Abstract
The TecA chlorobenzene dioxygenase and the TodCBA toluene dioxygenase exhibit substantial sequence similarity yet have different substrate specificities. Escherichia coli cells producing recombinant TecA enzyme dioxygenate and simultaneously eliminate a halogen substituent from 1,2,4,5-tetrachlorobenzene but show no activity toward benzene, whereas those producing TodCBA dioxygenate benzene but not tetrachlorobenzene. A hybrid TecA dioxygenase variant containing the large α-subunit of the TodCBA dioxygenase exhibited a TodCBA dioxygenase specificity. Acquisition of dehalogenase activity was achieved by replacement of specific todC1 α-subunit subsequences by equivalent sequences of the tecA1 α-subunit. Substrate transformation specificities and rates by E. coli resting cells expressing hybrid systems were analyzed by high-performance liquid chromatography. This allowed the identification of both a single amino acid and potentially interacting regions required for dechlorination of tetrachlorobenzene. Hybrids with extended substrate ranges were generated that exhibited activity toward both benzene and tetrachlorobenzene. The regions determining substrate specificity in (chloro)benzene dioxygenases appear to be different from those previously identified in biphenyl dioxygenases.
Aerobic degradation of aromatic compounds by bacteria is frequently initiated by non-heme iron-containing dioxygenases (22). These soluble multicomponent enzymes, which introduce two atoms of molecular oxygen into the aromatic ring and thereby activate it for subsequent cleavage, are classified into five groups according to the number of constituent components and the nature of the redox centers (3). Class IIB dioxygenases, such as the TecA chlorobenzene (4) and the TodCBA toluene dioxygenases (39), are comprised of a reductase and ferredoxin, which together serve as a short electron transport chain, and a catalytic terminal dioxygenase composed of a large α-subunit and small β-subunit with an (αβ)n configuration (22). Structural information about aromatic ring dioxygenases is very limited, and only recently has the terminal oxygenase component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 been crystallized (21).
α-Subunits of class IIB terminal dioxygenases contain a Rieske-type [2Fe-2S] iron-sulfur cluster (11, 28), an active-site non-heme mononuclear Fe(II) center (22), and the substrate binding site, which is assumed to be located in the vicinity of the activating iron (6). By exchanging subunits between different dioxygenase systems, several groups have shown that the α-subunit is responsible for substrate specificity (8–10, 26, 32, 34, 36). Further analyses of α-subunits subsequently identified a large C-terminal region of nitrotoluene dioxygenase of Pseudomonas sp. strain JS42 (26) and smaller elements of biphenyl dioxygenases from Pseudomonas pseudoalcaligenes KF707 and Pseudomonas sp. strain LB400 (19, 23) as being involved in the determination of substrate specificity.
The β-subunits of class IIB enzymes were reported to play a role in subunit association (6, 13, 24) and substrate recognition (12, 13), whereas some investigators excluded a direct involvement of the β-subunit in the determination of substrate specificity (26, 27, 34).
The substrate specificities of initial dioxygenases are crucial, because they often limit the range of compounds potentially degradable by the catabolic system. Because substituents often complicate mineralization, removal of one in the first step of a catabolic sequence is an advantageous mechanism that merits special attention. So far only one enzyme, the TecA chlorobenzene dioxygenase of Burkholderia sp. strain PS12, has been shown to dechlorinate a tetrachlorobenzene (4), and no dioxygenase able to transform higher chlorinated benzenes is known to date. Comprehension of the structural requirements for dechlorination is a prerequisite for the improvement of the catalytic properties of biocatalysts.
To identify the structural elements involved in dechlorination, we examined two class IIB enzymes (3) with complementary substrate specificities by exchanging equivalent polypeptide sequences. We report here the construction of an extensive number of chimeric dioxygenases and the analysis of their substrate specificities and transformation rates. This led to the identification of a single amino acid, as well as interacting regions required for dehalogenation of tetrachlorobenzene, and the generation of hybrid dioxygenases with extended substrate ranges.
MATERIALS AND METHODS
Strain and plasmids.
The host strain used in this study was Escherichia coli DH5α from Clontech. The cloning vectors used were pBluescript II KS(+) (Stratagene) and pCR2.1 (Invitrogen). The sources of the tecA1A2A3A4 chlorobenzene dioxygenase genes were plasmids pSTE3 and pSTE7 (4), and the source of the todC1C2BA toluene dioxygenase genes was plasmid pDTG601 (39).
DNA manipulations.
Standard procedures were performed as described by Sambrook et al. (30). Restriction enzymes were purchased from Amersham, Boehringer Mannheim, MBI Fermentas, New England Biolabs, and U.S. Biochemical Corp. T4 DNA ligase was purchased from New England Biolabs. Isopropyl-β-d-thiogalactopyranoside was obtained from Roth. Ampicillin was purchased from Sigma. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was obtained from Biomol. Oligonucleotides were obtained from Gibco Life Technologies. Taq and Pfu DNA polymerases were obtained from Boehringer Mannheim and Stratagene, respectively. Blunt-end fragments resulting from Pfu DNA polymerase amplification were A-tailed with Taq polymerase for subsequent cloning into the pCR2.1 T-vector system (Invitrogen) as described previously (31). Elution of DNA from agarose gels was performed with the QiaexII gel extraction kit (Qiagen). Plasmids were purified with a Qiawell 8 plasmid kit or a plasmid midi kit (Qiagen). Sequencing was done with the Applied Biosystems 373A DNA sequencer (Perkin-Elmer, Applied Biosystems) as described previously (18). Site-specific mutations were introduced by using splicing by overlap extension (SOE)-PCR (14) or with the Quikchange site-directed mutagenesis kit (Stratagene). The sequences of all de novo-synthesized DNA molecules and of the commercially obtained oligonucleotide primer sequences were confirmed by sequencing.
Oligonucleotides.
