Abstract
Neurological disorders are often debilitating conditions with no cure. The majority of current therapies are palliative rather than disease-modifying therefore new strategies for treating neurological disorders are greatly needed. mRNA-based therapeutics have great potential for treating such neurological diseases, however, challenges with delivery have limited their clinical potential. Lipid nanoparticles (LNPs) are a promising delivery vector for the brain given their safer toxicity profile and higher efficacy. Despite this, very little is known about LNP-mediated mRNA delivery into the brain. Here, we employ MC3-based LNPs and successfully deliver Cre mRNA and Cas9 mRNA/Ai9 sgRNA to the adult Ai9 mouse brain; greater than half of the entire striatum and hippocampus was found to be penetrated along the rostro-caudal axis by direct intracerebral injections of MC3 LNP mRNAs. MC3 LNP Cre mRNA successfully transfected cells in the striatum (~52% efficiency), and hippocampus (~49% efficiency). In addition, we demonstrate that MC3 LNP Cas9 mRNA/Ai9 sgRNA edited cells in the striatum (~7% efficiency) and hippocampus (~3% efficiency). Further analysis demonstrates that MC3 LNPs mediate mRNA delivery to multiple cell types including neurons, astrocytes and microglia in the brain. Overall, LNP-based mRNA delivery is effective in brain tissue and shows great promise for treating complex neurological disorders.
Introduction
mRNA-based therapeutics have excellent potential for treating neurological disorders, however, delivery challenges have limited their clinical impact (1–4). Lipid nanoparticle (LNP) and mRNA (LNP mRNA) complexes have great potential for transfecting brain tissue. LNP mRNA complexes have excellent biocompatibility and can transfect a variety of tissues and organs (5–9). In addition, several studies in the brain have been performed with lipid/mRNA complexes, which demonstrate the potential of lipid-based delivery vectors for transfecting the brain with mRNA (6, 7, 10). Kariko et al. (2001) demonstrated that lipofectin complexed luciferase mRNA can transfect brain tissue after intracerebral injections into rat brains (10). Lipofectin is a complex of the cationic lipid, N-(2,3-bis(oleyloq)propyl)-N,N,N-trimethylammonium chloride (DOTMA) and dioleoylphosphatidylethanolamine (DOPE) (11). Although lipofectin complexes mRNA via electrostatic and hydrophobic interactions, it lacks the PEG-lipid, is formulated via an aqueous process, is a larger size, and has a different structure than lipid nanoparticles (LNPs) (8, 11, 12). In addition, LNPs have an ionizable lipid, which is predominantly neutral at physiological pH, as opposed to the quaternary lipid DOTMA, and this allows them to have an excellent combination of in vivo efficacy and low toxicity (8, 13). As such, LNPs are promising delivery vectors for the brain (14, 15). Rungta et al. (2013) demonstrated that LNPs encapsulating siRNA have been injected into the brain after a direct intracerebral or intracerebroventricular (ICV) injection, and were able to efficiently knock down neuronal gene expression (16). Further studies by Tanaka et al. (2018) and Li et al. (2020) demonstrated that mRNA can also be delivered using LNPs after ICV injection into mouse brain tissue (6, 7). Tanaka et al. (2018) employed LNPs comprising thiol cleavable lipids to deliver luciferase mRNA and eGFP mRNA into the cerebroventricular region of the brain (6). Tanaka et al. (2018) demonstrated that LNPs can deliver eGFP mRNA into neurons and astrocytes post-ICV injections (6). More recently, Li et al. (2020) infused their LNPs, which contained Cre mRNA, into the lateral ventricle of adult Ai14 mice continuously from day 1 to day 4 using an osmotic pump connected to a brain cannula. As a result of Cre recombination, tdTomato expression was observed in the cells lining the injected lateral ventricle (hypothesized to be ependymal cells) and in the hippocampus, specifically in the stratum oriens of field CA1, CA2 and CA3 (7). Taken together, these pioneering studies have demonstrated that mRNA delivery by LNPs is possible after direct ICV injection into the brain. However, transfected cells appeared to be localized to the restricted brain regions, primarily to the cells lining the ventricle wall, with small levels of transfection in the brain tissue. This implicates that the tissue penetration of LNP mRNA complexes after an ICV injection is thought to be low for most potential therapeutic applications. In addition, the transfection distance and the delivery efficiency for mRNAs of larger proteins such as Cas9, which are considered to be significantly more challenging to deliver, were not fully investigated in these studies.
Intracerebral injections of LNP mRNA complexes represent an alternative approach for transfecting brain tissue. Intracerebral injections are clinically feasible, and are performed in a variety of clinical scenarios (17–23), as well as in various preclinical settings (4, 24–27). An intracerebral injection avoids the tissue barrier that blood-cerebrospinal fluid (CSF) barrier or blood-brain-barrier (BBB) represents, and consequently should lead to significantly higher brain transfection levels by specifically targeting local regions of the brain (4, 20, 22, 26, 27). In addition, a previous study estimated that human brain extracellular space has pores even larger than 200 nm in diameter, although more than 95% of all pores have diameters of 175 nm or smaller (28). In mice, nanoparticles densely coated with PEG measuring up to 200 nm in diameter were able to spread within brain tissue (28), a size range achievable for LNPs (9, 29). Notably, PEGylated polymer nanoparticles have previously been able to transfect DNA into brain tissue after an intracerebral injection; these nanoparticles were similar in size and surface chemistry as LNPs, suggesting that LNPs will be able to transfect brain tissue after intracerebral injection (28, 30).
CRISPR/Cas9 is one of the most widely used gene editing tools due to the simple design of the sgRNA giving it a combination of precision and ease of use. Studies employing CRISPR have already exhibited treatment efficiency in preclinical models of neurological disorders (31–36). Several pioneering studies have demonstrated that gene editing in the brain with Cas9 using nonviral delivery vehicles is possible. Specifically, Staahl et al. (2017) demonstrated that direct injection of the Cas9 protein fused to 6 SV40 nuclear localization signals (4xNLS-Cas9–2xNLS), complexed to sgRNA (4xNLS-Cas9–2xNLS RNPs) into the striatum and hippocampus of Ai14 mice was able to edit genes in brain tissue efficiently (37). Lee et al. (2018) further demonstrated that CRISPR-Gold which contains gold nanoparticles and Cas9 RNPs was able to successfully edit target gene and rescue increased repetitive behaviors in a mouse model for fragile X syndrome, a neurodevelopmental disorder (33). This was achieved by injecting directly into the striatum (33). More recently, there have been significant advancements made to delivering mRNA into the brain using nonviral delivery vehicles. A study by Abbasi et al. (2021) demonstrated that polycations based upon poly[N’-[N-(2-aminoethyl)-2-aminoethyl]aspartamide] (P[Asp(DET)]) were also capable of delivering mRNA expressing Cas9 along with sgRNA (38). These polyplexes were able to edit genes in the cortex after a direct intracerebral injection, using the Ai9 mouse model (38). Notably, there has been an effort of Cas9 mRNA/sgRNA delivery using LNPs in cultured cells by Li et al. (2020), however, Cas9 mRNA has not been tested using LNPs in the brain yet (7).
In this report, we demonstrate that LNP mRNA complexes can efficiently transfect brain tissue after an intracerebral injection into the striatum or hippocampus. We constructed LNPs containing 4-(dimethylamino)-butanoic acid, (10Z,13Z)-1-(9Z,12Z)-9,12-octadecadien-1-yl-10,13-nonadecadien-1-yl ester (DLin-MC3-DMA; MC3), named MC3 LNPs, and prepared them along with Cre mRNA or Cas9 mRNA/Ai9 sgRNA. We subsequently injected these LNPs into the striatum and hippocampus of Ai9 mice and then performed extensive analysis, which determined the transfection distance, total number of transfected cells, local delivery efficiency, and cell types transfected. We observed that MC3 LNP Cre mRNA transfected more than half of the entire striatum and hippocampus based on the rostro-caudal transfection distance analysis. Furthermore, we found that MC3-based LNPs transfected Cre mRNA to more than 1×105 brain cells with an approximate 52% and 49% local transfection efficiency in the striatum and hippocampus, respectively. In addition, MC3 LNPs also delivered Cas9 mRNA/Ai9 sgRNA and edited 1.5–3.3×103 cells with an approximate 7% and 3% local transfection efficiency in the striatum and hippocampus, respectively. These results demonstrate that MC3 LNPs were able to deliver mRNAs for larger size of proteins into the brain tissue. We further demonstrate that MC3 LNPs deliver mRNAs to multiple brain cell types namely neurons, astrocytes and microglia. Collectively, our results demonstrate that MC3 LNPs hold great potential for mRNA delivery in the brain while having both the transfection efficiency and tissue penetration properties needed for a variety of therapeutic applications.
Materials and Methods
Materials:
Paraformaldehyde (PFA) and D(+)-Sucrose were purchased from ACROS Organics™; Phosphate Buffered Saline (PBS) was purchased from Fisher BioReagents; Superfrost Plus Microscope Slides was purchased from Fisher Scientific; Tissue-Plus O.C.T. Compound was purchased from Fisher Healthcare; Isoflurane was purchased from Vetone; Prolong Gold Antifade Reagent was purchased from Invitrogen; Embedding Molds was purchased from ThermoSCIENTIFIC; DAPI (4,6-diamidino-2-phenylindole) was purchased from Invitrogen; Cre mRNA (L-7211) and Cas9 mRNA (L-7606) were purchased from TriLink BioTechnologies; Ai9 sgRNA were purchased from Integrated DNA Technologies; MC3 was purchased from MedKOO; DOPE and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol (DMG-PEG) are purchased from Avanti Polar Lipid; Cholesterol was purchased from Sigma Aldrich.
Antibodies:
The chicken IgY polyclonal RFP antibody (cat. #: 600-901-379S) was purchased from Rockland, the mouse IgG1 monoclonal NeuN antibody (cat. #: MAB377) from Millipore, the rabbit polyclonal GFAP antibody (cat. #: AB5804) from Sigma Aldrich, and the rabbit polyclonal Iba1 antibody (cat. #: 019-19741) from WAKO Chemicals. The Cy™3 AffiniPure Donkey Anti-Chicken IgY (cat. #: 703-165-155), Alexa Fluor® 647 AffiniPure Donkey Anti-Mouse IgG (cat. #: 715-605-151), Alexa Fluor® 488 AffiniPure Donkey Anti-Rabbit IgG (cat. #: 711-545-152), and were purchased from Jackson ImmunoResearch Laboratories, Inc.
