ABSTRACT
Bevacizumab plays an important role in the first and second line treatment for metastatic colorectal cancer (CRC). And induction of hypoxia and the tumors response to it plays an important role in determining the efficacy of antiangiogenic therapy while the connection between them remains unclear. Here, we found that lactate accumulated in the tumor environment of CRC and acted as substrates for histone lactylation, and this process was further induced by cellular enhanced glycolysis in hypoxia. We determined that CRC patients resistant to bevacizumab treatment presented with elevated levels of histone lactylation and inhibition of histone lactylation efficiently suppressed CRC tumorigenesis, progression and survival in hypoxia. Histone lactylation promoted the transcription of RUBCNL/Pacer, facilitating autophagosome maturation through interacting with BECN1 (beclin 1) and mediating the recruitment and function of the class III phosphatidylinositol 3-kinase complex, which had a crucial role in hypoxic cancer cells proliferation and survival. Moreover, combining inhibition of histone lactylation and macroautophagy/autophagy with bevacizumab treatment demonstrated remarkable treatment efficacy in bevacizumab-resistance patients-derived pre-clinical models. These findings delivered a new exploration and important supplement of metabolic reprogramming-epigenetic regulation, and provided a new strategy for improving clinical efficacy of bevacizumab in CRC by inhibition of histone lactylation.
Abbreviations: 2-DG: 2-deoxy-D-glucose; BECN1: beclin 1; CQ: chloroquine; CRC: colorectal cancer; DMOG: dimethyloxalylglycine; H3K18la: histone H3 lysine 18 lactylation; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; Nala: sodium lactate; PDO: patient-derived orgnoid; PDX: patient-derived xenograft; RUBCNL/Pacer: rubicon like autophagy enhancer; SQSTM1/p62: sequestosome 1.
KEYWORDS: Antiangiogenesis, autophagy, colorectal cancer, histone lactylation, hypoxia
Introduction
Colorectal cancer (CRC) is the third most common malignancy worldwide, accounting for nearly 8.5% of all cancer deaths [1]. Recurrence and distant metastasis are still the leading causes of treatment failure and death among CRC patients [2,3]. At diagnosis, 20%-30% of patients suffer from synchronous hepatic metastases and 50%-75% of all patients with CRC develop liver metastases [4,5]. An estimated 70–80% of cases with late-stage CRC are defined as initially unresectable, and their 5-year overall survival (OS) rate is less than 10% [6–9].
Currently, the application of doublet or triplet chemotherapy regimens with targeted therapies including antiangiogenic therapy has increased response rates to 60%-70%, increasing the survival of patients with metastatic CRC [10–12]. Bevacizumab, specifically targeting VEGFA (vascular endothelial growth factor A) to inhibit VEGF (vascular endothelial growth factor)-VEGFR (vascular endothelial growth factor receptor) signaling [13], plays an essential role in the first-line and second-line treatment for metastatic CRC [14,15]. Despite the improvement in clinical outcomes for metastatic CRC as a result of antiangiogenic therapy, the majority of patients manifest intrinsic or acquired resistance to treatment and the overall benefit is limited eventually [16,17]. Therefore, it is critical to explore the mechanisms that underlie the resistance of CRC cells to antiangiogenic therapy, which may improve the prognosis of metastatic CRC.
In solid tumors, intratumoral hypoxic regions caused by antiangiogenic therapy were found to be implicated in treatment resistance [18,19]. Hypoxia induces metabolic alterations of cancer cells as an adaptive response to nutrient deprivation, which influences susceptibility to hypoxia induced cell death during antiangiogenic therapy [20]. Enhanced glycolysis, One of the most critical metabolic alterations during hypoxia, resulting in augmented lactogenesis in cancer, is associated with diverse cellular processes [21]. Lactate, an abundant oncometabolite in the tumor environment generated during Warburg effect, triggers histone lysine lactylation as a new epigenetic modification which directly stimulates gene transcription from chromatin [22]. In recent years, histone lactylation, especially histone H3 lysine 18 lactylation (H3K18la), has been reported to regulate multitudinous biological processes such as oncogenesis [23], progression [24], tumor immune escape [25] and cancer cellular metabolism reprogramming [26].
In this study, we revealed that, induced by cellular enhanced glycolysis and consequent lactogenesis, histone lactylation levels were upregulated in CRC patients resistant to bevacizumab treatment. Inhibition of histone lactylation efficiently suppressed CRC tumorigenesis, progression and survival in hypoxia. Mechanistically, histone lactylation promoted the transcription of RUBCNL/Pacer (rubicon like autophagy enhancer), which was recognized as a promoter of autophagosome maturation through interaction with BECN1/Beclin 1, one of the main components of this pathway. We revealed that inhibition of histone lactylation enhanced the sensitivity of CRC cells to bevacizumab treatment in cell-based xenografts, patient-derived xenografts (PDXs) and patient-derived orgnoids (PDOs) models. Thus, our study suggested that inhibition of histone lactylation may be a novel strategy for improving clinical efficacy of bevacizumab in CRC.
Results
Colorectal cancer (CRC) resistance to bevacizumab treatment exhibited increased lactylation levels which was associated with poor survival in CRC patients
In order to explore the mechanisms that underlie the resistance of CRC cells to bevacizumab treatment, we collected CRC tissue specimens from fresh primary surgical specimens without any systemic treatment in the cohort previously described [27] and built up a biobank containing paraffin-embedded tumor section, extracted RNA, PDXs, PDOs and etc. (Figure 1A). Among them, patients with recurrent or metastatic tumors treated with first-line bevacizumab treatment whose clinical therapeutic response matched with the treatment evaluation in biobank (PDXs treated with bevacizumab, and PDOs treated with Dimethyloxalylglycine (DMOG) [28,29], an inhibitor of HIF prolylhydroxylase stabilizing and accumulating HIF1A/HIF-1α (hypoxia inducible factor 1 subunit alpha) protein in the nucleus under conditions of normoxia, which could mimic hypoxic environment caused by antiangiogenic therapy) were included (Table S1) and transcriptome sequencing was performed using pre-retained RNA (Figure 1B). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of upregulated genes of bevacizumab treated stable disease (SD)/progressive disease (PD) versus partial response (PR) revealed a significant enrichment in glycolysis signaling (Figure 1C). In the TCGA database, the genes associated with lactate production and secretion are highly expressed and positively correlated with VEGFA expression in CRC (Figure S1A-B), which suggested that bevacizumab resistance may be related to enhanced glycolysis and follow-by lactogenesis of cancer cells. To further investigate, enhanced glycolysis and lactogenesis were also found in paraffin-embedded tumor section derived from PDXs with bevacizumab resistance (Figure 1D, Figure S1C-D). Interestingly, the levels of lactylation and H3K18la, which were recognized as oncogenesis markers, obviously increased in patients with bevacizumab treated SD/PD compared with PR (Figure 1D-E, Figure S1C-D). And the increased levels of lactylation and H3K18la were further reflected in fresh surgical tumor tissues compared with the matched adjacent normal tissues from 10 random stage I-III CRC patients (Figure 1F). We examined the levels of lactylation and H3K18la in the CRC cohort and their conceivable clinical significance. Immunohistochemistry (IHC) staining showed higher levels of lactylation (Figure 1G) and H3K18la (Figure 1J) in CRC tissues than in adjacent normal tissues and high levels of lactylation (Figure 1H, Figure S1E) and H3K18la (Figure 1K, Figure S1F) were associated with shorter overall survival in patients with CRC. Moreover, in the SYSUCC cohort, based on the clinical response assessment, response rates were adversely associated with levels of lactylation (Figure 1I) and H3K18la (Figure 1L). These clinical data suggested that increased lactylation and H3K18la levels might confer resistance to bevacizumab and were associated with poor survival for patients with CRC.
Figure 1.
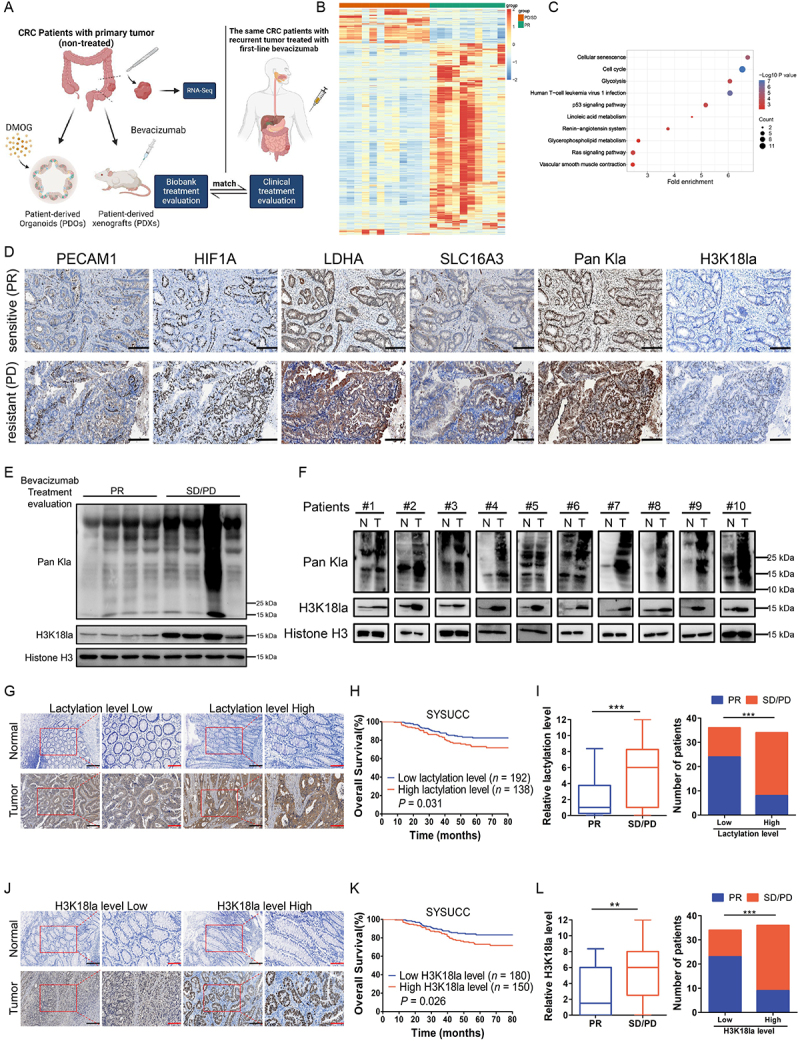
Colorectal cancer (CRC) resistance to bevacizumab treatment exhibited increased lactylation levels which was associated with poor survival in CRC patients.
