ABSTRACT
Macroautophagy/autophagy, is widely recognized for its crucial role in enabling cell survival and maintaining cellular energy homeostasis during starvation or energy stress. Its regulation is intricately linked to cellular energy status. In this review, covering yeast, mammals, and plants, we aim to provide a comprehensive overview of the understanding of the roles and mechanisms of carbon- or glucose-deprivation related autophagy, showing how cells effectively respond to such challenges for survival. Further investigation is needed to determine the specific degraded substrates by autophagy during glucose or energy deprivation and the diverse roles and mechanisms during varying durations of energy starvation.
Abbreviations: ADP: adenosine diphosphate; AMP: adenosine monophosphate; AMPK: AMP-activated protein kinase; ATG: autophagy related; ATP: adenosine triphosphate; ER: endoplasmic reticulum; ESCRT: endosomal sorting complex required for transport; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GD: glucose deprivation; GFP: green fluorescent protein; GTPases: guanosine triphosphatases; HK2: hexokinase 2; K phaffii: Komagataella phaffii; LD: lipid droplet; MAP1LC3/LC3: microtubule-associated protein1 light chain 3; MAPK: mitogen-activated protein kinase; Mec1: mitosis entry checkpoint 1; MTOR: mechanistic target of rapamycin kinase; NAD (+): nicotinamide adenine dinucleotide; OGD: oxygen and glucose deprivation; PAS: phagophore assembly site; PCD: programmed cell death; PtdIns3K: class III phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol-3-phosphate; ROS: reactive oxygen species; S. cerevisiae: Saccharomyces cerevisiae; SIRT1: sirtuin 1; Snf1: sucrose non-fermenting 1; STK11/LKB1: serine/threonine kinase 11; TFEB: transcription factor EB; TORC1: target of rapamycin complex 1; ULK1: unc-51 like kinase 1; Vps27: vacuolar protein sorting 27; Vps4: vacuolar protein sorting 4.
KEYWORDS: AMPK, autophagy, carbon starvation, energy metabolism, glucose starvation, Snf1
Introduction
Nutrient and energy metabolism are fundamental to basic biological and pathological activities and the ability to accommodate conditions where nutrients and energy are deficient is vital for survival. In response to nutrient and energy deprivation, organisms undergo metabolic adaptations by enhancing catabolic processes to generate energy and limiting anabolic processes to conserve energy expenditure [1]. The exploration of energy metabolism is particularly crucial when linked to its association with various diseases, particularly the metabolic reprogramming aspects of cancer or in ischemias. In-depth mechanistic investigations into energy metabolism offer valuable insights into such diseases and may lead to advancements in therapeutic interventions.
Autophagy is a key catabolic process in eukaryotes that occurs in response to metabolic and energy stress. Autophagy enables cells to efficiently break down diverse macromolecules, such as proteins, lipids and glycogen [2], thereby gleaning energy and nutrient resources for cells under starvation conditions [2,3]. Autophagy also plays a role in regulating cellular metabolic status by suppressing anabolism. When yeast cells are under starvation, ribosomal proteins are degraded to halt the energy-consuming process of protein synthesis [4]. In this way, energy deprivation-induced autophagy is widely considered significant as a survival strategy on both a cellular and organismal level [5,6]. Moreover, the failure to properly induce autophagy during glucose deprivation has been associated with poor prognosis in disease conditions such as ischemia and cancer [7,8]. Despite the growing focus on the interplay of autophagy and energy metabolism, there remains a lack of comprehensive reviews that clearly integrate the roles and mechanisms of autophagy triggered by carbon or glucose deprivation.
Here, we discuss recent advances in understanding the interplay between energy metabolism and autophagy. We particularly focus on the effect of carbon or glucose starvation on autophagy and note its physiological and pathological relevance. While our discussion is mainly on the pathways in yeast and mammalian cells, we also briefly discuss key mechanisms in plant cells. We also address unresolved key issues such as whether autophagy selectively targets specific substrates during glucose or energy deprivation. Furthermore, we advocate for rigorous studies to explore the precise mechanisms that directly impact autophagy during energy deprivation.
Autophagy Regulation During Carbon Deprivation
In the early 1960s, Ashford and Porter observed that the treatment of hepatocytes with glucagon increased the number of lysosomes containing cytoplasmic components [9]. These were later recognized as autolysosomes. In 1967, Deter et al., in their experiments on rat livers, then confirmed that autophagy could be induced by glucagon [10]. This observation was not further explored until early 1990s when Ohsumi’s group reported that the depletion of carbon nutrients could induce extensive autophagic degradation in the budding yeast Saccharomyces cerevisiae (S. cerevisiae) [11]. Since then, numerous studies using this yeast have shown that carbon depletion, including glucose deprivation, can induce macroautophagy [12–17], lipophagy (autophagic degradation of lipid droplets) [18,19], mitophagy (autophagic degradation of mitochondria) [20] or reticulophagy (autophagic degradation of the endoplasmic reticulum [ER]) [21]. The induction of autophagy by carbon starvation was also observed in plants, such as rice (Oryza sativa) [22], tobacco (Nicotiana tabacum) [23], Arabidopsis [24,25], and in mammalian cells [26–29]. Glucose deprivation-responsive autophagy shares many common key aspects with nitrogen starvation-induced autophagy, but it also has unique mechanisms.
Yeast can assimilate a wide range of carbon sources. While glucose is the most preferred, other sources such as glycerol, ethanol, acetate, and lactate are also utilized [30,31]. However, in the presence of glucose, the utilization of alternative carbon sources becomes repressed [32]. Based on this observation, several studies have explored the specific carbon starvation conditions that induce autophagy in yeast. The initial study was conducted using a mutant budding yeast strain unable to use glycerol as a carbon source [11]. In this mutant yeast strain, autophagy was induced in a standard growth medium where the only carbon source available was glycerol [11]. Moreover, several studies showed that glucose starvation induces macroautophagy [12,14,16] and lipophagy [33] in wild-type strains of budding yeast, although some studies suggested glucose starvation does not induce autophagy in budding yeast [15,34]. In fission yeast (S. pombe), either partial or complete glucose depletion could trigger autophagy [35].
While much evidence has been gained from studies using yeasts that were subjected to abrupt carbon starvation, this approach provided a somewhat artificial scenario, as changes would more usually occur gradually in natural environments. To address such shortcomings, Iwama and Ohsumi explored the processes of yeasts growing in media where energy sources were more gradually depleted [21]. For glucose-fermenting yeasts, such as S. cerevisiae, the process can be divided into 3 phases: the glucose-utilizing phase, the ethanol-utilizing (or lag) phase, and the ethanol-depleted phase [21]. Initially, yeast consumes glucose and diverts it to ethanol, acquiring adenosine triphosphate (ATP) from glycolysis in this stage. As proliferation continues, glucose becomes depleted and yeast cells begin to utilize ethanol (i.e. diauxic shift) to generate ATP in the mitochondria and initiate autophagy [36,37]. This phase is also known as the lag phase reflecting its growth status [21]. In the final or ethanol-depleted phase, cells enter stationary status (the period when the cell number stops increasing) and autophagy is activated to a greater extent [18,19]. Various cellular components undergo degradation in different stages. For instance, bulk autophagy, lipophagy, and reticulophagy are initiated during the ethanol-utilizing phase, whereas mitophagy is induced during the ethanol-depleted phase [21]. Methylotrophic yeasts such as Komagataella phaffii present contrasting features relating to their carbon metabolism since they do not ferment glucose. Lipophagy naturally occurs in K. phaffii after glucose depletion, releasing fatty acids to produce energy [38,39]. When the carbon source is shifted from glucose to methanol, K. phaffii undergoes lag phase autophagy, similar to S. cerevisiae [40].
