Abstract
The nuclear factor erythroid 2-related factor 2 (NRF2) transcription factor activates cytoprotective and metabolic gene expression in response to various electrophilic stressors. Constitutive NRF2 activity promotes cancer progression, whereas decreased NRF2 function contributes to neurodegenerative diseases. We used proximity proteomic analysis to define protein networks for NRF2 and its family members NRF1, NRF3 and the NRF2 heterodimer MAFG. A functional screen of co-complexed proteins revealed previously uncharacterized regulators of NRF2 transcriptional activity. We found that ZNF746 (also known as PARIS), a zinc finger transcription factor implicated in Parkinson’s disease, physically associated with NRF2 and MAFG, resulting in suppression of NRF2-driven transcription. ZNF746 overexpression increased oxidative stress and apoptosis in a neuronal cell model of Parkinson’s disease, phenotypes that were reversed by chemical and genetic hyperactivation of NRF2. This study presents a functionally annotated proximity network for NRF2 and suggests a link between ZNF746 overexpression in Parkinson’s disease and inhibition of NRF2-driven neuroprotection.
INTRODUCTION
Oxidative stress is caused by the unmitigated accumulation of reactive oxygen species (ROS), which damages DNA, RNA, protein, and lipids (1). Extensive oxidative damage contributes to many human pathologies, including neurodegenerative disease, cardiovascular disease, cancer, and aging (2, 3). Although elevated levels of ROS are harmful, homeostatic levels of ROS serve as a critical second messenger to govern many aspects of normal cell biology (4). NFE2-like bZIP transcription factor 2 (NFE2L2, referred to as NRF2) is a member of the cap ‘n’ collar (CNC) family of transcription factors and constitutes a primary cellular defense mechanism against oxidative stress. NRF2 drives a gene expression program of approximately 200 genes involved in redox homeostasis, metabolism, DNA repair, and cell survival and proliferation (5, 6).
In the absence of stress, NRF2 protein levels are kept low though ubiquitylation-dependent proteasomal degradation (7, 8) mediated by the Kelch-like ECH-associated protein 1 (KEAP1) homodimer and its associated E3 ubiquitin ligase scaffold, Cullin-3 (CUL3) (7, 9, 10). As the substrate adaptor within the complex, KEAP1 captures NRF2 by binding the DLG and ETGE motifs within NRF2 (11). In normal cells, oxidative, electrophilic, and metabolic stressors react with cysteine residues in KEAP1, suppressing NRF2 degradation. The current model suggests that cysteine modifications to the KEAP1 dimer suppresses the ubiquitylation or release of NRF2 to the proteasome, resulting in a catalytically “dead” KEAP1/CUL3 complex that cannot bind to additional NRF2 molecules (12–14). Thus, newly synthesized NRF2 escapes degradation and translocates into the nucleus. NRF2 forms a heterodimer with the small musculoaponeurotic fibrosarcoma (sMAF) transcription factors and binds to the CNC-sMaf binding element (CsMBE) in the promoters of target genes (15, 16).
In addition to NRF2, the CNC family of NFE2-like bZIP transcription factors includes NFE2L1 (referred to as NRF1) and NFE2L3 (referred to as NRF3). Whereas NRF2 protects against oxidative and electrophilic stress, NRF1 mediates the proteasomal bounce-back response upon inhibition of the proteasome (17). NRF1 localizes to the endoplasmic reticulum (ER) and is posttranslationally regulated by the ER-associated degradation (ERAD) pathway (18–20). In response to disruptions in protein homeostasis, NRF1 escapes the ER to activate gene expression (21). Although relatively little is known about the function of NRF3, it may complement NRF1 in maintaining proteasome homeostasis (22). Active NRF3 is reported to promote proliferation in colon and pancreatic cancer cell line models (23–25).
Because NRF2 functions to improve cellular health and viability in response to stress, NRF2 contributes to both cancer prevention and progression (6, 26, 27). In carcinogen-driven mouse models, NRF2-deficient mice show an increase tumor incidence and metastasis (28–30). However, after cancer initiation, NRF2 activity promotes cancer progression (29, 31). Mutational activation of NRF2 is common in cancers of the lung, head and neck, esophagus, and bladder (10). Gain-of-function mutations in NRF2 disrupt the DLG or ETGE motifs, including the E79Q mutation; loss-of-function mutations in KEAP1 are not localized to hotspots and variably disrupt protein function (10, 32). In many tumors lacking NRF2 or KEAP1 mutations, aberrant NRF2 activation is achieved through other mechanisms, including KEAP1 posttranslational modifications and altered protein-protein interactions that displace NRF2 from KEAP1 (10, 33). Regardless of the mechanism of activation, increased NRF2 signaling strongly correlates with poor patient prognosis and chemo- and radioresistance (34–37).
In contrast to cancer, NRF2 function is decreased in multiple neurodegenerative diseases (38). Parkinson’s disease is characterized by the progressive loss of dopaminergic neurons in the substantia nigra (39). Parkinson’s disease arises from the accumulation of α-synuclein protein aggregates, known as Lewy bodies, due to familial recessive mutations in PARKIN (PRKN), PINK1, DJ-1 (PARK7), and other genes (40). Oxidative stress and mitochondrial dysfunction are hallmarks of Parkinson’s disease that contribute to dopaminergic neuron cell death (41). As a critical modulator of the oxidative stress response, NRF2 is proposed to be a potential therapeutic target in Parkinson’s disease (42–44). Pharmacological activation of NRF2 in both the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced and α-synuclein mouse models of Parkinson’s disease prevents dopaminergic neuron cell death and attenuates Parkinson’s disease-associated behavioral symptoms (45–50). Reciprocally, NRF2 knockout (KO) mice demonstrate hypersensitivity to chemical and genetic drivers of Parkinson’s disease neurobiology (51–54). Collectively, these and other studies suggest that suppressed NRF2 signaling contributes to neuronal pathology and Parkinson’s disease.
In this study, we sought to gain new understanding of NRF2-driven biology by defining the protein-protein proximity networks for the NRF family and the NRF-heterodimerizing MAFG partner. Of the 59 high-confidence NRF2 proximal proteins, we selected ZNF746 for further study based on functional screening and disease annotations. We report that ZNF746 (also known as PARIS) bound to NRF2 and inhibited the activation of NRF2 target genes. ZNF746 expression increased ROS production and cell death, which were rescued by forced expression of NRF2. In healthy neurons, ZNF746 is phosphorylated by PINK1, ubiquitylated by PARKIN and subsequently degraded by the proteasome. In Parkinson’s disease, loss of PINK1 or PARKIN results in ZNF746 overexpression, leading to loss of mitochondrial biogenesis and eventual apoptosis (55, 56). Our results establish ZNF746 as a direct repressor of the NRF2/MAFG heterodimer, establishing a potential new mechanism for NRF2 loss, oxidative stress, and cell death in Parkinson’s disease.
RESULTS
Protein proximity networks for NRF1, NRF2, NRF3, and MAFG reveal unique transcriptional regulatory complexes
Immuno- and affinity protein purification followed by mass spectrometry (MS) is a powerful and unbiased approach to define protein complexes. However, false negatives are common, particularly for protein-protein interactions that are transient, low-affinity, or require a chromatin template. We used live cell miniTurbo-based promiscuous biotin labeling to define the proximity networks for NRF1, NRF2, NRF3, and MAFG (Fig. 1A) (57). Open reading frames for each ‘bait’ protein were cloned in frame with a V5 epitope tag fused to miniTurbo; N-terminal and C-terminal fusions were made for each bait protein. In addition to wild-type (WT) NRF2, we expressed miniTurbo-tagged NRF2E79Q, which is a cancer-derived constitutively active NRF2 mutant that is resistant to KEAP1-mediated ubiquitylation (10, 32). Naked miniTurbo (V5) and miniTurbo fused to a nuclear localization signal (NLS) or to a nuclear export signal (NES) served as negative controls for probabilistic scoring (Fig. 1A). Human embryonic kidney 293T (HEK293T) cells stably expressing each bait protein were treated with biotin before being subjected to Western blot analysis for biotinylated proximal proteins (fig. S1A, S1B). Cells expressing NRF2 and NRF2E79Q showed increased NQO1 protein levels, a canonical NRF2-induced gene, indicating that the miniTurbo fusion did not abrogate NRF2 function (fig. S1A).
Fig. 1. Protein proximity networks for NRF1, NRF2, NRF3, and MAFG reveal unique transcriptional regulatory complexes.
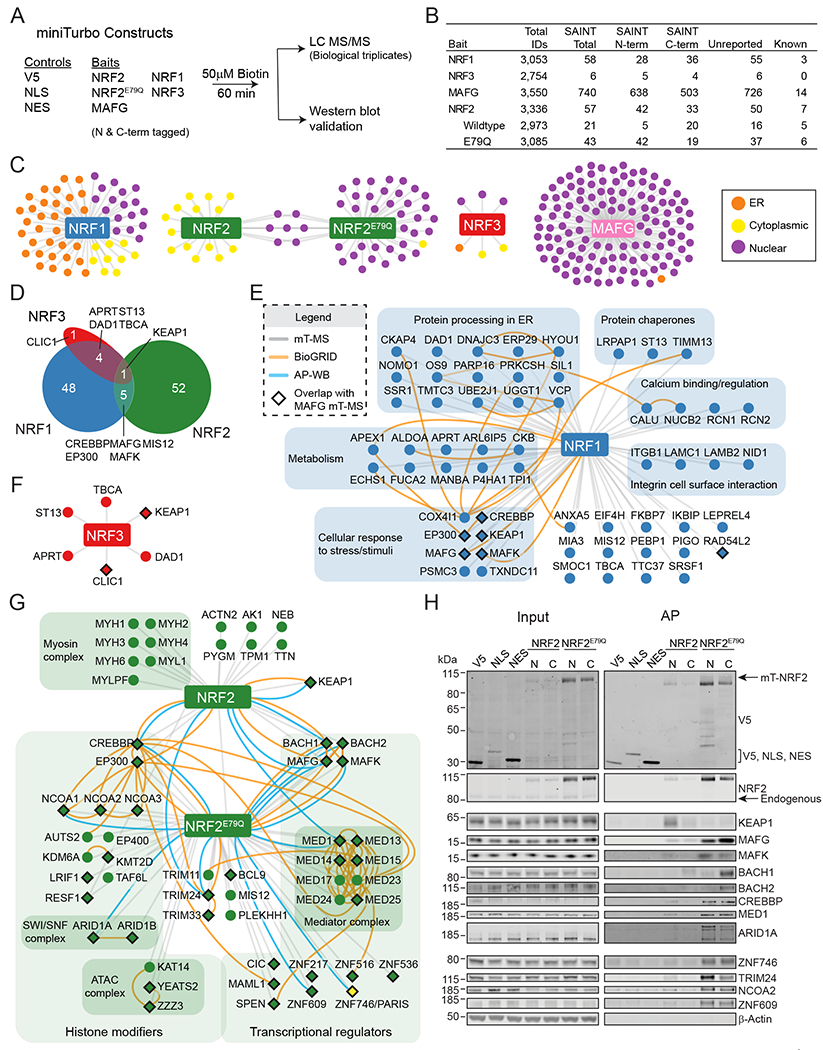
(A) Experimental design. HEK293T cells stably expressing N or C-terminally miniTurbo tagged control or bait proteins were treated with 50 μM biotin for 60 min. Biotinylated proteins were purified with streptavidin conjugated beads prior to MS or Western blot analysis. (B) Table showing the protein identifications for each bait protein from the MS analysis. Total IDs were probabilistically scored using SAINT. Known interactions were collected from the BioGRID database. (C) Protein proximity networks for NRF1, NRF2, NRF3, and MAFG with nodes colored based on cellular localization defined by GO analysis (orange=ER, yellow=cytoplasmic, and purple=nuclear). (D) Venn diagram depicting the overlap in proximal proteins that passed SAINT between NRF1, NRF2 (WT and NRF2E79Q combined), and NRF3. (E-G) Protein proximity networks for NRF1 (E), NRF3 (F), and NRF2 (G). Gray lines indicate bait-prey interactions that passed SAINT (n=3 biological replicates per bait). Orange lines indicate interactions collected from the BioGRID database and blue lines represent validated interactions by affinity purification (AP) followed by Western blot analysis. Diamonds depict the proteins that were also observed as prey in the MAFG network. (H) Western blot validation of select NRF2 proximal interactors identified by MS analysis. HEK293T cells stably expressing miniTurbo (mT)-tagged proteins were treated with 50 μM biotin for 1 h and biotinylated proteins affinity were purified using streptavidin beads. NLS= nuclear localization signal, NES= nuclear export signal, N= N-terminally-tagged baits, and C= C-terminally-tagged baits. Images are representative Western blots of n≥2 biological replicates per blotted protein. See also Figures S1 and S2.