Note: (A) Schematic description of biobank construction and CRC patients cohort administration. (B) Heatmap of 1,554 differentially expressed genes in bevacizumab treated SD/PD (red) and PR (green) CRC patients (1,285 downregulated and 269 upregulated in SD/PD versus PR (n = 12 and 10; all absolute log2 fold change > 0.5, false discovery rate (FDR) <10%); PD, progressive disease; SD, stable disease; and PR, partial response. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of top 1,000 significantly (FDR <5%) upregulated genes of SD/PD versus PR. (D) Representative Paraffin-embedded tumor sections derived from patient-derived xenografts (PDXs) mice (PDX #1-bevacizumab treated sensitive (PR) and PDX #6-bevacizumab treated resistant (PD)) were stained with PECAM1, HIF1A, LDHA, SLC16A3, Pan Kla or H3K18la antibodies (scale bar: 200 μm). (E) Western blotting analysis showing the levels of lactylation or H3K18la in 8 primary tumor tissues reserved before relapse from CRC patients who were treated with first-line bevacizumab treatment after recurrence, including 4 samples from patients with bevacizumab treated SD/PD and 4 samples from patients with bevacizumab treated PR. (F) Western blotting analysis of lactylation or H3K18la levels in 10 matched CRC tissues (T) and adjacent noncancerous tissues (N). (G and J) Two representative cases show high compared with low levels of lactylation (G) or H3K18la (J) in human CRC tumor tissues and adjacent normal tissues analyzed by immunohistochemistry (IHC) staining (scale bar: black, 200 μm; red, 100 μm;). (H and K) The overall survival curves of CRC patients in the SYSUCC cohort with low and high lactylation (H) or H3K18la (K) levels are generated using the Kaplan-Meier method and the log-rank test; SYSUCC, Sun Yat-sen University Cancer Center. (I and L) Statistics of relative lactylation (I) or H3K18la levels (L) in CRC patients with bevacizumab treated PR (n = 32) or bevacizumab treated SD/PD (n = 38) (left) and number of CRC patients with PR or SD/PD in low or high lactylation (I) or H3K18la levels (L) groups (right); **P ≤0.01, ***P ≤0.001, by Mann-Whitney test (left), or Chi-Square test (right).
Effects of histone lactylation inhibition on colorectal cancer cells proliferation and survival in hypoxia
Next, we set out to explore the biological functions of histone lactylation in CRC cell lines. Much higher levels of lactylation and H3K18la levels were observed in CRC cell lines compared with normal colon epithelial cell lines, especially in HCT116 and SW620 cells (Figure 2A). Then, HCT116 and SW620 cells were cultured in hypoxia (1% oxygen) compared with in normoxia and followed by western blotting analysis with antibodies against diverse forms of histone lactylation, which showed a certain level of H3K18la in normoxia and significantly increased level of H3K18la in hypoxia (Figure 2B). Combined with the fact that H3K18la is known to be an important cancer-related marker, we identified that H3K18la was a key histone lactylation modification in CRC cell lines. Herein, intracellular lactate production and histone lactylation were blocked by glycolytic inhibitors (2-deoxy-D-glucose [2-DG] and oxamate) or siRNAs for LDHA (lactate dehydrogenase A) (Figure S2A). Notably, glycolysis inhibitors achieved a significant dose-dependent decrease in intracellular lactate (Figure S2B), as well as global lactylation and H3K18la levels in CRC cells in hypoxia (Figure 2C) or in normoxia (Figure S2C). The same phenomenon was found after LDHA silencing, which could be partially restored by supplementing with sodium lactate (Nala) (Figure S2C). Next, we found that inhibition of lactylation and H3K18la levels suppressed CRC cells proliferation (Figure S2E-F) and colony formation ability (Figure S2G-H). At the meantime, hypoxia induced intracellular lactate production and increased lactylation and H3K18la in HCT116 and SW620 cells (Figure S2D, Figure 2D). In hypoxia, inhibition of lactylation and H3K18la levels also suppressed proliferation (Figure 2E-F) and colony formation (Figure 2G-H) ability of CRC cells and increased hypoxia induced apoptosis (Figure 2I-K) of CRC cells. Taken together, these results revealed that histone lactylation and H3K18la play a role in tumorigenesis and progression of CRC and inhibition of lactylation and H3K18la levels suppressed CRC cells proliferation and survival in hypoxia.
Figure 2.
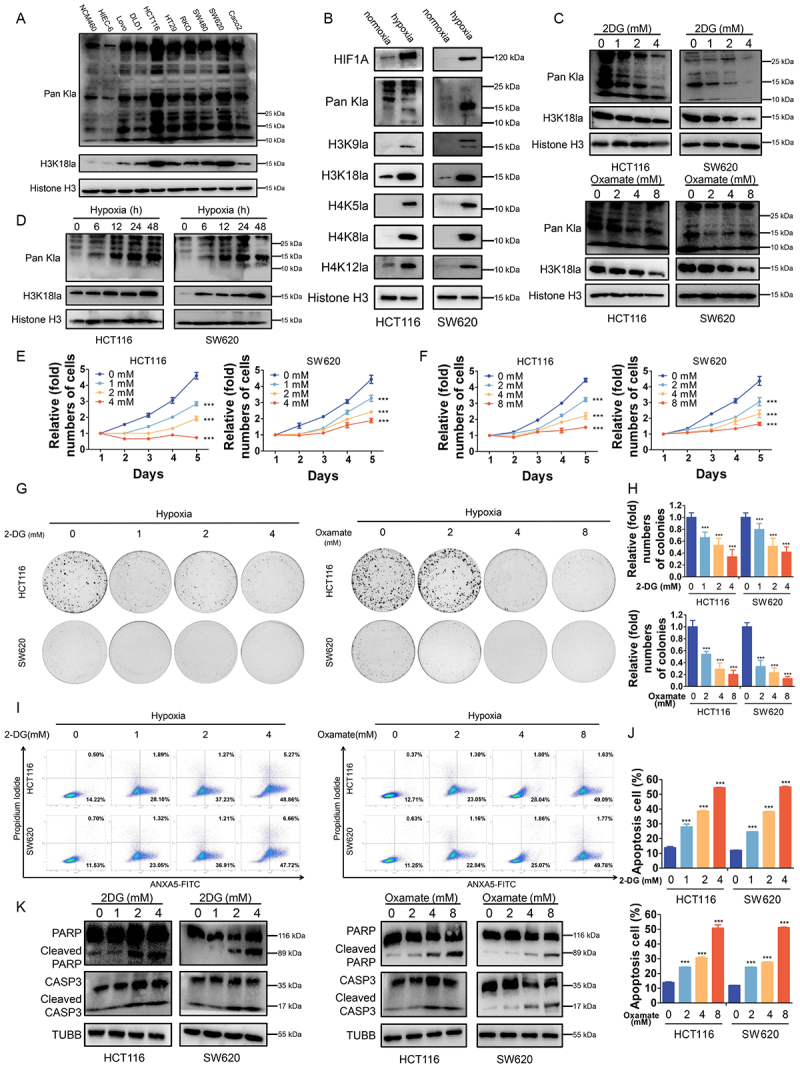
Effects of histone lactylation inhibition on colorectal cancer cells proliferation and survival in hypoxia.
Note: (A) Western blotting analysis of lactylation or H3K18la levels in normal colon epithelial cells and CRC cells. (B) Western blotting analysis of Pan- and site-specific histone lactylation in HCT116 and SW620 cells in normoxia compared with in hypoxia (1% oxygen, 24 h). (C) Lactylation and H3K18la levels were detected in HCT116 and SW620 cells cultured in different concentrations of 2-DG or oxamate for 24 h in hypoxia (1% oxygen) by western blot. (D) Lactylation and H3K18la levels were detected in HCT116 and SW620 cells cultured in hypoxia (1% oxygen) at the indicated time. (E and F) Proliferation of HCT116 and SW620 cells cultured in different concentrations of 2-DG (E) or oxamate (F) in hypoxia (1% oxygen) was analyzed using CCK8 assay. ***P ≤0.001. (G) Tumor growth of HCT116 and SW620 cells treated with different concentrations of 2-DG or oxamate in hypoxia (1% oxygen) was evaluated by colony formation assay. (H) Statistical analysis of the colony formation assay performed using HCT116 and SW620 cells treated with different concentrations of 2-DG or oxamate in hypoxia (1% oxygen). All of the experiments were performed in triplicate, and relative colony numbers are shown as means ± SD. ***P ≤0.001. (I) ANXA5/annexin V-FITC and PI staining showing apoptosis in HCT116 and SW620 cells cultured in different concentrations of 2-DG or oxamate in hypoxia (1% oxygen). (J) Quantification of apoptotic cells. ***P ≤0.001. (K) Western blotting analysis of cleaved CASP3 and cleaved PARP in in HCT116 and SW620 cells cultured in different concentrations of 2-DG or oxamate in hypoxia (1% oxygen).