The studies discussed above suggest that either abrupt or gradual depletion of available sources of carbon, or a shift from the preferable carbon source to alternative sources in yeast cells, induces autophagy. However, it is important to note that different induction conditions can result in varying consequences, since there are discernible differences between abrupt and gradual starvation processes. Following abrupt glucose starvation, ATP levels drop sharply and are only replenished gradually [41,42]. In such cases, autophagy can be a way to recover the energy pool. By contrast, gradual starvation does not induce such a sudden decline in ATP levels, at least until the entire carbon source is consumed. It is also noteworthy that, while the regulation of autophagy by carbon starvation has been clearly demonstrated in yeasts, studies on this topic in other organisms have been relatively limited [26–28,43,44].
Mechanisms of Carbon Starvation-Responsive Autophagy in Yeast
In yeast cells, the intracellular energy status is sensed by Snf1 (sucrose non-fermenting 1), which is homologous to mammalian AMP-activated protein kinase (AMPK) (Figure 1). As Snf1 responds to declining levels of glucose to regulate metabolic activities, it is reasonable to hypothesize that Snf1 May therefore play a role in the regulation of autophagy during energy deprivation. Supporting such a hypothesis, Adachi et al. showed that Snf1 is required for autophagy under carbon starvation conditions [15]. Seo et al. showed that Snf1 facilitates the translocation of Atg14 from the ER to the vacuole membrane, which is critical for acute glucose restriction-induced lipophagy [33].
Figure 1.
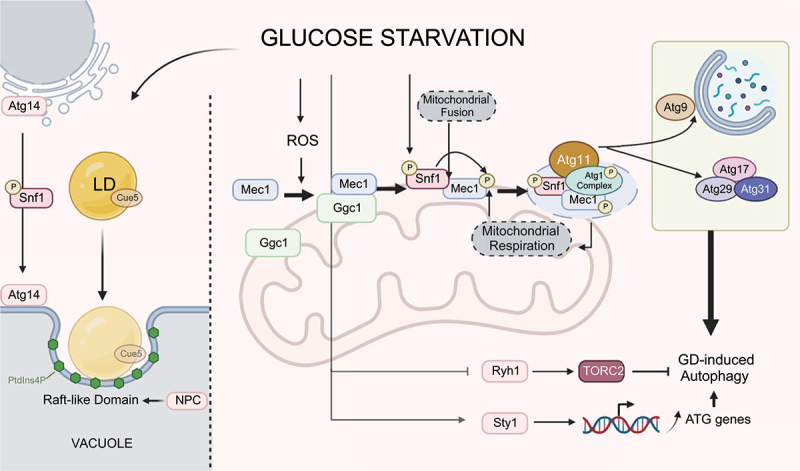
Molecular mechanisms underlying macroautophagy and lipophagy induced by glucose deprivation (GD) in yeasts. Under glucose starvation, Ggc1 recruits Mec1 to the mitochondrial surface, leading to the formation of Mec1 puncta in a process that requires ROS. Mitochondrial fusion facilitates the binding between Snf1 and Mec1, resulting in Mec1 phosphorylation by Snf1. The phosphorylation of Mec1 promotes the recruitment of Atg1 to the mitochondria. Atg11 mediates the interaction of Snf1 and Atg1. The Snf1-Mec1-Atg1 complex is necessary for maintaining mitochondrial respiration. In turn, mitochondrial respiration is required for glucose starvation-induced Mec1 phosphorylation. Atg11 also mediates the association of Atg17 with Atg29-Atg31 and facilitates the recruitment of Atg9 vesicles to the PAS. Additionally, glucose starvation suppresses the activity of Ryh1, a small GTPase, inhibiting TORC2 and enhancing autophagy. Glucose deprivation also enhances the activity of Sty1, a MAPK family member, to upregulate autophagy-related genes expression. During lipophagy, Snf1-dependant translocation of Atg14 from the ER to the vacuole is critical for the formation of the raft-like domain, which contains PtdIns4P. NPC/Niemann Pick type C proteins facilitate the expansion of the raft-like domain. Specifically in glucose starvation, Cue5, a selective aggrephagy receptor in K. phaffii, is degraded together with lipid droplets (LD, spherical organelles containing lipids) via autophagy.
In our studies, we have shown that Snf1 induces the recruitment of Atg1 to the mitochondria via binding and phosphorylating Mec1 (mitosis entry checkpoint 1, a homolog of mammalian ATR involved in the DNA damage response pathway), on the mitochondrial surface [12]. In this way, the Snf1-Mec1-Atg1 pathway was confirmed to act to maintain mitochondrial respiration. We also found that mitochondrial respiration is necessary for Mec1 phosphorylation by Snf1 and for glucose starvation-induced autophagy. In response to energy starvation, Mec1 forms puncta on the mitochondria by interacting with Ggc1 (GDP/GTP carrier 1), a mitochondrial transmembrane protein. Mec1 also directly interacts with Atg13 (an essential protein in autophagy initiation), its binding crucial for recruiting Atg13 to the phagophore assembly site (PAS) and inducing autophagy during glucose starvation [45].
We further elucidated that the mitochondrial fusion machinery is required for the interaction of Snf1 and Mec1 upon glucose deprivation [46]. These findings of the close relationship between mitochondria and glucose starvation-induced autophagy are consistent with a report by Adachi et al. [15] where the authors showed that inhibiting cytochrome C reductase led to suppression of autophagy induction during carbon starvation. Their study also showed that carbon deprivation-induced autophagy depends on Atg11 and Atg17 [15]. Upon energy starvation, Atg11 mediates the interaction between Snf1 and Atg1 and enables Snf1 to regulate Atg1 [13]. In addition, Atg11 facilitates the association of Atg17 with Atg29-Atg31 and the recruitment of Atg9 to the PAS [13]. All these steps are important for the formation of the PAS.
Corral-Ramos et al. recently showed that the TOR and MAP kinase pathways regulate autophagy in fission yeast during glucose starvation [35]. In nutrient-rich media, Ryh1, a small GTPase, activates the TOR complex 2 (TORC2) pathway [47]. Under glucose starvation, the inactivation of Ryh1 suppressed TORC2. This results in subsequent TORC1 inhibition and autophagy induction [35]. Their study also showed that autophagy is regulated by Sty1, a MAPK family member that controls the transcription of ATG genes, in fission yeast during glucose starvation.
Lang et al. also showed that autophagy initiation occurs during glucose starvation, evidencing this via the formation of Atg8 puncta. However, the completion of autophagy and the delivery of Atg8 to the vacuoles did not occur [34]. This study suggested that autophagy is suppressed by increased amino acids, as produced from endocytosis and vacuolar hydrolysis and induced by glucose starvation. However, Adachi and colleagues showed that glucose starvation did not induce Atg8 degradation in two yeast strains [15]. The discrepancy between these two studies may be due to differences in the glucose starvation conditions used, such as acute versus chronic deprivation. Further investigation is needed to clarify such discrepant results on the effect of glucose starvation on autophagy.
Mechanisms of Lipophagy in Yeast Stationary Phase
One of the other key mechanisms related to carbon starvation-induced autophagy involves the pathway that regulates lipophagy in the stationary phase of budding yeast, as occurs when the whole carbon source is depleted Figure 1. In such a condition, the size and number of lipid droplets (LDs, spherical organelles containing lipids surrounded by a protein-associated phospholipid monolayer), increase, and the LDs translocate from the perinuclear ER to the vacuolar lipid rafts to provide inner contents, such as sterol esters, to maintain the integrity of the lipid rafts [18]. In a feed-forward manner, the integrity of lipid rafts supports the delivery of LDs to the vacuoles during the stationary phase [18]. The molecular mechanisms underlying this process have been uncovered to some extent. Ncr1 (Niemann-pick type C Related 1) and Npc2 (Niemann Pick type C homolog 2), which are homologs of a protein that causes Niemann-Pick type C (NPC) disease, play a role in facilitating the formation and expansion of the lipid rafts and lipophagy during the stationary phase. This process likely occurs through the insertion of sterols into the lysosome membrane [19]. Additionally, Kurokawa et al. showed that Pik1 (phosphatidyl inositol kinase 1) and Stt4 (staurosporine and temperature sensitive 4), both phosphatidylinositol 4-kinases, produce phosphatidylinositol 4-phosphate (PtdIns4P) and facilitate the engulfment of LDs by PtdIns4P-positive raft-like domains during the stationary phase of budding yeast [48]. How those molecules regulate lipophagy during the stationary phase, and whether the observed lipophagy is due to carbon source deprivation, requires further investigation.