MC3 LNP synthesis and mRNA preparation:
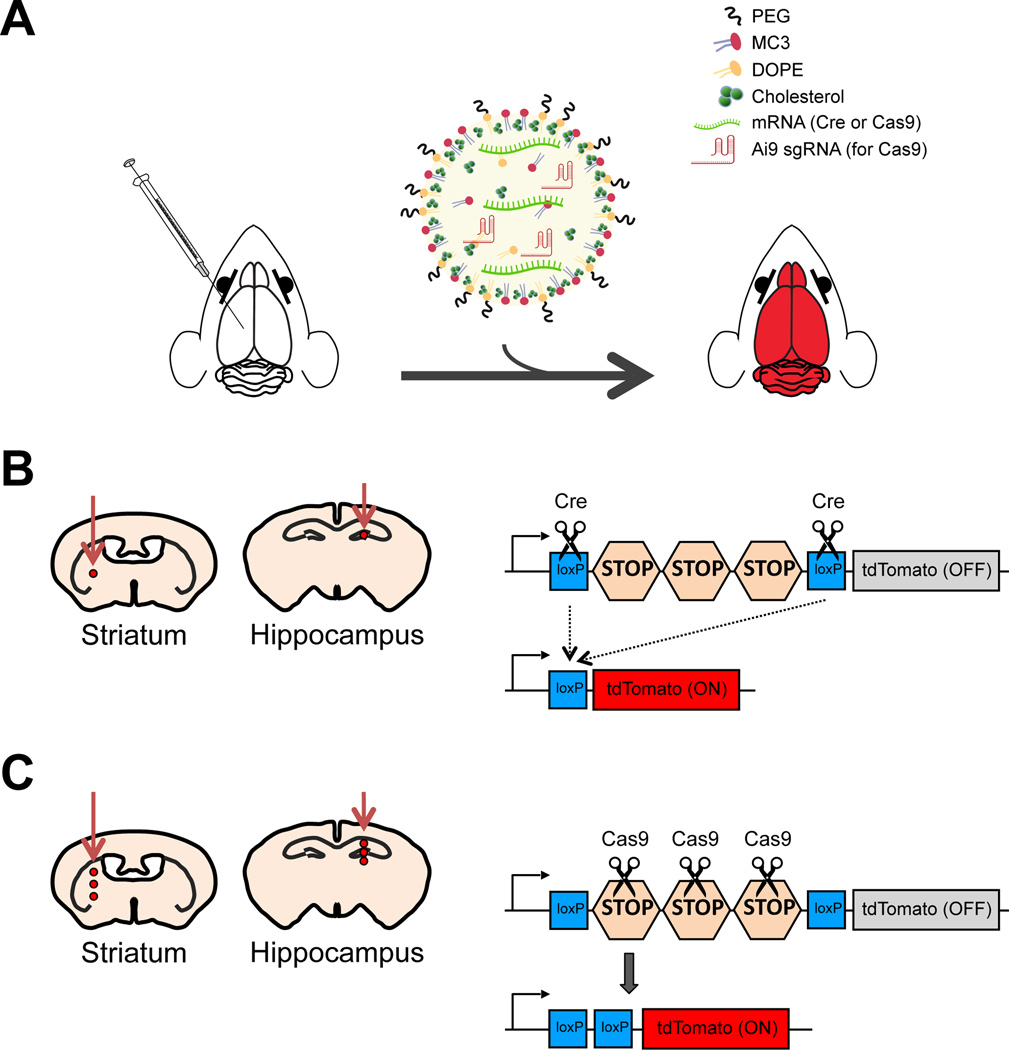
For MC3 LNP preparation, a stock solution (10 mg mL−1) of MC3 (MedKoo, cat. #: 555308), DOPE (Avanti Polar Lipids, cat. #: 850725), Cholesterol (Sigma Aldrich, cat. # C8667), and DMG-PEG (Avanti Polar Lipids, cat. #: 880151) was made by dissolving each lipid separately in ethanol and stored at −30 °C. Just before the LNP formation, the lipids were taken out and kept on ice and vortexed whenever necessary. The cholesterol solution was warmed up to dissolve the crystals which form during the cold storage. Then MC3, DOPE, cholesterol, and DMG-PEG were mixed in 36.8:23.8:38.2:1.2 molar ratios, respectively. RNAs 1 μg μL−1 (Cre mRNA, Cas9 mRNA/Ai9 sgRNA) were dissolved in 200 mM citrate buffer (pH 4) and mixed with MC3 LNPs in a 1:3 ratio (volume/volume). The resulting MC3 LNP mRNA complexes were gently vortexed and incubated at room temperature for 15 min for immediate use or stored at 4 °C for shipping and handling. Size distribution and polydispersity of MC3 LNPs with Cre mRNAs were determined by Malvern Zetasizer (Malvern) with an average size of 162 nm as shown in Figure S1. A schematic diagram of the formulation process for MC3 LNPs with mRNA is shown in Figure 1A. The Ai9 sgRNA targeting sequence which was used with Cas9 mRNA is AAGTAAAACCTCTACAAATG.
Figure 1. The scheme of the procedure.
(A) Schematic showing the injection procedure. MC3 LNPs containing either Cre mRNA or Cas9 mRNA/Ai9 sgRNA injected into the brain were able to deliver mRNA and induce gene recombination or editing in Ai9 mice. (B) Left: Scheme of the injection site for MC3 LNP Cre mRNA and controls (saline or Cre mRNA) in either the striatum or hippocampus of adult Ai9 mice. Saline, Cre mRNA (0.125 μg μL−1 or 0.250 μg μL−1) or MC3 LNP Cre mRNA (0.125 μg μL−1 or 0.250 μg μL−1) was injected into the striatum or hippocampus of Ai9 mice. Right: Scheme of Cre-mediated recombination of tdTomato gene. (C) Left: Schematic showing the injection site for MC3 LNP Cas9 mRNA/Ai9 sgRNA and controls (saline or Cas9 mRNA) in either the striatum or hippocampus of adult Ai9 mice. Saline, Cas9 mRNA (0.125 μg μL−1 or 0.250 μg μL−1) or MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.125 μg μL−1 or 0.250 μg μL−1) was injected into the striatum or hippocampus of Ai9 mice. Right: Scheme of Cas9-mediated editing of tdTomato gene.
Animal care and use:
Ai9 mice (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; Cre-dependent tdTomato reporter mice in C57BL/6J background) were obtained from Jackson Laboratory (stock #007909). The use and care of animals in this study follow the guidelines of the UTHSCSA Institutional Animal Care and Use Committee.
Stereotaxic injection:
Male and female Ai9 mice aged 2–3 months were used in this study. Animals were anaesthetized using isoflurane (4% for induction and 2% for maintaining during the surgery) and injected with either saline (as a control), Cre mRNA (with MC3 LNPs or without MC3 LNPs as a control; 2 different concentrations) or Cas9 mRNA/Ai9 sgRNA (with MC3 LNPs or without MC3 LNPs as a control; 2 different concentrations) into the striatum (AP: +0.50 mm, ML: +1.87 mm or −1.87 mm depending on the experiment) and hippocampus (AP: −2.56 mm, ML: +1.55 mm or −1.55 mm depending on the experiment) (Figure 1). The saline control was injected as a single injection (for striatum DV: −3.47 mm; for hippocampus DV: −2.00 mm) with a volume of 1.5 μL per injected site (0.5 μL min−1). The Cre mRNA control and MC3 LNP Cre mRNA were injected (DV for striatum: −3.47 mm; DV for hippocampus: −2.00 mm) with either low (0.125 μg μL−1) or high (0.250 μg μL−1) dosage with 1.5 μL per injection site (0.5 μL min−1, as a single injection; Figure 1B). The Cas9 mRNA/Ai9 sgRNA control and MC3 LNP Cas9 mRNA/Ai9 sgRNA were injected (DV for striatum: −3.47, −3.07 and −2.67 mm with a 0.4 mm interval; DV for hippocampus: −2.00, −1.85, and −1.70 mm with a 0.15 mm interval) with either low (0.125 μg μL−1) or high (0.250 μg μL−1) dosage with 2.1 μL (0.5 μL min−1, 0.7 uL per injection site X 3 injection sites while pulling out the needle; Figure 1C). Injections were performed using Hamilton Neuros syringe (Hamilton Company). After the infusion, the injector was left at the injection site for 5 min and then slowly withdrawn. A period of 21 days was given before the perfusion and brain analysis.
Sample processing:
The brain collection was performed 21 days after stereotaxic injection. Mice were anaesthetized by isoflurane and were perfused with ice-cold PBS followed by 4% PFA in PBS. The brains were postfixed for 4 hr in 4% PFA, washed once with PBS, and then cryoprotected by 30% sucrose in PBS at 4 °C for 2–3 days. After cryoprotection, the brains were embedded in O.C.T. compound, frozen, and then stored at −80 °C until further processing. The brain sections were obtained by cryostat (CM1860 UV; Leica Microsystems). Slices with striatum and hippocampus were sectioned on the coronal plane at 20 μm, mounted on the glass slides, and stored at −20 °C until further use. The sections for DAPI staining without further immunostaining were washed three times in PBS. After the washing, the sections were incubated with DAPI diluted in PBS (1:1500 of 5 μg μL−1 stock solution) for 10 min. Next, the sections were washed three times in PBS and mounted with Prolong Gold Antifade Reagent for imaging.
Immunostaining:
Every 3rd brain section collected from the striatum and the hippocampus was immunostained and used for analyses. The sections for immunostaining were washed three times in PBS and then antigen retrieval was performed by steaming in a citrate buffer (0.294% sodium citrate, 0.05% Tween 20 in distilled water, pH 6.0) for 15 min with subsequent cooling over ice for 10 min. The sections were rinsed in PBS and blocked (5% goat serum, 0.2% Triton X-100 in PBS) at room temperature for 1 hr. The sections were next incubated in the same blocking solution with primary antibodies (1:1000 chicken IgY anti-RFP, 1:500 mouse IgG1 anti-NeuN, 1:500 rabbit anti-GFAP, and 1:500 rabbit anti-Iba1) overnight at 4 °C. The sections were washed four times in PBS before incubation with secondary antibodies (1:500 Cy™3 AffiniPure Donkey Anti-Chicken IgY, 1:500 Alexa Fluor® 647 AffiniPure Donkey Anti-Mouse IgG, and 1:500 Alexa Fluor® 488 AffiniPure Donkey Anti-Rabbit IgG) for 2 hr. After the incubation with secondary antibodies the sections were washed four times in PBS, and then incubated with DAPI diluted in PBS (1:1500 of 5 μg μL−1 stock solution) for 10 min. Next, the sections were washed three times in PBS and mounted with Prolong Gold Antifade Reagent.