Across biological triplicate experiments, affinity purified biotinylated proteins were analyzed by label-free liquid chromatography–tandem mass spectrometry (LC-MS/MS). The resulting data were probabilistically scored against the controls using the SAINT (significance analysis of interactome) express algorithm (Fig. 1B and Data File S1) (58). Combining the N-terminal and C-terminal NRF2 and NRF2E79Q networks revealed 50 previously unreported ‘prey’ proteins and 7 co-complexed proteins previously identified in the BioGRID database (Fig. 1B) (59). The N- and C-terminal networks comprised shared and unique prey-proteins, which likely reflects the 10-35nm biotin labeling radius of the miniTurbo enzyme and higher-order structure of the NRF2 protein complex (Fig. 1B) (60, 61). For example, BACH1 and BACH2 preferentially co-complexed with C-terminal NRF2E79Q as compared to N-terminally tagged NRF2E79Q (Data File S1). Such comparisons provide crude yet valuable insight into protein complex structure.
As expected, proteins within the NRF2E79Q and MAFG networks localize primarily to the nuclear compartment, whereas proximal proteins to WT NRF2 localize to both the nucleus and cytoplasm (Fig. 1C, compare the purple and yellow nodes). NRF1 binds to the ER membrane and is released in response to cellular stress (62) and 38 of the 66 NRF1 proximal proteins are reported to localize to the endoplasmic reticulum (ER). These data confirm proper subcellular localization of the miniTurbo-tagged bait proteins. In total, 111 proteins were enriched within the NRF1, NRF2, and NRF3 networks. Of these, 52 were unique to NRF2 and 48 were specific to NRF1 (Fig. 1D). KEAP1 was the only protein found in all three proximity networks. The lack of common interactors highlights differences in the regulation, subcellular localization, and function for the NRF protein family.
We next considered each network individually, grouping ‘prey’ proteins based on functional ontology (Fig. 1E–G). Fifteen proteins within the NRF1 network contribute to protein processing within the ER, including VCP, a known regulator of NRF1 (63). The NRF1 network was also enriched for proteins involved in stress response, including the known interactors MAFG and MAFK (Fig. 1E, orange lines). Additional functional groups included proteins broadly involved in metabolism, Ca2+ binding/regulation, integrin signaling, and protein chaperones. The NRF3 network was comparatively small and contained only six proteins that passed SAINT thresholds (Fig. 1F). One of these interactors was DAD1, which assembles protein N-glycosylation complexes at the ER (Fig. 1F) (64). NRF3 is N-glycosylated at the ER; these data nominate DAD1 as the responsible oligosaccharyltransferase (65).
The WT NRF2 network contained primarily cytoplasmic proteins, including seven myosin proteins. As expected, KEAP1 was observed proximal to WT NRF2, but not to NRF2E79Q (Fig. 1G). All but a few of the NRF2E79Q proximal proteins are annotated with transcriptional regulatory and/or histone modification functions. Previously reported members of the NRF2 transcriptional complex were identified: MAFG, MAFK, BACH1 (66), CREBBP (67), EP300 (68), eight components of the Mediator complex (69) and ARID1A and ARID1B of the SWI/SNF complex (70) (Fig. 1G, orange lines). Consistent with the heterodimeric nature of the NRF2:sMAF complex, we observed more than 75% of the NRF2E79Q network within the MAFG proximal network (Fig. 1G, denoted with diamond-shaped nodes). Beyond the established NRF2 interacting proteins, 50 previously unreported proximal proteins were identified (Fig. 1G). For example, NRF2E79Q proximal proteins included three members of the ATAC complex, a group of proteins with histone acetyltransferase activity, and the nuclear receptor coactivators NCOA1/2/3 (Fig. 1G). The Notch signaling pathway transcription factor MAML1 and five zinc finger proteins were also enriched within the network.
To validate these interactions, NRF2 proximal proteins were purified with streptavidin beads and detected by Western blot analysis (Fig. 1G–H, S2A, blue lines). In line with the MS analysis, the majority of the proteins were enriched in the NRF2E79Q samples, likely due to the increased stability and expression of NRF2E79Q versus WT NRF2. Some ‘prey’ proteins were biotinylated by both the N- and C-terminus tagged samples (these included CREBBP, MED1, ARID1A, and ZNF746) whereas a subset were enriched in either the N-terminus samples or the C-terminus (Fig. 1H, S2A). In summary, these data provide unbiased and probabilistically scored proximity networks for NRF1, NRF2, NRF3, and MAFG.
Functional annotation of the NRF2 proximal protein network
To determine if NRF2 proximal proteins regulate NRF2-dependent transcription, we performed an arrayed siRNA screen against 30 of the interactors and 5 controls in H1299 lung adenocarcinoma cells harboring eYFP within the first intron of the endogenous NQO1 gene (71). These H1299-NQO1-eYFP cells also carry constitutively expressed mCherry for cell number normalization. Transient transfection with KEAP1 siRNAs or treatment with the KEAP1 small molecule inhibitor CDDOme resulted in NRF2-dependent eYFP expression (Fig. 2A–C). siRNAs specific for genes encoding the NRF2 proximal proteins were introduced into cells before treatment with DMSO or CDDOme. Live cell imaging and florescence quantitation revealed the impact of gene loss on eYFP expression (Fig. 2D, S3, Data File S2). Except for siRNAs targeting KEAP1, silencing of the NRF2 proximal proteins did not affect eYFP expression in DMSO treated cells (fig. S3). In the presence of CDDOme, silencing the expression of five proximal proteins significantly reduced eYFP intensity. Conversely, silencing six genes increased reporter activation (Fig. 2D). These ‘hits’ largely consisted of known NRF2 regulators, including BACH1 and members of the Mediator complex (66, 69). Loss of EP300 enhanced eYFP expression, despite being previously reported to activate the expression of NRF2 downstream targets (Fig. 2D) (67, 68). Together, this functionally annotated proximity network provides a valuable resource and mechanistic insight into NRF2 regulation.
Fig. 2. Functional annotation of the NRF2 proximal protein network.
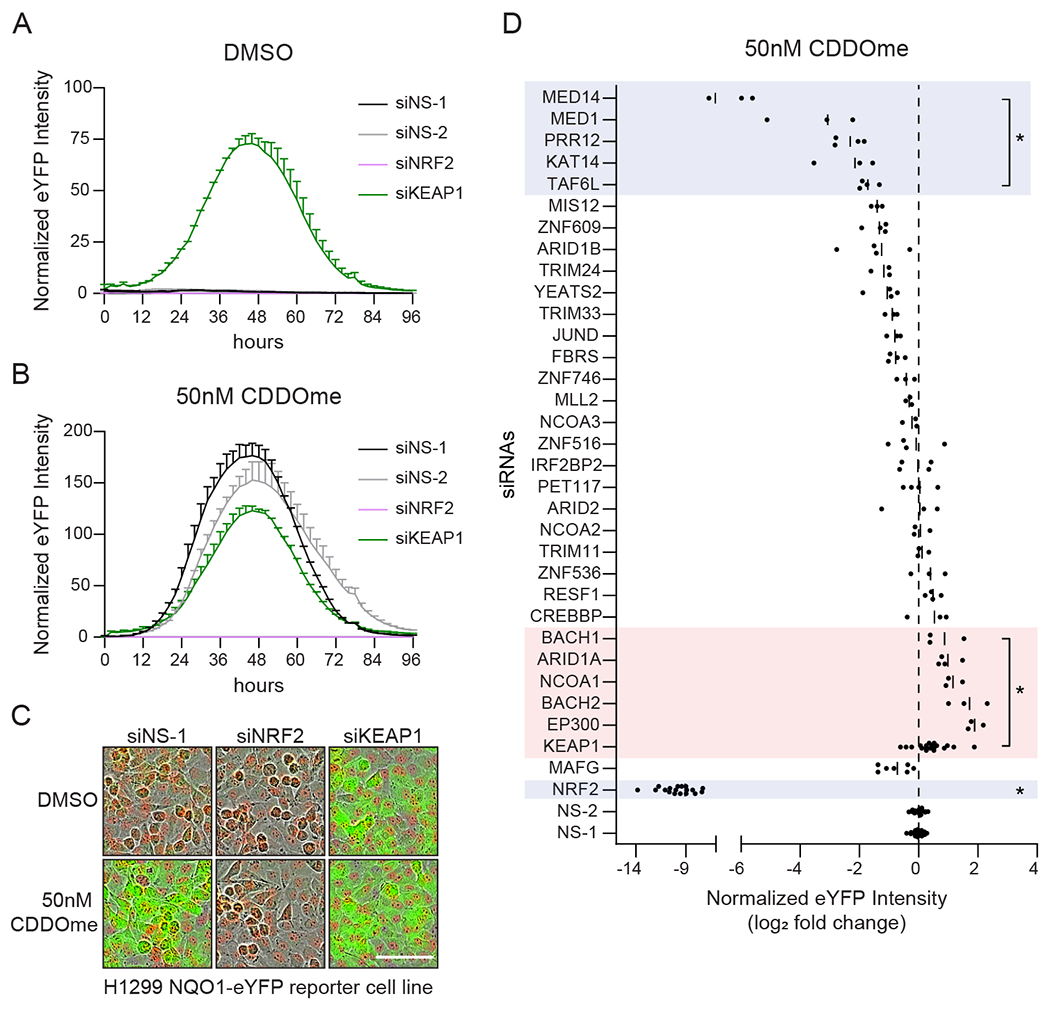
(A, B) Time course showing activation of the H1299 NQO1-eYFP reporter cell line for the control siRNAs used in the screen. H1299 NQO1-eYFP cells were transfected with siRNAs for 24 h prior to treatment with DMSO (A) or 50 nM CDDOme (B). eYFP intensity was normalized to cell number (mCherry positive). The graphs are representative of the controls used on each plate throughout the screen. (C) Representative images at the 48 h time point from (A) and (B). Scale bar, 100 μm. (D) H1299 NQO1-eYFP cells were transfected with siRNAs for 24 h prior to treatment with CDDOme. eYFP intensity was normalized to cell number and log2 fold change was calculated compared to average of siNS-1 and siNS-2. Data are represented by the mean of three or four biological replicates per siRNA. *, adjP≤0.05 by one-way ANOVA and Dunnett test for multiple comparisons compared to siNS-1 and siNS-2. See also Figure S3.
Loss of ZNF746 expression does not affect NRF2 signaling
From the NRF2 network, we chose ZNF746 for further exploration for several reasons. First, it is implicated in the molecular etiology of Parkinson’s disease (55, 56, 72). Second, it is reported to promote ROS production and proliferation in cancer cell lines (73–76). Finally, it has been suggested to be a NRF2 target gene and may serve in a negative feedback loop because it is a KRAB-domain containing transcriptional repressor (77, 78). Although siRNA silencing of ZNF746 did not affect NQO1-driven eYFP expression, we tested if siRNA or CRISPR knockout (KO) of ZNF746 regulated NRF2 governance of endogenous target genes. Transient knock-down of NRF2 in H1299 cells treated with CDDOme or PRL-295, a non-covalent inhibitor of the KEAP1-NRF2 interaction, resulted in a significant decrease in NRF2 and two NRF2 downstream target genes, NQO1 and HMOX1 (Fig. 3A, S4A) (79). ZNF746 silencing did not affect NRF2 or the expression of these target genes (Fig. 3A, S4A). For additional validation, we used CRISPR engineering to generate monoclonal ZNF746 knockouts in H1299 cells (fig. S4B). Again, deletion of ZNF746 did not alter expression of NRF2 or NRF2 targets in the presence of CDDOme or PRL-295 (Fig. 3B, S4C, S5A). Lastly, we silenced ZNF746 in two KEAP1 mutant lung cancer cell lines, A549 and H460, which constitutively express high levels of NRF2. ZNF746 silencing in these lines did not affect NRF2 target gene expression (Fig. 3C, 3D). Together, these data suggest that loss of ZNF746 expression does not alter NRF2 signaling in the lung cell lines tested.
Fig. 3. Loss of ZNF746 expression does not affect NRF2 signaling.