The designation, sequence (5′→3′), and priming direction of the oligonucleotide primers used for amplification of DNA fragments by PCR (29), SOE-PCR (14), and the Quikchange kit (Stratagene) are as follows: prSTB70 (forward), gcccgcgggctctatgcccattggc (SacII); prSTB71 (reverse), gacgtcggctctcttgacggaatcaagc (AatII); prSTB72 (forward), cgagctcggtgagaagacaatgaatc (SacI); prSTB73 (reverse), cctcggtgcggtcgagcatatggtc (NdeI); prSTB74 (forward), cgaagttctacatggaccatatgctcg (NdeI); prSTB75 (reverse), gtagctggtgacctttggccccatg (BstEII); prSTB76 (reverse), ggatgccatgtccggaccgtgttg (RsrII); prSTB77 (forward), caacacggtccggacatggcatcc (RsrII); prSTB104 (reverse), ctggcaggcctgccaggatgcc (StuI); prSTB105 (forward), ctgtaactggaaactcgccgcagagc (Phe211Leu); prSTB106 (forward), gagcagttttgctgggacatgtaccatg (Ser218Trp); prSTB107 (forward), gttttgcagcgacgcgtaccatgccg (Met220Ala); prSTB108 (forward), gtaccatgccgcgacgacctcgcatc (Gly224Ala); prSTB109 (forward), ccgggacgaccgcgcatctgtctgg (Ser227Ala); prSTB110 (forward), ctgtaactggaaagccgccgcagagc (Phe211Ala); prSTB111 (forward), gagcagttttgcgccgacatgtaccatg (Ser218Ala); prSTB114 (forward), caggcctgccagacggcgttgaactg (StuI); prSTB124 (reverse), gctctgcggcgagtttccagttacag (Phe211Leu); prSTB125 (reverse), catggtacatgtcccagcaaaactgctc (Ser218Trp); prSTB126 (reverse), cggcatggtacgcgtcgctgcaaaac (Met220Ala); prSTB127 (reverse), gatgcgaggtcgtcgcggcatggtac (Gly224Ala); prSTB128 (reverse), ccagacagatgcgcggtcgtcccgg (Ser227Ala); prSTB129 (reverse), gctctgcggcggctttccagttacag (Phe211Ala); prSTB130 (reverse), catggtacatgtcggcgcaaaactgctc (Ser218Ala); prSTB132 (reverse), ggtgacctttggccccatgatggcaagcagcagatcgggttcgccaataaagaagccac (BstEII); prSTB135 (forward), ggcaggcctgccagaagaccttgaaatggccgatc (StuI); prSTB136 (forward), ctggcaggcctgccggacggcgttg (StuI); and prSTB137 (forward), cgaaggtcaccagctactggacc (BstEII). Recognition sequences for the restriction enzymes indicated are underlined, and boldface letters indicate the triplets which were changed by the Quikchange system.
Resting cell assays.
E. coli strains were cultured (1% inoculum) in Luria-Bertani medium (30) containing 0.1 mg of ampicillin per ml and 1.0 mM isopropyl-thio-β-galactopyranoside in baffled round-bottom flasks at 30°C on a rotary shaker operated at 160 rpm, and cells were prepared as described previously (4). Briefly, after washing twice with assay buffer (10 mM glucose in 0.1 × M9 mineral medium), cell suspensions were concentrated to an A600 of 50, and then 1.0-ml samples were removed from the concentrate, shock-frozen in liquid nitrogen, and stored at −20°C for Western blot analysis and determination of whole-cell protein. The resting cell assay was started by dilution of concentrated cell suspension to an A600 of 2.0 in 10 ml of prewarmed assay buffer containing 0.5 mM substrate. To monitor product formation, samples were taken at regular intervals between 0 and 180 min and immediately shock-frozen in liquid nitrogen for subsequent analysis. None of the transformation products indicated in Table 1 was detected in control experiments with E. coli resting cells carrying pBluescript II KS(+). Standard deviations were below 50%, as determined from three independent transformation experiments each with E. coli (pSTO4), E. coli (pSTE7), and E. coli (pSTE81) cells.
TABLE 1.
Retention volumes and absorption maxima in HPLC analysis of dihydroxy intermediates formed from unchlorinated and chlorinated benzenesa
| Substrate | Proposed product | % [MetOH] used | RV (ml) | λmax (nm) |
|---|---|---|---|---|
| Benzene | cis-1,2-Dihydroxy-1,2-dihydrocyclohexa-3,5-diene | 18 | 3.1 | 262 |
| 1,2-Dichlorobenzene | 3,4-Dichloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene | 45 | 4.1 | 272 |
| 1,2,4,5-Tetrachlorobenzene | 3,4,6-Trichloro-1,2-dihydroxybenzene | 63 | 5.5 | 210 |
Supernatant fluid from E. coli resting cells carrying different dioxygenase systems was analyzed by HPLC for the formation of dihydroxylated compounds. Chromatographic and spectral properties of the proposed products derived from the substrates listed are shown. [MetOH], methanol used in the aqueous solvent system; RV, retention volume; λmax, wavelength of the absorption maximum.
Analysis of transformation products.
Dihydroxy compounds were analyzed with a reverse-phase high-performance liquid chromatography (HPLC) system equipped with autosampler and a sample cooler unit operated at 4°C (Shimadzu) on an SC 125 by 4.6-mm Lichrospher 100 RP8 5.0-μm column (Bischoff). The aqueous solvent system contained 0.1% ortho-phosphoric acid and 18 to 63% methanol at a 1.0-ml/min flow rate (Table 1). Shock-frozen samples were thawed and centrifuged (20 min, 4°C, 14,000 × g), and 20 μl of cell-free supernatant fluid was analyzed. Absorbance was monitored between 200 and 400 nm. The identities of cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene and 3,4,6-trichlorocatechol were confirmed by comparison with authentic standards, which were also used to calibrate HPLC analyses. Spectral data of 3,4-dichloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene were in agreement with those published previously (4).
Western blot analysis.
Immunologically detectable soluble α-subunits from individual constructs used for transformation experiments were visualized by Western blot analysis. Cell suspensions were thawed and diluted as required in the resting cell assays, ruptured on ice by ultrasonic pulses (six times at 10 s each, 100 W; Labsonic U; B. Braun) in the presence of 40 μg of DNAse I per ml and 20 μg of RNase A per ml, and centrifuged (40 min, 4°C, 130,000 × g). Supernatant fluid was subjected to electrophoresis on a sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gel (20). Extract of E. coli cells carrying pBluescript II KS(+) obtained in a similar fashion was used as negative control, and different volumes of a similar extract of E. coli (pSTE7) cells producing the wild-type TecA dioxygenase were used as internal standards on each gel.
Proteins were subsequently electrotransferred (30 min, 100 V) (30) in a Bio-Rad wet-blot apparatus onto a 0.2-μm-pore-diameter transblot nitrocellulose membrane (Bio-Rad). Immunological detection of α-subunit proteins was carried out with polyclonal affinity-purified antibodies raised against denatured α-subunit of the terminal BedC1 benzene dioxygenase component (38). The membrane was blocked (30 min, phosphate-buffered saline [PBS]–5% nonfat dry milk) (30) and incubated for 1 h with anti-BedC1 antibody (diluted 1:500 in PBS–2.5% nonfat dry milk), which was omitted from the solution in a conjugate-control experiment. After washing (three times for 10 min each in PBS–0.05% Tween 20), the membrane was incubated overnight at 4°C with peroxidase-conjugated Affinipure goat anti-rabbit immunoglobulin G (H+L) from Dianova (diluted 1:1,500 in PBS–0.05% Tween 20), which was found to be highly specific for anti-BedC1 antibody (data not shown). After removal of unbound antibody (three times for 10 min each, PBS–0.05% Tween 20), ECL (enhanced chemiluminescence) Western blotting detection reagents were added according to the manufacturer’s instructions (Amersham), and chemoluminescence was detected on BioMax XR film (Kodak). Polyclonal anti-BedC1 antibody was specific for α-subunit proteins (see Fig. 2) and immuno-cross-reactivity was observed only with a protein with a size of 57.6 ± 0.3 kDa, which was the only protein detected in E. coli cells carrying pBluescript II KS(+) (data not shown). Molecular weights of immunodetected proteins were determined with high-molecular-weight rainbow-colored protein markers (Amersham).