Imaging:
The stained brain sections were imaged using Zeiss Axio Observer 7 microscope with dry 10x, 20x or oil 40x objectives, and captured with camera Axiocam 503 mono (Carl Zeiss Microscopy GmbH). For each experiment, the exposure time was kept consistent to allow a comparative analysis between images within the experiment. Apotome.2 (Carl Zeiss Microscopy GmbH) was utilized to prevent scattered out-of-focus light and to improve resolution. Post-processing including Apotome processing or stitching processing were done using Zen 2.3 (Carl Zeiss Microscopy GmbH). For whole brain slide images, images were taken using the 10x objective and were stitched together. For the transfection distance analysis, every 3rd brain slice was stained with DAPI and the transfection distance was defined by identifying the brain slices with the presence of tdTomato and DAPI copositive (tdTomato+; DAPI+) cells. For determining the number of total tdTomato+; DAPI+ cells in the injected area, every 4th of DAPI-stained brain slice (every 12th brain slice per mouse) was imaged for analysis using the 20x objective, and then were stitched together. For determining the local transfection efficiency and for counting specific cell types (neurons, astrocytes or microglia), DAPI stained and/or immunostained brain slices (10–15 images from 2–3 mice, close to the injection site) were imaged with z-stack (10 μm thickness; 1 μm intervals) using 40x objective.
Image analysis:
Transfection distance was quantified by identifying the brain slices of the coronal sections with presence of tdTomato+; DAPI+ cells as defined in the previous section, and was calculated in the rostro-caudal axis. Specifically, the total number of stitched images containing tdTomato+; DAPI+ cells was then multiplied by 60 µm, the thickness of 3 brain slices, to obtain the transfection distance in the striatum or hippocampus given every 3rd brain slice was imaged as described above. For determining the number of total tdTomato+; DAPI+ cells, tdTomato+; DAPI+ cells were counted in every 4th DAPI stained brain slice (every 12th brain slice per mouse) through the entire rostro-caudal extent of the striatum or hippocampus. Then, the total number of cells counted from every 4th DAPI stained brain slice was multiplied by 12 to obtain the estimate of the total number of tdTomato+; DAPI+ cells in the striatum or hippocampus, as similar to previously reported methods (37). We analyzed whole brain area except the inner wall of ventricle to avoid the possibility that vectors were transported by ventricular CSF. The number of cells was counted using the ImageJ software (NIH).
Quantifications of local transfection efficiency and cell type-specific analyses were performed in defined Region of Interests (ROIs) of the heaviest transfected area of striatum or hippocampus, which was the identical size (219.74 × 165.71 μm) between all animals. Analyzed ROI images originated from 5 different brain sections per mouse, 10–15 images from 2–3 mice. The cells were quantified along the z-stack using the optical dissector to avoid the over/under-counting of superimposed cells. To analyze the percentage of edited cells in ROIs, tdTomato+ cells were counted and normalized with the number of DAPI+ cells. Total number of DAPI+ cells in Ai9 mice was also counted and presented. To determine the percentage of each cell type (neuron, astrocyte or microglia) among edited cells, tdTomato+; NeuN+, tdTomato+; GFAP+, and tdTomato+; Iba1+ in ROIs were counted and normalized by a number of tdTomato+ cells. Each cell count before normalization (non-normalized raw value) is also presented. All contrast/brightness parameters were kept identical between ROI images and the counting was performed as a blind experiment. The number of cells was counted using the ImageJ software (NIH).
Statistical analysis:
Statistical analyses were conducted with data acquired from a total of 5 images per animal, a total of 10–15 images per group. The data were analyzed using two-tailed permutation t-test with 10,000 permutations. The significance level was defined as P < 0.05. Statistical analyses were conducted using RVAideMemoire packages for R version 3.6.0. All data are presented as mean ± SEM.
Results and Discussion
1. MC3 LNP Cre mRNA complexes induce efficient Cre-mediated recombination in the striatum and hippocampus of Ai9 mice
Cationic lipid mRNA complexes have been injected intracerebrally and LNP mRNA complexes have been injected via the ICV route (6, 7, 10), however, LNP mRNA complexes have not been fully investigated for transfection efficiencies after intracerebral injections into the brain tissue. In addition, the tissue tropism and transfection efficiency of LNP mRNA complexes have not been investigated. In order to determine the transfection efficiency of MC3 LNPs in brain tissue, we injected MC3 LNP Cre mRNA into two brain regions, the striatum and hippocampus of Ai9 mice (Figure 1B). We selected MC3 as the ionizable lipid for our LNPs because of its well-characterized low level of toxicity, excellent clinical track record, and high efficiency (14, 39, 40). LNPs containing MC3 were formulated using a standard two-phase mixing protocol, where mRNA dissolved in citrate buffer (pH 4) was mixed with an ethanol solution containing the lipids, as similar to previously reported methods (39, 41). The mixing was performed by hand pipetting and generated MC3 LNP Cre mRNA of approximately 150–170 nm in diameter (Figure S1). Genetically engineered Ai9 mice have a fluorescent tdTomato gene containing upstream loxP-flanked STOP cassettes (42). Once the Cre recombinase activates the transgene, tdTomato is expressed and the gene-edited cells appear as tdTomato-positive cells (Figure 1). First, we injected Cre mRNA (0.125 μg μL−1) with MC3 LNPs (MC3 LNP Cre mRNA) into the brains (striatum or hippocampus) of Ai9 mice and collected the brains 21 days after the injection (uninjected group was used as a control). Our pilot in vivo experiment demonstrated that MC3 containing LNPs were stable and were able to transfect brain cells for at least 2 days after synthesis if stored at 4°C (Figure 2), therefore our experiments were performed within 2 days after LNP formulation.
Figure 2. Post-synthesis stability of MC3 LNPs.
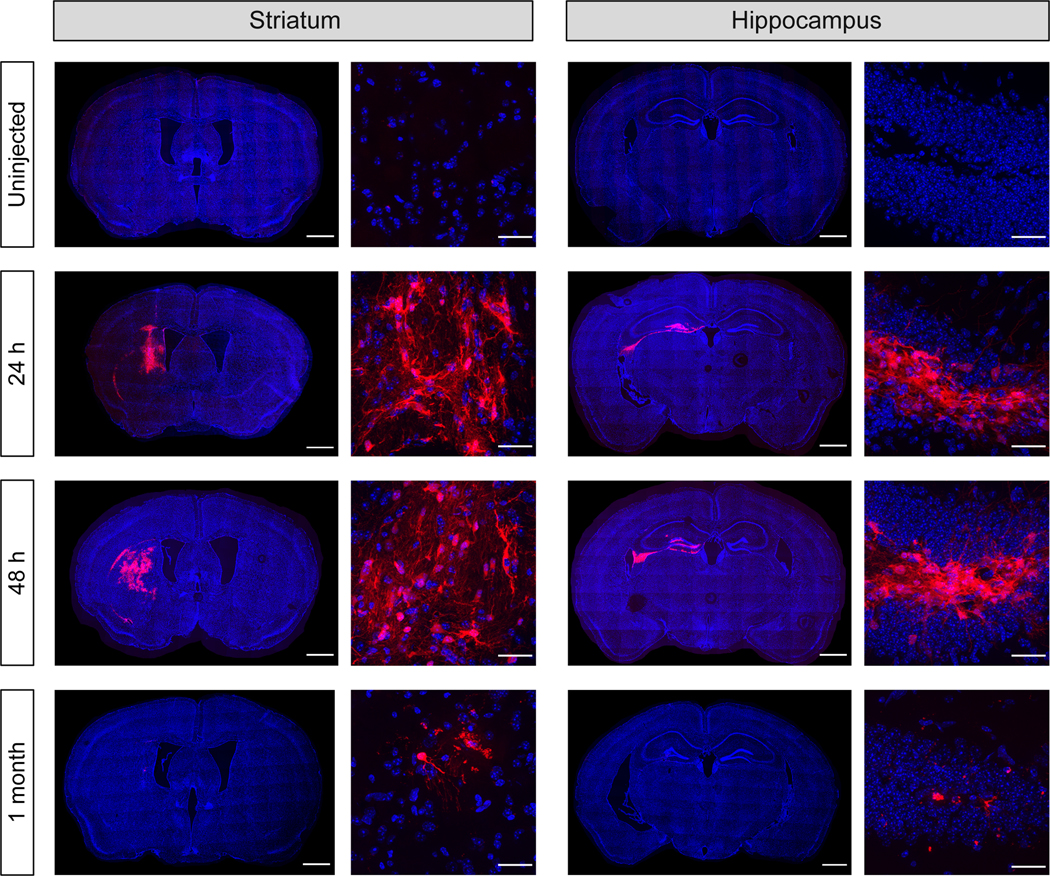
MC3 LNP Cre mRNA (0.125 μg μL−1; stored at 4°C) was injected 24 hr, 48 hr and 1 month after the MC3 LNP synthesis into the striatum (Left) or hippocampus (Right) of Ai9 mice. The presence of tdTomato (red) expression and DAPI (blue) nuclear staining were analyzed 21 days after the MC3 LNP Cre mRNA injection. Scale bars represent 100 μm (Left) for whole brain images and 30 μm (Right) for higher magnification images, respectively.