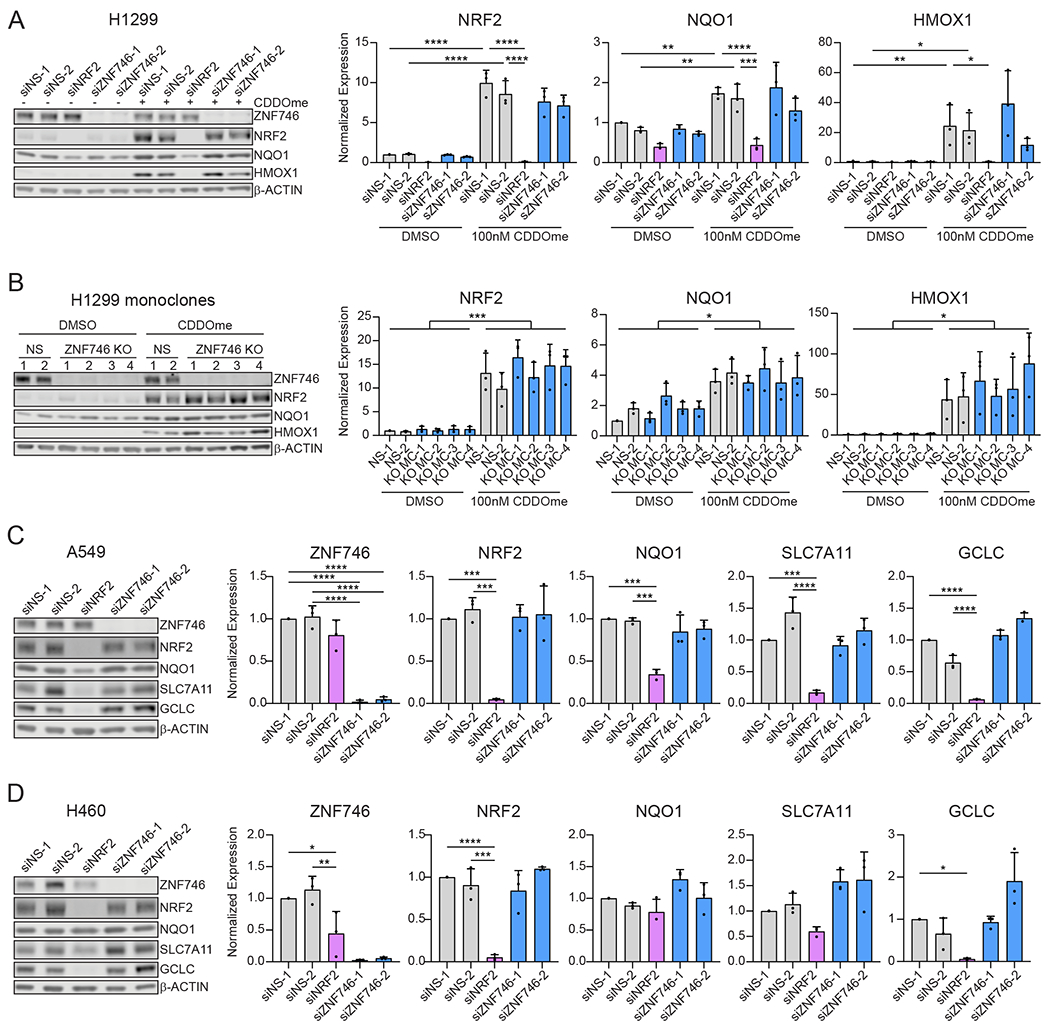
(A) H1299 cells were transfected with the indicated siRNAs for 48 h prior to treatment with 100 nM CDDOme for 16 h. NS= non-specific. The expression of each target was normalized to β-ACTIN. Data are presented as means ± SD. *, adjP<0.05; **, adjP<0.01; ***, adjP<0.001; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group). (B) Monoclonal NS and ZNF746 KO H1299 cells were treated with 100 nM CDDOme for 16 h before analysis by Western blot. The expression of each target was normalized to β-ACTIN. Data are presented as means ± SD. *, adjP<0.05; ***, adjP<0.001 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group). The indicated significant comparisons are between the CDDOme treatment and the respective DMSO control for each cell line. (C and D) A549 (C) and H460 (D) cells were transfected with the indicated siRNAs for 48 h. The expression of each target was normalized to β-ACTIN. Data are presented as means ± SD. *, adjP<0.05; **, adjP<0.01; ***, adjP<0.001; ****, adjP<0.0001 by one-way ANOVA and Šidák test for multiple comparisons (n=3 biological replicates per group). See also Figures S4 and S5.
ZNF746 overexpression inhibits the expression of NRF2 target genes
Overexpression of ZNF746 leads to oxidative stress in myoblasts and to loss of dopaminergic neurons in Parkinson’s disease models (55, 73). Therefore, we tested if ZNF746 overexpression impacted NRF2 target gene expression. As a control, we included the ZNF746C571A mutant which localizes to the zinc finger domain and prevents DNA binding (55). H1299 NQO1-eYFP cells stably expressing doxycycline-inducible (DI)-ZNF746 showed a dose dependent increase in ZNF746 protein levels (Fig. 4A). Overexpression of ZNF746 decreased NQO1-driven eYFP expression, whereas ZNF746C571A had no effect, suggesting that DNA binding is necessary for repression (Fig. 4B). Similarly, in parental H1299 cells, DI-ZNF746 expression significantly reduced endogenously expressed NQO1 protein levels in the presence of CDDOme or PRL-295 (Fig. 4C–D, S5B, S6A–B). ZNF746 did not repress the NRF2 target gene SLC7A11, suggesting potential specificity for ZNF746 regulation of NRF2.
Fig. 4. ZNF746 overexpression inhibits expression of NRF2 targets.
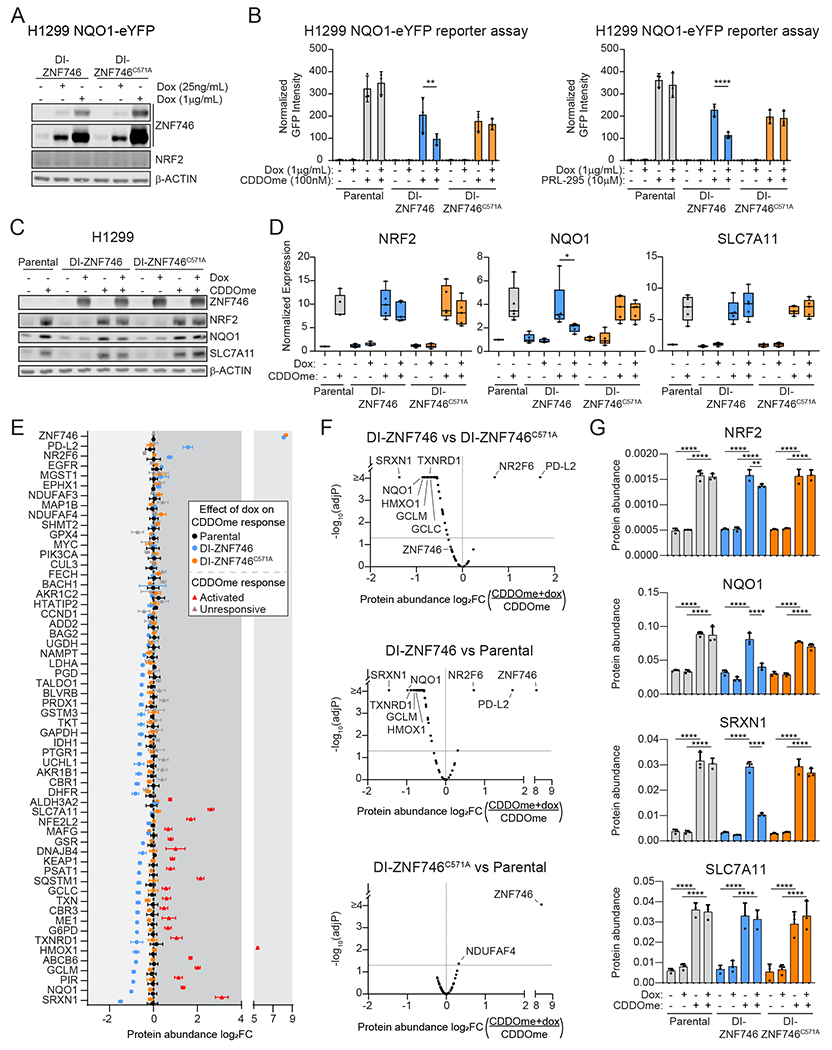
(A) H1299 NQO1-eYFP cells stably expressing doxycycline-inducible (DI) ZNF746 were treated with doxycycline for 48 h and analyzed by Western blot. (B) H1299 NQO1-eYFP cells stably expressing DI-ZNF746 were co-treated with 1 μg/mL doxycycline and 100 nM CDDOme or 10 μM PRL-295 and imaged over time. eYFP intensity was normalized to cell number (mCherry positive) and the peak eYFP time point was used from each replicate. Data are presented as means ± SD. **, adjP<0.01; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group). (C) H1299 cells stably expressing DI-ZNF746 were co-treated with 1 μg/mL doxycycline and 100 nM CDDOme for 24 h and analyzed by Western blot. Image is representative of n=5 biological replicates per group. (D) Quantification of protein expression normalized to β-ACTIN from (C). Data are presented as a boxplot with whiskers indicating the minimum and maximum values. *, adjP<0.05 by two-way ANOVA and Tukey test for multiple comparisons (n=5 biological replicates per group). (E) H1299 cells stably expressing DI-ZNF746 were co-treated with 1 μg/mL doxycycline and 100 nM CDDOme for 24 h. Protein abundance was quantified using targeted mass spectrometry against a library of defined NRF2 targets and controls. Log2 fold change (FC) (CDDOme+dox over CDDOme) was calculated for each protein and shown for parental, DI-ZNF746 and DI-ZNF746C571A cells. The CDDOme response (log2FC of CDDOme over DMSO) was calculated for each protein from the Parental sample only. Significance for the CDDOme response was calculated using two-way ANOVA and Šidák test for multiple comparisons (red dots indicate adjP<0.05). Data are presented as means ± SD (n=3 biological replicates per group). (F) Volcano plots depicting the difference in the log2FC protein abundance (CDDOme+dox over CDDOme) and adjP-value for the indicated comparisons from (E). Significance was calculated using two-way ANOVA and Tukey’s test for multiple comparisons (n=3 biological replicates per group). (G) Protein abundance of select proteins from (E). Data are presented as means ± SD. **, adjP<0.01; ****, adjP<0.0001 by two-way ANOVA and Tukey’s test for multiple comparisons (n=3 biological replicates per group). See also Figures S5 and S6.
To broadly investigate the consequence of ZNF746 overexpression on NRF2 signaling, we used an optimized internal standard parallel reaction monitoring (OIS-PRM) MS approach to quantify a predefined library of NRF2-related proteins (80). Our NRF2 panel consists of 227 peptides mapping to 90 proteins, including NRF2, ZNF746, MAFG, KEAP1. In total, 68 proteins are NRF2-related pathway members with 30 being transcriptional targets. The remaining proteins include immune cell markers, tumor suppressors, oncogenes and ‘housekeeping’ proteins. CDDOme treatment of parental H1299 cells significantly increased protein expression of 21 NRF2 target genes (Fig. 4E, red triangles). To determine the effect of ZNF746 and ZNF746C571A expression on the CDDOme response, we calculated the fold change of CDDOme treatment in the presence or absence of doxycycline for each cell line (Fig. 4E, Data File S3). Expression of ZNF746 repressed 16 of the 21 CDDOme-activated NRF2 targets. Dox treatment of ZNF746C571A and parental cells had no effect on the proteomic response to CDDOme (Fig. 4E, compare the blue circles to the black and orange circles). Direct comparisons of doxycycline treatment on CDDOme-induced activation between the samples showed a robust and significant decrease in specific NRF2 targets in ZNF746-expressing compared to ZNF746C571A-expressing cells and ZNF746-expressing compared to parental cells, but not for ZNF746C571A-expressing compared to parental cells (Fig. 4F, 4G S6C–J). Specifically, ZNF746 reduced the CDDOme induction of the protein abundance of NQO1 without affecting that of SLC7A11, corroborating the Western blot data (Fig. 4C–D, 4G and S6A, B). SRXN1, a regulator of the ROS detoxifying peroxiredoxin family, displayed the greatest decrease in protein abundance following ZNF746 expression (Fig. 4E–G). From the OIS-PRM data, ZNF746 induction reduced NRF2 protein levels in a statistically significant manner; however, the change was modest and was not supported by Western blot analysis (Fig. 4C, 4D, 4G and S6A–B). Unexpectedly, ZNF746 induced the expression of two proteins that function as immune checkpoints, NR2F6 and PD-L2 (Fig. 4E–F, S6H–I). ZNF746-induced expression of NR2F6 was reversed upon NRF2 activation by CDDOme, suggesting co-regulation of the immune cell response. In summary, ZNF746 overexpression suppressed most but not all NRF2 target genes.
NRF2 and ZNF746 co-complex and share common interactors
ZNF746 was among the targets initially validated to interact with NRF2 (Fig. 1H). To further confirm and study the interaction, we identified proteins proximal to ZNF746 and ZNF746C571A using miniTurbo biotinylation. Western blot analysis of HEK293T cells stably expressing each ‘bait’ protein confirmed biotinylation (fig. S7). Streptavidin purified protein complexes from miniTurbo-tagged ZNF746 cells contained ZNF777, a previously reported binding partner of ZNF746 (Fig. 5A, S2B) (81). To examine ZNF746-NRF2 interaction, cells were treated with CDDOme to stabilize NRF2 protein expression. Only after CDDOme treatment was NRF2 detected in protein complexes proximal to ZNF746, whereas NRF2 displayed reduced proximity to ZNF746C571A (Fig. 5A, S2B). Similarly, ZNF746 captured more MAFG protein as compared to ZNF746C571A. These data suggest that ZNF746 interacts with the NRF2/MAFG heterodimer specifically when bound to chromatin.