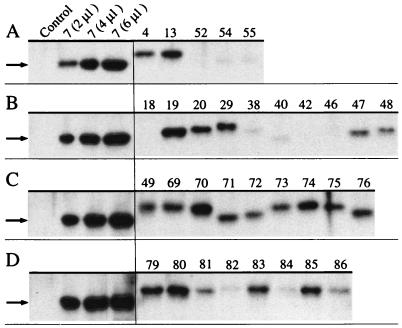
FIG. 2.
Western blot analysis of soluble α-subunit proteins. After electrophoretic separation of 4 μl of crude extracts of E. coli cells carrying different plasmids encoding dioxygenase systems, soluble α-subunits were detected with anti-BedC1 rabbit antibody (38), and bands corresponding to chimeric α-subunit proteins (50 to 51 kDa) were visualized on film. Eight microliters of E. coli (pBluescript II KS[+]) was used as a negative control, and 2, 4, and 6 μl of E. coli (pSTE7) crude cell extract served as internal standards on the first four lanes of each gel. Numbers at the top of each lane correspond to the number of the plasmid carried in E. coli according to Tables 2 and 3 and Fig. 3. The sizes of the wild-type proteins produced in E. coli (pSTE7) and E. coli (pSTO4) were 50.1 ± 0.3 (indicated by an arrow) and 51.3 ± 0.3 kDa, respectively, which is in close agreement with the deduced molecular masses of wild-type TecA (50.5 kDa) (4) and TodCBA (50.9 kDa) (39) dioxygenases. Small differences in the relative mobility of α-subunit proteins are assumed to be due to differences in size and amino acid composition of individual chimeras. The panel was composed from four different gels with Photoshop software (version 3.0; Adobe).
Substrate transformation rates of 1,2-dichlorobenzene, benzene, and 1,2,4,5-tetrachlorobenzene.
Substrate transformation rates were expressed as the amount of product formed per unit of time and amount of whole-cell protein (1,000 area units [AU] min−1 μg−1) measured at the wavelength of the corresponding absorption maximum after HPLC separation (Table 1). Rates were usually constant for at least 30 min (Fig. 1). The concentration of whole-cell protein was determined by boiling cell suspensions for 10 min in 100 mM NaOH according to the method of Bradford (5) with bovine serum albumin as the standard. The use of the same bacterial host and assay conditions allowed a direct comparison of the transformation rates of different dioxygenase systems for each substrate.
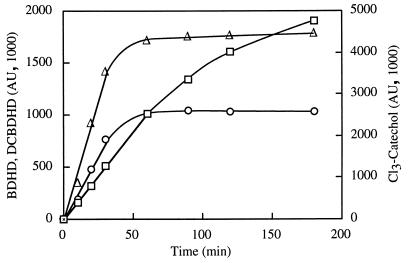
FIG. 1.
HPLC analysis of product formation. E. coli cells carrying dioxygenase genes were incubated with 0.5 mM substrate. At regular time intervals, samples were taken, and the supernatant fluids were analyzed by HPLC (Table 1) for product accumulation. The formation of 3,4-dichloro-cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene (DCBDHD [▵]) from 1,2-dichlorobenzene and cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene (BDHD [○]) from benzene by E. coli (pSTO4) and the formation of 3,4,6-trichlorocatechol (Cl3-catechol) from tetrachlorobenzene (□) by E. coli (pSTE7) are shown as a function of time.
Chemicals.
Benzene and 1,2-dichlorobenzene were purchased from Fluka, 1,2,4,5-tetrachlorobenzene was obtained from Aldrich, and cis-1,2-dihydroxy-1,2-dihydrocyclohexa-3,5-diene was from Sigma. All chemicals were of the highest purity available. HPLC-grade methanol was from Baker, and 3,4,6-trichlorocatechol was kindly provided by H.-A. Arfmann.
RESULTS AND DISCUSSION
To define the subunit, region, and amino acids which are responsible for dechlorination of tetrachlorobenzene, genetic elements were exchanged between the tecA and todCBA dioxygenase systems of Burkholderia sp. strain PS12 (4) and Pseudomonas putida F1 (39), respectively (Table 2). A large number of hybrids were prepared and analyzed in order to localize specificity determinants as precisely as possible and to investigate the combined effects of multiple determinants. The resulting hybrid dioxygenases were subsequently expressed in E. coli for analysis of the effects of the substitutions on their catalytic potential (Table 3). Because the final product concentration was not always highest for the substrate that was converted at the highest initial rate (data not shown), initial substrate transformation rates were determined (Fig. 1).
TABLE 2.
Hybrid plasmids constructed in this studya
| Type of plasmid | Relevant characteristics | Structure |
|---|---|---|
| pCR2.1 derivatives | ||
| pCR5 | todC1(III from SacII site)-tecA2(5′ sequence to AatII site) | 0.2-kb fragment PCR amplified from pSTO4 template with primer pair prSTB70/71 |
| pCR6 | tecA1(I) | 0.6-kb fragment PCR amplified from pSTE7 template with primer pair prSTB72/73 |
| pCR7 | tecA1(II) | 0.5-kb fragment PCR amplified from pSTE7 template with primer pair prSTB74/76 |
| pCR8 | tecA1(III)-tecA2(5′ sequence to AatII site) | 0.5-kb fragment PCR amplified from pSTE7 template with primer pair prSTB77/71 |
| pCR9 | todC1 (I+IIABCD) (with NdeI site) | 0.9-kb fragment SOE-PCR-amplified from pSTO4 template with primer pairs prSTB72/73 and prSTB74/75 |
| pCR13 | tecA1(IIA) | 0.15-kb fragment PCR amplified from pSTE7 template with primer pair prSTB74/104 |
| pCR14 | tecA1(IIB)-todC1(IICD) | 0.15-kb fragment PCR amplified from pSTO4 template with primer pair prSTB114/75 |
| pCR15 | todC1(IIABC)-tecA1(IID) | 0.3-kb fragment PCR amplified from pSTO4 template with primer pair prSTB74/132 |
| pCR20 | todC1(IIB)-tecA1(IICDE) | 0.3-kb fragment PCR amplified from pSTE7 template with primer pair prSTB135/76 |
| pCR21 | tecA1(IIBCD) | 0.15-kb fragment PCR amplified from pSTE7 template with primer pair prSTB136/75 |
| pCR22 | tecA1(IIE) | 0.15-kb fragment PCR amplified from pSTE7 template with primer pair prSTB137/76 |
| pCR23 | tecA1(IIBCDE) | 0.3-kb fragment PCR amplified from pSTE7 template with primer pair prSTB136/76 |
| pBluescript KS II(+) derivatives | ||
| pSTE10.1 | pBluescript KS II(+) lacking part of its multiple cloning site | pBluescript KS II(+) religated after removal of EcoRV-Ecl136II fragment |
| pSTE11 | todC1-todC2 (5′ sequence to HindIII site) | 1.6-kb HindIII fragment of pSTO4 into HindIII site of pSTE10.1 |
| pSTE12 | todC1-tecA2 (5′ sequence to AatII site) | 0.2-kb SacII-AatII fragment of pCR5 into SacII-AatII site of pSTE11 |
| pSTE14 | todC1 (with NdeI site) | 0.8-kb SacI-BstEII fragment of pCR9 into SacI-BstEII site of pSTE12 |
| pSTE15 | tecA1(I)-todC1(II+III) | 0.6-kb SacI-NdeI fragment of pCR6 into SacI-NdeI site of pSTE14 |
| pSTE16 | todC1(I)-tecA1(II)-todC1(III) | 0.4-kb NdeI-RsrII fragment of pCR7 into NdeI-RsrII site of pSTE14 |
| pSTE17 | todC1(I+II)-tecA1(III) | 0.5-kb RsrII-AatII fragment of pCR8 into RsrII-AatII site of pSTE14 |
| pSTE21 | todC1(Phe211Leu) | Quikchange mutation of pSTE14 with primer pair prSTB105/124 |