In budding yeast, the endosomal sorting complex required for the transport (ESCRT) complex plays a crucial role in lipophagy during glucose starvation. Oku et al. showed that Vps27 (vacuolar protein sorting 27), a component of the ESCRT complex, particularly its binding with clathrin, is necessary for the induction of lipophagy after a diauxic shift switching the carbon source from glucose to ethanol [36]. Vps4 (vacuolar protein sorting 4) of the ESCRT complex is also required for lipophagy during the stationary phase of budding yeast [18,49]. However, the role of the ESCRT complex in lipophagy requires further clarification, as Zhang et al. suggested that the ESCRT complex plays a negative role in lipophagy under cases of abrupt glucose limitation [50].
Regulation of Autophagy During Glucose Starvation in Mammalian Cells
In mammalian cells, the regulation of autophagy during glucose starvation is mediated by multiple linked pathways. These have been well summarized in other reviews [51,52]. The central mediator of the mechanism is AMPK (Figure 2). Glucose deprivation activates AMPK via increasing the relative ratio of AMP or ADP level to ATP level [53,54]. Additionally, one recent study confirmed fructose-1,6-bisphosphate (a glycolysis metabolite) and ALDO/aldolase mediate glucose sensing by AMPK [55]. The activated AMPK then phosphorylates ULK1, the protein kinase that triggers the initiation of autophagy [26]. AMPK also phosphorylates RPTOR/raptor [56], a component of MTORC1, and TSC2 (TSC complex subunit 2) [57–60], an upstream suppressor of MTORC1 signaling. Through RPTOR phosphorylation, AMPK can enhance autophagy induction. Autophagy can be enhanced under glucose-deficient conditions through the inhibition of MTORC1, independent of AMPK activation. Glucose limitation deactivates RRAG guanosine triphosphatases (GTPases) and thereby suppresses the recruitment of MTORC1 to the lysosome for its activation [61]. In cultured HEK293E cells, glucose deprivation was shown to induce the accumulation of DEPTOR (DEP domain-containing MTOR-interacting protein), which binds and inhibits MTORC1 and MTORC2 [62], enhancing autophagy [63]. In addition, glucose deficiency was shown to enhance the interaction between HK2 (hexokinase 2), an enzyme that catalyzes the production of glucose-6-phosphate during glycolysis, and MTORC1 [64], the binding of which suppresses MTORC1 activity and induces autophagy.
Figure 2.
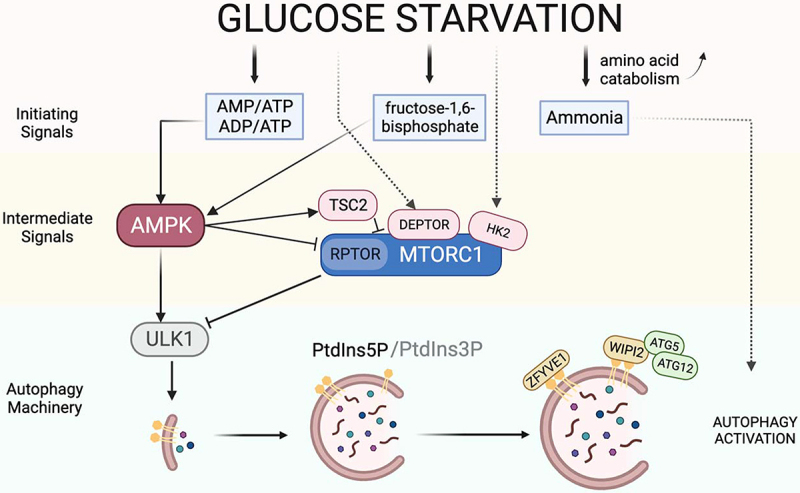
Schematic diagram illustrating the process of autophagy initiation in mammalian cells in response to glucose starvation. When glucose is deprived, the decrease in cellular AMP or ADP levels is detected by AMPK. A decrease in fructose-1,6-bisphosphate (FBP), a metabolic intermediate of glycolysis, can activate AMPK to induce autophagy. In addition, an increase in ammonia produced by amino acid catabolism can also trigger autophagy initiation. Activated AMPK can suppress MTORC1 via phosphorylating RPTOR (a component of MTORC1) and activating TSC2 (suppressor of MTORC1). Independent of AMPK, MTORC1 can also be inhibited by enhanced interaction with DEPTOR and HK2 under glucose starvation. PtdIns3P (a phospholipid in cell membranes) is required to initiate autophagy under nitrogen starvation, while PtdIns5P can substitute for PtdIns3P in glucose deprivation-induced autophagy. PtdIns5P mediates the recruitment of WIPI2 and ZFYVE1 to phagophores, maintaining ATG12–ATG5 conjugation and autophagy. Dashed lines indicate that the intermediate mechanisms are unclear.
In addition to the pathways discussed above, glucose deprivation has also been shown to induce autophagy by enhancing amino acid catabolism that leads to the accumulation of cellular ammonia. The accumulation of ammonia has been shown to induce autophagy in both mouse embryonic fibroblasts (MEFs) and in a human bone osteosarcoma epithelial cell line [28,65]. Cheong et al. showed that autophagy could be induced in the absence of ULK1 and ULK2 through the accumulation of ammonia during prolonged glucose starvation [28]. Another non-canonical autophagy pathway induced by glucose starvation involves phosphatidylinositol 5-phosphate (PtdIns5P) [66]. While phosphatidylinositol-3-phosphate (PtdIns3P)-mediated recruitment of autophagic effectors such as WIPI (WD repeat domain, phosphoinositide interacting) proteins are known to be essential for autophagy induction in response to amino acid starvation [67], McAlpine et al. and Vicinanza et al. showed in cultured MEFs and HeLa cells that glucose deprivation-induced autophagy does not require PtdIns3P or WIPI proteins, but instead requires PtdIns5P [29,66]. Upon glucose deprivation, PtdIns5P mediates the recruitment of WIPI2 and ZFYVE1/DFCP1 (double FYVE domain-containing protein) to phagophores, maintaining ATG12–ATG5 conjugation and autophagy, even in PtdIns3P-depleted cells [66]. These findings suggest that glucose starvation induces autophagy in mammalian cells via multiple pathways. Such multiple pathways may have evolved to ensure homeostasis in mammalian cells and to adapt to the complex and drastically changing environments encountered during evolution.
Autophagy During Low Energy in Plants
As sessile organisms, plants extensively rely on autophagy to deal with inevitable existential challenges [68,69]. Autophagy-deficient Arabidopsis thaliana (A. thaliana) mutants have shown early senescence under darkness or sugar deprivation [24,70–72]. Darkness, during which the carbohydrate pool is depleted in plants, is widely used to create carbon-starvation conditions, though light signaling might suppress autophagy independently of carbon starvation [70,72,73].