Fig. 5. NRF2 and ZNF746 co-complex and share common interactors.
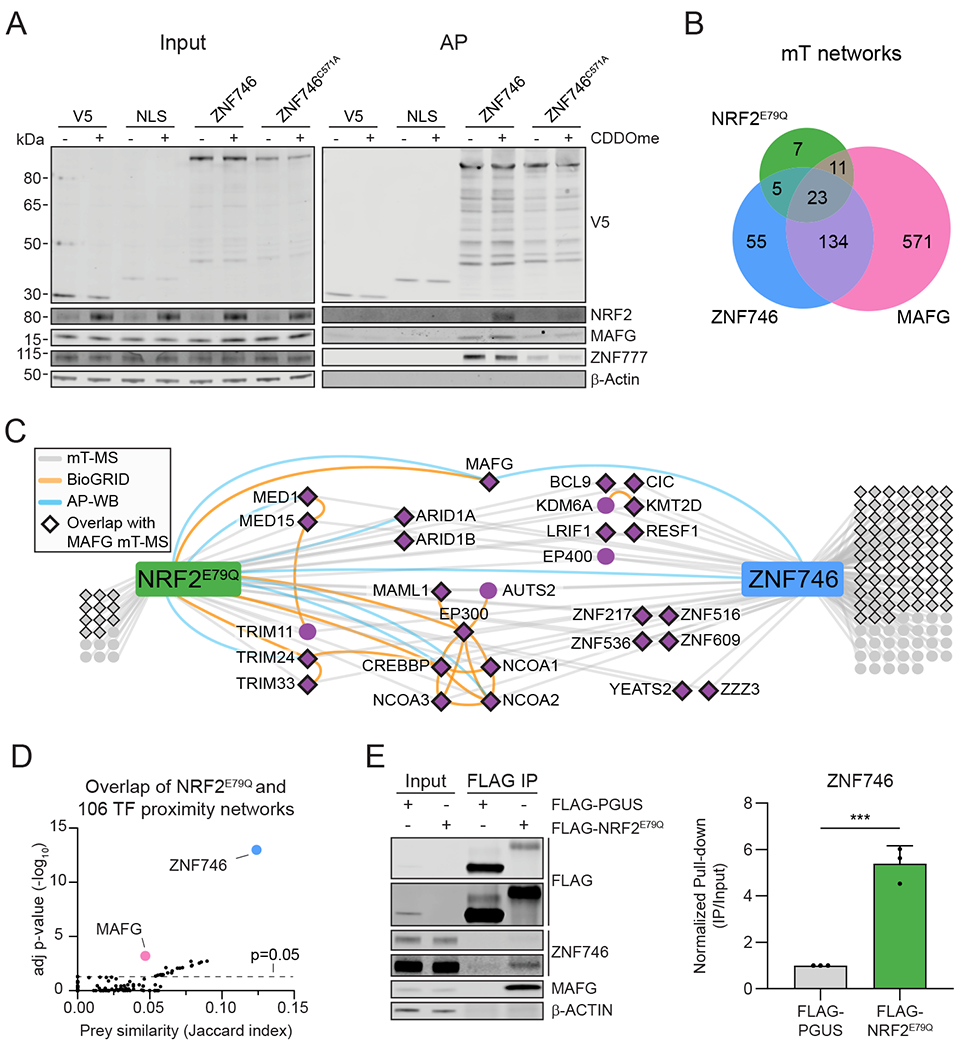
(A) HEK293T cells stably expressing miniTurbo (mT)-tagged proteins were treated with 100 nM CDDOme for 16 h before being incubated with 50 μM biotin for 1 h. Biotinylated proteins were affinity purified using streptavidin beads and analyzed by SDS-PAGE. Images are representative Western blots (n=3 biological replicates per group). (B) Venn diagram showing the overlap between the NRF2E79Q, ZNF746, and MAFG proximity networks. (C) Protein proximity network for ZNF746 and NRF2E79Q generated from HEK293T cells stably expressing miniTurbo-tagged proteins. Gray lines indicate bait-prey interactions identified in the MS analysis that passed SAINT (n≥2 biological replicates per bait). Orange lines indicate interactions collected from the BioGRID database and blue lines represent validated interactions by affinity purification (AP) followed by Western blot analysis. Diamonds depict the prey proteins that were also observed in the MAFG network. (D) Overlap of NRF2E79Q and 106 transcription factor proximity networks. P-value was calculated using the hypergeometric test. Jaccard index indicates prey similarity. (E) Representative Western blot from FLAG-immunoprecipitations from HEK293T cells. IPs were analyzed by SDS-PAGE and probed with the indicated antibodies. The quantified data are presented as means ± SD. ***, adjP<0.001 by unpaired t-test (n=3 biological replicates per group). See also Figures S2 and S7.
Using MS analysis, we defined the miniTurbo-based ZNF746 proximity network and compared it to the NRF2E79Q and MAFG networks (Data File S1). Sixty percent of the NRF2E79Q network overlapped with that for ZNF746 (Fig. 5B–C). To determine the specificity and importance of this overlap, we compared the NRF2 network with a proximity proteomic dataset containing 102 transcription factors (82). The ZNF746 network was the most significantly similar to the NRF2 network (Fig. 5D, Data File S4). Although the MAFG network also significantly overlapped with the NRF2 network, the Jaccard similarity index was reduced compared to ZNF746, possibly because of the large size of the MAFG network (Fig. 5B, D).
To investigate if NRF2 and ZNF746 co-complexed in an orthogonal assay, we immunopurified FLAG tagged NRF2E79Q from HEK293T cells and used immunoblotting to detect associated proteins. Endogenously expressed ZNF746 significantly co-purified with FLAG-NRF2E79Q as compared to the FLAG-PGUS control (Fig. 5E). Together, these data suggest NRF2, ZNF746, and MAFG co-complex within the nucleus.
NRF2 activation rescues ZNF746 induced ROS production and cell death
Our data showed that ZNF746 bound to the NRF2:MAFG protein complex and inhibited NRF2-driven transcription. Because NRF2 is central to the oxidative stress response, we next tested if ZNF746 expression affected cellular levels of ROS. Dox-induced ZNF746 expression significantly increased ROS, which was not observed with ZNF746C571A expression (Fig. 6A). Pharmacological activation of NRF2 by CDDOme or treatment with the antioxidant N-acetyl-cysteine (NAC) suppressed ZNF746-driven ROS production (Fig. 6A). Similarly, genetic activation of the NRF2 pathway through co-expression of NRF2E79Q prevented the induction of ROS (Fig. 6B). Sustained elevated levels of ROS can lead to cell death. Indeed, we observed an increase in cell death starting at 48 h after ZNF746 induction (Fig. 6C). Chemical or genetic activation of NRF2 curtailed the number of apoptotic cells (Fig. 6D–H). Expression of ZNF746C571A did not induce cell death, suggesting the ZNF746 transcriptional response is necessary for its induction of ROS and cell death (Fig. 6C–H).
Fig. 6. NRF2 activation rescues ZNF746 induced ROS elevation and cell death.
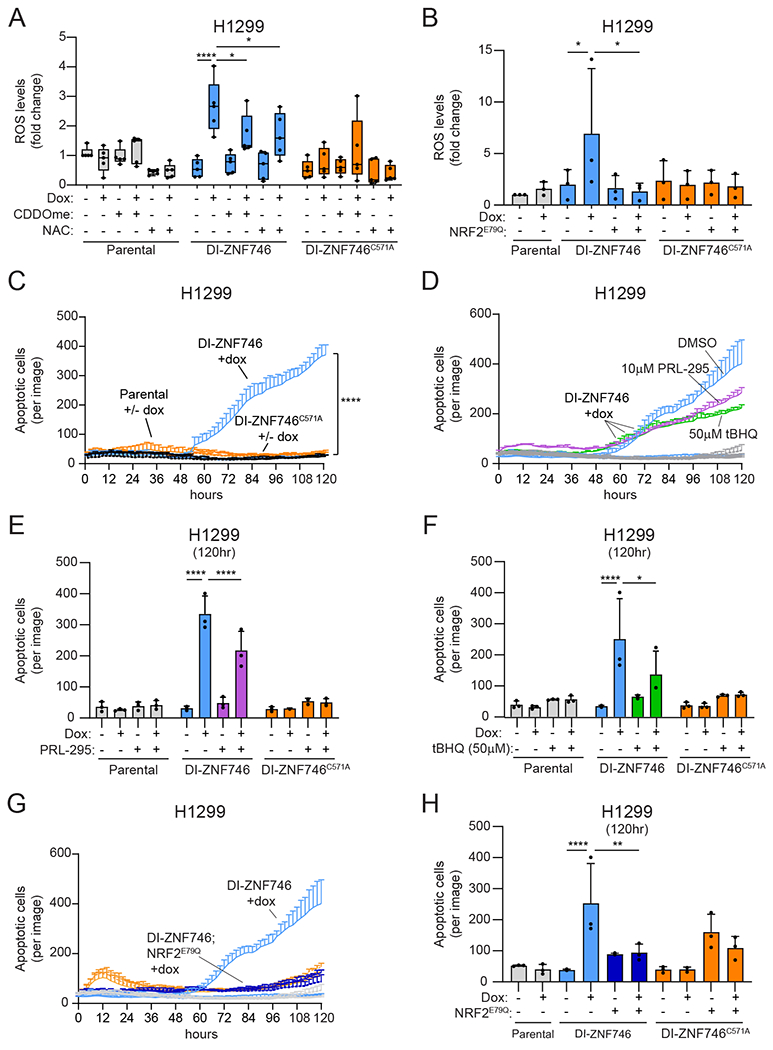
(A) H1299 cells stably expressing doxycycline-inducible (DI) ZNF746 were co-treated with 0.5 μg/mL doxycycline and 100 nM CDDOme or 3 mM NAC for 48 h. ROS was quantified using CellROX. Data are presented as a boxplot with whiskers indicating the minimum and maximum values. *, adjP<0.05; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=5 biological replicates per group). (B) H1299 cells stably expressing DI-ZNF746 and NRF2E79Q were treated with 1 μg/mL doxycycline. ROS was quantified using CellROX. The quantified data are presented as means ± SD. *, adjP<0.05 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group). (C) H1299 cells stably expressing DI-ZNF746 were treated with 0.5 μg/mL doxycycline and cell death was quantified over time using Cytotox-Red. The time course shown is representative of biological triplicates per group and the statistics were calculated from n=3 biological replicates per group at the 96 h time point. ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons. (D) H1299 cells stably expressing DI-ZNF746 were co-treated with 0.5 μg/mL doxycycline and 10 μM PRL-295 or 50 μM tBHQ. Cell death was quantified using Cytotox-Red. The time course shown is representative of biological triplicates per group. (E and F) Quantification of cell death at 120 h time point from (D). The quantified data are presented as means ± SD. *, adjP<0.05; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group). (G) H1299 cells stably expressing DI-ZNF746 and NRF2E79Q were treated with 0.5 μg/mL doxycycline. Cell death was quantified using Cytotox-Red. The time course shown is representative of biological triplicates per group. (H) Quantification of cell death at 120 h time point from (G). The quantified data are presented as means ± SD. **, adjP<0.01; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group).
ZNF746 promotes elevated ROS and cell death in a neuronal cell model
ZNF746/PARIS was discovered as a substrate for PINK1 and PARKIN, which together target ZNF746 to the proteasome for degradation (55, 56). Loss of PARKIN or PINK1 in models of Parkinson’s disease increases ZNF746 protein levels and subsequent transcriptional reprogramming. Current models suggests that ZNF746 upregulation in Parkinson’s disease leads to the repression of PGC-1α and mitochondrial biogenesis and ultimately the death of dopaminergic neurons (55, 56). Our data demonstrated that ZNF746 directly repressed NRF2 signaling, leading to the production of ROS and cell death. Therefore, we tested whether ZNF746 governs NRF2 transcriptional activity and biology in SH-SY5Y cells, a commonly used neuronal cell line model for Parkinson’s disease (83). Using OIS-PRM analysis to report protein-level quantitation of NRF2 and its target genes, we found that doxycycline-induced ZNF746 significantly repressed CDDOme induction of NRF2 targets compared to ZNF746C571A (Fig. 7A–G, S8A–I; Data File S3). A modest decrease in NRF2 protein levels was observed in SH-SY5Y cells (Fig. 7B). Some of the top repressed proteins included NQO1 and SRXN1, which agrees with the H1299 dataset (Fig. 7C, 7D). However, SLC7A11 protein abundance was significantly decreased in SH-SY5Y cells which remained unchanged in H1299 cells (Fig. 7E). Collectively, nine CDDOme-activated proteins were repressed by ZNF746 in both SH-SY5Y and H1299 cells (Fig. 7G). The proteins unique to each cell line were largely driven by cell line specific activation of target genes by CDDOme (Fig. 7G). Two immune cell markers, NR2F6 and PD-L2, were induced by ZNF746 in H1299 cells (Fig. 5E, S6H, S6I). In SH-SY5Y cells, ZNF746 also increased NR2F6 protein levels and co-treatment with CDDOme suppressed this induction (fig. S8H). However, the expression of PD-L2 remained unchanged in SH-SY5Y cells following ZNF746 induction (fig. S8I). Western blot analysis confirmed the reduction of NRF2 and its targets NQO1 and SLC7A11 (Fig. 7H, S5C, S8J). In summary, ZNF746 repressed the activation of NRF2 target genes in neuronal SH-SY5Y cells.