| pSTE22 | todC1(Ser218Trp) | Quikchange mutation of pSTE14 with primer pair prSTB106/125 |
| pSTE23 | todC1(Met220Ala) | Quikchange mutation of pSTE14 with primer pair prSTB107/126 |
| pSTE24 | todC1(Gly224Ala) | Quikchange mutation of pSTE14 with primer pair prSTB108/127 |
| pSTE25 | todC1(Ser227Ala) | Quikchange mutation of pSTE14 with primer pair prSTB109/128 |
| pSTE26 | todC1(Phe211Ala) | Quikchange mutation of pSTE14 with primer pair prSTB110/129 |
| pSTE27 | todC1(Ser218Ala) | Quikchange mutation of pSTE14 with primer pair prSTB111/130 |
| pSTE28 | todC1(I)-tecA1(IIA)-todC1(IIBCDE+III) | 0.15-kb NdeI-StuI fragment of pCR13 into NdeI-StuI site of pSTE14 |
| pSTE30 | todC1(with NdeI site)-tecAaAbAcAd | 1.5-kb HindIII-AatII fragment of pSTE14 into HindIII-AatII site of pSTE7 |
| pSTE39 | todC1(I)-tecA1(IIAB)-todC1(IICDE+III) | 0.15-kb StuI-BstEII fragment of pCR14 into StuI-BstEII site of pSTE28 |
| pSTE41 | todC1(I)-tecA1(IIABCD)-todC1(IIE+III) | 0.15-kb StuI-BstEII fragment of pCR21 into StuI-BstEII site of pSTE28 |
| pSTE43 | todC1(I)-tecA1(IIAD)-todC1(IIBCE+III) | 0.15-kb StuI-BstEII fragment of pCR15 into StuI-BstEII site of pSTE28 |
| pSTE53 | todC1(I)-tecA1(II+III) | 0.4-kb NdeI-RsrII fragment of pSTE16 into NdeI-RsrII site of pSTE17 |
| pSTE61 | todC1(I)-tecA1(IIACDE)-todC1(IIB+III) | 0.3-kb StuI-RsrII fragment of pCR20 into StuI-RsrII site of pSTE28 |
| pSTE62 | todC1(I)-tecA1(IIAE)-todC1(IIBCD+III) | 0.15-kb BstEII-RsrII fragment of pCR22 into BstEII-RsrII site of pSTE28 |
| pSTE63 | todC1(I+IIACDE)-tecA1(IIB)-todC1(III) | 0.15-kb StuI-BstEII fragment of pCR14 into StuI-BstEII site of pSTE14 |
| pSTE64 | todC1(I+IIAE)-tecA1(IIBCD)-todC1(III) | 0.15-kb StuI-BstEII fragment of pCR21 into StuI-BstEII site of pSTE14 |
| pSTE65 | todC1(I+IIABCE)-tecA1(IID)-todC1(III) | 0.15-kb StuI-BstEII fragment of pCR15 into StuI-BstEII site of pSTE14 |
| pSTE66 | todC1(I+IIAB)-tecA1(IICDE)-todC1(III) | 0.3-kb StuI-RsrII fragment of pCR20 into StuI-RsrII site of pSTE14 |
| pSTE67 | todC1(I+IIABCD)-tecA1(IIE)-todC1(III) | 0.15-kb BstEII-RsrII fragment of pCR22 into BstEII-RsrII site of pSTE14 |
| pSTE68 | todC1(I+IIA)-tecA1(IIBCDE)-todC1(III) | 0.3-kb StuI-RsrII fragment of pCR23 into StuI-RsrII site of pSTE14 |
| Expressing dioxygenase genes | ||
| pSTO4 | todC1-C2BA | 4.1-kb EcoRI-BamHI fragment of pDTG601 into EcoRI-BamHI site of pBluescript II KS(+) |
| pSTE13 | todC1-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE12 into HindIII-AatII site of pSTE7 |
| pSTE18 | tecA1(I)-todC1(II+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE15 into HindIII-AatII site of pSTE7 |
| pSTE19 | todC1(I)-tecA1(II)-todC1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE16 into HindIII-AatII site of pSTE7 |
| pSTE20 | todC1(I+II)-tecA1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE17 into HindIII-AatII site of pSTE7 |
| pSTE29 | todC1(I)-tecA1(IIA)-todC1(IIBCDE+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE28 into HindIII-AatII site of pSTE7 |
| pSTE38 | tecA1(I+IIA)-todC1(IIBCDE)-tecA1(III)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE28 into NdeI-BstEII site of pSTE55 |
| pSTE40 | tecA1(I+IIAB)-todC1(IICDE)-tecA1(III)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE39 into NdeI-BstEII site of pSTE55 |
| pSTE42 | tecA1(I+IIABCD)-todC1(IIE)-tecA1(III)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE41 into NdeI-BstEII site of pSTE55 |
| pSTE46 | tecA1(I+IIAD)-todC1(IIBCE)-tecA1(III)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE43 into NdeI-BstEII site of pSTE55 |
| pSTE47 | todC1(I)-tecA1(IIAB)-todC1(IICDE+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE39 into HindIII-AatII site of pSTE7 |
| pSTE48 | todC1(I)-tecA1(IIABCD)-todC1(IIE+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE41 into HindIII-AatII site of pSTE7 |
| pSTE49 | todC1(I)-tecA1(IIAD)-todC1(IIBCE+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE43 into HindIII-AatII site of pSTE7 |
| pSTE52 | tecA1(I+II)-todC1(III)-tecA2A3A4 | 0.9-kb NdeI-AatII fragment of pSTE16 into NdeI-AatII site of pSTE18 |
| pSTE54 | todC1(I)-tecA1(II+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE53 into HindIII-AatII site of pSTE7 |
| pSTE55 | tecA1(I)-todC1(II)-tecA1(III)-tecA2A3A4 | 0.9-kb NdeI-AatII fragment of pSTE17 into NdeI-AatII site of pSTE18 |
| pSTE69 | todC1(I)-tecA1(IIACDE)-todC1(IIB+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE61 into HindIII-AatII site of pSTE7 |
| pSTE70 | todC1(I)-tecA1(IIAE)-todC1(IIBCD+III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE62 into HindIII-AatII site of pSTE7 |
| pSTE71 | todC1(I+IIACDE)-tecA1(IIB)-todC1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE63 into HindIII-AatII site of pSTE7 |
| pSTE72 | todC1(I+IIAE)-tecA1(IIBCD)-todC1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE64 into HindIII-AatII site of pSTE7 |
| pSTE73 | todC1(I+IIABCE)-tecA1(IID)-todC1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE65 into HindIII-AatII site of pSTE7 |
| pSTE74 | todC1(I+IIAB)-tecA1(IICDE)-todC1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE66 into HindIII-AatII site of pSTE7 |
| pSTE75 | todC1(I+IIABCD)-tecA1(IIE)-todC1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE67 into HindIII-AatII site of pSTE7 |
| pSTE76 | todC1(I+IIA)-tecA1(IIBCDE)-todC1(III)-tecA2A3A4 | 1.5-kb HindIII-AatII fragment of pSTE68 into HindIII-AatII site of pSTE7 |
| pSTE79 | todC1(Phe211Leu)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE21 into NdeI-BstEII site of pSTE30 |
| pSTE80 | todC1(Ser218Trp)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE22 into NdeI-BstEII site of pSTE30 |
| pSTE81 | todC1(Met220Ala)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE23 into NdeI-BstEII site of pSTE30 |
| pSTE82 | todC1(Gly224Ala)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE24 into NdeI-BstEII site of pSTE30 |
| pSTE83 | todC1(Ser227Ala)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE25 into NdeI-BstEII site of pSTE30 |
| pSTE84 | todC1(Phe211Ala)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE26 into NdeI-BstEII site of pSTE30 |
| pSTE85 | todC1(Ser218Ala)-tecA2A3A4 | 0.3-kb NdeI-BstEII fragment of pSTE27 into NdeI-BstEII site of pSTE30 |
| pSTE86 | todC1(Met220Ala)-todC2BA | 1.0-kb EcoRI-BspEI fragment of pSTE81 into EcoRI-BspEI site of pSTO4 |
The relevant gene regions correspond to those defined in Fig. 3.