Several works have contributed to our understanding of the autophagy mechanism in plants during carbon starvation. In A. thaliana, ATI1 (ATG8-interacting protein 1) and ATI2 bind to Atg8 [44]. Located in the ER network and plastids in favorable growth conditions, ATI1 transports cargo proteins, such as chloroplast proteins, to the vacuole in response to carbon starvation [43,44]. Wu et al. showed that ATI1 and ATI2 regulate reticulophagy in A. thaliana in response to carbon starvation [74]. In this process, ATI1 and ATI2 recruit and carry MSBP1 (Membrane Steroid Binding Protein 1) from the ER to the vacuoles for degradation [74]. MSBP1 is a scaffold protein that stabilizes P450 enzymes involved in the biosynthesis of lignin, a complex biopolymer, as a main component of the plant cell wall [74,75]. Such a result implies that the function of P450 enzymes might be regulated by the reticulophagy under carbon starvation.
Chloroplasts, the site where the core metabolic processes occur in photoautotrophs, are also degraded and utilized as a major energy source for survival during energy-limited conditions such as sugar starvation [76]. According to a model proposed by Ishida et al., chloroplasts in A. thaliana are decomposed and transported into specialized autophagic vesicles called Rubisco-containing bodies when individual leaves are shaded from light [77]. A similar model has been proposed in rice (Oryza sativa) where recycling of chloroplastic proteins was observed during darkness, which is consistent with the finding in A. thaliana [78].
In plants, programmed cell death is intertwined with autophagy during carbon starvation [79]. One study, conducted on both potatoes and tobacco, suggested that the translocation of VPE (vacuolar processing enzyme) from the ER to the central vacuole via autophagy results in programmed cell death in response to carbon starvation [80]. However, the role of autophagy in programmed cell death is controversial. Phillips et al. showed that autophagy-deficient A. thaliana is prone to programmed cell death under carbon starvation [72]. Further study is necessary to clarify whether autophagy plays a positive or a negative role in programmed cell death during carbon starvation in plants.
Autophagy in Supplying Energy and Regulating Cellular Survival During Energy Stress
One way autophagy maintains energy homeostasis and supports cell survival during glucose starvation is to mobilize nutrient reserves, such as LDs. For instance, glucose deprivation-induced autophagy enhances the number of LDs in mammalian cells that are subjected to lipolysis thus resulting in the production of ATP [81]. Kaushik et al. showed that chaperone-mediated autophagy facilitates lipolysis during glucose starvation via the degradation of PLIN2 (perilipin 2) and PLIN3 [82]. Lipophagy has been shown to contribute to energy production in budding yeast under glucose restriction conditions [17,33]. In mice, depleting an autophagy gene specifically in the liver or kidney prevented fasting-induced LD formation and ketogenesis in those tissues [83,84]. This suggested that autophagy is necessary for the metabolic response to fasting in mice. Other studies have further confirmed the critical role of autophagy in supplying energy for animal survival during fasting. For example, mouse neonates have been shown to depend on autophagy for survival due to the limited availability of food after birth [85]. During this neonatal period, autophagy breaks down glycogen and proteins, releasing glucose and amino acids as energy sources, thus supporting neonatal survival [6,61,86,87].
Besides degrading lipids, glycogen, and proteins, autophagy can also facilitate cells to adapt to glucose deprivation through regulating energy demand or nutrient intake. For example, starvation-induced hypothalamic autophagy was shown to upregulate the level of AGRP (agouti related neuropeptide), which promotes food intake in mice [88]. AGRP levels are regulated by fatty acids generated from the autophagic degradation of lipids in neurons. Oh et al. also showed that the hypothalamic AMPK-mediated autophagy pathway can increase food intake in the mouse by upregulating NPY (neuropeptide Y) expression and downregulating POMC/pro-opiomelanocortin-α expression during glucose shortage conditions [89]. However, the specific mechanism of how autophagy can affect NPY and POMC/pro-opiomelanocortin-α expression remains unknown.
While fasting is a common condition that subjects living organisms to energy stress, exercise can also cause a corresponding energy stress challenge in organisms. Exercise has been shown to induce autophagy in organs associated with glucose homeostasis, including the liver, pancreas and muscle [90]. In response to exercise, autophagy may play roles in replenishing energy or adjusting energy demand. For example, He et al. showed that exercise-induced autophagy facilitates the muscle to uptake glucose during exercise [90]. Liu et al. showed that endurance exercise and glucose deprivation can facilitate the interaction of Toll-like receptor 9 with BECN1 resulting in muscle AMPK activation [91].
In addition to its pro-survival roles, autophagy has been shown to promote immunogenic cell death [92]. In their study, Pietrocola et al. showed that autophagy induction through treating with protein acetylation inhibitors, which mimics the effects of caloric restriction, enhances the immune response against tumor cells in mice. Autophagy enhances the secretion of ATP, a hallmark of immunogenic cell death, which then triggers the immune response [92,93]. These studies suggest that starvation-induced autophagy plays complex roles in regulating biological processes related to energy stress and cellular survival.
Energy Deprivation and Autophagy in Human Health and Disease
The proper regulation of autophagy during energy deprivation is closely related to many human diseases [94,95]. In the following section, we discuss three disease conditions (cerebral ischemia, myocardial ischemia, and cancer) where cells experience glucose deprivation and their response involves autophagy. These examples highlight the potential impact of dysregulated autophagy during glucose deprivation on human health.
Cerebral ischemia
In the brain, restricted blood supply can lead to the development of ischemia and oxygen-glucose deprivation (OGD). An increasing number of studies have suggested that autophagy might play crucial roles in cell survival under stress conditions [96,97]. Wang et al. showed that overexpression of NAMPT (nicotinamide phosphoribosyltransferase), a metabolic enzyme involved in the biosynthesis of nicotinamide adenine dinucleotide, enhances autophagy in the neurons via inhibiting MTOR, thus presenting a neuroprotective role [98]. However, autophagy does not appear to remain active throughout the period of cerebral ischemia. In cases of cerebral ischemia, TFEB (transcription factor EB) expression is both increased, and shows increased activity of translocation into the nucleus. This leads to autophagy activation, which alleviates the effects of ischemic injury [99]. Specifically, OGD-induced upregulation of calcineurin facilitates the translocation of TFEB. As the ischemic condition persists, the nuclear TFEB levels decrease and autophagy is reduced, exacerbating the extent of ischemic damage. In one recent study, Xu et al. showed that the expression of α7nAChR (alpha7 nicotinic acetylcholine receptor) is reduced in a mouse model of ischemic stroke [100]. Application of an agonist of the receptor was then shown to have a protective effect against the ischemic stroke condition, likely via the AMPK-MTORC1-autophagy pathway.
In contrast, some studies have suggested that the suppression of autophagy, rather than its enhancement, plays a beneficial role in cerebral ischemia [101,102]. Zhang et al. showed that autophagy inhibition during OGD decreased the area of cerebral tissue infarction in mice [103]. Using pharmacological inhibitors of autophagy, Qin et al. showed that OGD-induced autophagy may at least partly contribute to ischemic injury of astrocytes in rats [104]. In cell culture experiments, Pei et al. discovered a microRNA (small noncoding RNA), miR-190b, that inhibits autophagy and neuronal apoptosis during OGD [105]. One possible explanation is that excessive autophagy may aggravate neural deficits and the maintaining of moderate autophagy may therefore be a key to protecting brain tissue during cerebral ischemia.
Myocardial ischemia
Myocardial ischemia is a pathological condition characterized by the interruption of blood perfusion in the heart, resulting in reduced glucose and oxygen supply to cardiomyocytes and the failure of normal heart function. Enhancing autophagy has been regarded as a protective mechanism for myocardial ischemia [7]. In rat or mouse models, acute myocardial infarction enhancing autophagy via rapamycin treatment or starvation resulted in decreased myocardial infarction size [106,107]. According to another study in pigs, chronic coronary ischemia increased autophagy in the ischemic myocardium, which was reversely correlated with cellular apoptosis, the direct cause of infarction necrosis [108].