Fig. 7. ZNF746 promotes elevated ROS and cell death in a neuronal cell model.
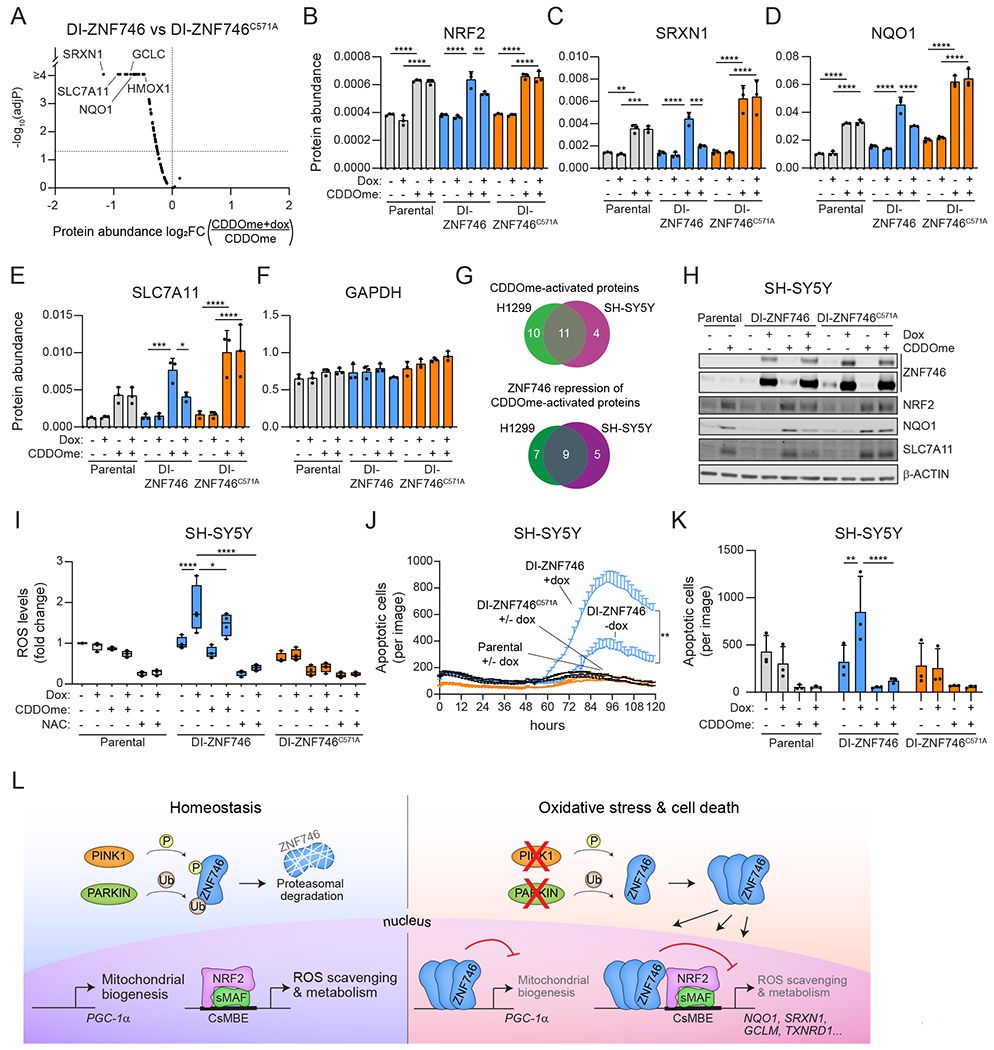
(A) SH-SY5Y cells stably expressing doxycycline-inducible (DI) ZNF746 were co-treated with 2 μg/mL doxycycline and 100 nM CDDOme for 24 h. Protein abundance was quantified using targeted mass spectrometry against a library of defined NRF2 targets and controls. Volcano plot depicting the difference in the log2FC protein abundance (CDDOme+dox over CDDOme) and adjP-value for DI-ZNF746 compared to DI-ZNF746C571A. Significance was calculated using two-way ANOVA and Tukey’s test for multiple comparisons (n=3 biological replicates per group). (B-F) Protein abundance of select targets from ‘A’. Data are presented as means ± SD. *, adjP<0.05; **, adjP<0.01; ***, adjP<0.001; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group). (G) Venn diagram of the overlap of proteins significantly repressed by ZNF746 between the H1299 and SH-SY5Y OIS-PRM datasets. (H) SH-SY5Y cells stably expressing DI-ZNF746 were co-treated with 2 μg/mL doxycycline and 100 nM CDDOme for 24 h before analysis by Western blot. Representative blot from five biological replicates per group. (I) SH-SY5Y cells stably expressing DI-ZNF746 were co-treated with 2 μg/mL dox and 100 nM CDDOme or 3 mM NAC for 48 h. ROS was quantified using CellROX. Data are presented as a boxplot with whiskers indicating the minimum and maximum values. *, adjP<0.05; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=4 biological replicates per group). (J) SH-SY5Y cells stably expressing DI-ZNF746 were treated with 2 μg/mL doxycycline and cell death was quantified using Cytotox-Red. Cells were treated with 10μM menadione after 48 hours. The time course shown is representative of n=4 biological replicates and the statistics were calculated from n=4 biological replicates per group at 96 h time point. Data are presented as means ± SD. **, adjP<0.01 by two-way ANOVA and Tukey test for multiple comparisons. (K) SH-SY5Y cells stably expressing DI-ZNF746 were co-treated with 2 μg/mL doxycycline and 100 nM CDDOme and cell death was quantified using Cytotox-Red. The quantified data are presented as means ± SD. **, adjP<0.01; ****, adjP<0.0001 by two-way ANOVA and Tukey test for multiple comparisons (n=3 biological replicates per group). (L) Proposed model in which elevated ZNF746 protein levels repress expression of both PGC-1α and NRF2 targets leading to dysfunction of mitochondrial biogenesis and redox homeostasis. Inhibition of both pathways ultimately leads to cell death and together may act as coordinated targets of ZNF746/PARIS to promote Parkinson’s disease. See also Figures S5, S8, and S9.
Last, we examined the impact of ZNF746 on oxidative stress and cell death in SH-SY5Y cells. ZNF746 expression increased the levels of ROS, and stabilization of NRF2 by CDDOme treatment dampened ROS accumulation (Fig. 7I). Cell death also significantly increased with ZNF746 induction in the presence of a sub-phenotypic dose of menadione, a small molecule that increases ROS (Fig. 7J, S9). In the absence of menadione, ZNF746 did not affect cell death. CDDOme-mediated activation of NRF2 significantly reduced the number of apoptotic cells following ZNF746 induction (Fig. 7K). Together, these data support a model wherein elevated levels of ZNF746 directly represses NRF2 target gene expression, leading to an increase in ROS and cell death (Fig. 7L). As such, we speculate that ZNF746 stabilization in PINK1 or PARKIN mutant neurons drives mitochondrial dysfunction, oxidative stress, and cell death through the concerted transcriptional suppression of PGC-1α and NRF2 target genes.
DISCUSSION
In general terms, NRF2 transcriptional activity is increased in cancer and suppressed during aging and in neurodegenerative disease. The goal of this study was to identify uncharacterized regulators of NRF2 to better understand disease mechanisms and highlight new therapeutic opportunities. We present a functionally annotated NRF2 proximity interactome with comparative analyses to NRF1, NRF3, MAFG and ZNF746 proximity networks. We found that ZNF746 co-complexed with NRF2 and MAFG on chromatin, resulting in the suppression of NRF2-driven gene transcription and subsequent oxidative stress and apoptosis. ZNF746/PARIS is a PINK1 and PARKIN substrate that is overexpressed in Parkinson’s disease. Based on our data, we posit that restructuring of the chromatin-bound NRF2 protein interaction network is one mechanism that contributes to NRF2 suppression in neurodegenerative disease (Fig. 7L).
Parkinson’s disease is characterized at the cellular level by oxidative stress, mitochondrial dysfunction, neuroinflammation, and accumulation of protein aggregates. The NRF2 transcriptional program functions to mitigate many of these stressors, and indeed NRF2 activation generally suppresses the molecular and pathophysiological hallmarks of Parkinson’s disease in several model systems (42, 43). Multiple mechanisms have been proposed to explain the observed decrease in NRF2 activity during organismal aging and in neurodegeneration, including Parkinson’s disease (38). Our data from H1299 lung cancer cells and SHSY-5Y neuroblastoma cells demonstrates that NRF2 transcriptional activity is suppressed by ZNF746. An important limitation of this study is that it remains unknown whether ZNF746 directly suppresses NRF2 signaling in physiologically relevant Parkinson’s models, such as iPS-derived dopaminergic neurons, mouse models, or Drosophila (84). Related to this, it is interesting that siRNA and CRISPR-mediated knockout of ZNF746 did not activate NRF2-target gene expression (Fig. 3A–D, S4A, S4C). Whether experimental model systems that express high levels of ZNF746, such as PINK1 or PARKIN mutant neurons, display a ZNF746 loss-of-function impact on NRF2 remains to be tested. We hypothesize that suppression of NRF2 by ZNF746 will exacerbate molecular events associated with Parkinson’s disease onset and progression in these models (Fig. 7L).
The underlying molecular mechanism of ZNF746/PARIS in Parkinson’s disease has been investigated by the Dawson laboratory (55, 56, 72, 85–87). ZNF746 protein accumulates in cells lacking PARKIN or PINK1, resulting in loss of PGC-1α expression and subsequent decrease in mitochondrial biogenesis, leading to the progressive loss of dopaminergic neurons. ZNF746 mRNA levels are also increased in sporadic forms of Parkinson’s disease (88). Our data offer an alternative, PGC-1α-independent mechanism of action for ZNF746. Although we did not observe altered PGC-1α expression following ZNF746 gain-of-function or loss-of-function, it is possible that ZNF746 suppression of both PGC-1α and NRF2 contributes to Parkinson’s disease pathogenesis (Fig. 7L). Continued studies that disentangle NRF2 from PGC-1α are needed. For example, although our data showed that chemical and genetic activation of NRF2 rescued ZNF746-driven ROS and cell death, they do not prove that repression of NRF2 signaling is necessary for these ZNF746-induced phenotypes. We speculate that point mutations localized to the ZNF746:NRF2 binding interface will relieve ZNF746-driven NRF2 suppression without globally impacting ZNF746 targets, such as PGC-1α. There are additional complexities to consider. PGC-1α coordinates expression of antioxidant genes such as GCLC and SOD2 and may directly regulate NRF2 expression (89–93). In a putative signaling loop, the PGC-1α promoter contains CsMBE binding sites, although whether NRF2 binds and regulates PGC-1α has not been reported (94). Furthermore, the PINK1 promoter contains functional CsMBEs that are bound and transcriptionally regulated by NRF2 (94).
To investigate the global impact of ZNF746 on gene expression, the Dawson laboratory profiled ZNF746 binding sites in SH-SY5Y cells and performed gene set enrichment analysis on the top 600 ChIP-seq peaks found within promoters (95). The NRF2 oxidative stress response pathway was the third ranked pathway using the BioCarta Metabolic Pathway Analysis algorithm. Although these data do not explicitly show that ZNF746 binds to CsMBEs, they indicate that ZNF746 binds the promoter regions of NRF2 target genes, supporting our study. Of these 600 peaks, 21 genes overlapped with our OIS-PRM library, 13 of which were altered by ZNF746 expression, including GCLM, GCLC, CBR3, and PRDX1 (Fig. 4E–F, 7A, S8A, Data File S3) (95). Together, these data suggest that ZNF746 binds the promoters of NRF2 target genes and inhibits their activation.
Within the same ZNF746 ChIP-seq dataset, Yazar et al. found a ZNF746 binding site in the NRF2 promoter, suggesting direct regulation of NRF2 expression (95). However, we observed that ZNF746 loss did not alter NRF2 protein abundance in the lung cell lines tested (Fig. 3A–D, S4A, C). ZNF746 overexpression slightly decreased NRF2 levels in SH-SY5Y cells but did not impact NRF2 levels in H1299 cells (Fig. 7B, 7H, S8J and Fig. 4C, 4D, 4G). Whether the modest decrease in NRF2 protein levels impacts SH-SY5Y biology remains to be determined. Conversely, increased ZNF746 mRNA has been reported in two NRF2 transcriptional profiling studies (77, 78). If NRF2 directly binds and activates the ZNF746 promoter, these data would suggest a negative feedback loop for ZNF746 and NRF2. However, in our study, ZNF746 expression was unchanged by NRF2 silencing or activation in all tested cell lines, except for in H460 cells (Fig. 3D). Undoubtedly, yet-to-be-defined contextual features may contribute to ZNF746-NRF2 co-regulation.