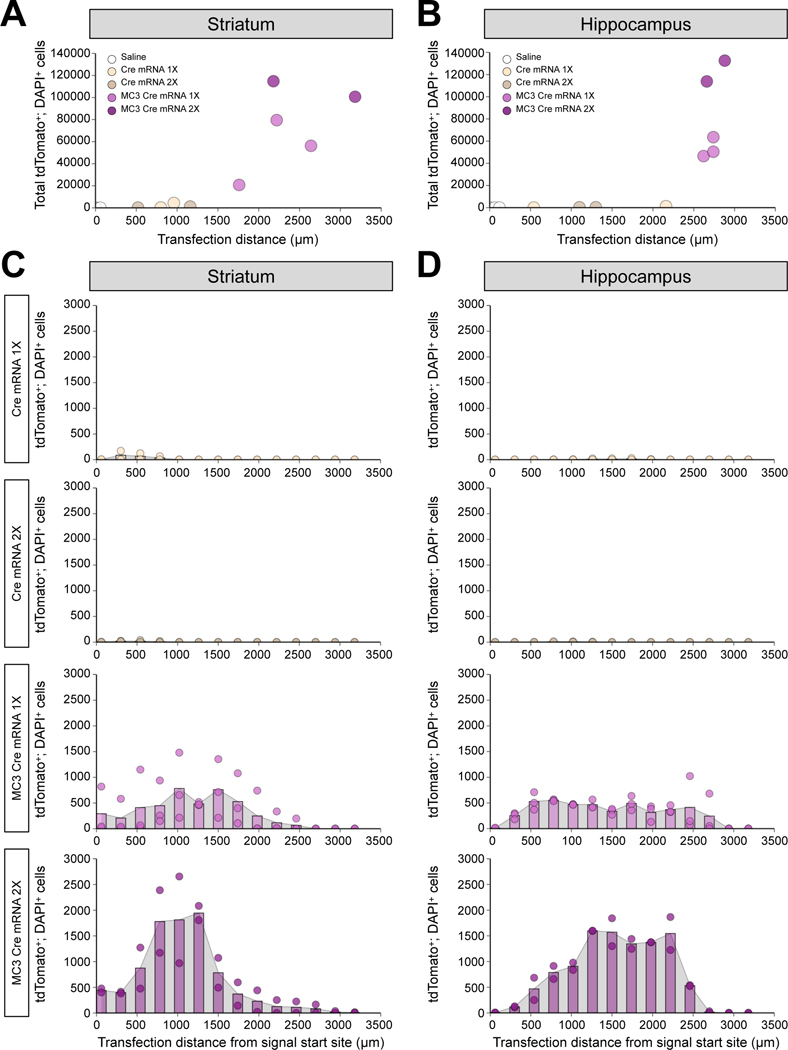
After demonstrating that MC3 LNP Cre mRNA was able to transfect brain cells, next we checked the transfection efficiency of MC3 LNP Cre mRNA by determining the transfection distance and the total number of transfected cells. We injected saline, low (0.125 μg μL−1) or high (0.250 μg μL−1) dose of Cre mRNA with or without MC3 LNPs into the brains (striatum or hippocampus) of Ai9 mice and collected the brains 21 days after the injection (saline and Cre mRNA without MC3 LNPs-injected group were used as controls). Brain slices were sectioned on the coronal plane at a thickness of 20 μm and were DAPI stained before imaging. For determining the transfection distance, we identified brain slices containing tdTomato+; DAPI+ cells and counted as described in the method ‘Image Analysis Section’. The transfection distance for low and high doses of Cre mRNA were 880 ± 80 μm and 840 ± 320 μm, respectively, and for low and high doses of MC3 LNP Cre mRNA were 2,207 ± 254 μm, and 2,680 ± 500 μm in striatum, respectively (Figure 3A). In hippocampus, the transfection distance for low and high doses of Cre mRNA were 1,350 ± 810 μm and 1,200 ± 100 μm, respectively, and for low and high doses of MC3 LNP Cre mRNA were 2,700 ± 40 μm and 2,770 ± 110 μm, respectively (Figure 3B). Further, for determining total Cre-recombined cells, tdTomato+; DAPI+ cells were counted in every 4th DAPI stained brain slice (every 12th brain slice per mouse) (Shown as histogram in Figure 3C–3D) and multiplied by 12 to obtain the estimate of the total number of tdTomato+; DAPI+ cells (Figure 3A–3B) as described in the method ‘Image Analysis Section’. As a result, the total tdTomato+; DAPI+ cells for low and high doses of Cre mRNA were 2,316 ± 2,040 cells and 672 ± 468 cells, respectively, and for low and high doses of MC3 LNP Cre mRNA were 52,160 ± 17,075 cells and 107,742 ± 7,050 cells in striatum, respectively (Figure 3A). In hippocampus, the total tdTomato+; DAPI+ cells for low and high doses of Cre mRNA were 648 ± 576 cells and 324 ± 12 cells, respectively, and for low and high doses of MC3 LNP Cre mRNA were 53,484 ± 5,158 cells and 123,210 ± 9,366 cells, respectively (Figure 3B). We did not observe a noticeable number of tdTomato+; DAPI+ cells by saline injection as a baseline control both in striatum and hippocampus. Taken together, these results demonstrate that MC3 LNPs deliver Cre mRNAs efficiently after intracerebral injection into striatum and hippocampus. Notably, high dose of MC3 LNP Cre mRNA transfected approximately 2-fold more cells than low dose of MC3 LNP Cre mRNA did in both striatum and hippocampus while low and high doses of MC3 LNP Cre mRNA showed similar transfection distance.
Figure 3. Transfection distance and efficiency after MC3 LNP-mediated Cre mRNA delivery in striatum and hippocampus.
(A, B) Transfection distance and total tdTomato+; DAPI+ cells in striatal injected samples (A), and hippocampal injected samples (B). (C, D) Quantification of tdTomato+; DAPI+ cells in every 12th brain slice in striatal injected samples (C), and hippocampal injected brain samples (D). Saline (n = 3 mice), low dose of Cre mRNA (0.125 μg μL−1; Cre mRNA 1X; n = 2 mice), high dose of Cre mRNA (0.250 μg μL−1; Cre mRNA 2X; n = 2 mice), low dose of MC3 LNP Cre mRNA (0.125 μg μL−1; MC3 Cre mRNA 1X; n = 3 mice), and high dose of MC3 LNP Cre mRNA (0.250 μg μL−1; MC3 Cre mRNA 2X; n = 2 mice).
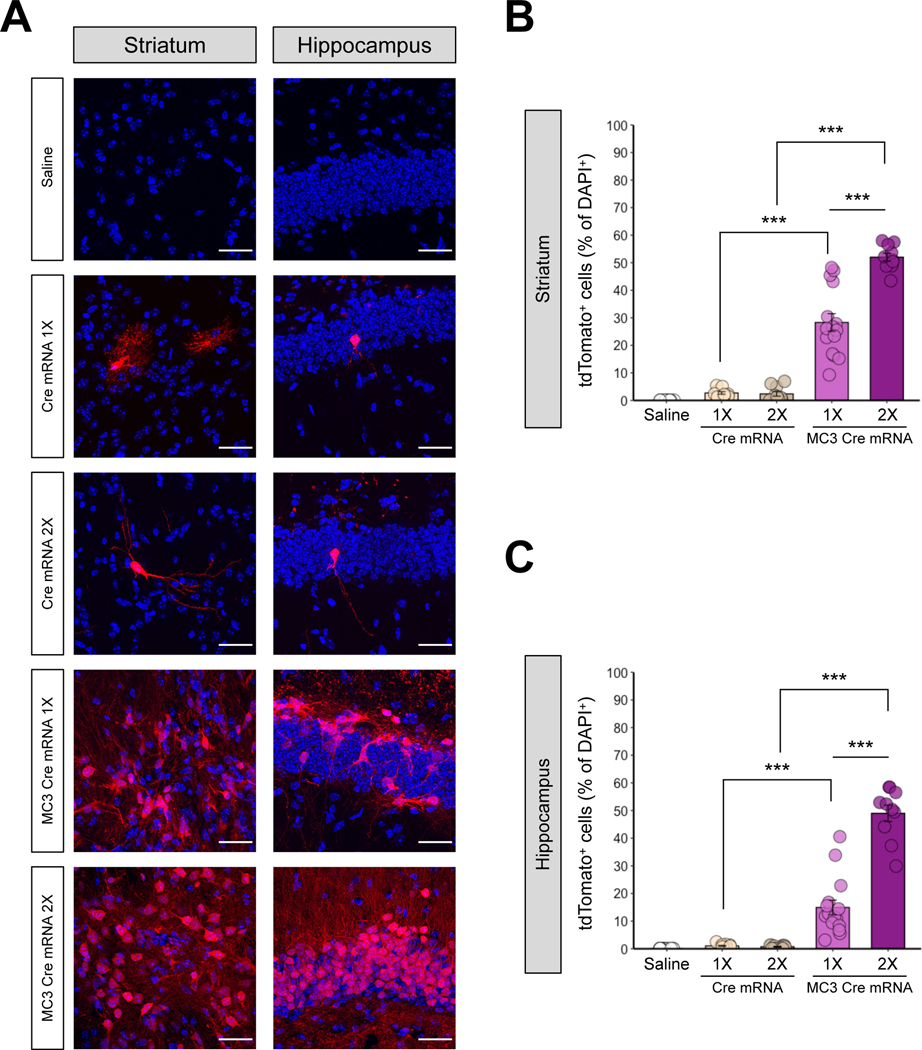
Next, we further analyzed the local transfection efficiency in the highest transfected areas of the striatum and hippocampus, determined by the result from Figure 3. As a result, Ai9 mice injected with MC3 LNP Cre mRNA had significantly increased tdTomato-positive cells in comparison to control groups (saline or Cre mRNA without MC3 LNPs) in the striatum (Figure 4A Left panel and Figure 4B) and hippocampus (Figure 4A Right panel and Figure 4C). The dose of the MC3 LNP Cre mRNA had a significant effect on the brain local transfection efficiency and we observed significantly higher levels of Cre-mediated recombination at the high dose of MC3 LNP Cre mRNA (0.250 μg μL−1), compared with the low dose of MC3 LNP Cre mRNA (0.125 μg μL−1), approximately 53.0% versus 28.0% efficiency in the striatum (Figure 4B). Similarly, the Cre-mediated recombination was significantly higher (~49.0% efficiency) with the high dose as compared with the low dose of MC3 LNP Cre mRNA (~14.9% efficiency) in the hippocampus (Figure 4C). We did not observe the dose-dependent effect with control injections of Cre mRNA without MC3 LNPs in both brain regions, a similar finding to Figure 3. Given that we occasionally observed significant differences in the total number of cells (DAPI+) between treatments in the striatum and the hippocampus (Figure S2A), the Cre-recombined (tdTomato+) cells were normalized by the number of DAPI+ cell.
Figure 4. MC3 LNPs can deliver Cre mRNA and enhance Cre-mediated recombination in mouse striatum and hippocampus.