Further evidence of the beneficial effect of autophagy in myocardial ischemia was provided by a study by Sun et al. in which the authors showed glucose starvation enhanced autophagy in cultured primary cardiomyocytes [109]. This autophagy was further increased by CDKN1B/p27 and played a pro-survival role by inhibiting apoptosis. Matsui et al. showed evidence that autophagy may play a protective role during myocardial ischemia via an AMPK-dependent mechanism [110]. In line with these findings, Sciarretta et al. showed that glucose starvation inactivates RHEB-MTORC1 signaling thus resulting in induction of autophagy that protects cardiomyocytes [111]. The beneficial effect of glucose starvation on cardiomyocytes during OGD might be at least in part due to the upregulation of many autophagy-related genes and involving Gabarapl1 and Atg12, via FOXO transcription factors [112].
Despite the beneficial effects of autophagy on cardiac cell survival, some studies have suggested that autophagy might have a counteractive effect. Troncoso et al. showed that IGF1 (insulin-like growth factor 1) suppresses autophagy in cardiomyocytes but enhances cell survival during prolonged serum and glucose deprivation [113]. Aki et al. observed that overexpression of PtdIns3K led to the accumulation of autophagic vacuoles and subsequent cardiomyocyte death during glucose starvation [114]. Taken together, these studies suggest that autophagy can have both beneficial and harmful effects on cardiomyocyte survival during glucose starvation or myocardial ischemia. Kriel et al. proposed that the extent of autophagy or an autophagy flux threshold might determine whether autophagy leads to cell death or survival [115]. It is important to note that autophagic flux was not thoroughly evaluated in several of the studies discussed above. Thus, further investigation is necessary to make definitive conclusions.
Cancer
Tumors are characterized by disorganized vasculature and uncontrolled rapid proliferation of cells. This can lead to inadequate delivery of nutrients, including glucose, and an increased demand for energy [116]. Adapting to such a metabolic status, tumor cells exhibit a preference for glycolysis over oxidative phosphorylation to produce energy even under aerobic conditions (known as the Warburg effect). This leads to their increased sensitivity to a reduction in glucose levels compared to untransformed cells [117,118].
Targeting autophagy in glucose-deficient tumor cells has been tested as a potential therapeutic strategy for cancer treatment. Li et al. showed that during glucose deprivation, ACSS (acyl-CoA synthetase short-chain family member) is translocated into the nucleus and promotes the transcription of lysosomal and autophagy genes by its binding to TFEB and by facilitating its translocation to the respective gene promoter regions [119]. The authors used a xenograft tumor model to demonstrate that the ability of ACSS to translocate into the nucleus is important in the induction of autophagy and results in a reduction in brain tumor growth. Studies on breast or pancreatic cancer cells have also shown that activating autophagy can promote cell survival under conditions of glucose deprivation [120,121]. Walker et al. showed that autophagy-mediated degradation of SQSTM1/p62 (sequestosome 1) stabilizes NFE2L2/NRF2 (nuclear factor, erythroid derived 2, like 2), a key transcription factor involved in antioxidative response, thus protecting breast cancer cells during glucose deprivation [120]. Meng et al. showed that glucose deprivation-induced degradation of GPX1 (glutathione peroxidase 1), an antioxidant enzyme, activates autophagy and inhibits pancreatic cancer cell death [121].
Although autophagy inhibition has been considered a potential therapeutic strategy for cancer, it should be noted that autophagy also plays a role in suppressing cancer cell growth. One example of the negative effects of autophagy on cancer cell growth is related to the tumor suppressor gene TP53/p53, where the mutant form of TP53 is oncogenic. Rodriguez et al. showed that glucose restriction induces binding between the mutant TP53 and BECN1, a core protein in the autophagy machinery, resulting in the activation of autophagic degradation of the mutant TP53 and subsequent tumor cell death [8]. Pietrocola et al. demonstrated that autophagy can also have negative effects on lung tumor growth [92]. Using a mouse lung cancer model, the authors showed that caloric restriction mimic mice showed autophagic-dependent depletion of regulatory T cells from the tumor. This resulted in enhanced anticancer immunosurveillance [92]. Taken together, autophagy triggered in energy-stressed cancer cells can exert both therapeutic and pathogenic effects. Such dual effects are likely due to multiple downstream pathways with different functions involved in responses to stress conditions. Further understanding of the mechanisms underlying the effects of autophagy could be beneficial toward the development of better cancer therapeutics.
Outstanding Questions
As discussed above, autophagy regulation during glucose or energy deprivation has unique aspects compared to nitrogen and other starvation-induced autophagy. Despite the significant importance of the topic, there are still many controversial issues and unresolved molecular mechanisms regarding the regulation and roles of autophagy during glucose starvation or energy deprivation. Several noteworthy questions related to this are discussed below.
Cells recognize glucose and nitrogen starvation as distinct stresses and cope with them in different ways to ensure cell survival and homeostasis. It is currently unclear whether autophagy specifically degrades certain substrates during glucose or energy deprivation. The only known example of a glucose starvation-specific autophagy substrate is Cue5, a receptor for aggrephagy (selective autophagy of ubiquitinated protein aggregates), that is degraded via autophagy during glucose starvation in K. phaffii (methylotrophic yeast) [39]. Cue5 accumulates on LDs and is degraded along with LDs via lipophagy in the stationary phase, thereby generating energy for the yeast cells. A global-scale proteomics approach has been useful in identifying proteins degraded during amino acid starvation [122]. A similar approach could be useful in identifying proteins degraded during glucose starvation at a global scale.
Although AMPK is a main player in the pathway that regulates autophagy in response to glucose starvation, other molecules have also been identified to regulate autophagy independently of AMPK during glucose starvation. Some examples of such proteins include Sty1 and Ryh1 in fission yeast as well as HK2 and RRAG GTPases in mammalian cells [35,47,61,64], as discussed above. It is possible that many more pathways, distinct from AMPK, could independently regulate autophagy during glucose starvation. For instance, Mec1, involved in DNA repair, has been found to regulate autophagy specifically in response to glucose starvation in budding yeast [12]; Prl1 has been found to specifically regulate lipophagy during glucose deprivation in K. phaffii [38]; and PtdIns5P has been identified as essential for autophagy during glucose deprivation, but shown to be dispensable for nitrogen starvation-induced autophagy in human cancer cells [66]. It is possible that more genes regulating glucose starvation-dependent autophagy, may be identified in the future.
Glucose starvation induces changes in gene expression and alters the function of many cellular factors. This makes it difficult to tease apart the specific mechanisms that directly affect autophagy. Additionally, the cellular response that occurs via autophagy during glucose starvation can also vary depending on the duration of glucose starvation or the cell type, potentially representing discrepancies among studies. Achieving a more comprehensive understanding of the molecular mechanisms governing autophagy under glucose starvation or energy deprivation, coupled with rigorous experimental validation of published works, holds the potential to resolve controversies within the literature and unlock novel insights into this pivotal cellular process for stress response.
