507. doi: 10.1371/journal.pone.0192507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang G, Shen X, Gong Y, Dong Z, Zhao X, Shen W, et al. Antibacterial properties of Acinetobacter baumannii phage Abp1 endolysin (PlyAB1). BMC Infect Dis. 2014;14: 681. doi: 10.1186/s12879-014-0681-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thummeepak R, Kitti T, Kunthalert D, Sitthisak S. Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage ØABP-01 endolysin (LysABP-01) in combination with colistin. Front Microbiol. 2016;7: 1402. doi: 10.3389/fmicb.2016.01402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y, Xu D, Wang L, Qu M, Li F, Tan Z, et al. Characterization of a broad-spectrum endolysin LysSP1 encoded by a Salmonella bacteriophage. Appl Microbiol Biotechnol. 2021;105: 5461–5470. doi: 10.1007/s00253-021-11366-z [DOI] [PubMed] [Google Scholar]
- 37.Ni P, Wang L, Deng B, Jiu S, Ma C, Zhang C, et al. Characterization of a lytic bacteriophage against Pseudomonas syringae pv. actinidiae and its endolysin. Viruses. 2021;13: 631. doi: 10.3390/v13040631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lood R, Winer BY, Pelzek AJ, Diez-Martinez R, Thandar M, Euler CW, et al. Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother. 2015;59: 1983–1991. doi: 10.1128/AAC.04641-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Düring K, Porsch P, Mahn A, Brinkmann O, Gieffers W. The non‐enzymatic microbicidal activity of lysozymes. FEBS Lett. 1999;449: 93–100. doi: 10.1016/s0014-5793(99)00405-6 [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Li G, Wan F, Liu P, Du F, Shao Y, et al. Virulence characteristics and drug resistance of the prevalent capsule types in Acinetobacter baumannii. Microb Drug Resist. 2023;29: 274–279. doi: 10.1089/mdr.2022.0310 [DOI] [PubMed] [Google Scholar]
- 41.Bae JY, Jun KI, Kang CK, Song KH, Choe PG, Bang JH, et al. Efficacy of intranasal administration of the recombinant endolysin SAL200 in a lethal murine Staphylococcus aureus pneumonia model. Antimicrob Agents Chemother. 2019;63: e02009–18. doi: 10.1128/AAC.02009-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raz A, Serrano A, Hernandez A, Euler CW, Fischetti VA. Isolation of phage lysins that effectively kill Pseudomonas aeruginosa in mouse models of lung and skin infection. Antimicrob Agents Chemother. 2019;63: e00024–19. doi: 10.1128/AAC.00024-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doehn JM, Fischer K, Reppe K, Gutbier B, Tschernig T, Hocke AC, et al. Delivery of the endolysin cpl-1 by inhalation rescues mice with fatal pneumococcal pneumonia. J Antimicrob Chemother. 2013;68: 2111–2117. doi: 10.1093/jac/dkt131 [DOI] [PubMed] [Google Scholar]
- 44.Jun SY, Jung GM, Yoon SJ, Choi YJ, Koh WS, Moon KS, et al. Preclinical safety evaluation of intravenously administered SAL200 containing the recombinant phage endolysin SAL-1 as a pharmaceutical ingredient. Antimicrob Agents Chemother. 2014;58: 2084–2088. doi: 10.1128/AAC.02232-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jun SY, Jang IJ, Yoon S, Jang K, Yu KS, Cho JY, et al. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob Agents Chemother. 2017;61: e02629–16. doi: 10.1128/AAC.02629-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDS-PAGE and western blot images for Fig 2. Lane 1, molecular size marker; lane 2, purified LysAB1245; lane 3, expression sample; lane 4, washed sample; lane 5, unbound sample.
(PDF)
The values used to build the graphs included the means and standard deviations.
(PDF)
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.