(A) tdTomato (red) expression and DAPI (blue) nuclear staining were analyzed 21 days after the saline, Cre mRNA or MC3 LNP Cre mRNA injection in the striatum (Left), and the hippocampus (Right). Cre mRNA or MC3 LNP Cre mRNA was injected with two doses: 0.125 μg μL−1 (1X) or 0.250 μg μL−1 (2X). The scale bar represents 30 μm. (B, C) Quantification of the percentage of tdTomato+ cells among DAPI+ cells in the striatum (permutation one-way ANOVA: F(4,55) = 129.20, P < 0.001) (B), and the hippocampus (permutation one-way ANOVA: F(4,55) = 110.80, p < 0.001) (C). All data are presented as mean ± SEM. ** p < 0.01, *** p < 0.001 by post hoc permutation t-test. Post hoc p values were calculated between Cre mRNA (0.125 μg μL−1 or 0.250 μg μL−1) and MC3 LNP Cre mRNA (0.125 μg μL−1 or 0.250 μg μL−1) as well as between dosages 0.125 μg μL−1 vs. 0.250 μg μL−1 within Cre mRNA or MC3 LNP Cre mRNA. Saline (n = 15 images from 3 mice), low dose of Cre mRNA (0.125 μg μL−1; Cre mRNA 1X; n = 10 images from 2 mice), high dose of Cre mRNA (0.250 μg μL−1; Cre mRNA 2X; n = 10 images from 2 mice), low dose of MC3 LNP Cre mRNA (0.125 μg μL−1; MC3 Cre mRNA 1X; n = 15 images from 3 mice), and high dose of MC3 LNP Cre mRNA high dose (0.250 μg μL−1, MC3 Cre mRNA 2X; n = 10 images from 2 mice).
Taken together, we observed that MC3 LNP Cre mRNA penetrated approximately more than 1/2 of the entire striatum or hippocampus (the entire hilus of the dentate gyrus) based on the transfection area of the highest transfected brain section (Figure 2), as well as based on the rostro-caudal transfection distance analysis (Figure 3). Next, we also demonstrated that MC3 LNP Cre mRNA transfected more than 1×105 brain cells (Figure 3) with approximately 53% and 49% of the local transfection efficiency in the highest transfected area of striatum and hippocampus, respectively (Figure 4). Notably, the concentration of LNP mRNA complexes that we used in our study were significantly lower than the previously reported doses used for ICV injections while the transfected brain regions were significantly larger than that observed with an ICV injection (7). These results suggest that MC3 LNP Cre mRNA can successfully transfect brain cells after intracerebral injection.
2. Cre mRNAs delivered by MC3 LNPs generate Cre-mediated gene recombination in neurons, astrocytes, and microglia
Next, we analyzed the cellular identity of the transfected cells with MC3 LNPs containing Cre mRNA by analyzing tdTomato expression in neurons, astrocytes, and microglia in the highest transfected areas of the striatum and hippocampus, determined by the result from Figure 3. The Ai9 mouse brains were injected with a high dose of MC3 LNP Cre mRNA (0.250 μg μL−1), sectioned and stained for the neuronal marker: neuronal nuclear protein (NeuN), astrocyte marker: glial fibrillary acidic protein (GFAP), and microglia marker: ionized calcium-binding adaptor molecule 1 (Iba1). The co-localization of tdTomato-positive neurons (tdTomato+; NeuN+), astrocytes (tdTomato+; GFAP+) or microglia (tdTomato+; Iba1+) demonstrated that approximately 33.7% were neurons, 47.9% were astrocytes, and 2.5% were microglia among Cre-recombined cells (tdTomato+) in the striatum respectively (Figure 5A and 5B, see Figure S3 for Supporting Information of non-normalized raw values). In the hippocampus, approximately 78.7% of the transfected cells were neurons, 6.0% were astrocytes, and 0.4% were microglia (Figure 5C and 5D, see Figure S4 for Supporting Information of non-normalized raw values). Overall, Cre-mediated recombination by MC3 LNP Cre mRNA delivery was observed slightly more in astrocytes compared to neurons in the striatum while Cre-mediated recombination was observed predominantly in the neuronal population in the hippocampus. Taken together, MC3 LNP Cre mRNA is capable to induce Cre-mediated recombination in various brain cell types including neurons, astrocytes, and microglia with different transfection efficiencies between striatum and hippocampus.
Figure 5. The cell type specificity after the injection of MC3 LNP Cre mRNA in the striatum and hippocampus of Ai9 mice.
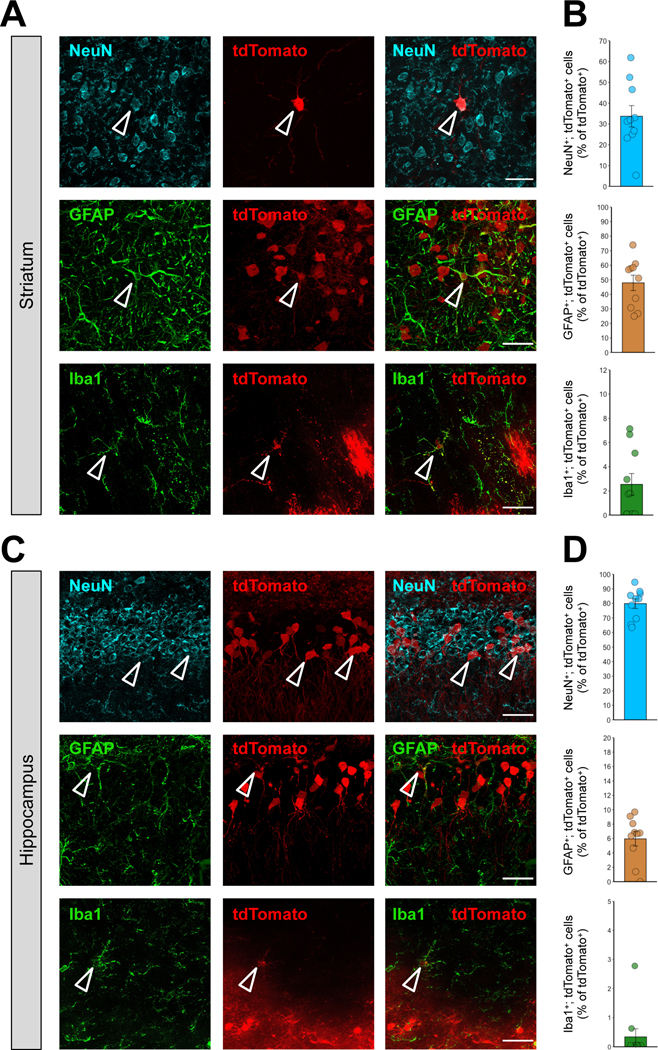
(A, C) Immunostaining of tdTomato+ (red) and either NeuN+ (cyan), GFAP+ (green) or Iba1+ (green) cells 21 days after stereotaxic injection of MC3 LNP Cre mRNA (0.250 μg μL−1) in the striatum (A), and the hippocampus (C). The scale bar represents 30 μm. (B, D) Quantification of tdTomato+; NeuN+, tdTomato+; GFAP+ or tdTomato+; Iba1+ cells among total tdTomato+ cells (%) in the MC3 LNP Cre mRNA (0.250 μg μL−1)-injected striatum (B), and hippocampus (D). All data are presented as mean ± SEM. MC3 LNP Cre mRNA (0.250 μg μL−1; n = 10 images from 2 mice).
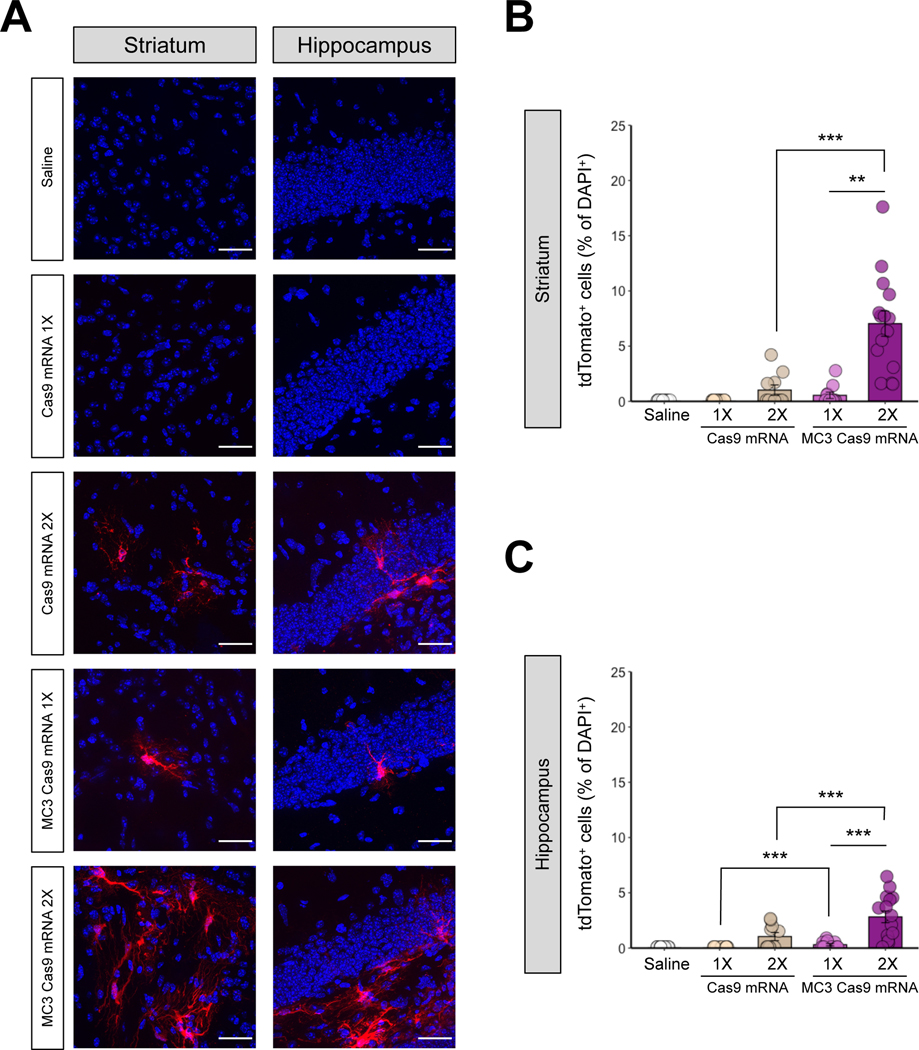
3. MC3 LNP Cas9 mRNA/Ai9 sgRNA complexes edit genes in the striatum and hippocampus of Ai9 mice
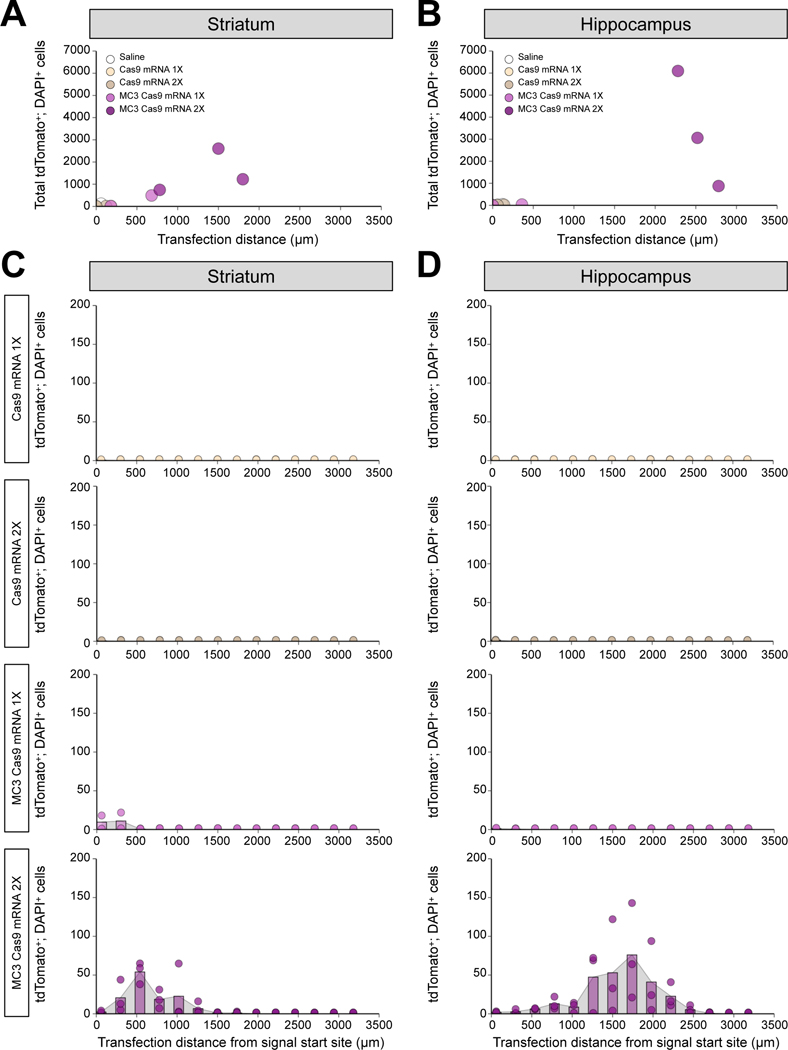
Cas9 mRNA and sgRNA have never been tested in brain tissue using LNPs and we, therefore, investigated whether MC3 LNPs can deliver Cas9 mRNA/Ai9 sgRNA to Ai9 mouse brains after intracerebral injection. The sgRNA used in these experiments targets the STOP sequence upstream of the fluorescent tdTomato gene and enables tdTomato gene expression, resulting in fluorescence (Figure 1C) as we previously reported (43). MC3 LNP Cas9 mRNA/Ai9 sgRNA were stereotaxically injected into the striatum and the hippocampus at low (0.125 μg μL−1) or high (0.250 μg μL−1) dose (saline and Cas9 mRNA/Ai9 sgRNA without MC3 LNPs-injected group were used as controls). Mice were sacrificed 21 days after the injections and the presence of tdTomato-positive cells was determined. In Figure 6A–6B, we quantified transfection distance and total tdTomato+; DAPI+ cells among different doses of Cas9 mRNA/Ai9 sgRNA with or without MC3 LNPs. We demonstrated that the transfection distance for low and high doses of Cas9 mRNA/Ai9 sgRNA were 90 ± 90 μm and 60 ± 60 μm, respectively, and for low and high doses of MC3 LNP Cas9 mRNA/Ai9 sgRNA were 430 ± 250 μm, and 1,360 ± 303 