Acknowledgements
We sincerely thank Prof. Do-Hyung Kim who has served as the editor. We thank the other members of the lab, especially Yingcong Chen, Weijing Yao and Yuting Chen, for their discussion in preparing this review. Figures were created using BioRender. This research was supported by the National Key Research and Development Program of China (2021YFC2600104) to Liqin Zhang, and National Natural Science Foundation of China (Grant No: 32122028, 92254307, and 32070739), Zhejiang Provincial Natural Science Foundation of China under Grant No.LR21C070001 to Cong Yi.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Funding Statement
The work was supported by the National Key Research and Development Program of China [2021YFC2600104]; the National Science Foundation of China [32122028, 32070739, and 91754107]; and Zhejiang Provincial Natural Science Foundation of China [LR21C070001].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Huang K, Fingar DC.. Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol. 2014. Dec;36:79–90. doi: 10.1016/j.semcdb.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].He C. Balancing nutrient and energy demand and supply via autophagy. Curr Biol. 2022 Jun 20;32(12):R684–r696. doi: 10.1016/j.cub.2022.04.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010 Dec 3;330(6009):1344–1348. doi: 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kraft C, Deplazes A, Sohrmann M, et al. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008. May;10(5):602–610. doi: 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- [5].Karsli-Uzunbas G, Guo JY, Price S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014. Aug;4(8):914–927. doi: 10.1158/2159-8290.CD-14-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004 Dec 23;432(7020):1032–1036. doi: 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- [7].Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006 Oct 6;281(40):29776–29787. doi: 10.1074/jbc.M603783200 [DOI] [PubMed] [Google Scholar]
- [8].Rodriguez OC, Choudhury S, Kolukula V, et al. Dietary downregulation of mutant p53 levels via glucose restriction: mechanisms and implications for tumor therapy. Cell Cycle. 2012 Dec 1;11(23):4436–4446. doi: 10.4161/cc.22778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Bio. 1962. Jan;12(1):198–202. doi: 10.1083/jcb.12.1.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Bio. 1967. Nov;35(2):C11–6. doi: 10.1083/jcb.35.2.C11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Takeshige K, Baba M, Tsuboi S, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Bio. 1992. Oct;119(2):301–311. doi: 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yi C, Tong J, Lu P, et al. Formation of a Snf1-Mec1-Atg1 Module on mitochondria governs energy deprivation-induced autophagy by regulating mitochondrial respiration. Dev Cell. 2017 Apr 10;41(1):59–71.e4. doi: 10.1016/j.devcel.2017.03.007 [DOI] [PubMed] [Google Scholar]
- [13].Yao W, Li Y, Wu L, et al. Atg11 is required for initiation of glucose starvation-induced autophagy. Autophagy. 2020. Dec;16(12):2206–2218. doi: 10.1080/15548627.2020.1719724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Miller-Fleming L, Antas P, Pais TF, et al. Yeast DJ-1 superfamily members are required for diauxic-shift reprogramming and cell survival in stationary phase. Proc Natl Acad Sci U S A. 2014 May 13;111(19):7012–7017. doi: 10.1073/pnas.1319221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Adachi A, Koizumi M, Ohsumi Y. Autophagy induction under carbon starvation conditions is negatively regulated by carbon catabolite repression. J Biol Chem. 2017 Dec 1;292(48):19905–19918. doi: 10.1074/jbc.M117.817510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yokota H, Gomi K, Shintani T. Induction of autophagy by phosphate starvation in an Atg11-dependent manner in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2017 Jan 29;483(1):522–527. doi: 10.1016/j.bbrc.2016.12.112 [DOI] [PubMed] [Google Scholar]
- [17].Weber CA, Sekar K, Tang JH, et al. β-Oxidation and autophagy are critical energy providers during acute glucose depletion in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2020 Jun 2;117(22):12239–12248. doi: 10.1073/pnas.1913370117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang CW, Miao YH, Chang YS. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Bio. 2014 Aug 4;206(3):357–366. doi: 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tsuji T, Fujimoto M, Tatematsu T, et al. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. Elife. 2017 Jun 7;6:e25960. doi: 10.7554/eLife.25960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zheng L, Shu WJ, Li YM, et al. The Paf1 complex transcriptionally regulates the mitochondrial-anchored protein Atg32 leading to activation of mitophagy. Autophagy. 2020. Aug;16(8):1366–1379. doi: 10.1080/15548627.2019.1668228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Iwama R, Ohsumi Y. Analysis of autophagy activated during changes in carbon source availability in yeast cells. J Biol Chem. 2019 Apr 5;294(14):5590–5603. doi: 10.1074/jbc.RA118.005698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen MH, Liu LF, Chen YR, et al. Expression of alpha-amylases, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. Plant J. 1994. Nov;6(5):625–636. doi: 10.1046/j.1365-313X.1994.6050625.x [DOI] [PubMed] [Google Scholar]
- [23].Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996. Aug;111(4):1233–1241. doi: 10.1104/pp.111.4.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hanaoka H, Noda T, Shirano Y, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002. Jul;129(3):1181–1193. doi: 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Izumi M, Wada S, Makino A, et al. The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 2010. Nov;154(3):1196–1209. doi: 10.1104/pp.110.158519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011. Feb;13(2):132–141. doi: 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gammoh N, Florey O, Overholtzer M, et al. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol. 2013. Feb;20(2):144–149. doi: 10.1038/nsmb.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cheong H, Lindsten T, Wu J, et al. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011 Jul 5;108(27):11121–11126. doi: 10.1073/pnas.1107969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McAlpine F, Williamson LE, Tooze SA, et al. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 2013. Mar;9(3):361–373. doi: 10.4161/auto.23066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zaman S, Lippman SI, Zhao X, et al. How Saccharomyces responds to nutrients. Ann Rev Genet. 2008;42(1):27–81. doi: 10.1146/annurev.genet.41.110306.130206 [DOI] [PubMed] [Google Scholar]
- [31].Turcotte B, Liang XB, Robert F, et al. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010. Feb;10(1):2–13. doi: 10.1111/j.1567-1364.2009.00555.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kayikci Ö, Nielsen J, Bolotin-Fukuhara M. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015. Sep;15(6):fov068. doi: 10.1093/femsyr/fov068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Seo AY, Lau PW, Feliciano D, et al. AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. Elife. 2017 Apr 10;6. doi: 10.7554/eLife.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lang MJ, Martinez-Marquez JY, Prosser DC, et al. Glucose starvation inhibits autophagy via vacuolar hydrolysis and induces plasma membrane internalization by down-regulating recycling. J Biol Chem. 2014 Jun 13;289(24):16736–16747. doi: 10.1074/jbc.M113.525782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Corral-Ramos C, Barrios R, Ayté J, et al. TOR and MAP kinase pathways synergistically regulate autophagy in response to nutrient depletion in fission yeast. Autophagy. 2022. Feb;18(2):375–390. doi: 10.1080/15548627.2021.1935522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Oku M, Maeda Y, Kagohashi Y, et al. Evidence for ESCRT- and clathrin-dependent microautophagy. J Cell Bio. 2017 Oct 2;216(10):3263–3274. doi: 10.1083/jcb.201611029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dickinson JR, Schweizer M, editors. The metabolism and molecular physiology of Saccharomyces cerevisiae. New York: Taylor & Francis; 1999. [Google Scholar]