In addition to our work, several prior studies have reported NRF2 interaction networks. First, integrating public data with predictions of protein domain-domain interactions nominated 125 NRF2 interacting proteins (96). Second, a focused yeast 2-hybrid screen of 187 proteins with follow-up validation revealed 27 NRF2 interactions (97). Third, FLAG epitope-based immunopurified NRF2 protein complexes have been analyzed by MS, though unabridged data have not been reported (98, 99). From these prior studies, we observed only seven shared discoveries (KEAP1, MAFG, MAFK, EP300, CBP, BACH2 and TRIM33). The lack of concordance is likely due to differences in technology (yeast two-hybrid and immunoprecipitation compared to proximity labeling) and cell models (yeast compared to mammalian cell lines). Overlap between immunoprecipitation and proximity labeling is often low, given fundamental differences in live-cell proximity relationships and the stability of protein-protein interactions during detergent extraction and in post-lysis washing (100).
Our functionally annotated proximity network offers new insights in NRF2 regulation. Knockdown of EP300, an activator of NRF2, or NCOA1, a protein with histone acetyltransferase activity, increased NRF2 activity (Fig. 2D) (67). Conversely, loss of the histone acetyltransferase KAT14 reduced NRF2 activity. BACH1 is an inhibitor of NRF2-dependent target gene expression, such as HMOX1; however, limited studies have investigated BACH2 (66). Our data showed that BACH2 may function similarly to BACH1 and repress NRF2 signaling (Fig. 2D). PRR12 and TAF6L are poorly understood and appear necessary for NRF2 reporter activation. Genomic alterations to PRR12 results in complications to neurodevelopment and TAF6L belongs to the family of GNAT histone acetyltransferases that activate gene transcription (101, 102). Collectively, we view these data as a valuable resource to understand NRF2 regulation. The expanded networks for MAFG, NRF1 and NRF3 may similarly empower future research.
MATERIALS AND METHODS
Cell Culture
Cell lines were maintained in a humidified incubator at 37°C with 5% CO2. Cell line identities were validated by short tandem repeat analysis (LabCorp, Genetica Cell Line Testing) and cultures were regularly tested for mycoplasma contamination using the MycoAlert mycoplasma detection kit (Lonza). HEK293T cells (ATCC CRL-3216; homo sapiens) were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. H1299 (ATCC CRL-5803; homo sapiens), H460 (ATCC HTB-177; homo sapiens), and A549 (ATCC CCL-185; home sapiens) cells were cultured in RPMI-1640 supplemented with 10% FBS and 1% penicillin–streptomycin. The H1299 NQO1-eYFP reporter cells (170407PL1A2-NQO1, homo sapiens) contain eYFP engineered into intron 1 the NQO1 gene and were a kind gift from Uri Alon and the Kahn Protein Dynamics group (71). SH-SY5Y cells (RRID:CVCL_0019, homo sapiens) were a kind gift from Jason Yi (WUSTL) and cultured in DMEM:F12 supplemented with 10% FBS and 1% penicillin–streptomycin.
Cloning
The miniTurbo constructs were assembled by Gateway multisite cloning as previously described (103). Briefly, ORFs were cloned into entry vectors using the primers listed in Data File S5. The miniTurbo constructs were obtained from Addgene (#107170, 107172, and 107174) (57). UBC promoter driven miniTurbo tagged proteins at the N- or C-terminus were generated utilizing the Protein Expression Laboratory-Combinatorial cloning platform (PEL-CCP) (a kind gift from Dominic Esposito (Frederick National Laboratory for Cancer Research)). The NRF2E79Q and ZNF746C571A point mutations were generated using PCR-based mutagenesis followed by Gibson cloning. The pDONR221 backbone was digested with BsrGI and the point mutations were amplified using the primer sequences in Data File S5. The backbone and PCR products were ligated together using Gibson Assembly Master Mix (NEB). sgRNAs for CRISPR knockout were cloned into pLenti-DECKO-Cas9 backbone (a kind gift from Sebla Kutluay (WUSTL)). Forward and reverse oligos containing the sgRNA and Gibson homology sequences (Data File S5) were annealed together and ligated into BsmBI-digested pLenti-DECKO-Cas9 using Gibson Assembly Master Mix. ZNF746 and ZNF746C571A were cloned into the doxycycline-inducible vector pCW57.1 (Addgene # 41393) by Gateway cloning. Plasmids generated were validated by Sanger sequencing and/or next-gen sequencing.
Lentivirus production and transduction
Recombinant lentivirus was produced in HEK293T cells using polyethylenimine (PEI) combined with a vector containing the gene of interest, psPAX2 packaging (Addgene #12260) and VSV-G envelope plasmids (Addgene #12259). Recipient cells were transduced for 24 h with 8 μg/ml polybrene and cultured in media containing the appropriate antibiotic to select for cells expressing the gene of interest.
Immunoblotting
Cell lines were grown to 70-80% confluence and lysed in RIPA (10% glycerol, 50mM Tris-HCl pH 7.4, 150mM NaCl, 2mM EDTA, 0.1% SDS, 1% NP40, 0.2% sodium deoxycholate) containing protease and phosphatase inhibitors (Thermo Scientific) and benzonase (Santa Cruz). Protein concentrations were determined using the BCA Protein Assay Kit. Equal amounts of protein were loaded onto Bis-Tris gels and separated by SDS-PAGE using MOPS running buffer. Separated proteins were transferred to nitrocellulose membranes, blocked in 5% milk, and incubated with primary antibodies overnight at 4°C. Washed membranes were incubated for 45 min at room temperature in secondary antibody solution (LI-COR IRDye 680, 800; 1:10,000 in 5% milk), imaged on an Odyssey® CLx and analyzed using Image Studio Software. Antibodies were used at the following dilutions: β-actin (MilliporeSigma #A5316, RRID: AB_476743, 1:5000), ARID1A (Cell Signaling Technology #12354, RRID: AB_2637010, 1:1000), BACH1 (Bethyl #A303-057A, RRID: AB_10890550, 1:2000), BACH2 (Bethyl #A305-502A, RRID: AB_2891429, 1:2000), CREBBP (Cell Signaling Technology #7389, AB_2616020, 1:1000), GCLC (Abcam #ab190685, RRID: AB_2889925, 1:1000), HMOX1 (Abcam #ab13243, AB_299790, 1:1000), KEAP1 (Protein Tech #10503-2-AP, RRID: AB_2132625, 1:500), MAFG (Abcam #ab154318, RRID: AB_2934223, 1:1000), MAFK (Abcam #ab50322, RRID: AB_881008, 1:1000), MED1 (Bethyl #A700-037, RRID: AB_2891837, 1:1000), NCOA2 (Bethyl #A300-346A, RRID: AB_185562, 1:2000), NRF2 (Cell Signaling Technology #20733, RRID: AB_2934224, 1:1000), NQO1 (Novus #NB200-209, RRID: AB_10002706, 1:1000), SLC7A11 (Cell Signaling Technology #12691, RRID: AB_2687474, 1:1000), TRIM24 (Bethyl #A300-815A, RRID: AB_577246, 1:1000), V5 (Invitrogen #R960-25, RRID: AB_2556564, 1:2000), ZNF609 (MilliporeSigma #HPA040742, RRID: AB_10963581, 1:1000), ZNF746 (Protein Tech #24543-1-AP, RRID: AB_2879598, 1:1000), ZNF777 (Bethyl #A301-881A, RRID: AB_1309807, 1:1000). To detect biotinylated proteins, blots were incubated with IRDye 680LT Streptavidin (LI-COR #926-68031) for 45 min at room temperature.
Affinity purification of biotinylated proteins
HEK293T cells stably expressing miniTurbo-tagged proteins were treated with 50 μM biotin for 1 h. Cells were then lysed in RIPA containing protease and phosphatase inhibitors and benzonase. Protein concentrations were determined using the BCA Protein Assay Kit and equal amounts of lysates (5-10 mg for Western blotting analysis and 19 mg for MS analysis) were incubated with 30 μL of packed streptavidin beads (Cytiva) overnight at 4°C on a rotator. The beads were washed 2x with WB1 (2% SDS), 1x with WB2 (50 mM HEPES pH 8.0, 500 mM NaCl, 1 mM EDTA, 1.0% Triton-X, and 0.1% sodium deoxycholate), 1x with WB3 (50 mM HEPES pH 8.0, 250 mM NaCl, 1 mM EDTA, 0.5% NP40, and 0.5% sodium deoxycholate), 1x with WB4 (50 mM HEPES pH 8.0, 50 mM NaCl), and 3x with 50 mM ammonium bicarbonate. Purified biotinylated proteins were either processed for mass spectrometry (see below) or eluted for Western blot by incubating the beads in 2x LDS buffer (Invitrogen) containing 10% β-mercaptoethanol and 1 mM biotin at 70°C for 10 min.
Immunoprecipitation analysis
HEK293T cells were transfected with FLAG-tagged constructs for 24 h and lysed with RIPA buffer. Lysates were incubated with FLAG conjugated beads (MilliporeSigma #F2426) overnight at 4°C. Beads were washed 3x with RIPA buffer and eluted in 2x LDS sample buffer and 50mM DTT at 70°C for 10 min. Eluted proteins were analyzed by Western blot.
Mass spectrometry sample preparation, data acquisition, raw data processing, and analysis for miniTurbo-based proximity proteomics
The streptavidin beads containing purified biotinylated proteins were resuspended in 0.1% Rapigest (Waters), reduced with 5 mM DTT at 60°C for 30 min, alkylated with 50 mM chloroacetamide at room temp for 20 min, and on-bead digested with 2.5 μg of trypsin (Promega) at 37°C for 18 h. Trypsin was neutralized with 50 mM HCl incubated at 37°C for 45 min. Samples were centrifuged at 400×g for 2 min to remove the beads and then at 21,000×g for 5 min to remove insoluble cleaved Rapigest. The peptides were dried in a speed vac and resuspended in 0.5% TFA. The samples were de-salted using C18 spin columns (Pierce) and residual detergent was removed using ethyl acetate. Peptides were dried in a speed vac and resuspended in 2% acetonitrile, 0.1% formic acid.
Trypsinized peptides were loaded onto a μPAC™ Trapping column (PharmaFluidics) and separated on μPAC™ columns (PharmaFluidics). The miniTurbo-tagged NRF1/2/3 samples were separated on a 200 cm column operated at 30°C using a 100 min gradient from 2% to 25% buffer B, which was followed by a 20 min gradient from 25% to 35% buffer B, flowing at 300 nL/min. The miniTurbo-tagged ZNF746 samples were separated on a 50 cm column operated at 30°C using a 4 min gradient from 2% to 7% buffer B flowing at 1.0 μL/min, then a 106 min gradient from 7% to 25% buffer B flowing at 300 nL/min, and lastly a 5 min gradient from 25% to 30% buffer B flowing at 300 nL/min. Mass spectrometry analysis was performed on an Orbitrap Eclipse (Thermo Fisher Scientific) operated in data-dependent acquisition mode.
The miniTurbo-tagged NRF1/2/3 MS1 scans were acquired in the Orbitrap at 120k resolution, with a 250% normalized automated gain control (AGC) target, auto max injection time, and a 375-1500 m/z scan range. Both the linear ion trap and the Orbitrap were used for the miniTurbo-tagged NRF1/2/3 MS2 scans. MS2 targets with a charge of +2 or +3, and ≥90% precursor fit at either a 0.8 m/z or 0.4 m/z-wide isolation window were fragmented by collision induced dissociation (CID) with a 35% normalized collision energy and scanned in the linear ion trap at the widest window width that passed the thresholds. Remaining MS2 targets with charge from +2 to +6, ≥5e4 intensity and ≥10% precursor fit at isolation widths of 1.6 m/z, 0.8 m/z, or 0.4 m/z wide were fragmented with higher-energy collision dissociation at 30% normalized collision energy and scanned in the Orbitrap at 15k resolution at the widest window width that passed the thresholds. MS2 AGC targets and maximum injection times were set to standard and auto for their respective analyzers. Dynamic exclusion was set to 60 seconds. Acquisition was performed with a 2.7 second cycle time.
The miniTurbo-tagged ZNF746 MS scan methods were as follows. The MS1 scans were acquired in the Orbitrap at 240k resolution, with a 250% normalized automated gain control (AGC) target, auto max injection time, and a 375-2000 m/z scan range. MS2 targets were filtered for charges states 2-7, with a dynamic exclusion of 60 seconds, and were accumulated using a 0.7 m/z quadrupole isolation window. MS2 scans were performed in the ion trap at a turbo scan rate following CID with a 35% normalized collision energy. MS2 scans used a 100% normalized AGC target and 35 ms max injection time.