Histone lactylation activated RUBCNL transcription in colorectal cancer
To reveal the regulatory role of histone lactylation in gene expression, we performed chromatin immunoprecipitation sequencing (ChIP-seq) using anti-H3K18la antibodies and found obvious enrichment in promoter regions (Figure 3A, Figure S3A). KEGG analysis of H3K18la-specific genes enriched in tumor-related and Macroautophagy/autophagy pathways (Figure 3B). By overlapping the gene sets among ChIP-seq, RNA-seq (Figure 1B) and TCGA database, 7 differentially expressed genes were identified (Figure 3C). Among them, RUBCNL, an autophagy enhancer gene, was identified to have a marked enrichment of H3K18la peaks in the promoter position (Figure 3D) and was positively correlated with the genes associated with lactate production and secretion in CRC in TCGA database (Figure S3C). Moreover, ChIP-qPCR assays confirmed that H3K18la was enriched at the RUBCNL promoter, which was reduced by glycolytic inhibitors or siRNA for EP300, a histone lactylation writer (Figure 3E-F). As expected, mRNA level of HIF1A, LDHA and SLC16A3 (solute carrier family 16 member 3) expression were significantly increased followed by increased mRNA level of RUBCNL expression in hypoxia (Figure 3G). Increased protein level of RUBCNL expression was also found in hypoxia or in cells cultured in different concentrations of lactate (Figure 3H-I). On the contrary, decreased protein level of H3K18la and RUBCNL expression were found after treated with siRNA for EP300 (E1A binding protein p300) or EP300 inhibitor, A-485 (Figure S3D). Both mRNA (Figure S3E-F) and protein (Figure 3J-K) levels of RUBCNL expression were increased after treated with DMOG and were decreased after treated with glycolysis inhibitors, which could be partially restored by supplementing with Nala. IHC staining also showed that RUBCNL was highly expressed in cancer and a poor prognostic factor for CRC (Figure 3L-M, Figure S3G), and its expression was negatively correlated with the efficacy of bevacizumab (Figure 3N). Moreover, expression levels of RUBCNL were positively associated with levels of lactylation and H3K18la (Figure S3H). Collectively, these data suggested that the transcription of RUBCNL is positively regulated by H3K18la.
Figure 3.
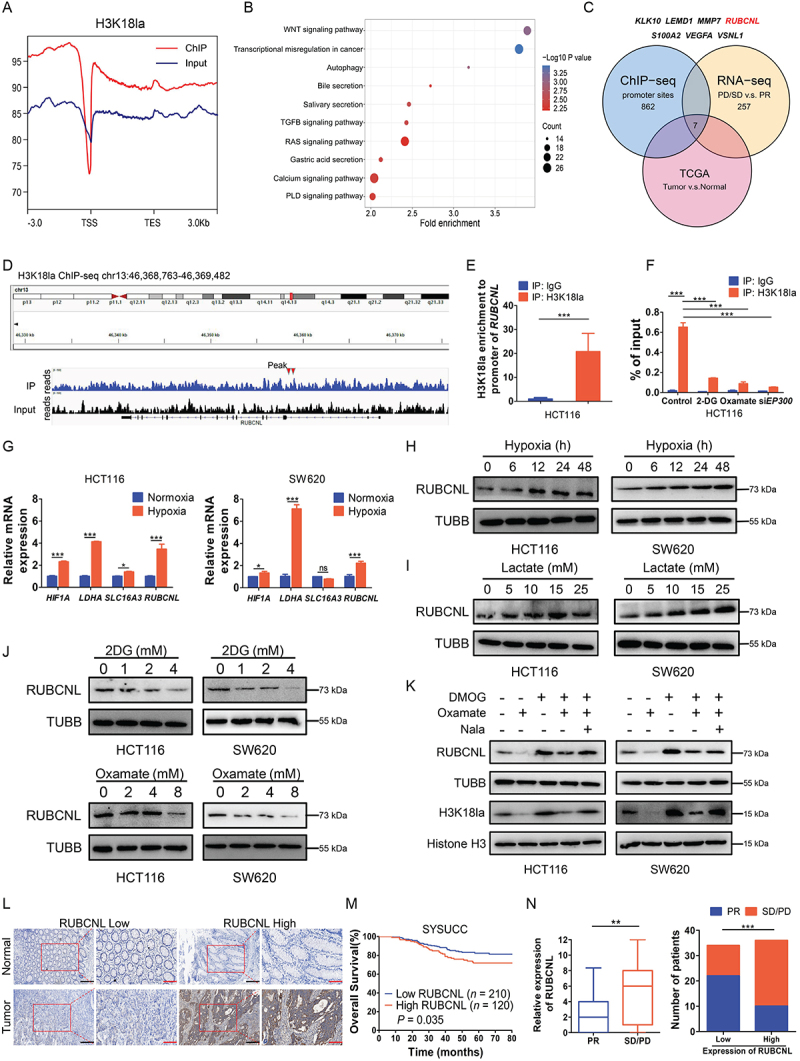
Histone lactylation activated RUBCNL transcription in colorectal cancer.
Note: (A) Distribution of H3K18la sites relative to translation start site (TSS). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of H3K18la peaks. (C) Strategy to identify potential specific downstream targets of H3K18la modification. (D) Representative IGV tracks showing enriched H3K18la modification in RUBCNL promotor by ChIP-seq. Arrows are the H3K18la peaks at the RUBCNL promotor. (E) DNA fragments from HCT116 cells were immunoprecipitated with the H3K18la-specific antibody and analyzed by qPCR using the indicated primers. ***P ≤0.001. (F) ChIP-qPCR analysis for H3K18la status at the RUBCNL promotor of HCT116 cells treated with 4 mM 2-DG, 8 mM oxamate or transiently transfected with siRNAs targeting EP300 for 24 h. ***P ≤0.001. (G) Expression of HIF1A, LDHA, SLC16A3 and RUBCNL were detected in HCT116 and SW620 cells in normoxia or hypoxia (1% oxygen) for 24 h by RT-qPCR. ns, no significance, *P ≤0.05, ***P ≤0.001. (H) Western blotting analysis of RUBCNL expression in HCT116 and SW620 cells cultured in hypoxia (1% oxygen) at the indicated time. (I) Western blotting analysis of RUBCNL expression in HCT116 and SW620 cells cultured in different concentrations of lactate for 24 h. (J) Western blotting analysis of RUBCNL expression in HCT116 and SW620 cells cultured in different concentrations of 2-DG or oxamate for 24 h. (K) Western blotting analysis of RUBCNL expression and H3K18la levels in HCT116 and SW620 cells treated with 2 mM DMOG, 8 mM oxamate or 5 mM Nala for 24 h. (L) Two representative cases show high compared with low RUBCNL expression in human CRC tumor tissues and adjacent normal tissues analyzed by immunohistochemistry (IHC) staining (scale bar: black, 200 μm; red, 100 μm;). (M) The overall survival curves of CRC patients in the SYSUCC cohort cohort with low and high RUBCNL expression are generated using the Kaplan-Meier method and the log-rank test. (N) Statistics of relative RUBCNL expression in CRC patients with bevacizumab treated PR (n = 32) or bevacizumab treated SD/PD (n = 38) (left) and number of CRC patients with PR or SD/PD in low or high RUBCNL expression groups (right); **P ≤0.01, ***P ≤0.001, by Mann-Whitney test (left), or Chi-Square test (right).
RUBCNL, as a novel oncogene in colorectal cancer, plays a crucial role in cancer cells proliferation and survival in hypoxia in vitro and in vivo
In light of our above findings, since RUBCNL can be up-regulated by H3K18la, we next explored its function in CRC. We first established stable RUBCNL knockdown and over expressing cells from the HCT116 and SW620 CRC cell lines (Figure 4A,4M). We found that RUBCNL acted as an oncogene in CRC cell lines (Figure S4A-C) and silencing RUBCNL expression suppressed proliferation (Figure 4B) and colony formation (Figure 4C-D) ability of CRC cells in hypoxia and increased hypoxia induced apoptosis (Figure 4E-G) of CRC cells. To further verify the role of RUBCNL, we subcutaneously injected stable shNC and shRUBCNL HCT116 cell lines into nude mice to establish xenograft mouse models. When tumors reached about 100 mm3, mice were randomized into four groups and treated with or without bevacizumab. The tumors with shRUBCNL were smaller and weighed less than those with shNC and the efficacy of bevacizumab was significantly enhanced in the shRUBCNL group compared with that in the shNC group (Figure 4H-J). Silencing RUBCNL expression combined with bevacizumab treatment significantly reduced the growth rate of CRC tumors, as shown by IHC staining (Figure 4K-L). While RUBCNL overexpression enhanced the ability of proliferation (Figure 4N) and colony formation (Figure 4O, Figure S4G) treated with oxamate in hypoxia and decreased hypoxia induced apoptosis (Figure S4H, Figure 2P-Q) of CRC cells. To further determine whether RUBCNL increased the survival of CRC cells in hypoxia through enhancing autophagy pathway, CRC cells with RUBCNL overexpression were treated with chloroquine (CQ), a classic autophagy inhibitor, and showed a decreased ability of proliferation (Figure S4I) and colony formation (Figure S4J-K). Taken together, these data indicated that RUBCNL acts as an oncogene and confers to bevacizumab resistance in CRC.
Figure 4.