μm in striatum, respectively (Figure 6A). In hippocampus, the transfection distance for low and high doses of Cas9 mRNA/Ai9 sgRNA were 0 μm and100 ± 40 μm, respectively, and for low and high doses of MC3 LNP Cas9 mRNA/Ai9 sgRNA were 180 ± 180 μm and 2527 ± 144 μm, respectively (Figure 6B). Further, we demonstrate that the total tdTomato+; DAPI+ cells as for low and high doses of Cas9 mRNA/Ai9 sgRNA were 6 ± 6 cells and 6 ± 6 cells, respectively, and for low and high doses of MC3 LNP Cas9 mRNA/Ai9 sgRNA were 252 ± 240 cells and 1524 ± 558 cells in striatum-injected samples, respectively (Figure 6A; Shown as histogram in Figure 6C). In the hippocampus, the total tdTomato+; DAPI+ cells for low and high doses of Cas9 mRNA/Ai9 sgRNA were 0 cells and 18 ± 6 cells, respectively, and for low and high doses of MC3 LNP Cas9 mRNA/Ai9 sgRNA were 18 ± 18 cells and 3,344 ± 1,514 cells, respectively (Figure 6B; Shown as histogram in Figure 6D). Taken together, MC3 LNPs were able to deliver Cas9 mRNA/Ai9 sgRNA in the brain after intracerebral injection. Notably, high dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA elevated transfection distance and tdTomato+; DAPI+ cells in the striatum and more noticeably in the hippocampus. Approximately a 10-fold increase in transfection distance and approximately a 200-fold increase in total tdTomato+; DAPI+ cells in high dose group of MC3 LNP Cas9 mRNA/Ai9 sgRNA compared to low dose group of MC3 LNP Cas9 mRNA/Ai9 sgRNA were observed.
Figure 6. Transfection distance and efficiency after MC3 LNP-mediated Cas9 mRNA/Ai9 sgRNA delivery in striatum and hippocampus.
(A, B) Transfection distance and total tdTomato+; DAPI+ cells in striatal injected samples (A), and hippocampal injected samples (B). (C, D) Quantification of tdTomato+; DAPI+ cells in every 12th brain slice in striatal injected samples (C), and hippocampal injected samples (D). Saline (n = 3 mice), low dose of Cas9 mRNA/Ai9 sgRNA (0.125 μg μL−1; Cas9 mRNA 1X; n = 2 mice), high dose of Cas9 mRNA/Ai9 sgRNA (0.250 μg μL−1; Cas9 mRNA 2X; n = 2 mice), low dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.125 μg μL−1; MC3 Cas9 mRNA 1X; n = 2 mice), and high dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.250 μg μL−1; MC3 Cas9 mRNA 2X; n = 3 mice).
Next, we further analyzed the local transfection efficiency in the highest transfected areas of the striatum and hippocampus, determined by the result from Figure 6. In the striatum, the number of edited cells (tdTomato+) in mice injected with the low dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA was not significantly different to the control composed of mice injected with saline or injected with the same dose of Cas9 mRNA/Ai9 sgRNA without MC3 LNPs (Figure 7A Left panel and Figure 7B), demonstrating that no significant gene-editing occurred. However, after the injection of the high dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA, we observed significantly increased number of tdTomato+ cells compared with the same dose of Cas9 mRNA/Ai9 sgRNA without MC3 LNPs (Figure 7B). The gene-editing efficiency increased approximately 12.8-fold (from ~0.5% to ~7.0% efficiency) with the high dose compared with the low dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA injection, while we did not observe any significant changes between two doses in Cas9 mRNA/Ai9 sgRNA without MC3 LNPs (Figure 7B). In the hippocampus, we observed significantly increased gene-editing efficiency with both low and high doses of MC3 LNP Cas9 mRNA/Ai9 sgRNA, as compared to the same dose of Cas9 mRNA/Ai9 sgRNA without MC3 LNPs (Figure 7A Right panel and Figure 7C). Figure 7C demonstrated a significantly increased number of tdTomato+ cells among all DAPI+ cells in the high dose treatment of MC3 LNP Cas9 mRNA/Ai9 sgRNA (~2.8% efficiency) compared to the low dose treatment of MC3 LNP Cas9 mRNA/Ai9 sgRNA (~0.3% efficiency), while we did not observe any significant changes between two doses in Cas9 mRNA/Ai9 sgRNA without MC3 LNPs (Figure 7C). We did not see any significant difference in total DAPI-positive cell numbers in the striatum between any groups (Figure S2B Left panel). However, in the hippocampus, we observed differences in DAPI+ cells between Cas9 mRNA/Ai9 sgRNA and MC3 LNP Cas9 mRNA/Ai9 sgRNA in both concentrations (Figure S2B Right panel). Nevertheless, all counted Cas9-edited cells were normalized by the total number of DAPI+ cells and therefore the results were not affected by the number of cells.
Figure 7. MC3 LNPs can deliver Cas9 mRNA with Ai9 sgRNA to promote gene editing in mouse striatum and hippocampus in a dose-dependent manner.
(A) tdTomato (red) expression and DAPI (blue) nuclear staining were analyzed 21 days after the saline, Cas9 mRNA/Ai9 sgRNA or MC3 LNP Cas9 mRNA/Ai9 sgRNA injection in the striatum (Left), and the hippocampus (Right). Cas9 mRNA/Ai9 sgRNA or MC3 LNP Cas9 mRNA/Ai9 sgRNA was injected with two doses: 0.125 μg μL−1 (1X) or 0.250 μg μL−1 (2X). The scale bar represents 30 μm. (B, C) Quantification of the percentage of tdTomato+ cells among DAPI+ in the striatum (permutation one-way ANOVA: F(4,55) = 23.36, p < 0.001) (B), and the hippocampus (permutation one-way ANOVA: F(4,55) = 15.29, p < 0.001) (C). All data are presented as mean ± SEM. ** p < 0.01, *** p < 0.001 by post hoc permutation t-test. Post hoc p values were calculated between Cas9 mRNA/Ai9 sgRNA (0.125 μg μL−1 or 0.250 μg μL−1) and MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.125 μg μL−1 or 0.250 μg μL−1) as well as between dosages 0.125 μg μL−1 vs. 0.250 μg μL−1 within Cas9 mRNA/Ai9 sgRNA or MC3 LNP Cas9 mRNA/Ai9 sgRNA. Saline (n = 15 images from 3 mice), Cas9 mRNA/Ai9 sgRNA low dose (0.125 μg μL−1; Cas9 mRNA 1X; n = 10 images from 2 mice), Cas9 mRNA/Ai9 sgRNA high dose (0.250 μg μL−1; Cas9 mRNA 2X; n = 10 images from 2 mice), MC3 LNP Cas9 mRNA/Ai9 sgRNA low dose (0.125 μg μL−1; MC3 Cas9 mRNA 1X; n = 10 images from 2 mice), and MC3 LNP Cas9 mRNA/Ai9 sgRNA high dose (0.250 μg μL−1; MC3 Cas9 mRNA 2X; n = 15 images from 3 mice).