Raw MS data files were processed by MaxQuant (version 2.0.1.0) using the UniProtKB/SwissProt human canonical sequence database (downloaded March 2021) (104). The following parameters were used: specific tryptic digestion with up to two missed cleavages, fixed carbamidomethyl modification, variable modifications for protein N-terminal acetylation, methionine oxidation and biotin, match between runs, and label-free quantification. Unique and razor peptides were used for protein quantification. Each bait was given a replicate number >1 away from all other baits in ensure match between runs occurred only for replicates of the same bait. The confidence of bait-prey protein interactions compared to the controls (V5, NLS, NES) were scored using SAINTexpress (v3.6.3) with an L-value of 3 for NRF1, 2, and 3 dataset and an L-value of 2 for the ZNF746 and MAFG dataset (58). Proteins that passed the following bait-specific thresholds were included in downstream analysis. NRF2 (SAINT≥0.42, BFDR ≤0.28), NRF1 (SAINT≥0.44, BFDR ≤0.26), NRF3 (SAINT≥0.44, BFDR ≤0.26), ZNF746 (SAINT≥0.7, BFDR ≤0.05), and MAFG (SAINT≥0.7, BFDR ≤0.05). Common BioID contaminants deposited in the BioID CRAPome database were filtered out of the dataset (105). Multi-validated prey-prey interactions from at least two separate experiments deposited in the BioGRID database were included (59). Protein networks were generated in Cytoscape (v 3.9.1) (106). Specifically for the MAFG localization network, an additional threshold of average spectral counts ≥20 was included for visualization purposes (Fig. 1C).
To determine the network similarities between NRF2, ZNF746, and MAFG, we performed Jaccard index-based overlap analysis against a published proximity proteomic dataset containing 102 transcription factors (82). Lists of interacting proteins were generated for NRF2, ZNF746, and MAFG as described above and SAINT>0.7 for the reference dataset. For simplicity, protein groups of more than one gene were reduced to include only the first gene in the list. The Jaccard index was calculated based on the lists of interactions and P-values representing the similarity of baits were determined using a hypergeometric test.
Mass spectrometry sample preparation, data acquisition, and raw data processing and analysis for OIS-PRM for NRF2 targets
Cells were lysed in urea lysis buffer (8 M urea, 1 mM EDTA, 50 mM Tris pH 8.0, and 70 mM NaCl) containing protease and phosphatase inhibitors and benzonase. Protein concentrations were determined using the BCA Protein Assay Kit for equal protein loading. Samples were reduced with 5 mM DTT at 37°C for 45 min and alkylated with 50 mM chloroacetamide at room temp for 20 min. Lysates were first digested with 20 mAu LysC at 30°C for 2 h and then with 20 μg of trypsin (Promega) per 1 mg of protein at 37°C for 18 h. Lysates were diluted to 3 M urea prior to LysC digestion and 1.5 M urea prior to trypsin digestion with 50 mM Tris pH 8.0. Trypsin was neutralized with 1% formic acid and high spin cleared. The peptides were de-salted using Strata-X 10 mg/mL columns (Phenomenex), dried in a speed vac, and resuspended in 2% acetonitrile, 0.1% formic acid. Endogenous peptides were injected at 1 ug/uL with 150 fmol stable isotopically labeled (SIL) peptides per injection. In sum, 218 SIL peptides were obtained in array-purity from Vivitide and were synthesized with 15N and 13C-labeled lysine and arginine residues.
Tryptic peptides were separated by reverse phase nano-HPLC using an Ultimate 3000 RSLCnano System (Thermo Fisher) coupled to a 25 cm x 75 um i.d. EASY-Spray™ HPLC column (Thermo Scientific) packed with 2 μm C18 particles and heated to 40°C. For peptide separation and elution, solvent A was 0.1% FA in water and Solvent B was 0.1% FA in ACN. Samples were loaded by a user defined program for a 1 μL full-loop injection. The gradient was 4% B at 1 min, 15.4% B at 31 min, 24.5% B at 46 min, and 98% B at 48 min. Each method included a wash step with three ramps between 2% and 98% solvent B, which was followed by 24 minutes of re-equilibration at 2% B and 300 nL/min flow. During the gradients, the flow was 250 nL/min.
Internal standard triggered-parallel reaction monitoring (IS-PRM) methods leverage SIL internal standard peptides to direct efficient acquisition of endogenous, unlabeled peptides (107). We implemented a customized IS-PRM algorithm through the Thermo Scientific™ Tribrid™ IAPI (80, 108). Once per cycle, the method looks for MS1 detection of any features that match the expected m/z of one of the SIL peptides to within 5 ppm. MS1 detection at sufficient intensity triggers a subsequent “watch” scan. Detection of at least five of six characteristic transitions to within 20 ppm confirms the presence of a SIL peptide. Confirmation triggers a high-resolution “quant” scan targeting the endogenous counterpart.
MS data were generated using an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Scientific). For all IS-PRM runs, Orbitrap MS1 scans used an AGC target of 1.2e6, resolution of 120K at 200 m/z, and a maximum injection time of 50 ms. Fragmentation was carried out by HCD with a 30% normalized collision energy and a precursor isolation width of 1 Th. MS2 watch scans were carried out with an AGC target of 5e5, maximum injection time (maxIT) of 11ms, and Orbitrap resolution of 7.5k. We specified an AGC target of 5e5, a maxIT of 246 ms, and an Orbitrap resolution at 120k for all MS2 quant scans.
IS-PRM identification was based on the six most abundant transitions for each peptide excluding precursor, y1, y2, and b1 ions within a mass tolerance window of 40ppm for the H1299 dataset and 20ppm for the SH-SY5Y dataset. For each run, an active IAPI instance generated a text file consisting of all centroided MS2 scans with their respective retention time and precursor m/z annotations. Custom python scripts were used to analyze the data as follows. The three most abundant transitions were used for quantification, but spectral angle contrast between light and heavy transition areas was used to exclude noisy or interfered transitions from consideration. Peak area ratios obtained by IS-PRM were normalized by global extraction from PRM (GXPRM) as previously proposed (109). Briefly, the intensities of commonly identified peptides that were co-isolated with the targeted peptides are used to derive a multiplicative normalization factor for each sample. Peptide abundances were summarized to the protein level by their geometric mean.
Insufficient or inadequate quantification of endogenous peptides resulted in missing values at the protein level (n=3 biological replicates; Data File S3). The missing values were imputed with the lowest observed log2 protein abundance for each protein across the H1299 and SH-SY5Y datasets. The imputed data was then used for all downstream analyses. Proteins that contained missing values across the majority of samples and replicates were not included in downstream analyses or plotted.
siRNA transfections
The sequences and catalog numbers for each siRNA are listed in Data File S5. 2nM of siRNAs for each target was complexed with RNAiMax (Invitrogen) in Opti-MEM following the manufacturer’s recommendations. Cells were collected for Western blot analysis 72 h after siRNA transfections.
Arrayed siRNA screen
H1299 NQO1-eYFP cells were seeded in a 48-well plate with two wells per siRNA on each plate across biological triplicate or quadruplicate (Data File S2). The next day, the cells were transfected with the indicated siRNA in a pooled format (3 siRNAs per well; Data File S5). 24 h after transfection, DMSO or 50 nM CDDOme was added and then imaged using the Incucyte® S3 Life Cell Analysis System. Phase and fluorescent images were taken every 2 h for 4 days. The H1299 NQO1-eYFP cells stably express constitutive nuclear mCherry and eYFP under control of the endogenous NQO1 promoter. Normalized NQO1-eYFP intensity was calculated by dividing eYFP intensity by red objects (number of cells). The average of the duplicate wells was taken for each biological replicate.
The nonspecific siRNA controls (NS-1 and NS-2) were included on each plate to control for batch effects. The siRNAs targeting the canonical NRF2 pathway (NRF2, KEAP1, and MAFG) were included on the majority of the plates to confirm NQO1-eYFP response. The fold change was calculated by dividing each target by the average of the NS-1 and NS-2 normalized eYFP intensity value from the same plate (Data File S2).
CRISPR/Cas9 gene editing
H1299 cells were transduced with pLenti-Cas9, which co-expresses Cas9 and the sgRNA (GCTCTACAAGCACGTGATG) that targets exon 3 of ZNF746 (CCDS55180). After transduction, cells were selected in 1 μg/mL puromycin. Monoclonal populations were generated for both non-specific and ZNF746 sgRNAs by single cell dilutions in 96-well plates. Genomic KO was confirmed by Sanger sequencing using ZNF746_screen-B-F and ZNF746_screen-B-R (Data File S5).
ROS quantification
ROS levels were measured using CellROX™ Green Reagent (Invitrogen #C10444) following the manufacturer’s recommendations. Briefly, cells were seeded in duplicate wells per biological replicate and treated with doxycycline for 48 to 72 h. 5 μM CellROX was added to each well and incubated at 37°C for 1 h. Cells were washed with PBS and imaged using the Incucyte® S3 Life Cell Analysis System. Total green intensity was normalized to cell confluence and duplicate wells were averaged together for each biological replicate. SH-SY5Y cells were treated with 10 μM menadione for 6 h prior to addition of CellROX. For rescue experiments, cells were co-treated with doxycycline and CDDOme or NAC.
Cell death quantification
Cell death was measured using Incucyte® Cytotox Red Dye (Sartorius #4632) following the manufacturer’s recommendations. Briefly, cells were seeded in duplicate wells per biological replicate. The following day, cells were changed to media containing doxycycline and 250 nM Cytotox Red Dye. Cells were then imaged every 2 h using the Incucyte® S3 Life Cell Analysis System. The total number of red cells was counted for each image and duplicate wells were averaged together for each biological replicate. SH-SY5Y cells were treated with 10 μM menadione 48 h after the addition of doxycycline/Cyotox Red and immediately placed back in the Incucyte device for imaging. For rescue experiments, cells were co-treated with doxycycline and Cytotox Red and PRL-295 or tBHQ.
Quantification and Statistical Analysis
Statistical parameters and the number of biological replicates for each experiment are reported in the respective figure legends with a minimum of three biological replicates used. Each plot includes points for individual biological replicates and mean ± SD error bars unless otherwise specified. GraphPad Prism 9 software was used for statistical analysis. One-way or two-way ANOVA was performed to assess significance and post hoc comparisons were employed to correct for multiple comparisons with the specific test indicated in the figure legend. For visual clarity, we only showed pertinent statistical comparisons in the graphs.
Supplementary Material
Acknowledgments:
We thank Dr. Kobayashi (Doshisha University) for sharing the NRF3 plasmid and Dr. Terry Moore (University of Illinois at Chicago) for sharing the PRL-295 compound.