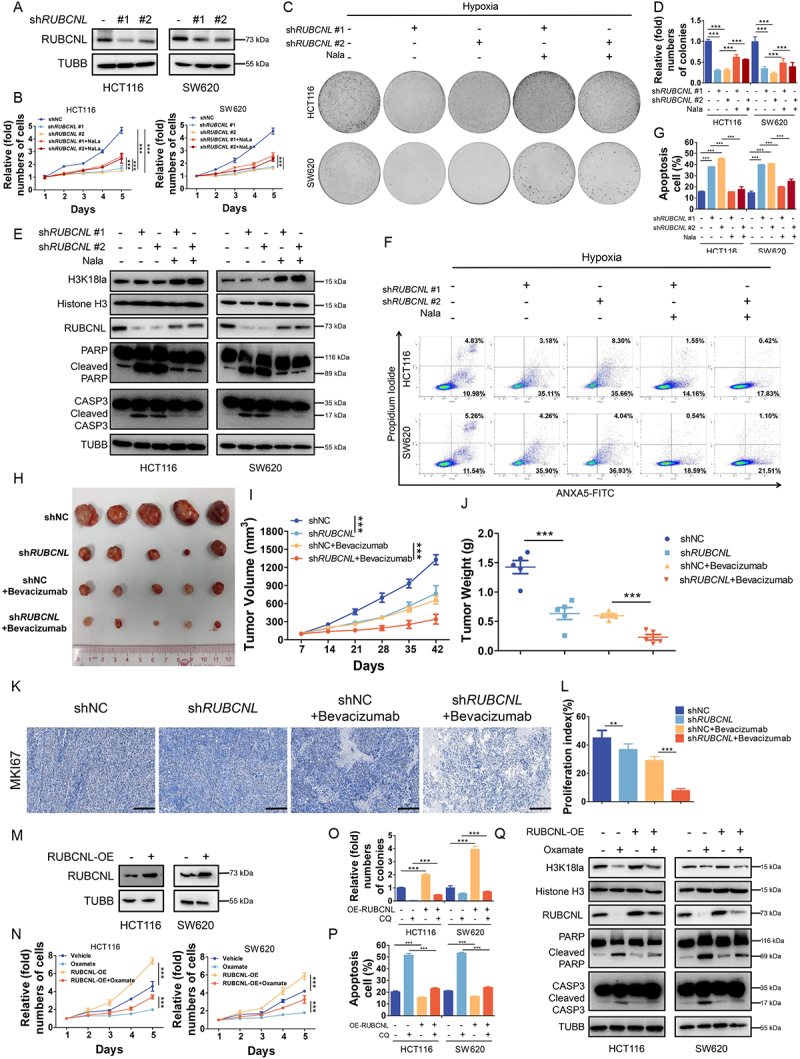
Histone lactylation promoted colorectal cancer cells survival and proliferation in hypoxia through RUBCNL.
Note: (A) Western blotting analysis of RUBCNL in HCT116 and SW620 cells transfected with shRUBCNL. (B) Proliferation of HCT116 and SW620 cells after RUBCNL silencing and supplemented with 5 mM sodium lactate (Nala) in hypoxia (1% oxygen) was analyzed using CCK8 assay. ***P ≤0.001. (C) Tumor growth of HCT116 and SW620 cells after RUBCNL silencing and supplemented with 5 mM Nala in hypoxia (1% oxygen) was evaluated by colony formation assay. (D) Statistical analysis of the colony formation assay performed using HCT116 and SW620 cells after RUBCNL silencing and supplemented with 5 mM Nala in hypoxia (1% oxygen). All of the experiments were performed in triplicate, and relative colony numbers are shown as means ± SD. ***P ≤0.001. (E) Western blotting analysis of cleaved CASP3 and cleaved PARP in HCT116 and SW620 cells after RUBCNL silencing and supplemented with 5 mM Nala in hypoxia (1% oxygen). (F) ANXA5/annexin V-FITC and PI staining showing apoptosis in HCT116 and SW620 cells after RUBCNL silencing and supplemented with 5 mM Nala in hypoxia (1% oxygen). (G) Quantification of apoptotic cells. ***P ≤0.001. (H) Photograph and comparison of tumor sizes in different groups. Bevacizumab, 5 mg/kg every 3 days. (I) Growth curves of tumors formed by indicated cells. ***P ≤0.001. (J) Tumor weights in the indicated groups. ***P ≤0.001. (K) Representative images of IHC staining of MKI67/Ki 67 in mouse tumors. (L) Quantification of proliferation index in the indicated xenograft tumors, proliferation index was determined using the percentage of MKI67-positive cells. **P ≤0.01, ***P ≤0.001. (M) Western blotting analysis of RUBCNL in HCT116 and SW620 cells transfected with RUBCNL-expressing plasmid. (N) Proliferation of HCT116 and SW620 cells with or without RUBCNL overexpression treated with 8 mM oxamate in hypoxia (1% oxygen) was analyzed using CCK8 assay. ***P ≤0.001. (O) Statistical analysis of the colony formation assay performed using HCT116 and SW620 cells with or without RUBCNL overexpression treated with 8 mM oxamate in hypoxia (1% oxygen). All of the experiments were performed in triplicate, and relative colony numbers are shown as means ± SD. ***P ≤0.001. (P) Quantification of apoptotic cells. ***P ≤0.001. (Q) Western blotting analysis of cleaved CASP3 and cleaved PARP in HCT116 and SW620 cells with or without RUBCNL overexpression treated with 8 mM oxamate in hypoxia (1% oxygen).
RUBCNL promoted autophagasome maturation through mediating the recruitment and function of the PtdIns3K complex
Next, we set out to investigate exactly how RUBCNL increases the survival of cancer cells in hypoxia and confers bevacizumab resistance through autophagy pathway in CRC. CRC cells with stable RUBCNL knockdown showed a decreased ratio of MAP1LC3/LC3 (microtubule associated protein 1 light chain 3 beta)-II:I and accumulation of SQSTM1/p62 (sequestome 1), indicating a suppression of autophagic flux (Figure S5A), which could be partially restored by supplementing with Nala (Figure 5A). While CRC cells with stable RUBCNL overexpression showed an increased ratio of LC3-II:I and degradation of SQSTM1, indicating a promotion of autophagic flux (Figure S5B), which could be suppressed by adding oxamate (Figure S5C, Figure 5B). Combined with the results of expression levels of RUBCNL and H3K18la shown in Figures 4E,Q we considered that H3K18la promoted autophagy mainly by facilitating the expression of RUBCNL, rather than directly regulated by H3K18la. To further assess the role of RUBCNL in autophagy, we tested the effects of RUBCNL on autophagic flux using the mRFP-GFP-LC3 adenovirus with or without DMOG in RUBCNL knockdown or overexpressing CRC cells. The data showed that RUBCNL knockdown cells significantly blocked autophagosome maturation, as measured by the mRFP+ GFP− puncta in both DMOG treated and untreated cells, while RUBCNL overexpression enhanced autophagosome maturation (Figure 5C-D). Furthermore, using transmission electron microscopy, we observed significant accumulation of autophagosome in RUBCNL knockdown cells while autolysosome in RUBCNL overexpression cells, under both DMOG treated and untreated conditions, confirming that RUBCNL mediates autophagasome maturation.
Figure 5.
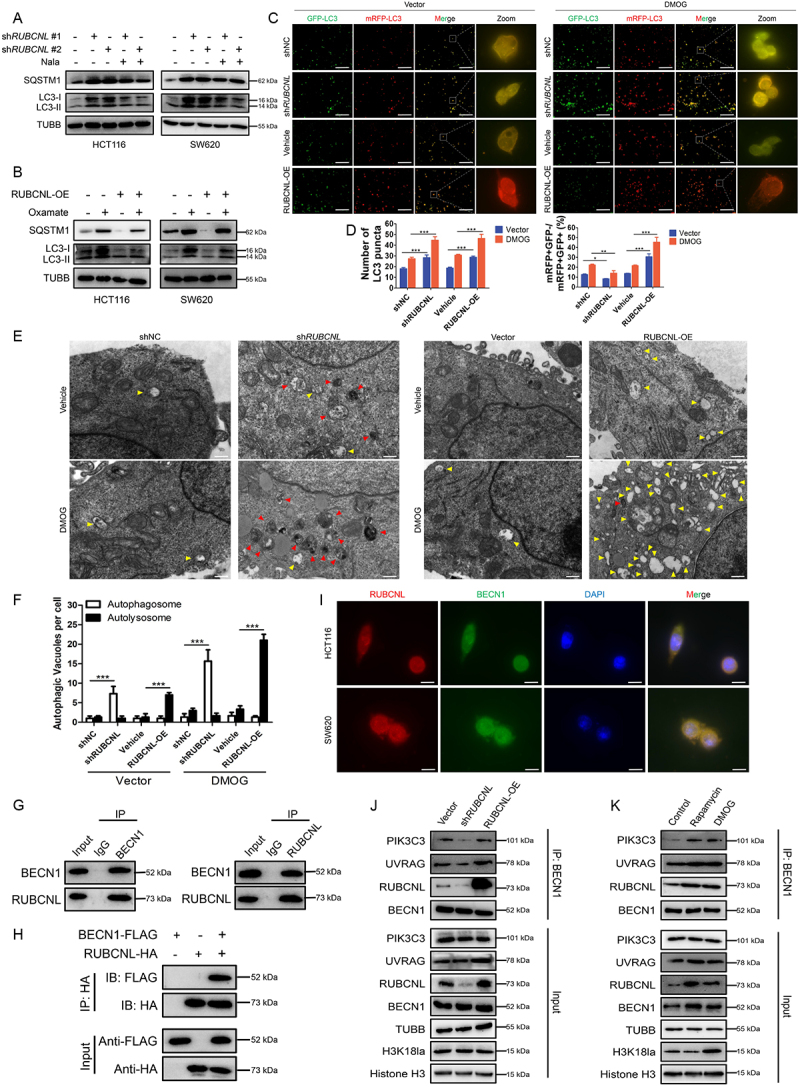
RUBCNL promoted autophagasome maturation through mediating the recruitment and function of PtdIns3K complex.