- [38].Kumar R, Rahman MA, Nazarko TY. Nitrogen starvation and stationary phase lipophagy have distinct molecular mechanisms. Int J Mol Sci. 2020 Nov 29;21(23):9094. doi: 10.3390/ijms21239094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kumar R, Shroff A, Nazarko TY. Komagataella phaffii Cue5 piggybacks on lipid droplets for its vacuolar degradation during stationary phase lipophagy. Cells. 2022 Jan 10;11(2):215. doi: 10.3390/cells11020215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yamashita S, Yurimoto H, Murakami D, et al. Lag-phase autophagy in the methylotrophic yeast pichia pastoris. Genes Cells. 2009. Jul;14(7):861–870. doi: 10.1111/j.1365-2443.2009.01316.x [DOI] [PubMed] [Google Scholar]
- [41].Ashe MP, De Long SK, Sachs AB, et al. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000. Mar;11(3):833–848. doi: 10.1091/mbc.11.3.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aoh QL, Hung CW, Duncan MC, et al. Energy metabolism regulates clathrin adaptors at the trans-Golgi network and endosomes. Mol Biol Cell. 2013. Mar;24(6):832–847. doi: 10.1091/mbc.e12-10-0750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Michaeli S, Honig A, Levanony H, et al. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014. Oct;26(10):4084–4101. doi: 10.1105/tpc.114.129999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Honig A, Avin-Wittenberg T, Ufaz S, et al. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell. 2012. Jan;24(1):288–303. doi: 10.1105/tpc.111.093112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yao W, Li Y, Chen Y, et al. Mec1 regulates PAS recruitment of Atg13 via direct binding with Atg13 during glucose starvation-induced autophagy. Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2215126120. doi: 10.1073/pnas.2215126120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu C, Yao W, Kai W, et al. Mitochondrial fusion machinery specifically involved in energy deprivation-induced autophagy. Front Cell Dev Biol. 2020;8:221. doi: 10.3389/fcell.2020.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hatano T, Morigasaki S, Tatebe H, et al. Fission yeast Ryh1 GTPase activates TOR complex 2 in response to glucose. Cell Cycle. 2015;14(6):848–856. doi: 10.1080/15384101.2014.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kurokawa Y, Konishi R, Yoshida A, et al. Microautophagy in the yeast vacuole depends on the activities of phosphatidylinositol 4-kinases, Stt4p and Pik1p. Biochim Biophys Acta Biomembr. 2020 Nov 1;1862(11):183416. doi: 10.1016/j.bbamem.2020.183416 [DOI] [PubMed] [Google Scholar]
- [49].Toulmay A, Prinz WA. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Bio. 2013 Jul 8;202(1):35–44. doi: 10.1083/jcb.201301039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang A, Meng Y, Li Q, et al. The endosomal sorting complex required for transport complex negatively regulates Erg6 degradation under specific glucose restriction conditions. Traffic. 2020. Jul;21(7):488–502. doi: 10.1111/tra.12732 [DOI] [PubMed] [Google Scholar]
- [51].Li Y, Chen Y. AMPK and autophagy. In: Qin Z-H, editor. Autophagy: biology and diseases: basic Science. Singapore: Springer Singapore; 2019. pp. 85–108. doi: 10.1007/978-981-15-0602-4_4 [DOI] [Google Scholar]
- [52].Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011 Sep 2;13(9):1016–1023. doi: 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Salt IP, Johnson G, Ashcroft SJ, et al. AMP-activated protein kinase is activated by low glucose in cell lines derived from pancreatic beta cells, and may regulate insulin release. Biochem J. 1998 Nov 1;335(Pt 3):533–9. doi: 10.1042/bj3350533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Oakhill JS, Chen ZP, Scott JW, et al. β-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK). Proc Natl Acad Sci U S A. 2010 Nov 9;107(45):19237–19241. doi: 10.1073/pnas.1009705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang CS, Hawley SA, Zong Y, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017 Aug 3;548(7665):112–116. doi: 10.1038/nature23275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008 Apr 25;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Inoki K, Li Y, Xu T, et al. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003 Aug 1;17(15):1829–1834. doi: 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003 Nov 26;115(5):577–590. doi: 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- [59].Lee MN, Ha SH, Kim J, et al. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol Cell Biol. 2009. Jul;29(14):3991–4001. doi: 10.1128/MCB.00165-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zheng M, Wang YH, Wu XN, et al. Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat Cell Biol. 2011. Mar;13(3):263–272. doi: 10.1038/ncb2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Efeyan A, Zoncu R, Chang S, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013 Jan 31;493(7434):679–683. doi: 10.1038/nature11745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009 May 29;137(5):873–886. doi: 10.1016/j.cell.2009.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011 Oct 21;44(2):304–316. doi: 10.1016/j.molcel.2011.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Roberts DJ, Tan-Sah VP, Ding EY, et al. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol Cell. 2014 Feb 20;53(4):521–533. doi: 10.1016/j.molcel.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Eng CH, Yu K, Lucas J, et al. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010 Apr 27;3(119):ra31. doi: 10.1126/scisignal.2000911 [DOI] [PubMed] [Google Scholar]
- [66].Vicinanza M, Korolchuk VI, Ashkenazi A, et al. PI(5)P regulates autophagosome biogenesis. Mol Cell. 2015 Jan 22;57(2):219–234. doi: 10.1016/j.molcel.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010. May;6(4):506–522. doi: 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- [68].Ren C, Liu J, Gong Q. Functions of autophagy in plant carbon and nitrogen metabolism. Front Plant Sci. 2014;5:301. doi: 10.3389/fpls.2014.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Araújo WL, Tohge T, Ishizaki K, et al. Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011. Sep;16(9):489–498. doi: 10.1016/j.tplants.2011.05.008 [DOI] [PubMed] [Google Scholar]
- [70].Thompson AR, Doelling JH, Suttangkakul A, et al. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005. Aug;138(4):2097–2110. doi: 10.1104/pp.105.060673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005. May;42(4):535–546. doi: 10.1111/j.1365-313X.2005.02397.x [DOI] [PubMed] [Google Scholar]
- [72].Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008. Mar;178(3):1339–1353. doi: 10.1534/genetics.107.086199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yang C, Shen W, Yang L, et al. HY5-HDA9 module transcriptionally regulates Plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol Plant. 2020 Mar 2;13(3):515–531. doi: 10.1016/j.molp.2020.02.011 [DOI] [PubMed] [Google Scholar]
- [74].Wu J, Michaeli S, Picchianti L, et al. ATI1 (ATG8-interacting protein 1) and ATI2 define a plant starvation-induced reticulophagy pathway and serve as MSBP1/MAPR5 cargo receptors. Autophagy. 2021. Nov;17(11):3375–3388. doi: 10.1080/15548627.2021.1872886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Gou M, Ran X, Martin DW, et al. The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nat Plants. 2018. May;4(5):299–310. doi: 10.1038/s41477-018-0142-9 [DOI] [PubMed] [Google Scholar]
- [76].Izumi M, Nakamura S, Li N. Autophagic turnover of chloroplasts: its roles and regulatory mechanisms in response to sugar starvation. Front Plant Sci. 2019;10:280. doi: 10.3389/fpls.2019.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ishida H, Wada S. Autophagy of whole and partial chloroplasts in individually darkened leaves: a unique system in plants? Autophagy. 2009. Jul;5(5):736–737. doi: 10.4161/auto.5.5.8568 [DOI] [PubMed] [Google Scholar]
- [78].Izumi M, Hidema J, Wada S, et al. Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 2015. Apr;167(4):1307–1320. doi: 10.1104/pp.114.254078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Üstün S, Hafrén A, Hofius D. Autophagy as a mediator of life and death in plants. Curr Opin Plant Biol. 2017. Dec;40:122–130. doi: 10.1016/j.pbi.2017.08.011 [DOI] [PubMed] [Google Scholar]