TABLE 3.
Expression of soluble dioxygenase α-subunits and corresponding transformation activities of E. coli cells carrying plasmids with wild-type and hybrid dioxygenase genesa
| Plasmidb | Transformation rate (1,000 AU min−1 μg−1)c
|
||
|---|---|---|---|
| D | B | T | |
| Set 1 | |||
| pSTE7 | 5.0 | <0.05 | 20 |
| pSTO4 | 30 | 15 | 0.2 |
| pSTE13 | 15 | 20 | 0.2 |
| Set 2 | |||
| pSTE52 | 1.0 | <0.05 | 0.5 |
| pSTE54 | 1.5 | <0.05 | 1.5 |
| pSTE55 | 5.0 | 6.5 | <0.05 |
| Set 3 | |||
| pSTE18 | <0.05 | <0.05 | <0.05 |
| pSTE19 | 5.0 | <0.05 | 2.5 |
| pSTE20 | 5.0 | 5.0 | <0.05 |
| Set 4 | |||
| pSTE38 | 0.1 | <0.05 | <0.05 |
| pSTE40 | 0.5 | <0.05 | <0.05 |
| pSTE42 | 6.0 | 0.4 | 4.0 |
| pSTE46 | 0.8 | <0.05 | <0.05 |
| Set 5 | |||
| pSTE29 | 1.0 | <0.05 | <0.05 |
| pSTE47 | 4.0 | <0.05 | <0.05 |
| pSTE48 | 7.0 | <0.05 | 0.4 |
| pSTE49 | 8.0 | <0.05 | <0.05 |
| pSTE69 | 10 | <0.05 | 0.5 |
| pSTE70 | 0.2 | <0.05 | <0.05 |
| Set 6 | |||
| pSTE71 | 8.0 | 15 | 0.1 |
| pSTE72 | 4.5 | 2.5 | <0.05 |
| pSTE73 | 8.0 | 5.0 | <0.05 |
| pSTE74 | 3.0 | 0.3 | <0.05 |
| pSTE75 | 0.5 | 0.4 | <0.05 |
| pSTE76 | 2.5 | 0.3 | <0.05 |
| Set 7 | |||
| pSTE79 | 10 | 10 | <0.05 |
| pSTE80 | 12 | 1.0 | <0.05 |
| pSTE81 | 15 | 0.6 | 2.0 |
| pSTE86 | 40 | 0.5 | 1.0 |
| pSTE82 | 7.5 | 5.0 | <0.05 |
| pSTE83 | 5.0 | 7.5 | 0.2 |
| pSTE84 | 4.0 | 0.8 | <0.05 |
| pSTE85 | 15 | 12 | <0.05 |
Transformation experiments were performed with resting E. coli cells carrying the plasmids listed.
The sets of constructs refer to those defined in Fig. 3. pSTE7 represents the wild-type TecA system, and pSTO4 represents the wild-type TodCBA system; all other plasmids specify chimeric dioxygenases.
Transformation rates are expressed as the amount of product formed per time and amount of whole-cell protein (1,000 AU min−1 μg−1) from the linear range determined by HPLC analysis. D, B, and T represent the transformation activities of 1,2-dichlorobenzene, benzene, and 1,2,4,5-tetrachlorobenzene, respectively.
Expression of α-subunits.
The concentrations of immunodetectable soluble α-subunit in recombinant bacteria differed significantly (Fig. 2). These differences may be caused by lower expression of recombinant α-subunit genes (37), or conformational changes in hybrid dioxygenases, leading to precipitation as inclusion bodies, accelerated degradation by cellular proteases, or less efficient transfer of ferrous iron cofactor into the active site by the cellular machinery of E. coli (16). A different response of the antibody to the various constructs analyzed cannot completely be excluded. However, because a polyclonal antibody which generally recognizes different epitopes on a protein was used, such an effect would be insufficient to explain the drastically different signals obtained.
Role of dioxygenase α-subunits in substrate specificity.
A comparison of recombinant wild-type dioxygenases showed that TecA chlorobenzene dioxygenase dioxygenolytically dechlorinates tetrachlorobenzene but fails to attack benzene (4), whereas the TodCBA toluene dioxygenase (pSTE7 and pSTO4, respectively, in Table 3) displays converse activities. Because 1,2-dichlorobenzene was transformed by both systems, it was used to detect active recombinant enzyme.
The substrate specificity of class IIB enzymes is usually determined by the α-subunit (8–10, 19, 23, 26, 27, 32, 34, 36) and we assumed that this applies also to the TecA and TodCBA dioxygenases. The tecA1 α-subunit gene from chlorobenzene dioxygenase was therefore replaced with the corresponding subunit gene (todC1) of toluene dioxygenase, resulting in construct pSTE13 (Table 2), which produced active TodC1::TecA2A3A4 hybrid dioxygenase in E. coli (Table 3). The substrate specificity of the hybrid enzyme is identical to that of wild-type toluene dioxygenase, with a similar transformation efficiency for benzene and undetectable dehalogenase activity (Table 3). These results confirm that the α-subunit is responsible for substrate specificity and indicate that conformation, subunit association, and electron flow are not significantly affected by exchange of α-subunits between the two enzymes. The results are also in agreement with the observation that functional and stable heterodimers and tetramers of TodC1::BphA2 polypeptides are formed in cells expressing hybrid dioxygenase genes (13). Restoration of dehalogenase activity was subsequently investigated by introduction of smaller tecA1 elements into this hybrid system in order to identify the elements responsible for dehalogenation.
Role of region II in dechlorination.