Note: (A and B) Western blot analysis of LC3-II:I and SQSTM1 expression in HCT116 and SW620 cells after RUBCNL silencing and supplemented with 5 mM Nala (A) or with or without RUBCNL overexpression treated with 8 mM oxamate (B). (C and D) mRFP-GFP-LC3 was expressed in HCT116 cells transfected with shRUBCNL or RUBCNL-expressing plasmid, respectively. Cells were treated with DMSO (left panel) or 2 mM DMOG (right panel) for 6 h. The number of LC3 puncta was analyzed by fluorescence microscope (C). GFP-negative mRFP-positive (GFP-mRFP+) puncta, which indicate autolysosomes, were quantified and are summarized in (D) (scale bar: 100 μm). *P ≤0.05, **P ≤0.01, ***P ≤0.001. (E and F) HCT116 cells transfected with shRUBCNL or RUBCNL-expressing plasmid, respectively, were treated with DMSO or 2 mM DMOG for 6 h and then analyzed by transmission electron microscopy (E) (scale bar: 0.5 μm). Arrows indicate autophagic vacuole (red arrows, autophagosome; yellow arrows, autolysosome). The autophagic vacuole per cross-sectioned cell under EM was calculated and is summarized in (F). ***P ≤0.001. G. Endogenous interaction between BECN1 and RUBCNL was determined using co-IP with anti-BECN1 or anti-RUBCNL antibodies in HCT116 cells. (H) Exogenous interaction between BECN1 and RUBCNL was determined using co-IP with anti-FLAG or anti-HA antibodies in HEK 293T cells co-transfected with BECN1-FLAG and RUBCNL-HA. (I) Immunofluorescence staining showing colocalization of endogenous RUBCNL (red) and BECN1 (green) in HCT116 and SW620 cells. The nucleus is labeled by DAPI (blue) (scale bar: 20 μm). (J) Endogenous PtdIns3K complexes were IP from cells with Vector, shRUBCNL, or RUBCNL overexpression using anti-BECN1 antibody, and then they were analyzed by western blot. (K) Endogenous PtdIns3K complexes were IP from control cells and cells treated with 200 nM rapamycin or 2 mM DMOG using anti-BECN1 antibody, and they were analyzed by western blot.
We searched for RUBCNL-interacting proteins by immunoprecipitation (IP) with anti-RUBCNL beads (Figure S5A). Mass spectrometry from the BioGRID database showed that RUBCNL may interact with BECN1, a component of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex which mediates vesicle-trafficking processes, and that was confirmed by western blotting analysis after IP. A Co-IP assay was performed using endogenous proteins in HCT116 cells or exogenous proteins in HEK 293T cells transfected with BECN1-FLAG and RUBCNL-HA. And an interaction between BECN1 and RUBCNL was detected (Figure 5G-H). In addition, immunofluorescence was used to observe the colocalization status of BECN1 and RUBCNL (Figure 5I). We further examined their association by immunoprecipitating endogenous BECN1 complexes in RUBCNL wild type, knockdown and overexpression HCT116 cells. As shown in Figure 5J, RUBCNL knockdown decreased the association among BECN1, UVRAG (UV radiation resistance associated), and PIK3C3/VPS34 (phosphatidylinositol 3-kinase catalytic subunit type 3) while RUBCNL overexpression increased the association. Moreover, the association between the RUBCNL and PtdIns3K complex was enhanced in CRC cells treated with DMOG or rapamycin (an autophagy agonist) (Figure 5K). Collectively, the data suggested that RUBCNL promoted autophagasome maturation through mediating the recruitment and function of PtdIns3K complex and H3K18la promoted autophagy mainly by facilitating the expression of RUBCNL and subsequent cellular processes.
Histone lactylation inhibition enhanced antitumor effects of bevacizumab in colorectal cancer
Previously, several studies implicated autophagy as an essential mechanism of resistance to antiangiogenic therapies and autophagy inhibitors may help prevent resistance to anti-angiogenic therapy in cancer [19,30,31]. However, the efficacy of autophagy inhibitors is still limited. Herein, we found that either target inhibition of histone lactylation by oxamate or inhibition of autophagy by CQ could significantly suppress proliferation (Figure 6A) and induce apoptosis (Figure 6B-D) of CRC cells treated with DMOG, and these effects were reinforced by combination. To further verify, we tested the effects of inhibition of histone lactylation or autophagy in PDXs and PDOs resistant to bevacizumab. In PDXs, we found that inhibition of histone lactylation or autophagy combined with bevacizumab treatment significantly reduced the growth of CRC tumors than bevacizumab treatment alone, as shown by H&E and MKI67/Ki 67 IHC staining (Figure 6E) and significant reduced PDX tumor growth and tumor weight (Figure 6F-G, Figure S6A-B). The same results were found in bevacizumab resistant PDOs. When the combination was assessed on matrix-embedded human colorectal cancer-derived organoid cultures, synergy between DMOG and oxamate, CQ or both was maintained (Figure 6H-I, Figure S6C). Bright field images and paraffin-embedded tumor sections stained with H&E of PDOs showed an outstanding synergistic antitumor effect (Figure 6J). Furthermore, HCT116 cell lines were injected into nude mice to establish xenograft mouse models and also showed a synergistic antitumor effect of combining bevacizumab with inhibition of histone lactylation and autophagy, which was evident by significantly reduced tumor growth and tumor weight (Figure 6K-M). The combination of bevacizumab with inhibition of histone lactylation and autophagy resulted in proliferation suppression and more apoptosis by comparison with bevacizumab treatment alone, as determined by IHC (Figure 6N-P). These data collectively showed that inhibition of histone lactylation enhanced antitumor effects of bevacizumab in colorectal cancer.
Figure 6.
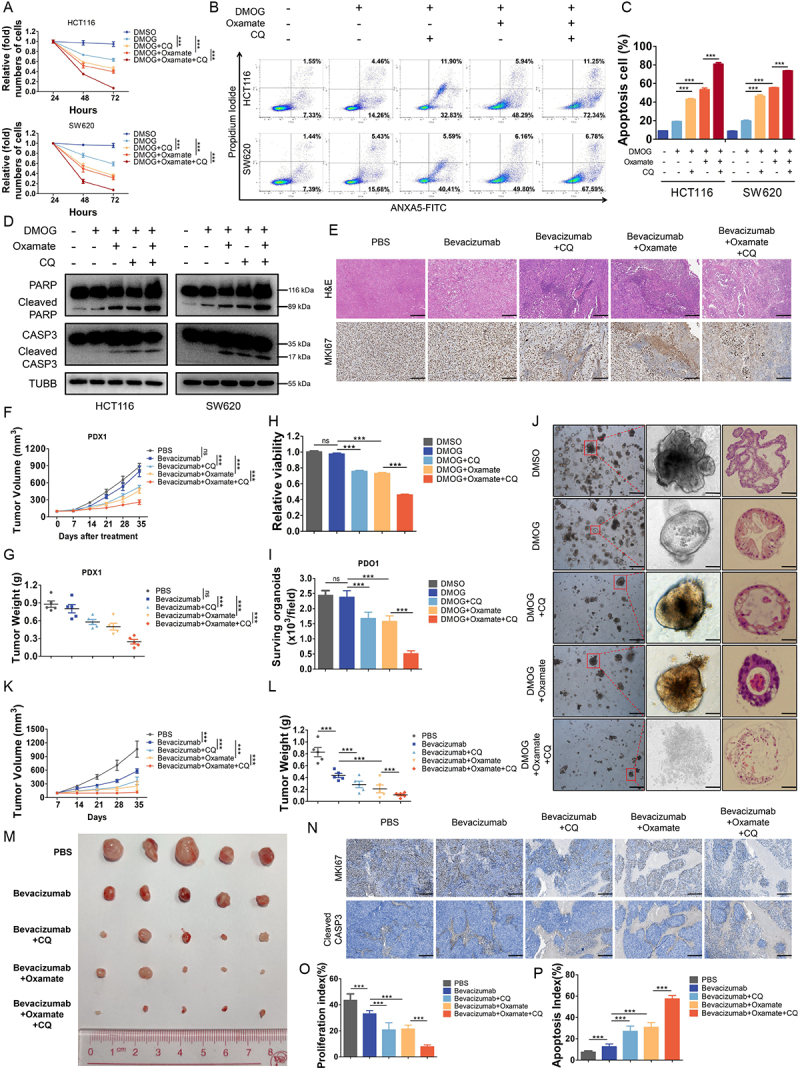
Histone lactylation inhibition enhanced antitumor effects of bevacizumab in colorectal cancer.
Notes: (A) Proliferation of HCT116 and SW620 cells treated with 2 mM DMOG, 5 μM chloroquine (CQ), 8 mM oxamate or indicated combination was analyzed using CCK8 assay. ***P ≤0.001. (B) ANXA5/annexin V-FITC and PI staining showing apoptosis in HCT116 and SW620 cells treated with 2 mM DMOG, 5 μM CQ, 8 mM oxamate or indicated combination. (C) Quantification of apoptotic cells. ***P ≤0.001. (D) Western blotting analysis of cleaved CASP3 and cleaved PARP in HCT116 and SW620 cells treated with 2 mM DMOG, 5 μM CQ, 8 mM oxamate or indicated combination. (E) Paraffin-embedded tumor sections derived from the patient-derived xenograft (PDX) mice treated with bevacizumab (5 mg/kg every 3 days), CQ (3.5 mg/kg, every day), oxamate (500 mg/kg, every day) or indicated combination were stained with H&E or MKI67 antibodies (scale bar: 200 μm). (F) Growth curves of PDX #1 tumors in the indicated groups. ns, no significance, ***P ≤0.001. (G) Tumor weights of PDX #1 in the indicated groups. ns, no significance, ***P ≤0.001. (H) Quantification viability of patient-derived organoids (PDOs) (#1, #2, #3) treated with 2 mM DMOG, 5 μM CQ, 8 mM oxamate or indicated combination. ns, no significance, ***P ≤0.001. (I) Quantification of the number of viable living cells per well of PDO #1. ns, no significance, ***P ≤0.001. (J) Representative bright field images and paraffin-embedded tumor sections stained with H&E of PDOs (scale bar: left panel, 200 μm; middle and right panel, 50 μm). (K) Growth curves of tumors formed by indicated cells and treated with bevacizumab (5 mg/kg every 3 days), CQ (3.5 mg/kg, every day), oxamate (500 mg/kg, every day) or indicated combination. ***P ≤0.001. (L) Tumor weights in the indicated groups. ***P ≤0.001. (M) Photograph and comparison of tumor sizes in the indicated groups. (N) Representative images of IHC staining of MKI67 or cleaved CASP3 in mouse tumors. (O and P) Quantification of proliferation index (O) and apoptosis index (P) in the indicated groups, proliferation index was determined using the percentage of MKI67-positive cells and apoptosis index was determined using the percentage of cleaved CASP3-positive cells. ***P ≤0.001.