- [80].Teper-Bamnolker P, Danieli R, Peled-Zehavi H, et al. Vacuolar processing enzyme translocates to the vacuole through the autophagy pathway to induce programmed cell death. Autophagy. 2021. Oct;17(10):3109–3123. doi: 10.1080/15548627.2020.1856492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Roa-Mansergas X, Fadó R, Atari M, et al. CPT1C promotes human mesenchymal stem cells survival under glucose deprivation through the modulation of autophagy. Sci Rep. 2018 May 3;8(1):6997. doi: 10.1038/s41598-018-25485-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015. Jun;17(6):759–770. doi: 10.1038/ncb3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Takagi A, Kume S, Kondo M, et al. Mammalian autophagy is essential for hepatic and renal ketogenesis during starvation. Sci Rep. 2016 Jan 6;6(1):18944. doi: 10.1038/srep18944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Li Y, Chao X, Yang L, et al. Impaired fasting-induced adaptive lipid droplet Biogenesis in liver-specific Atg5-deficient mouse liver is mediated by persistent nuclear factor-like 2 activation. Am J Pathol. 2018. Aug;188(8):1833–1846. doi: 10.1016/j.ajpath.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shelley HJ, Bassett JM, Milner RD. Control of carbohydrate metabolism in the fetus and newborn. Br Med Bull. 1975. Jan;31(1):37–43. doi: 10.1093/oxfordjournals.bmb.a071239 [DOI] [PubMed] [Google Scholar]
- [86].Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202(9):631–638. doi: 10.1016/j.prp.2006.04.001 [DOI] [PubMed] [Google Scholar]
- [87].Schiaffino S, Hanzlíková V. Autophagic degradation of glycogen in skeletal muscles of the newborn rat. J Cell Bio. 1972. Jan;52(1):41–51. doi: 10.1083/jcb.52.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kaushik S, Rodriguez-Navarro JA, Arias E, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011 Aug 3;14(2):173–183. doi: 10.1016/j.cmet.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Oh TS, Cho H, Cho JH, et al. Hypothalamic AMPK-induced autophagy increases food intake by regulating NPY and POMC expression. Autophagy. 2016. Nov;12(11):2009–2025. doi: 10.1080/15548627.2016.1215382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].He C, Bassik MC, Moresi V, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012 Jan 18;481(7382):511–515. doi: 10.1038/nature10758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liu Y, Nguyen PT, Wang X, et al. TLR9 and beclin 1 crosstalk regulates muscle AMPK activation in exercise. Nature. 2020. Feb;578(7796):605–609. doi: 10.1038/s41586-020-1992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Pietrocola F, Pol J, Vacchelli E, et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell. 2016 Jul 11;30(1):147–160. doi: 10.1016/j.ccell.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Martins I, Wang Y, Michaud M, et al. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ. 2014. Jan;21(1):79–91. doi: 10.1038/cdd.2013.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mizushima N. Autophagy: process and function. Genes Dev. 2007 Nov 15;21(22):2861–2873. doi: 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- [95].Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008 Jan 11;132(1):27–42. doi: 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wang P, Shao BZ, Deng Z, et al. Autophagy in ischemic stroke. Prog Neurobiol. 2018. Apr-May;163-164:98–117. [DOI] [PubMed] [Google Scholar]
- [97].Ajoolabady A, Wang S, Kroemer G, et al. Targeting autophagy in ischemic stroke: from molecular mechanisms to clinical therapeutics. Pharmacol Ther. 2021. Sep;225:107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wang P, Guan YF, Du H, et al. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy. 2012. Jan;8(1):77–87. doi: 10.4161/auto.8.1.18274 [DOI] [PubMed] [Google Scholar]
- [99].Liu Y, Xue X, Zhang H, et al. Neuronal-targeted TFEB rescues dysfunction of the autophagy-lysosomal pathway and alleviates ischemic injury in permanent cerebral ischemia. Autophagy. 2019. Mar;15(3):493–509. doi: 10.1080/15548627.2018.1531196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Xu ZQ, Zhang JJ, Kong N, et al. Autophagy is involved in neuroprotective effect of Alpha7 nicotinic acetylcholine receptor on ischemic stroke. Front Pharmacol. 2021;12:676589. doi: 10.3389/fphar.2021.676589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Rami A, Kögel D. Apoptosis meets autophagy-like cell death in the ischemic penumbra: Two sides of the same coin? Autophagy. 2008. May;4(4):422–426. doi: 10.4161/auto.5778 [DOI] [PubMed] [Google Scholar]
- [102].Wang JF, Mei ZG, Fu Y, et al. Puerarin protects rat brain against ischemia/reperfusion injury by suppressing autophagy via the AMPK-mTOR-ULK1 signaling pathway. Neural Regen Res. 2018. Jun;13(6):989–998. doi: 10.4103/1673-5374.233441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zhang X, Yan H, Yuan Y, et al. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013. Sep;9(9):1321–1333. doi: 10.4161/auto.25132 [DOI] [PubMed] [Google Scholar]
- [104].Qin AP, Liu CF, Qin YY, et al. Autophagy was activated in injured astrocytes and mildly decreased cell survival following glucose and oxygen deprivation and focal cerebral ischemia. Autophagy. 2010. Aug;6(6):738–753. doi: 10.4161/auto.6.6.12573 [DOI] [PubMed] [Google Scholar]
- [105].Pei X, Li Y, Zhu L, et al. Astrocyte-derived exosomes transfer miR-190b to inhibit oxygen and glucose deprivation-induced autophagy and neuronal apoptosis. Cell Cycle. 2020. Apr;19(8):906–917. doi: 10.1080/15384101.2020.1731649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Aisa Z, Liao GC, Shen XL, et al. Effect of autophagy on myocardial infarction and its mechanism. Eur Rev Med Pharmacol Sci. 2017. Aug;21(16):3705–3713. [PubMed] [Google Scholar]
- [107].Kanamori H, Takemura G, Goto K, et al. Autophagy limits acute myocardial infarction induced by permanent coronary artery occlusion. Am J Physiol Heart Circ Physiol. 2011. Jun;300(6):H2261–71. doi: 10.1152/ajpheart.01056.2010 [DOI] [PubMed] [Google Scholar]
- [108].Yan L, Vatner DE, Kim SJ, et al. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005 Sep 27;102(39):13807–13812. doi: 10.1073/pnas.0506843102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Sun X, Momen A, Wu J, et al. p27 protein protects metabolically stressed cardiomyocytes from apoptosis by promoting autophagy. J Biol Chem. 2014 Jun 13;289(24):16924–16935. doi: 10.1074/jbc.M113.542795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007 Mar 30;100(6):914–922. doi: 10.1161/01.RES.0000261924.76669.36 [DOI] [PubMed] [Google Scholar]
- [111].Sciarretta S, Zhai P, Shao D, et al. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012 Mar 6;125(9):1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009 Oct 9;284(41):28319–28331. doi: 10.1074/jbc.M109.024406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Troncoso R, Vicencio JM, Parra V, et al. Energy-preserving effects of IGF-1 antagonize starvation-induced cardiac autophagy. Cardiovasc Res. 2012 Feb 1;93(2):320–329. doi: 10.1093/cvr/cvr321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Aki T, Yamaguchi K, Fujimiya T, et al. Phosphoinositide 3-kinase accelerates autophagic cell death during glucose deprivation in the rat cardiomyocyte-derived cell line H9c2. Oncogene. 2003 Nov 20;22(52):8529–8535. doi: 10.1038/sj.onc.1207197 [DOI] [PubMed] [Google Scholar]
- [115].Kriel J, Loos B. The good, the bad and the autophagosome: exploring unanswered questions of autophagy-dependent cell death. Cell Death Differ. 2019. Mar;26(4):640–652. doi: 10.1038/s41418-018-0267-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- [117].Hsu PP, Sabatini DM. Cancer cell metabolism: warburg and beyond. Cell. 2008 Sep 5;134(5):703–707. doi: 10.1016/j.cell.2008.08.021 [DOI] [PubMed] [Google Scholar]
- [118].Goldsmith J, Levine B, Debnath J. Autophagy and cancer metabolism. Methods Enzymol. 2014;542:25–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Li X, Yu W, Qian X, et al. Nucleus-Translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol Cell. 2017 Jun 1;66(5):684–697.e9. doi: 10.1016/j.molcel.2017.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Walker A, Singh A, Tully E, et al. Nrf2 signaling and autophagy are complementary in protecting breast cancer cells during glucose deprivation. Free Radic Biol Med. 2018 May 20;120:407–413. doi: 10.1016/j.freeradbiomed.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Meng Q, Xu J, Liang C, et al. GPx1 is involved in the induction of protective autophagy in pancreatic cancer cells in response to glucose deprivation. Cell Death Dis. 2018 Dec 11;9(12):1187. doi: 10.1038/s41419-018-1244-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gretzmeier C, Eiselein S, Johnson GR, et al. Degradation of protein translation machinery by amino acid starvation-induced macroautophagy. Autophagy. 2017 Jun 3;13(6):1064–1075. doi: 10.1080/15548627.2016.1274485 [DOI] [PMC free article] [PubMed] [Google Scholar]