To determine which part of the α-subunit is responsible for dechlorination, regions I, II, and III of todC1 in the TodC1::TecA2A3A4 hybrid system were sequentially or pairwise replaced by equivalent regions of tecA1 (Table 2 and Fig. 3, sets 2 and 3). Comparison of transformation rates (Table 3) of the resulting active hybrid enzymes showed that the presence of the middle region, TecA1-II, is sufficient for dechlorination of tetrachlorobenzene and that in contrast, the presence of region TodC1-II correlates with benzene transformation (Fig. 3). These results differ from those obtained with biphenyl (8, 19, 23) dioxygenases, in which the C-terminal regions (approximately III and IV in Fig. 4A) of the corresponding α-subunits are responsible for substrate specificity.
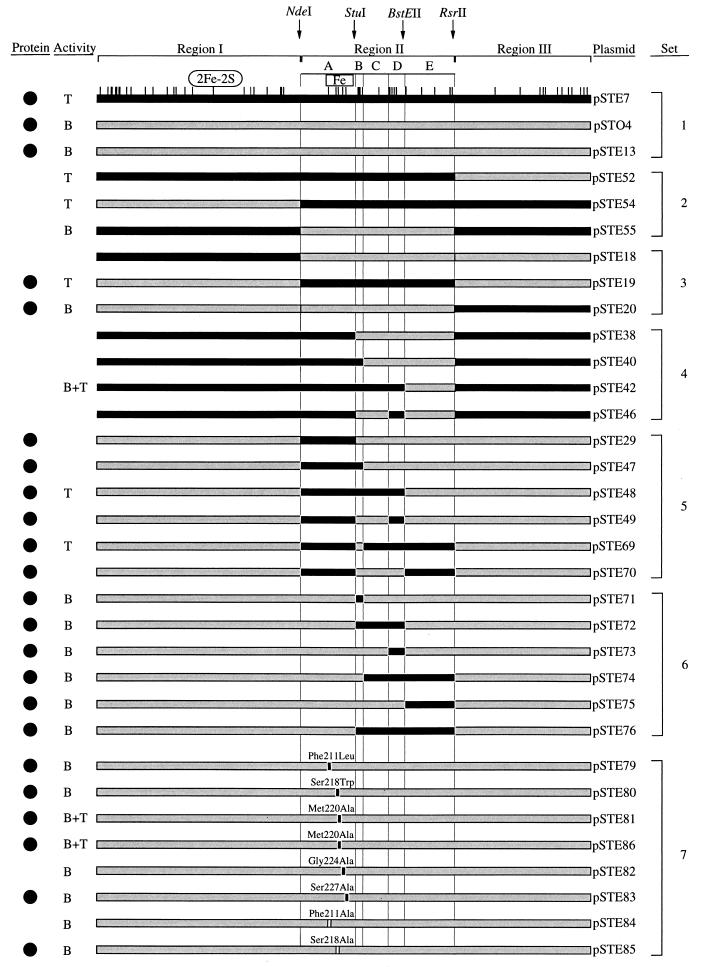
FIG. 3.
Constructs of dioxygenase α-subunit proteins. Plasmids pSTE7 (tecA1A2A3A4) and pSTO4 (todC1C2BA) carry wild-type dioxygenase systems. Hybrid systems were constructed with the chimeric α-subunits in the tecA2A3A4 background, except for plasmid pSTE86, which contains the todC1C2BA background (Table 2). The regions I of the two enzymes differ by 24 amino acid residues, subregions IIA differ by 5, IIB differ by 4, IIC differ by 1, IID differ by 5, IIE differ by 5, and III differ by 12. The positions at which the amino acids differ between TecA1 and TodC1 are shown on top of pSTE7 α-subunit. Black bars indicate fragments of TecA1 origin, grey bars indicate fragments of TodC1 origin, and white boxed amino acids in pSTE84 and pSTE85 were present in neither TecA1 nor in TodC1. Regions I, II, and III and subregions IIA, IIB, IIC, IID, and IID are delineated by black vertical lines. Relevant restriction sites are indicated. The position of the putative Rieske-type [2Fe-2S] iron-sulfur cluster (11, 28) in region I is indicated as an oval. The putative mononuclear iron-coordination motif Glu214-Xaa3–4-Asp219-Xaa2-His222-Xaa4–5-His228 (17) is shown as a boxed area. Solid circles (•) indicate E. coli cells expressing soluble α-subunit protein of TecA1 at a significant level compared to the TecA1 wild-type level. With the exception of E. coli (pSTE18), all constructs transformed 1,2-dichlorobenzene. In addition to 1,2-dichlorobenzene, E. coli resting cells carrying the corresponding plasmid transformed benzene (B), tetrachlorobenzene (T), or all three substrates (B+T) with activities above the detection limit (see Table 3).
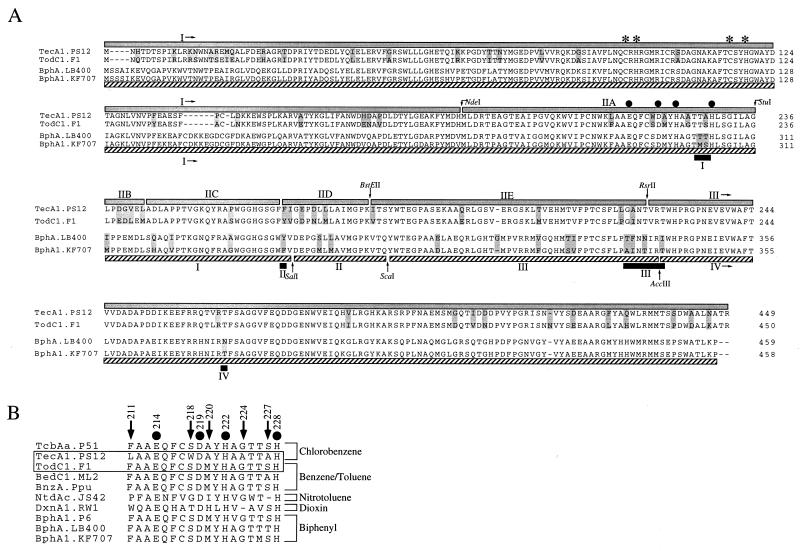
FIG. 4.