Discussion
It is well known that cancer cells reprogram metabolic processes in order to support rapid proliferation, invasion and several cellular processes, as one of the key hallmarks of cancer. Lactate, an abundant energy source and metabolic byproduct in the tumor environment generated during Warburg effect, has been identified to be associated with tumorigenesis and multiple biological functions [32] and has recently been found to trigger histone lysine lactylation as a novel epigenetic modification stimulating gene transcription directly [22]. In the present study, we found that lactate accumulated in the tumor environment of CRC and acted as substrates for histone lactylation, and this process was further induced by cellular enhanced glycolysis in hypoxia. We identified that CRC patients resistant to bevacizumab treatment presented with elevated level of histone lactylation and inhibition of histone lactylation efficiently suppressed CRC tumorigenesis, progression and survival in hypoxia (Figure 7). These results were further confirmed in several pre-clinical models. These findings might provide a new strategy for improving clinical efficacy of bevacizumab in CRC by inhibition of histone lactylation.
Figure 7.
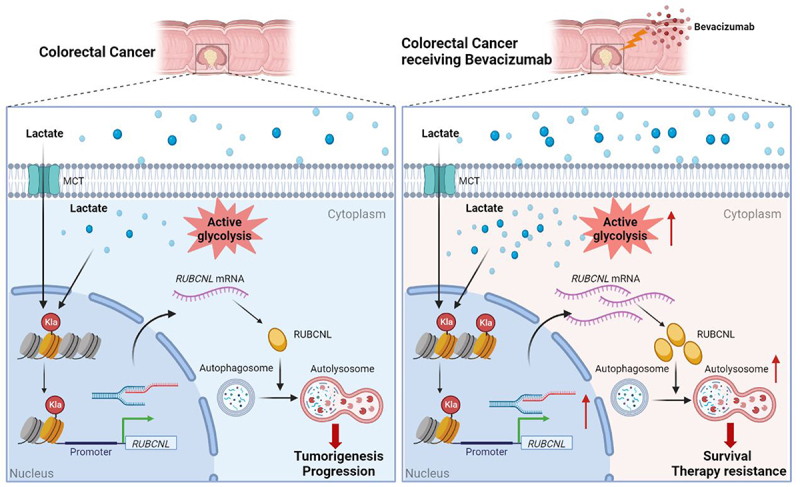
In colorectal cancer cells, high histone lactylation level induced by aerobic glycolysis promotes the transcription of RUBCNL, which enhances autophagy through promoting autophagasome maturation, and contributes to colorectal cancer tumorigenesis and progression. When receiving bevacizumab, the glycolysis of cancer cell in hypoxic regions may be further enhanced, followed by higher levels of histone lactylation and RUBCNL is transcriptionally upregulated by histone lactylation, which contributes to colorectal cancer cells survival and therapy resistance.
Post-translational modifications of histones act as a key role in regulating gene expression modulating a variety of biological processes by acting as docking sites for recruiting chromatin modulators [33,34]. Dysregulation of various histone modifications is frequently identified in several human diseases including cancer [35], highlighting the importance of histone-based gene regulation. In recent years, lactate-triggered histone lysine lactylation, serving as a novel type of histone modification, has been identified to induce the expression of genes related to tumorigenesis, progression and proliferation of cancer cells and the expression of homeostatic genes during M1 macrophage polarization caused by infection or hypoxia [36]. Here, we for the first time revealed that histone lactylation could drive tumorigenesis and progression of CRC and, in hypoxia, facilitate cancer cell survival, conferring resistance to bevacizumab treatment. Our study delivered a new exploration and important supplement of metabolic reprogramming-epigenetic regulation and provided a new tract for dealing with clinical treatment failure in cancer.
In solid tumors, the contribution of hypoxia to the resistance to antiangiogenic therapy has been established in decades [37,38]. And induction of hypoxia and the tumors response to it plays an important role in determining the efficacy of antiangiogenic therapy [39–41]. A clinical trial showed outstanding treatment efficiency of a hypoxia-activated prodrug (Evofosfamide) combined with bevacizumab in patients with bevacizumab-refractory glioblastoma [42,43]. However, the connection between hypoxia and resistance to antiangiogenic therapy is still unclear. Both hypoxic cancer cells stimulate blood vessel sprouting (angiogenesis) and hypoxia-inducible factors (HIFs)-mediated cellular responses that favor the selection of resistant cells adapted to the hypoxic microenvironment were identified to be the two most promising mechanisms [44,45].
Cancer cells induce multiple oncogenic signaling pathways to survive when the tumor microenvironment rapidly changes, such as in hypoxia, under chemotherapy, and under nutrient deprivation. Autophagy, an intracellular lysosomal-dependent degradation system for cell survival in response to extracellular and intracellular stress [46], was reported as one of the key properties of cancer cells to maintain cellular homeostasis [47–49]. However, the role of autophagy in tumorigenesis and drug resistance remains controversial and the precise molecular mechanisms remain unclear. Researchers found that efficacy of antiangiogenic therapy is partially dependent on the ability of cancer cells survival in hypoxic stress [19], which could be augmented by cancer-related autophagy [20,50,51]. Autophagy inhibition was found to be a novel way of increasing the efficacy of antiangiogenic agents [30,31,52]. Here, we revealed that histone lactylation promoted the transcription of RUBCNL, facilitating autophagosome maturation through interacting with BECN1 and mediating the recruitment and function of PtdIns3K complex, which played a crucial role in cancer cell proliferation and survival in hypoxia. Inhibition of histone lactylation enhanced the sensitivity of CRC cells to bevacizumab treatment, owning comparable treatment efficiency to autophagy inhibitor. Moreover, inhibiting RUBCNL expression through inhibition of histone lactylation has a synergistic inhibitory effect on autophagy together with CQ. Combining inhibition of histone lactylation and autophagy with bevacizumab treatment demonstrated remarkable treatment efficacy in bevacizumab-resistance patients-derived pre-clinical models (PDXs, PDOs).
In summary, in colorectal cancer cells, high histone lactylation level induced by aerobic glycolysis promotes the transcription of RUBCNL, which enhances autophagy through promoting autophagasome maturation, and contributes to colorectal cancer tumorigenesis and progression. When receiving bevacizumab, the glycolysis of hypoxic cancer cells may be further enhanced, followed by higher levels of histone lactylation and RUBCNL is transcriptionally upregulated by histone lactylation, which contributes to colorectal cancer cells survival and therapy resistance. Our study suggested that inhibition of histone lactylation may be a novel strategy for increasing the efficacy of bevacizumab in CRC, which demonstrated remarkable treatment efficacy in bevacizumab-resistance PDXs and PDOs.
Materials and methods
Patients and specimens
We obtained 330 CRC tissue specimens from fresh surgical specimens between November 2007 and May 2015 at Sun Yat-sen University Cancer Center (SYSUCC) and 150 CRC tissue specimens between October 2015 and October 2019 at The Sixth Affiliated Hospital of Sun Yat-sen University (SYSU-6thAH). And 70 patients among them received bevacizumab as first-line systemic therapy at SYSUCC after tumor progression. Bevacizumab (5 mg/kg) was administered intravenously on the first day every 2 weeks combined with chemotherapy. All patients underwent an assessment of tumor status at baseline and every 6 weeks after chemotherapy by abdominal/pelvic/chest computerized tomography (CT) or magnetic resonance imaging (MRI) and the best response was classified as a complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD) according to response evaluation criteria in solid tumors (RECIST) standard 1.1 as previously described [27]. Overall survival was defined as the time from the date of surgery to the date of death from any cause or latest follow-up. Informed consent for the use of the imaging and clinical data was obtained from the patients before first-line treatment. The study was approved by the Institutional Research Ethics Committee of Sun Yat-sen University Cancer Center (Guangzhou, China, approval number: B2022-025-01) and The Sixth Affiliated Hospital of Sun Yat-sen University (Guangzhou, China, approval number: 2022ZSLYEC–116).
Cell lines and cell cultures
The CRC cells (SW480, SW620, RKO, Caco2, DLD1, Lovo, HT29 and HCT116), normal colon epithelial cells (NCM460 and HIEC-6) and HEK293T were purchased from American Type Culture Collection (CCL-228, CCL-227, CRL-2577, HTB-37, CCL-221, CCL-229, HTB-38, CCL-247, CRL-1831, CRL-3266, CRL-3216) and cultured in complete medium as recommended, supplemented with 100 μg/mL streptomycin, 100 IU/mL penicillin, and 10% fetal bovine serum (Gibco, 10099141) at 37°C in 5% CO2. All cell lines used in our study were authenticated by STR profiling and negatively tested for mycoplasma contamination before experiments.
Quantification of lactate
Lactate levels were measured by using a colorimetric lactate assay according to the manufacturer’s protocol (Solarbio, YZ-K-LATE).