4. MC3 LNP-mediated delivery of Cas9 mRNA/Ai9 sgRNA into the brain leads to Cas9-mediated gene editing in neurons, astrocytes, and microglia of Ai9 mice
Next, we also determined the MC3 LNP Cas9 mRNA/Ai9 sgRNA-mediated gene editing efficiency in the various cell types in the brain by analyzing the tdTomato-positive neurons, astrocytes, and microglia in the highest transfected areas of striatum and hippocampus, determined by the result from Figure 6. Ai9 mice were injected with the high dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.250 μg μL−1) and after 21 days the brain sections were stained for neurons (NeuN+), astrocytes (GFAP+), and microglia (Iba1+). The co-localization of tdTomato-positive neurons (tdTomato+; NeuN+), astrocytes (tdTomato+; GFAP+) or microglia (tdTomato+; Iba1+) in the striatum demonstrated that among all edited (tdTomato+) cells, approximately 1.5% were neurons, 79.4% were astrocytes, and 2.9% were microglia (Figure 8A and 8B, see Figure S5 for Supporting Information of non-normalized raw values). In the hippocampus. we found that among the cells edited by MC3 LNP Cas9 mRNA/Ai9 sgRNA, approximately 45.1% were neurons, 44.5% were astrocytes, and 4.2% were microglia (Figure 8C and 8D, see Figure S6 for Supporting Information of non-normalized raw values). Overall, in the striatum, a high dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA induced gene editing predominantly in astrocytes. On the contrary, both neurons and astrocytes comprise similarly of the gene-edited cell populations in the hippocampus. In conclusion, MC3 LNPs-mediated Cas9 mRNA/Ai9 sgRNA delivery can induce gene editing in neurons, astrocytes, and microglia with different efficiencies as we observed with MC3 LNPs-mediated Cre-mRNA delivery.
Figure 8. The cell type specificity after the injection of MC3 LNP Cas9 mRNA with Ai9 sgRNA in the striatum and hippocampus of Ai9 mice.
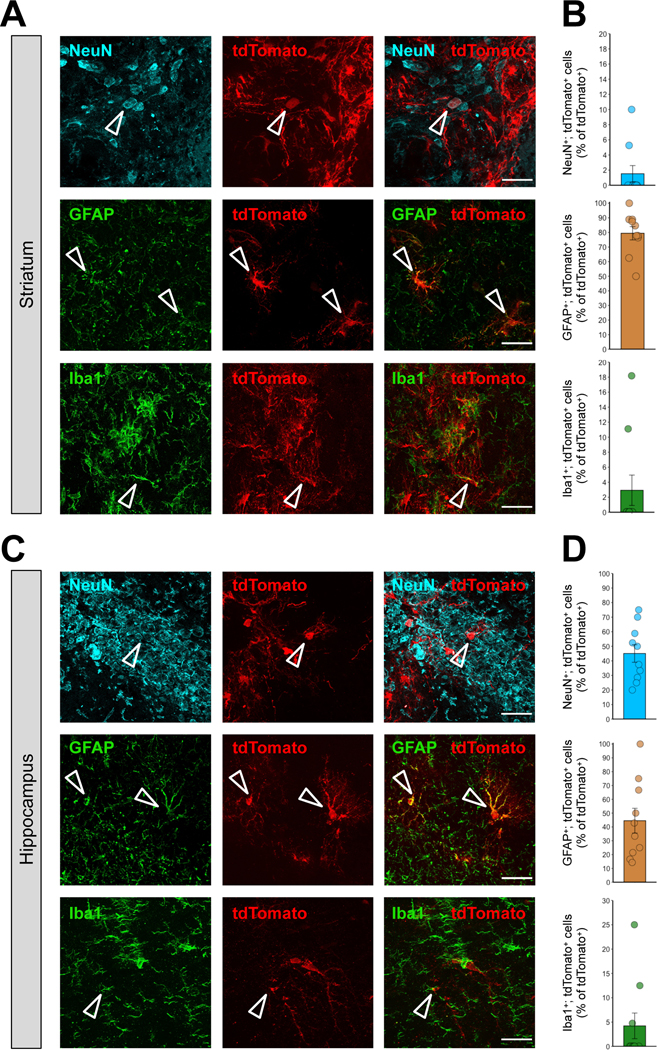
(A, C) Immunostaining of tdTomato+ (red) and either NeuN+ (cyan), GFAP+ (green) or Iba1+ (green) cells 21 days after stereotaxic injection of MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.250 μg μL−1) in the striatum (A), and the hippocampus (C). The scale bar represents 30 μm. (B, D) Quantification of (B) tdTomato+; NeuN+, tdTomato+; GFAP+ or tdTomato+; Iba1+ cells among total tdTomato+ cells (%) in the MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.250 μg μL−1)-injected striatum (B), and hippocampus (D). All data are presented as mean ± SEM. MC3 LNP Cas9 mRNA/Ai9 sgRNA (0.250 μg μL−1; n = 10 images from 2 mice).
5. Concluding remarks
LNP-mediated mRNA delivery has shown great therapeutic potential for treating a wide array of diseases due to its safe toxicity profile and transfection efficiency in vivo (8, 15). However, very little is known about LNPs-mediated mRNA delivery in the brain. Here we demonstrate successful delivery of Cre mRNA by MC3 LNPs into the striatum and hippocampus of Ai9 mice via robust manifestation of Cre-mediated recombination in the brain. Notably, our results indicate that a single intracerebral injection of MC3 LNP mRNAs can penetrate more than half of the striatum and hippocampus. We also demonstrate that the delivery of Cas9 mRNA/Ai9 sgRNA by MC3 LNPs results in successful gene editing in the striatum and the hippocampus of Ai9 mice. MC3 LNP Cas9 mRNA/Ai9 sgRNA demonstrates similar or better local transfection efficiency in the striatum and the hippocampus compared to previously demonstrated results using Cas9 RNPs (37, 43). This implicates that MC3 LNPs can deliver mRNAs for larger size proteins such as Cas9 into the brain. Moreover, we show that the recombination or editing efficiencies are dependent on the concentration of the Cre mRNA or Cas9 mRNA/Ai9 sgRNA being introduced to the brain. Our extensive quantification result of total transfected cell number implicates that transfected cell numbers are dose-dependent (Figure S7), however, the transfection distance between lower and higher doses was similar, giving important insight for potential clinical therapeutic strategies. In addition, we demonstrate that MC3 LNPs can deliver mRNAs to a variety of cell types in the brain including neurons, astrocytes and microglia. Higher efficiencies were found in neurons and astrocytes compared to microglia. In conclusion, our investigation reveals that the MC3 LNPs have great potential for delivering various sizes of mRNA after intracerebral injection to the target brain regions. Furthermore, our attempt to deliver Cas9 mRNA/sgRNA in vivo will open up new therapeutic opportunities and possibilities.
Our results demonstrate that Cas9-mediated gene editing efficiency was considerably lower than that of Cre-mediated gene recombination; the transfection efficiency between MC3 LNP Cas9 mRNA/Ai9 sgRNA and MC3 LNP Cre mRNA demonstrates a larger difference in the total number of transfected cells and the local transfection efficiency than in the transfection distance. The cause of different transfection efficiency between MC3 LNP Cas9 mRNA/Ai9 sgRNA and MC3 LNP Cre mRNA is hypothesized to be the temporal spacing between Cas9 mRNA expression and complexation of the sgRNA with Cas9 protein, resulting in the sgRNA being degraded in the cytoplasm before complexation with Cas9 protein can occur. We also observe that high dose of MC3 LNP Cas9 mRNA/Ai9 sgRNA improves gene editing compared to low dose, which implicates that prolonged expression of Cas9 mRNA/Ai9 sgRNA could increase gene editing efficiency. As such, increasing sgRNA stability in vivo could be the way forward. Overall, MC3 LNP-mediated mRNA delivery via intracerebral injection has great promise for treating a wide array of complex conditions.
Supplementary Material
Acknowledgement
This work was supported by the NIH R01MH125979, NIH R21AG072423, the UT Rising STARs award, and the Simons Foundation Autism Research Initiative (SFARI) pilot award (#574967) to H.Y.L; NIH UG3NS115599, NIH R33AI119115, NIH R33DA048444, NIH R01EB029320, NIH R01MH125979, NIH 1UM1AI164559, NIH U19NS132303, CIRM DISC2-14097-0, the BAKAR Spark award, the Innovative Genomics Institute, the CRISPR-Cures grant, and the Cystic Fibrosis Foundation to N.M. The authors thank Rodrigo Gonzales-Rojas for comments on the manuscript.
Footnotes
Supporting Information
The Supporting Information is available free of charge:
(a) Size distribution and polydispersity index of MC3 LNP mRNA complexes (Figure S1); (b) quantification of total number of DAPI+, tdTomato+, NeuN+, Iba1+ and GFAP+ cells as well as total number of tdTomato+; NeuN+, tdTomato+; GFAP+ and tdTomato+; Iba1+ cells in the striatum and hippocampus of Ai9 mice after the injection with MC3 LNP Cas9 mRNA/sgRNA (0.250 μg μL−1) or with MC3 LNP Cre mRNA (0.250 μg μL−1) (Figures S2-S6); (c) quantification of tdTomato+; DAPI+ cells normalized by injection concentration of MC3 LNP mRNA (Figure S7)
Competing interests
The authors declare no competing interests.