Protein sequence alignment of α-subunits of selected class IIB dioxygenases. Amino acid alignment of α-subunits of chlorobenzene, toluene, and biphenyl class IIB dioxygenases is shown as follows: TecA1.PS12, chlorobenzene dioxygenase of Burkholderia sp. strain PS12 (4); TodC1.F1, toluene dioxygenase of P. putida F1 (39); BphA.LB400, biphenyl dioxygenase of Pseudomonas sp. LB400 (7); BphA1.KF707, biphenyl dioxygenase of P. pseudoalcaligenes KF707 (33). The assignments of regions, subregions, and restriction sites used in this study are indicated by grey boxes above the alignment, and those used in the study of Kimura et al. (19) are indicated below the alignment by dashed boxes, whereas regions specified by Mondello et al. (23) are shown as black boxes (A). Additional sequences used in the alignment of the putative iron ligand region were as follows: TcbAa.P51, chlorobenzene dioxygenase of Pseudomonas sp. strain P51 (37); BedC1.ML2, benzene dioxygenase of P. putida ML2 (35); BnzA.Ppu, benzene dioxygenase of P. putida (15); BphA1.P6, biphenyl dioxygenase of Rhodococcus globerulus P6 (2); NtdAc.JS42, 2-nitrotoluene dioxygenase of Pseudomonas sp. strain JS42 (25); DxnA1.RW1, dioxin dioxygenase of Sphingomonas sp. strain RW1 (1) (B). Differences between TecA1 and TodC1 and BphA and BphA1, respectively, are indicated by shaded amino acids. The numbering of the position of amino acid residues in the putative active-site iron liganding region IIA refers to the TecA1 sequence. Arrows indicate amino acids and their positions, which are different between TecA1 and TodC1 in subregion IIA. Solid circles show putative active-site mononuclear iron ligands (17). Asterisks indicate the conserved cysteines and histidines, putative ligands of the Rieske-type [2Fe-2S] iron-sulfur cluster (11, 28), with the consensus sequence Cys-Xaa-His16–17-Xaa-Cys-Xaa2-Xaa-His (22).
Importance of the putative iron ligand region IIA.
Although neither the location nor the structure of the substrate binding site of benzene dioxygenases has yet been elucidated, it is assumed to be in the neighbourhood of the non-heme ferrous prosthetic group (6). Mutagenesis studies have suggested that histidines His222 and His228 of the benzene dioxygenase α-subunit are iron ligands (6), which is consistent with site-directed mutagenesis studies of TodC1 protein by Jiang (17), who proposed that the motif Glu214-Xaa3–4-Asp219-Xaa2-His222-Xaa4–5-His228, which is also conserved in TecA1 (Fig. 4B), is involved in mononuclear iron coordination.
We therefore postulated that amino acid differences in the subregion IIA containing the iron ligands (Fig. 4B) may contribute to the observed differences in substrate specificity. Introduction of subregion TecA1-IIA (pSTE29) almost abolished enzyme activity (Table 3), so in addition to subregion TecA1-IIA, other interacting subregions must be necessary for a restoration of dehalogenase activity.
Various combinations of subregions IIB, IIC, IID, and IIE either together with TecA1-IIA (Fig. 3, set 5) or, as control, without TecA1-IIA (Fig. 3, set 6) were used to substitute equivalent regions of TodC1. The results indicate that the simultaneous presence of the subregions TecA1-IIA, -IIC, and -IID is sufficient for restoration of dehalogenase activity (Fig. 3, pSTE48 and pSTE69), suggesting critical interactions between these polypeptide sequences.
Single amino acid substitutions.
Since region TecA1-IIA is crucially involved in dehalogenation, but insertion into TodC1 results in an inactive enzyme, we decided to make individual amino acid substitutions which may perturb the protein less (Fig. 3, set 7). Sequence comparison revealed that the amino acids in region IIA of TecA1 and TodC1 differ in positions 211, 218, 220, 224, and 227, of which only Ala220 is conserved in the two chlorobenzene dioxygenases TecA (4) and TcbA (37) (Fig. 4B). Of a number of substitutions made, only one, Met220Ala (Fig. 3, pSTE81), led to restoration of dehalogenase activity to wild-type levels (Table 3). Substitution of methionine by alanine, which has a smaller side chain, may facilitate access of tetrachlorobenzene to the active-site iron. Because E. coli (pSTE81) cells additionally transformed benzene (Table 3), a biocatalyst has been generated with an extended substrate range. Replacement of two other large amino acids (Phe211 and Ser218) by alanine in the putative active-site iron ligand region (Fig. 3, pSTE84 and pSTE85, respectively) did not lead to restoration of dehalogenase activity.
A single amino acid substitution in region II was sufficient to restore dehalogenase activity, whereas replacements including the entire region II in most cases (e.g., pSTE29) did not (Fig. 3). Moreover, those hybrids, despite being active with 1,2-dichlorobenzene, mostly did not display any activity with benzene, indicating the possibility of negative interactions between upstream and downstream sequence elements.
The dioxygenase β-subunits.
Because the β-subunits of toluate-1,2-dioxygenase and of toluene and biphenyl dioxygenases have been suggested to be involved in substrate specificity (12, 13), we investigated the influence of the β-subunit on transformation specificity and efficiency by introduction of the Met220Ala substitution into the TodCBA wild-type system (Fig. 3 [pSTE86]). E. coli (pSTE86) cells were capable of dechlorinating tetrachlorobenzene (Table 3). Thus, the β-subunits of the (chloro)benzene dioxygenases studied here are not directly involved in the control of substrate specificity, which is consistent with the indications of other investigators that the β-subunits of 2-nitrotoluene, 2,4-dinitrotoluene, and biphenyl dioxygenases are not determinants of substrate specificity (26, 27, 34).
In conclusion, a system for assessment of the catalytic performance of hybrid dioxygenases was established and was used to identify interacting polypeptide elements and a single amino acid involved in dechlorination of tetrachlorobenzene. Moreover, this study has yielded new chlorobenzene dioxygenases with wider substrate spectra. Exchange of polypeptide segments between the α-subunit of the nondehalogenating Tod benzene dioxygenase and that of the dehalogenating Tec tetrachlorobenzene dioxygenase localized the dehalogenation potential to region IIA, a region comprising the ligands of the mononuclear ferrous iron of the active site of the enzyme and containing only five amino acid differences between the two enzymes. Sequential exchange of these individual amino acids identified amino acid residue 220 in the α-subunit of the dioxygenase as critical for dehalogenation. Since the bulkier methionine is located at this position in nondehalogenating Tod dioxygenase and the less bulky alanine is present in the dehalogenating Tec dioxygenase, it seems likely that the larger halogenated substrate is sterically hindered by the methionine from entering the catalytic site of the enzyme. Region IIA is located in the middle of the α-subunit. Recent studies of the α-subunits of LB400 and KF707 biphenyl dioxygenases showed that differences in substrate specificity and regioselectivity can also be attributed to a single amino acid exchange (19, 23). However, the location of these residues is closer to the C-terminal end of the α-subunit polypeptide and distant from the putative active-site iron ligands. The regions critical for substrate specificity in the (chloro)benzene dioxygenases studied here and biphenyl dioxygenases thus seem to be distinct.
ACKNOWLEDGMENTS
This work was supported by contract BIO4-CT972040 of the BIOTECH program of the EC.
We thank Silke Backhaus for sequencing support and Anke Peterseim for valuable assistance. We are indebted to Christiane Beckmann and Michael Tesar for excellent advice on immunological techniques, and we gratefully acknowledge Jean Armengaud and Michael Klemba for critically reading the manuscript. K.N.T. expresses gratitude to the Fonds der Chemischen Industrie for generous support.