Western blot analysis
Cells and tissues were lysed with RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40 [Beyotime, P0013F], 0.1% SDS, 0.5% sodium deoxycholate [Sigma-Aldrich, 302-95-4], 1 mM EDTA, and 10% glycerol) and centrifuged at 13,000 × g for 30 min at 4°C. Equal amounts of total protein lysates were subjected to 10% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked in 5% BSA (Biosharp, BS114-500 g) in TBS (Biosharp, BL602A) containing 0.1% Tween-20 (Beyotime, ST825-100 ml) for 1 h and then probed with the appropriate specific primary antibody overnight at 4°C. Primary antibodies used in this study are used in this study are anti-Pan Kla (Jingjie, PTM-1401RM), anti-H3K18la (Jingjie, PTM-1406RM), anti-H3K9la (Jingjie, PTM-1419RM), anti-H4K8la (Jingjie, PTM-1415RM), anti-H4K12la (Jingjie, PTM-1411RM), anti-HIF1A (Abcam, ab179483), anti-Histone H3 (Cell Signaling Technology, 4499), anti-LDHA (Proteintech, 19987–1-AP), anti-PARP (Cell Signaling Technology, 9532), anti-CASP3 (Cell Signaling Technology, 9662), anti-RUBCNL (Abcam, ab246954), anti-EP300 (Abcam, ab275378), anti-SQSTM1 (Cell Signaling Technology, 8025), anti-LC3 (Cell Signaling Technology, 3868), anti-FLAG tag (Proteintech, 60002–1-Ig), anti-HA tag (Proteintech, 51064–2-AP), anti-BECN1 (Proteintech, 11306–1-AP), anti-PIK3C3/VPS34 (Cell Signaling Technology, 4263), anti-UVRAG (Cell Signaling Technology, 13115) and anti-TUBB/β-tubulin (Proteintech, 10068–1-AP). Membranes were washed and incubated for 1 h at room temperature with certain HRP-linked secondary antibodies (Cell Signaling Technology, 7076).
Immunohistochemistry
Immunohistochemistry (IHC) analysis using paraffin embedded CRC specimens was conducted following standard manufacturer’s protocols as described previously. Primary antibodies anti-PECAM1/CD31 (Proteintech, 11265–1-AP), anti-HIF1A (Abcam, ab179483), anti-LDHA (Proteintech, 19987–1-AP), anti-SLC16A3 (Abcam, ab244385), anti-Pan Kla (Jingjie, PTM-1401RM), anti-H3K18la (Jingjie, PTM-1406RM) and anti-RUBCNL (Abcam, ab246954) were used for IHC staining. IHC staining was evaluated by two independent gastrointestinal pathologists blinded to the patients’ characteristics and clinical outcomes. Final IHC score was calculated based on both the extent and the intensity of staining. The staining extent that scored according to the percentage of positively stained cells ranged from 0 to 3 (0, 0–5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%), although the intensity of staining was scored as 0 (negative staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The Cutoff Finder program was used to determine the optimal cutoff for Pan Kla, H3K18la and RUBCNL expression. Specimens with the final scores ≥ 4 were defined as high expression, and specimens with the final scores < 4 were defined as low expression.
Immunofluorescent staining
Adherent cells were fixed with methanol and then gently washed with PBS (Biosharp, BL-601A) twice. Then, the cells were incubated with permeabilization buffer (eBioscience, 00-8333-56) for 10 min at 4°C. After blocking in 5% BSA in TBS/Tween-20 for 1 h at room temperature, the cells were incubated with primary antibodies against RUBCNL and BECN1 at 4°C overnight. The secondary antibody was marked with fluorescence (Abcam, ab150080 and ab150113) in the dark, and DAPI was applied. The images were captured using an Olympus® Fluoview FV1000 microscope (Japan), and the microscope software FV10-ASW 4.0 was used to separate each multispectral image.
The primary CRC cells prepared as described were transiently transfected with mRFP-GFP-LC3 adenovirus (Hanbio Co. Ltd., HBAP2100001) according to the manufacturer’s protocol. Subsequently, the treated cells were observed under Olympus® Fluoview FV1000 microscope.
Cell transfection and lentivirus production
The small interfering RNA (siRNA) targeting LDHA and EP300 were synthesized by RiboBio (Guangzou, China). Transfection was conducted using jetPRIME reagent (Polyplus, 101000046) as recommended. For knockdown or overexpression of RUBCNL, negative control or target gene shRNAs or over expressing plasmids were co-transfected into HEK293T with pHelper and pEnv. The virus was harvested from the transfected HEK293T cells, and the target cells were then infected with the viral supernatants for 2 consecutive days. Stable CRC cell lines were selected by treating with puromycin (2 μg/mL) for 10 days.
CCK-8 assay
For the proliferation assay, cells were seeded in 96-well plates (2000 cells per well during normoxia or 1 × 104 cells per well during hypoxia) and treated with corresponding processes. At the indicated time point, CCK8 (DOJINDO, CCK8) was added into the wells and incubated with cells according to the product manual. The absorbance was detected at a wavelength of 450 nm, and growth curves were generated to determine the growth rates.
Colony formation assay
Cells were seeded in 6-well plates (1000 cells per well) with 3 replicates and were then cultured for 14 days with complete medium at 37°C with 5% CO2. Cells were then fixed with methanol and stained with crystal violet, and the colonies of each well were counted.
Apoptosis assay
For cell apoptosis assay, CRC cells treated with corresponding processes were harvested and stained with ANXA5/annexin V-FITC and propidiumiodide (PI) using an Annexin V-FITC/PI staining kit (BD Pharmingen™, 556547). After incubation at room temperature for 15 min, the cells were analyzed by flow cytometry.
Coimmunoprecipitation
Coimmunoprecipitation was carried out with the Pierce™ Classic Magnetic IP/Co-IP kit (Thermo, 88804) according to the manufacturer’s protocol. Cells were lysed in lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 2 mM MgCl2, 2 mM EGTA) with protease inhibitor and phosphatase inhibitor cocktail (Thermo, 78446) on ice. Cell lysates were centrifuged to remove insoluble materials. Antibodies incubated with beads overnight at 4°C. The beads were washed repeatedly, and bound proteins were analyzed by western blotting.
Chromatin immunoprecipitation sequencing
The ChIP assay was performed using a Pierce™ Magnetic ChIP Kit (Thermo Fisher Scientific, 26157) according to the manufacturer’s instructions. Briefly, after cross-linking and chromatin digestion, digested chromatin was incubated with 5 μg anti-H3K18la-ChIP Grade (Jingjie, PTM-1427RM) or anti-IgG (Cell Signaling Technology, 3900) antibodies at 4°C overnight. Protein A/G magnetic beads (MCE, HY-K0202) were then added into the lysate the following morning and incubated for another 4 h. Purified DNA fragments were constructed and added to ChIP-seq libraries, amplified, and sequenced on an HiSeq 2500 platform (Illumina). The primers used for the qPCR analysis in this study are listed in supplementary materials Table S3.
RNA isolation, quantitative real-time PCR (RT-qPCR) and RNA sequencing
Total RNA was extracted using TRIzol reagent (Invitrogen, 15596026). The RNA purity was assessed using the ND-1000 Nanodrop. Each RNA sample had an A260:A280 ratio above 1.8 and an A260:A230 ratio above 2.0. Complementary DNA (cDNA) was generated using the PrimeScript RT reagent kit (TaKaRa Bio, RR037A) according to the manufacturer’s instructions. The cDNA was used for qPCR using SYBR Green PCR master mix (Life Technologies) on a CFX96 real-time PCR detection system (Bio-Rad). The libraries were paired-end sequenced (PE150, sequencing reads were 150 bp) on an Illumina HiSeq3000 at Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The clean reads were obtained after removal of reads containing adapter, ploy-N and at low quality from raw data. HISAT2 was used to align the clean reads to the human reference genome hg19 with default parameters. HTSeq was subsequently employed to convert aligned short reads into read counts for each gene model. Differential expression was assessed by DE sequencing read counts as input. The Benjamini-Hochberg multiple test correction method was enabled. Differentially expressed genes were chosen according to the criteria of p-value <0.05. KEGG analysis was used for gene functional annotation.
Transmission electron microscopy
The treated CRC cells were incubated with a pre-chilled 2% glutaraldehyde solution for 2 h at 4°C to fix the cell pellet. The cells were stained with 2% uranyl acetate solution for 2 h and then dehydrated in 50%, 70%, 90%, and 100% acetone. The cells were embedded in spurr embedding kit (KYD bio, 14300), and ultrathin sections were prepared for observation under an electron microscope (HITACHI).
Organoid culture
Tumors were obtained by surgical resection and processed for organoid culture within 24 h. Tumor tissue was cut into small pieces and enzymatically digested using digestion buffer (RPMI 1640 medium, 10% FBS, 1% penicillin-streptomycin, 4 mg/mL collagenase (Sigma-Aldrich, C5138) and 2 mg/mL deoxyribonuclease (Sigma-Aldrich, 18047019) before embedding in Matrigel (Corning, 356231). After solidification for 30 min at 37°C, cells embedded in Matrigel were overlaid with IntestiCult OGM Human (Stem Cell, 06010) supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL) and 10 mM Y-27632 (Sigma-Aldrich, 5092280001) at 37°C supplemented with 5% CO2. Organoids were passaged approximately every week by incubating in TrypLE Express (Gibco, 12604013) for 30 min at 37°C to dissociate organoids into single cells and replating them in fresh Matrigel. Organoids < passage 30 were used in experiments. On the second day after seeding organoids were drugged. At 10 d after drugging, organoids were imaged with the Olympus® I×73microscope. After image acquisition, organoid viability was accessed with Cell Titer Glo 3D Cell Viability assay (Promega, G9682–1) according to the manufacturer’s protocol on the SpectraMax i3× plate reader.
In vivo therapeutic study
CRC cell line-based xenograft (CDX) and patient-derived xenograft (PDX) models were performed. All mice were housed in specific-pathogen-free conditions. All procedures, following the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978), were approved by the Sun Yat-sen University Animal Care and Use Committee (L102012022050E, Guangzhou, China). Six to 8-week-old female BALB/c nude mice were purchased from The Vital River Laboratory (Beijing, China). RUBCNL-knockdown or control HCT116 cells (5 × 105) were suspended in 100 μl PBS and subcutaneously injected to the left flank of each nude mouse (5 mice/group, randomly divided into indicated groups). One week later, the mice were intraperitoneally injected with indicated treatment.