References
- (1).Damase TR; Sukhovershin R; Boada C; Taraballi F; Pettigrew RI.Cooke JP. The Limitless Future of RNA Therapeutics, Front Bioeng Biotechnol 2021, 9, 628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Qin S; Tang X; Chen Y; Chen K; Fan N; Xiao W; Zheng Q; Li G; Teng Y; Wu M.Song X. mRNA-based therapeutics: powerful and versatile tools to combat diseases, Signal Transduct Target Ther 2022, 7, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Anthony K. RNA-based therapeutics for neurological diseases, RNA Biol. 2022, 19, 176–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Peng H; Guo X; He J; Duan C; Yang M; Zhang X; Zhang L; Fu R; Wang B; Wang D; Chen H; Xie M; Feng P; Dai L; Tang X.Luo J. Intracranial delivery of synthetic mRNA to suppress glioblastoma, Mol Ther Oncolytics 2022, 24, 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Pardi N; Tuyishime S; Muramatsu H; Kariko K; Mui BL; Tam YK; Madden TD; Hope MJ.Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes, J Control Release 2015, 217, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tanaka H; Nakatani T; Furihata T; Tange K; Nakai Y; Yoshioka H; Harashima H.Akita H. In Vivo Introduction of mRNA Encapsulated in Lipid Nanoparticles to Brain Neuronal Cells and Astrocytes via Intracerebroventricular Administration, Mol Pharm 2018, 15, 2060–2067. [DOI] [PubMed] [Google Scholar]
- (7).Li Y; Jarvis R; Zhu K; Glass Z; Ogurlu R; Gao P; Li P; Chen J; Yu Y; Yang Y.Xu Q. Protein and mRNA Delivery Enabled by Cholesteryl-Based Biodegradable Lipidoid Nanoparticles, Angew Chem Int Ed Engl 2020, 59, 14957–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hou X; Zaks T; Langer R.Dong Y. Lipid nanoparticles for mRNA delivery, Nat Rev Mater 2021, 6, 1078–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Di J; Du Z; Wu K; Jin S; Wang X; Li T.Xu Y. Biodistribution and Non-linear Gene Expression of mRNA LNPs Affected by Delivery Route and Particle Size, Pharm Res 2022, 39, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kariko K; Keller JM; Harris VA; Langer DJ.Welsh FA. In vivo protein expression from mRNA delivered into adult rat brain, J Neurosci Methods 2001, 105, 77–86. [DOI] [PubMed] [Google Scholar]
- (11).Mahato RI; Kawabata K; Nomura T; Takakura Y.Hashida M. Physicochemical and pharmacokinetic characteristics of plasmid DNA/cationic liposome complexes, J Pharm Sci 1995, 84, 1267–1271. [DOI] [PubMed] [Google Scholar]
- (12).Palmer LR; Chen T; Lam AM; Fenske DB; Wong KF; MacLachlan I.Cullis PR. Transfection properties of stabilized plasmid-lipid particles containing cationic PEG lipids, Biochim Biophys Acta 2003, 1611, 204–216. [DOI] [PubMed] [Google Scholar]
- (13).Han X; Zhang H; Butowska K; Swingle KL; Alameh MG; Weissman D.Mitchell MJ. An ionizable lipid toolbox for RNA delivery, Nat Commun 2021, 12, 7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tam YY; Chen S.Cullis PR. Advances in Lipid Nanoparticles for siRNA Delivery, Pharmaceutics 2013, 5, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Satapathy MK; Yen TL; Jan JS; Tang RD; Wang JY; Taliyan R.Yang CH. Solid Lipid Nanoparticles (SLNs): An Advanced Drug Delivery System Targeting Brain through BBB, Pharmaceutics 2021, 13, 1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rungta RL; Choi HB; Lin PJ; Ko RW; Ashby D; Nair J; Manoharan M; Cullis PR.Macvicar BA. Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain, Mol Ther Nucleic Acids 2013, 2, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Freed CR; Greene PE; Breeze RE; Tsai WY; DuMouchel W; Kao R; Dillon S; Winfield H; Culver S; Trojanowski JQ; Eidelberg D.Fahn S. Transplantation of embryonic dopamine neurons for severe Parkinson’s disease, N Engl J Med 2001, 344, 710–719. [DOI] [PubMed] [Google Scholar]
- (18).Kunwar S; Prados MD; Chang SM; Berger MS; Lang FF; Piepmeier JM; Sampson JH; Ram Z; Gutin PH; Gibbons RD; Aldape KD; Croteau DJ; Sherman JW; Puri RK.Cintredekin Besudotox Intraparenchymal Study, G. Direct intracerebral delivery of cintredekin besudotox (IL13-PE38QQR) in recurrent malignant glioma: a report by the Cintredekin Besudotox Intraparenchymal Study Group, J Clin Oncol 2007, 25, 837–844. [DOI] [PubMed] [Google Scholar]
- (19).Ma Y; Tang C; Chaly T; Greene P; Breeze R; Fahn S; Freed C; Dhawan V.Eidelberg D. Dopamine cell implantation in Parkinson’s disease: long-term clinical and (18)F-FDOPA PET outcomes, J Nucl Med 2010, 51, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Chaichana KL; Pinheiro L.Brem H. Delivery of local therapeutics to the brain: working toward advancing treatment for malignant gliomas, Ther Deliv 2015, 6, 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chang Z; Mao G; Sun L; Ao Q; Gu Y.Liu Y. Cell therapy for cerebral hemorrhage: Five year follow-up report, Exp Ther Med 2016, 12, 3535–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gernert M; Feja M. Bypassing the Blood-Brain Barrier: Direct Intracranial Drug Delivery in Epilepsies, Pharmaceutics 2020, 12, 1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Duerinck J; Schwarze JK; Awada G; Tijtgat J; Vaeyens F; Bertels C; Geens W; Klein S; Seynaeve L; Cras L; D’Haene N; Michotte A; Caljon B; Salmon I; Bruneau M; Kockx M; Van Dooren S; Vanbinst AM; Everaert H; Forsyth R.Neyns B. Intracerebral administration of CTLA-4 and PD-1 immune checkpoint blocking monoclonal antibodies in patients with recurrent glioblastoma: a phase I clinical trial, J Immunother Cancer 2021, 9, e002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Bilang-Bleuel A; Revah F; Colin P; Locquet I; Robert JJ; Mallet J.Horellou P. Intrastriatal injection of an adenoviral vector expressing glial-cell-line-derived neurotrophic factor prevents dopaminergic neuron degeneration and behavioral impairment in a rat model of Parkinson disease, Proc Natl Acad Sci U S A 1997, 94, 8818–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gey L; Gernert M.Loscher W. Continuous bilateral infusion of vigabatrin into the subthalamic nucleus: Effects on seizure threshold and GABA metabolism in two rat models, Neurobiol Dis 2016, 91, 194–208. [DOI] [PubMed] [Google Scholar]
- (26).Gao Y; Geng L; Chen VP.Brimijoin S. Therapeutic Delivery of Butyrylcholinesterase by Brain-Wide Viral Gene Transfer to Mice, Molecules 2017, 22, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Noh JE; Oh SH; Park IH.Song J. Intracerebral Transplants of GMP-Grade Human Umbilical Cord-Derived Mesenchymal Stromal Cells Effectively Treat Subacute-Phase Ischemic Stroke in a Rodent Model, Front Cell Neurosci 2020, 14, 546659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nance EA; Woodworth GF; Sailor KA; Shih TY; Xu Q; Swaminathan G; Xiang D; Eberhart C.Hanes J. A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue, Sci Transl Med 2012, 4, 149ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen S; Tam YY; Lin PJ; Leung AK; Tam YK.Cullis PR. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration, J Control Release 2014, 196, 106–112. [DOI] [PubMed] [Google Scholar]
- (30).Mastorakos P; Zhang C; Berry S; Oh Y; Lee S; Eberhart CG; Woodworth GF; Suk JS.Hanes J. Highly PEGylated DNA Nanoparticles Provide Uniform and Widespread Gene Transfer in the Brain, Adv Healthc Mater 2015, 4, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gaj T; Ojala DS; Ekman FK; Byrne LC; Limsirichai P.Schaffer DV. In vivo genome editing improves motor function and extends survival in a mouse model of ALS, Sci Adv 2017, 3, eaar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yang S; Chang R; Yang H; Zhao T; Hong Y; Kong HE; Sun X; Qin Z; Jin P; Li S.Li XJ. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease, J Clin Invest 2017, 127, 2719–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lee B; Lee K; Panda S; Gonzales-Rojas R; Chong A; Bugay V; Park HM; Brenner R; Murthy N.Lee HY. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours, Nat Biomed Eng 2018, 2, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gyorgy B; Loov C; Zaborowski MP; Takeda S; Kleinstiver BP; Commins C; Kastanenka K; Mu D; Volak A; Giedraitis V; Lannfelt L; Maguire CA; Joung JK; Hyman BT; Breakefield XO.Ingelsson M. CRISPR/Cas9 Mediated Disruption of the Swedish APP Allele as a Therapeutic Approach for Early-Onset Alzheimer’s Disease, Mol Ther Nucleic Acids 2018, 11, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ekman FK; Ojala DS; Adil MM; Lopez PA; Schaffer DV.Gaj T. CRISPR-Cas9-Mediated Genome Editing Increases Lifespan and Improves Motor Deficits in a Huntington’s Disease Mouse Model, Mol Ther Nucleic Acids 2019, 17, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ricci R; Colasante G. CRISPR/dCas9 as a Therapeutic Approach for Neurodevelopmental Disorders: Innovations and Limitations Compared to Traditional Strategies, Dev Neurosci 2021, 43, 253–261. [DOI] [PubMed] [Google Scholar]
- (37).Staahl BT; Benekareddy M; Coulon-Bainier C; Banfal AA; Floor SN; Sabo JK; Urnes C; Munares GA; Ghosh A.Doudna JA. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes, Nat Biotechnol 2017, 35, 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Abbasi S; Uchida S; Toh K; Tockary TA; Dirisala A; Hayashi K; Fukushima S.Kataoka, K. Co-encapsulation of Cas9 mRNA and guide RNA in polyplex micelles enables genome editing in mouse brain, J Control Release 2021, 332, 260–268. [DOI] [PubMed] [Google Scholar]
- (39).Kulkarni JA; Myhre JL; Chen S; Tam YYC; Danescu A; Richman JM.Cullis PR. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA, Nanomedicine 2017, 13, 1377–1387. [DOI] [PubMed] [Google Scholar]
- (40).Cullis PR; Hope MJ. Lipid Nanoparticle Systems for Enabling Gene Therapies, Mol Ther 2017, 25, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Long J; Yu C; Zhang H; Cao Y; Sang Y; Lu H; Zhang Z; Wang X; Wang H; Song G; Yang J.Wang S. Novel Ionizable Lipid Nanoparticles for SARS-CoV-2 Omicron mRNA Delivery, Adv Healthc Mater 2023, e2202590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Madisen L; Zwingman TA; Sunkin SM; Oh SW; Zariwala HA; Gu H; Ng LL; Palmiter RD; Hawrylycz MJ; Jones AR; Lein ES.Zeng HA robust and high-throughput Cre reporting and characterization system for the whole mouse brain, Nat. Neurosci. 2010, 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Li J; Tuma J; Han H; Kim H; Wilson RC; Lee HY.Murthy N. The Coiled-Coil Forming Peptide (KVSALKE)(5) Is a Cell Penetrating Peptide that Enhances the Intracellular Delivery of Proteins, Adv Healthc Mater 2022, 11, e2102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.