REFERENCES
- 1. Armengaud, J., B. Happe, and K. N. Timmis. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954–3966. [DOI] [PMC free article] [PubMed]
- 2.Asturias J A, Diaz E, Timmis K N. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from gram-negative bacteria. Gene. 1995;156:11–18. doi: 10.1016/0378-1119(94)00530-6. [DOI] [PubMed] [Google Scholar]
- 3.Batie C J, Ballou D P, Correll C J. Phthalate dioxygenase reductase and related flavin-iron-sulphur containing electron transferases. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Boca Raton, Fla: CRC Press; 1992. pp. 544–554. [Google Scholar]
- 4.Beil S, Happe B, Timmis K N, Pieper D H. Genetic and biochemical characterization of the broad spectrum chlorobenzene dioxygenase from Burkholderia sp. strain PS12: dechlorination of 1,2,4,5-tetrachlorobenzene. Eur J Biochem. 1997;247:190–199. doi: 10.1111/j.1432-1033.1997.00190.x. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Butler C S, Mason J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. In: Poole R K, editor. Advances in microbial physiology. London, United Kingdom: Academic Press; 1997. pp. 47–84. [DOI] [PubMed] [Google Scholar]
- 7.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson B D, Mondello F J. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl Environ Microbiol. 1993;59:3858–3862. doi: 10.1128/aem.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa K, Hirose J, Hayashida S, Nakamura K. Efficient degradation of trichloroethylene by a hybrid aromatic ring dioxygenase. J Bacteriol. 1994;176:2121–2123. doi: 10.1128/jb.176.7.2121-2123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa K, Hirose J, Suyama A, Zaiki T, Hayashida S. Gene components responsible for discrete substrate specificity in the metabolism of biphenyl (bph operon) and toluene (tod operon) J Bacteriol. 1993;175:5224–5232. doi: 10.1128/jb.175.16.5224-5232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurbiel R J, Doan P E, Gassner G T, Macke T J, Case D A, Ohnishi T, Fee J A, Ballou D P, Hoffman B M. Active site structure of Rieske-type proteins: electron nuclear double resonance studies of isotopically labeled phthalate dioxygenase from Pseudomonas cepacia and Rieske protein from Rhodobacter capsulatus and molecular modeling studies of a Rieske center. Biochemistry. 1996;35:7834–7845. doi: 10.1021/bi960380u. [DOI] [PubMed] [Google Scholar]
- 12.Harayama S, Rekik M, Timmis K N. Genetic analysis of a relaxed substrate specificity aromatic ring dioxygenase, toluate 1,2-dioxygenase, encoded by TOL plasmid pWW0 of Pseudomonas putida. Mol Gen Genet. 1986;202:226–234. doi: 10.1007/BF00331641. [DOI] [PubMed] [Google Scholar]
- 13.Hirose J, Suyama A, Hayashida S, Furukawa K. Construction of hybrid biphenyl (bph) and toluene (tod) genes for functional analysis of aromatic ring dioxygenases. Gene. 1994;138:27–33. doi: 10.1016/0378-1119(94)90779-x. [DOI] [PubMed] [Google Scholar]
- 14.Horton R M. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 15.Irie S, Doi S, Yorifuji T, Takagi M, Yano K. Nucleotide sequencing and characterization of the genes encoding benzene oxidation enzymes of Pseudomonas putida. J Bacteriol. 1987;169:5174–5179. doi: 10.1128/jb.169.11.5174-5179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahng D, Wood T K. Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1994;60:2473–2482. doi: 10.1128/aem.60.7.2473-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang H, Parales R E, Lynch N A, Gibson D T. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol. 1996;178:3133–3139. doi: 10.1128/jb.178.11.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlson U, Rojo F, van Elsas J D, Moore E. Genetic and serological evidence for the recognition of four pentachlorophenol-degrading bacterial strains as a species of the genus Sphingomonas. Syst Appl Microbiol. 1995;18:539–548. [Google Scholar]
- 19.Kimura N, Nishi A, Goto M, Furukawa K. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J Bacteriol. 1997;179:3936–3943. doi: 10.1128/jb.179.12.3936-3943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee K, Kauppi B, Parales R E, Gibson D T, Ramaswany S. Purification and crystallization of the oxygenase component of naphthalene dioxygenase in native and selenomethionine-derivatized forms. Biochem Biophys Res Commun. 1997;241:553–557. doi: 10.1006/bbrc.1997.7863. [DOI] [PubMed] [Google Scholar]
- 22.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 23.Mondello F J, Turcich M P, Lobos J H, Erickson B D. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63:3096–3103. doi: 10.1128/aem.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parales J V, Kumar A, Parales R E, Gibson D T. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene. 1996;181:57–61. doi: 10.1016/s0378-1119(96)00462-3. [DOI] [PubMed] [Google Scholar]
- 26.Parales J V, Parales R E, Resnick S M, Gibson D T. Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the α subunit of the oxygenase component. J Bacteriol. 1998;180:1194–1199. doi: 10.1128/jb.180.5.1194-1199.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parales R E, Emig M D, Lynch N A, Gibson D T. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J Bacteriol. 1998;180:2337–2344. doi: 10.1128/jb.180.9.2337-2344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rieske J S, Maclennan D H, Coleman R. Isolation and properties of an iron-protein from the (reduced coenzyme Q)-cytochrome C reductase complex of the respiratory chain. Biochem Biophys Res Commun. 1964;15:338–344. [Google Scholar]
- 29.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sánchez A, Bullejos M, Burgos M, Jiménez R, Díaz R. An alternative to blunt-end ligation for cloning DNA fragments with incompatible ends. Trends Genet. 1996;12:44. doi: 10.1016/s0168-9525(96)90051-7. [DOI] [PubMed] [Google Scholar]
- 32.Suyama A, Iwakiri R, Kimura N, Nishi A, Nakamura K, Furukawa K. Engineering hybrid pseudomonads capable of utilizing a wide range of aromatic hydrocarbons and of efficient degradation of trichloroethylene. J Bacteriol. 1996;178:4039–4046. doi: 10.1128/jb.178.14.4039-4046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 34.Tan H M, Cheong C M. Substitution of the ISP alpha subunit of biphenyl dioxygenase from Pseudomonas results in a modification of the enzyme activity. Biochem Biophys Res Commun. 1994;204:912–917. doi: 10.1006/bbrc.1994.2546. [DOI] [PubMed] [Google Scholar]
- 35.Tan H M, Tang H Y, Joannou C L, Abdel-Wahab N H, Mason J R. The Pseudomonas putida ML2 plasmid-encoded genes for benzene dioxygenase are unusual in codon usage and low in G+C content. Gene. 1993;130:33–39. doi: 10.1016/0378-1119(93)90343-2. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Garnon J, Labbe D, Bergeron H, Lau P C. Sequence and expression of the bpdC1C2BADE genes involved in the initial steps of biphenyl/chlorobiphenyl degradation by Rhodococcus sp. M5. Gene. 1995;164:117–122. doi: 10.1016/0378-1119(95)00448-f. [DOI] [PubMed] [Google Scholar]
- 37.Werlen C, Kohler H P, van der Meer J R. The broad substrate chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase of Pseudomonas sp. strain P51 are linked evolutionarily to the enzymes for benzene and toluene degradation. J Biol Chem. 1996;271:4009–4016. doi: 10.1074/jbc.271.8.4009. [DOI] [PubMed] [Google Scholar]
- 38.Zamanian M, Mason J R. Benzene dioxygenase in Pseudomonas putida: subunit composition and immuno-cross-reactivity with other aromatic dioxygenases. Biochem J. 1987;244:611–616. doi: 10.1042/bj2440611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]