Funding:
National Institute of Health grant R01CA244236 (MBM)
T32 training grant T32CA009547-34 (KML)
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE (110) partner repository with the dataset identifiers PXD043182 and PXD043248 and are publicly available as of the date of publication. This paper does not contain original code. Any additional information or unique reagents required to reanalyze the data reported in this paper is available from M.B.M. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes:
- 1.Schieber M, Chandel NS, ROS function in redox signaling and oxidative stress. Curr Biol 24, R453–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sies H, Berndt C, Jones DP, Oxidative Stress. Annu Rev Biochem 86, 715–748 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Hayes JD, Dinkova-Kostova AT, Tew KD, Oxidative Stress in Cancer. Cancer Cell 38, 167–197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sies H, Jones DP, Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat Rev Mol Cell Biol 21, 363–383 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Zhu M, Fahl WE, Functional characterization of transcription regulators that interact with the electrophile response element. Biochem Biophys Res Commun 289, 212–219 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Rojo de la Vega M, Chapman E, Zhang DD, NRF2 and the Hallmarks of Cancer. Cancer Cell 34, 21–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi A et al. , Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24, 7130–7139 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rada P et al. , SCF/beta-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31, 1121–1133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh K, Mimura J, Yamamoto M, Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal 13, 1665–1678 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Cloer EW, Goldfarb D, Schrank TP, Weissman BE, Major MB, NRF2 Activation in Cancer: From DNA to Protein. Cancer Res 79, 889–898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong KI et al. , Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol 27, 7511–7521 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hast BE et al. , Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res 74, 808–817 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloer EW et al. , p62-Dependent Phase Separation of Patient-Derived KEAP1 Mutations and NRF2. Mol Cell Biol 38, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baird L, Yamamoto M, The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol Cell Biol 40, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh K et al. , An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236, 313–322 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T, Takahashi J, Yamamoto M, Molecular Basis of the KEAP1-NRF2 Signaling Pathway. Mol Cells 46, 133–141 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radhakrishnan SK et al. , Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38, 17–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radhakrishnan SK, den Besten W, Deshaies RJ, p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. Elife 3, e01856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sha Z, Goldberg AL, Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol 24, 1573–1583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Lucocq JM, Yamamoto M, Hayes JD, The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. Biochem J 408, 161–172 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koizumi S et al. , The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waku T et al. , NFE2L1 and NFE2L3 Complementarily Maintain Basal Proteasome Activity in Cancer Cells through CPEB3-Mediated Translational Repression. Mol Cell Biol 40, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi A, Waku T, New addiction to the NRF2-related factor NRF3 in cancer cells: Ubiquitin-independent proteolysis through the 20S proteasome. Cancer Sci 111, 6–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chowdhury A et al. , Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci Rep 7, 12494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H et al. , Elevated expression of NFE2L3 predicts the poor prognosis of pancreatic cancer patients. Cell Cycle 17, 2164–2174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang XJ et al. , Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29, 1235–1243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torrente L, DeNicola GM, Targeting NRF2 and Its Downstream Processes: Opportunities and Challenges. Annu Rev Pharmacol Toxicol 62, 279–300 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Ramos-Gomez M et al. , Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A 98, 3410–3415 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao S, Rojo de la Vega M, Chapman E, Ooi A, Zhang DD, The effects of NRF2 modulation on the initiation and progression of chemically and genetically induced lung cancer. Mol Carcinog 57, 182–192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer AK et al. , Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PLoS One 6, e26590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeBlasi JM et al. , Distinct Nrf2 Signaling Thresholds Mediate Lung Tumor Initiation and Progression. Cancer Res, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibata T et al. , Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A 105, 13568–13573 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hast BE et al. , Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res 73, 2199–2210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan L et al. , NFE2L2 mutations enhance radioresistance in head and neck cancer by modulating intratumoral myeloid cells. Cancer Res, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pillai R, Hayashi M, Zavitsanou AM, Papagiannakopoulos T, NRF2: KEAPing Tumors Protected. Cancer Discov 12, 625–643 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binkley MS et al. , KEAP1/NFE2L2 Mutations Predict Lung Cancer Radiation Resistance That Can Be Targeted by Glutaminase Inhibition. Cancer Discov 10, 1826–1841 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata T et al. , NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia 13, 864–873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidlin CJ, Dodson MB, Madhavan L, Zhang DD, Redox regulation by NRF2 in aging and disease. Free Radic Biol Med 134, 702–707 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poewe W et al. , Parkinson disease. Nat Rev Dis Primers 3, 17013 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Funayama M, Nishioka K, Li Y, Hattori N, Molecular genetics of Parkinson’s disease: Contributions and global trends. J Hum Genet 68, 125–130 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dias V, Junn E, Mouradian MM, The role of oxidative stress in Parkinson’s disease. J Parkinsons Dis 3, 461–491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bento-Pereira C, Dinkova-Kostova AT, Activation of transcription factor Nrf2 to counteract mitochondrial dysfunction in Parkinson’s disease. Med Res Rev 41, 785–802 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Zgorzynska E, Dziedzic B, Walczewska A, An Overview of the Nrf2/ARE Pathway and Its Role in Neurodegenerative Diseases. Int J Mol Sci 22, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson DA, Johnson JA, Nrf2--a therapeutic target for the treatment of neurodegenerative diseases. Free Radic Biol Med 88, 253–267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi JW et al. , Optimization of Vinyl Sulfone Derivatives as Potent Nuclear Factor Erythroid 2-Related Factor 2 (Nrf2) Activators for Parkinson’s Disease Therapy. J Med Chem 62, 811–830 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Jazwa A et al. , Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid Redox Signal 14, 2347–2360 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Lee JA et al. , Activation of the Nrf2 signaling pathway and neuroprotection of nigral dopaminergic neurons by a novel synthetic compound KMS99220. Neurochem Int 112, 96–107 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Kim S et al. , Nrf2 activator via interference of Nrf2-Keap1 interaction has antioxidant and anti-inflammatory properties in Parkinson’s disease animal model. Neuropharmacology 167, 107989 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Moreira S et al. , Nrf2 activation by tauroursodeoxycholic acid in experimental models of Parkinson’s disease. Exp Neurol 295, 77–87 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Williamson TP, Johnson DA, Johnson JA, Activation of the Nrf2-ARE pathway by siRNA knockdown of Keap1 reduces oxidative stress and provides partial protection from MPTP-mediated neurotoxicity. Neurotoxicology 33, 272–279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahuja M et al. , Distinct Nrf2 Signaling Mechanisms of Fumaric Acid Esters and Their Role in Neuroprotection against 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine-Induced Experimental Parkinson’s-Like Disease. J Neurosci 36, 6332–6351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen PC et al. , Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: Critical role for the astrocyte. Proc Natl Acad Sci U S A 106, 2933–2938 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burton NC, Kensler TW, Guilarte TR, In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology 27, 1094–1100 (2006). [DOI] [PubMed] [Google Scholar]
- 54.A A et al. , alpha-Syn overexpression, NRF2 suppression, and enhanced ferroptosis create a vicious cycle of neuronal loss in Parkinson’s disease. Free Radic Biol Med 192, 130–140 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin JH et al. , PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell 144, 689–702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee Y et al. , PINK1 Primes Parkin-Mediated Ubiquitination of PARIS in Dopaminergic Neuronal Survival. Cell Rep 18, 918–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Branon TC et al. , Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teo G et al. , SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J Proteomics 100, 37–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oughtred R et al. , The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci 30, 187–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.May DG, Scott KL, Campos AR, Roux KJ, Comparative Application of BioID and TurboID for Protein-Proximity Biotinylation. Cells 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim DI et al. , Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A 111, E2453–2461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Crouch DH, Yamamoto M, Hayes JD, Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J 399, 373–385 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsuchiya Y et al. , Dual regulation of the transcriptional activity of Nrf1 by beta-TrCP- and Hrd1-dependent degradation mechanisms. Mol Cell Biol 31, 4500–4512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roboti P, High S, The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J Cell Sci 125, 3474–3484 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Nouhi Z, Chevillard G, Derjuga A, Blank V, Endoplasmic reticulum association and N-linked glycosylation of the human Nrf3 transcription factor. FEBS Lett 581, 5401–5406 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Wiel C et al. , BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell 178, 330–345 e322 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Sun Z, Chin YE, Zhang DD, Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol 29, 2658–2672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganner A et al. , The acetyltransferase p300 regulates NRF2 stability and localization. Biochem Biophys Res Commun 524, 895–902 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Sekine H et al. , The Mediator Subunit MED16 Transduces NRF2-Activating Signals into Antioxidant Gene Expression. Mol Cell Biol 36, 407–420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song S et al. , Loss of SWI/SNF Chromatin Remodeling Alters NRF2 Signaling in Non-Small Cell Lung Carcinoma. Mol Cancer Res 18, 1777–1788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen AA et al. , Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Jo A et al. , PARIS farnesylation prevents neurodegeneration in models of Parkinson’s disease. Sci Transl Med 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bae JH et al. , ZNF746/PARIS overexpression induces cellular senescence through FoxO1/p21 axis activation in myoblasts. Cell Death Dis 11, 359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim B et al. , Inhibition of ZNF746 suppresses invasion and epithelial to mesenchymal transition in H460 non-small cell lung cancer cells. Oncol Rep 31, 73–78 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Jung JH et al. , Zinc finger protein 746 promotes colorectal cancer progression via c-Myc stability mediated by glycogen synthase kinase 3beta and F-box and WD repeat domain-containing 7. Oncogene 37, 3715–3728 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Kim H et al. , ZNF746/PARIS promotes the occurrence of hepatocellular carcinoma. Biochem Biophys Res Commun 563, 98–104 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Levings DC, Wang X, Kohlhase D, Bell DA, Slattery M, A distinct class of antioxidant response elements is consistently activated in tumors with NRF2 mutations. Redox Biol 19, 235–249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okazaki K et al. , Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non-small cell lung cancers. Nat Commun 11, 5911 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lazzara PR et al. , Isoquinoline Kelch-like ECH-Associated Protein 1-Nuclear Factor (Erythroid-Derived 2)-like 2 (KEAP1-NRF2) Inhibitors with High Metabolic Stability. J Med Chem 63, 6547–6560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wamsley NT et al. , Targeted proteomic quantitation of NRF2 signaling and predictive biomarkers in HNSCC. Mol Cell Proteomics, 100647 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Al Chiblak M, Steinbeck F, Thiesen HJ, Lorenz P, DUF3669, a “domain of unknown function” within ZNF746 and ZNF777, oligomerizes and contributes to transcriptional repression. BMC Mol Cell Biol 20, 60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goos H et al. , Human transcription factor protein interaction networks. Nat Commun 13, 766 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xicoy H, Wieringa B, Martens GJ, The SH-SY5Y cell line in Parkinson’s disease research: a systematic review. Mol Neurodegener 12, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cobb MM, Ravisankar A, Skibinski G, Finkbeiner S, iPS cells in the study of PD molecular pathogenesis. Cell Tissue Res 373, 61–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar M et al. , Defects in Mitochondrial Biogenesis Drive Mitochondrial Alterations in PARKIN-Deficient Human Dopamine Neurons. Stem Cell Reports 15, 629–645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pirooznia SK et al. , PARIS induced defects in mitochondrial biogenesis drive dopamine neuron loss under conditions of parkin or PINK1 deficiency. Mol Neurodegener 15, 17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brahmachari S et al. , Parkin interacting substrate zinc finger protein 746 is a pathological mediator in Parkinson’s disease. Brain 142, 2380–2401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alieva A et al. , Potential Biomarkers of the Earliest Clinical Stages of Parkinson’s Disease. Parkinsons Dis 2015, 294396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.St-Pierre J et al. , Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127, 397–408 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Olmos Y et al. , Mutual dependence of Foxo3a and PGC-1alpha in the induction of oxidative stress genes. J Biol Chem 284, 14476–14484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aquilano K et al. , p53 orchestrates the PGC-1alpha-mediated antioxidant response upon mild redox and metabolic imbalance. Antioxid Redox Signal 18, 386–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deng X et al. , The Nrf2/PGC1alpha Pathway Regulates Antioxidant and Proteasomal Activity to Alter Cisplatin Sensitivity in Ovarian Cancer. Oxid Med Cell Longev 2020, 4830418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choi HI et al. , PGC-1alpha attenuates hydrogen peroxide-induced apoptotic cell death by upregulating Nrf-2 via GSK3beta inactivation mediated by activated p38 in HK-2 Cells. Sci Rep 7, 4319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murata H et al. , NRF2 Regulates PINK1 Expression under Oxidative Stress Conditions. PLoS One 10, e0142438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yazar V, Kang SU, Ha S, Dawson VL, Dawson TM, Integrative genome-wide analysis of dopaminergic neuron-specific PARIS expression in Drosophila dissects recognition of multiple PPAR-gamma associated gene regulation. Sci Rep 11, 21500 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Papp D et al. , The NRF2-related interactome and regulome contain multifunctional proteins and fine-tuned autoregulatory loops. FEBS Lett 586, 1795–1802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Poh J et al. , A functionally defined high-density NRF2 interactome reveals new conditional regulators of ARE transactivation. Redox Biol 37, 101686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen D et al. , NRF2 Is a Major Target of ARF in p53-Independent Tumor Suppression. Mol Cell 68, 224–232 e224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meng C et al. , The deubiquitinase USP11 regulates cell proliferation and ferroptotic cell death via stabilization of NRF2 USP11 deubiquitinates and stabilizes NRF2. Oncogene 40, 1706–1720 (2021). [DOI] [PubMed] [Google Scholar]
- 100.Lambert JP, Tucholska M, Go C, Knight JD, Gingras AC, Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics 118, 81–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chowdhury F et al. , Haploinsufficiency of PRR12 causes a spectrum of neurodevelopmental, eye, and multisystem abnormalities. Genet Med 23, 1234–1245 (2021). [DOI] [PubMed] [Google Scholar]
- 102.Seruggia D et al. , TAF5L and TAF6L Maintain Self-Renewal of Embryonic Stem Cells via the MYC Regulatory Network. Mol Cell 74, 1148–1163 e1147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wall VE, Garvey LA, Mehalko JL, Procter LV, Esposito D, Combinatorial assembly of clone libraries using site-specific recombination. Methods Mol Biol 1116, 193–208 (2014). [DOI] [PubMed] [Google Scholar]
- 104.Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Mellacheruvu D et al. , The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10, 730–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shannon P et al. , Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gallien S, Kim SY, Domon B, Large-Scale Targeted Proteomics Using Internal Standard Triggered-Parallel Reaction Monitoring (IS-PRM). Mol Cell Proteomics 14, 1630–1644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wamsley NT et al. , Targeted proteomic quantitation of NRF2 signaling and predictive biomarkers in HNSCC. bioRxiv, 2023.2003.2013.532474 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chambers AG, Sweet SMM, Chain D, Kim YJ, Global Extraction from Parallel Reaction Monitoring to Quantify Background Peptides for Improved Normalization and Quality Control in Targeted Proteomics. Anal Chem 93, 13434–13440 (2021). [DOI] [PubMed] [Google Scholar]
- 110.Perez-Riverol Y et al. , The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE (110) partner repository with the dataset identifiers PXD043182 and PXD043248 and are publicly available as of the date of publication. This paper does not contain original code. Any additional information or unique reagents required to reanalyze the data reported in this paper is available from M.B.M. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.