Immunocompromised NOD.SCID mice were obtained from GemPharmatech (China) and used to establish patient-derived xenograft models. The PDX models were initially generated using fresh tumor samples from patients with CRC that were subcutaneously implanted into the dorsal flank of mice as the first generation (F0). Once an appropriate volume was reached, the tumors were excised, divided into equal pieces, and subcutaneously implanted into mice as the second generation (F1). When the tumors became palpable, the tumor-bearing mice were divided randomly into 5 groups with indicated treatment. Tumor growth curve was measured every 3 days for 4–5 weeks and the volume of tumor was calculated as length × Width2 ×1/2. At the end of the study, we surgically removed the tumors from the sacrificed mice.
Statistical analysis
Statistical analyses were performed using SPSS version 24.0 software. All data were presented as the mean ± SD. Statistical tests used in this study included the two-tailed Student’s t test, χ2 test and log-rank test. P value < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We greatly appreciate the help from all of our colleagues in the Department of Colorectal Surgery at Sun Yat-sen University Cancer Center who were involved in administering the treatments in the current study.
Funding Statement
This study was funded by grants from the National Natural Science Foundation of China (no. 82003051, no. 82072606, no. 82103273, no. 82202947 and no. 82203626), Guangdong Basic and Applied Basic Research Foundation (2021A1515110509 and 2019A1515110144).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15548627.2023.2249762
Data availability statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www. researchdata.org.cn).
References
- [1].Sung H, Ferlay J, Siegel RL, et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA A Cancer J Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601 [DOI] [PubMed] [Google Scholar]
- [3].Van Cutsem E, Cervantes A, Adam R, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–1422. doi: 10.1093/annonc/mdw235 [DOI] [PubMed] [Google Scholar]
- [4].Jönsson K, Gröndahl G, Salö M, et al. Repeated liver resection for colorectal liver metastases: a comparison with primary liver resections concerning perioperative and long-term outcome. Gastroenterol Res Pract. 2012;2012:568214. doi: 10.1155/2012/568214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rahbari NN, Reissfelder C, Schulze-Bergkamen H, et al. Adjuvant therapy after resection of colorectal liver metastases: the predictive value of the MSKCC clinical risk score in the era of modern chemotherapy. BMC Cancer. 2014;14(1):174. doi: 10.1186/1471-2407-14-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tzeng CW, Aloia TA.. Colorectal liver metastases. J Gastrointest Surg. 2013;17(1):195-201; quiz p.201–2. doi: 10.1007/s11605-012-2022-3 [DOI] [PubMed] [Google Scholar]
- [7].Chiche L. Prise en charge chirurgicale des métastases hépatiques des cancers colorectaux [Surgical management of hepatic metastases from colorectal cancer]. J Chir (Paris). 2003;140(2):77–89. [PubMed] [Google Scholar]
- [8].Maeda Y, Shinohara T, Nagatsu A, et al. Long-term outcomes of conversion hepatectomy for initially unresectable colorectal liver metastases. Ann Surg Oncol. 2016;23 Suppl 2(S2):S242–8. doi: 10.1245/s10434-015-4460-0 [DOI] [PubMed] [Google Scholar]
- [9].Maughan TS, Adams RA, Smith CG, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377(9783):2103–2114. doi: 10.1016/S0140-6736(11)60613-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–2019. doi: 10.1200/JCO.2010.33.5091 [DOI] [PubMed] [Google Scholar]
- [11].Venook AP, Niedzwiecki D, Lenz HJ, et al. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: arandomized clinical trial. JAMA. 2017;317(23):2392–2401. doi: 10.1001/jama.2017.7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tomasello G, Petrelli F, Ghidini M, et al. FOLFOXIRI plus bevacizumab as conversion therapy for patients with initially unresectable metastatic colorectal cancer: a systematic review and pooled analysis. JAMA Oncol. 2017;3(7):e170278. doi: 10.1001/jamaoncol.2017.0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hurwitz H. Integrating the anti-VEGF-A humanized monoclonal antibody bevacizumab with chemotherapy in advanced colorectal cancer. Clin Colorectal Cancer. 2004;4(Suppl 2):S62–8. doi: 10.3816/CCC.2004.s.010 [DOI] [PubMed] [Google Scholar]
- [14].Botrel TEA, Clark LGO, Paladini L, et al. Efficacy and safety of bevacizumab plus chemotherapy compared to chemotherapy alone in previously untreated advanced or metastatic colorectal cancer: a systematic review and meta-analysis. BMC Cancer. 2016;16(1):677. doi: 10.1186/s12885-016-2734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Benson AB, Venook AP, Al-Hawary MM, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19(3):329–359. doi: 10.6004/jnccn.2021.0012 [DOI] [PubMed] [Google Scholar]
- [16].Vasudev NS, Reynolds AR. Anti-angiogenic therapy for cancer: current progress, unresolved questions and future directions. Angiogenesis. 2014;17(3):471–494. Erratum in: Angiogenesis 17(3):495-7. doi: 10.1007/s10456-014-9420-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Garcia J, Hurwitz HI, Sandler AB, et al. Bevacizumab (avastin®) in cancer treatment: a review of 15 years of clinical experience and future outlook. Cancer Treat Rev. 2020;86:102017. doi: 10.1016/j.ctrv.2020.102017 [DOI] [PubMed] [Google Scholar]
- [18].Cao D, Hou M, Guan YS, et al. Expression of HIF-1alpha and VEGF in colorectal cancer: association with clinical outcomes and prognostic implications. BMC Cancer. 2009;9(1):432. doi: 10.1186/1471-2407-9-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hu YL, DeLay M, Jahangiri A, et al. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012;72(7):1773–1783. doi: 10.1158/0008-5472.CAN-11-3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Selvakumaran M, Yao KS, Feldman MD, et al. Antitumor effect of the angiogenesis inhibitor bevacizumab is dependent on susceptibility of tumors to hypoxia-induced apoptosis. Biochem Pharmacol. 2008;75(3):627–638. doi: 10.1016/j.bcp.2007.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tennant DA, Durán RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10(4):267–277. doi: 10.1038/nrc2817 [DOI] [PubMed] [Google Scholar]
- [22].Zhang D, Tang Z, Huang H, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yu J, Chai P, Xie M, et al. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021;22(1):85. doi: 10.1186/s13059-021-02308-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang J, Luo L, Zhao C, et al. A positive feedback loop between inactive VHL-triggered histone lactylation and PDGFRβ signaling drives clear cell renal cell carcinoma progression. Int J Biol Sci. 2022;18(8):3470–3483. doi: 10.7150/ijbs.73398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xiong J, He J, Zhu J, et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82(9):1660–1677.e10. doi: 10.1016/j.molcel.2022.02.033 [DOI] [PubMed] [Google Scholar]
- [26].Yang Z, Yan C, Ma J, et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat Metab. 2023;5(1):61–79. doi: 10.1038/s42255-022-00710-w [DOI] [PubMed] [Google Scholar]
- [27].Lu ZH, Peng JH, Wang FL, et al. Bevacizumab with preoperative chemotherapy versus preoperative chemotherapy alone for colorectal cancer liver metastases: a retrospective cohort study. Medicine (Baltimore). 2016;95(35):e4767. doi: 10.1097/MD.0000000000004767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cummins EP, Berra E, Comerford KM, et al. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci U S A. 2006;103(48):18154–18159. doi: 10.1073/pnas.0602235103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Milkiewicz M, Pugh CW, Egginton S. Inhibition of endogenous HIF inactivation induces angiogenesis in ischaemic skeletal muscles of mice. J Physiol. 2004;560(Pt 1):21–26. doi: 10.1113/jphysiol.2004.069757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Selvakumaran M, Amaravadi RK, Vasilevskaya IA, et al. Autophagy inhibition sensitizes colon cancer cells to antiangiogenic and cytotoxic therapy. Clin Cancer Res. 2013;19(11):2995–3007. doi: 10.1158/1078-0432.CCR-12-1542 [DOI] [PubMed] [Google Scholar]
- [31].Guo XL, Li D, Sun K, et al. Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma. J Mol Med (Berl). 2013;91(4):473–483. doi: 10.1007/s00109-012-0966-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Feng Y, Xiong Y, Qiao T, et al. Lactate dehydrogenase A: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018;7(12):6124–6136. doi: 10.1002/cam4.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- [34].Dunn RK, Kingston RE. Gene regulation in the postgenomic era: technology takes the wheel. Mol Cell. 2007;28(5):708–714. doi: 10.1016/j.molcel.2007.11.022 [DOI] [PubMed] [Google Scholar]
- [35].Chi P, Allis CD, Wang GG. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen AN, Luo Y, Yang YH, et al. Lactylation, a novel metabolic reprogramming code: current status and prospects. Front Immunol. 2021;12:688910. doi: 10.3389/fimmu.2021.688910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6(8):583–592. doi: 10.1038/nrc1893 [DOI] [PubMed] [Google Scholar]
- [38].Sorensen AG, Emblem KE, Polaskova P, et al. Increased survival of glioblastoma patients who respond to antiangiogenic therapy with elevated blood perfusion. Cancer Res. 2012;72(2):402–407. Erratum in: Cancer Res;72(2). doi: 10.1158/0008-5472.CAN-11-2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4(6):437–447. doi: 10.1038/nrc1367 [DOI] [PubMed] [Google Scholar]
- [40].Höckel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93(4):266–276. doi: 10.1093/jnci/93.4.266 [DOI] [PubMed] [Google Scholar]
- [41].Keunen O, Johansson M, Oudin A, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci U S A. 2011;108(9):3749–3754. doi: 10.1073/pnas.1014480108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Brenner A, Zuniga R, Sun JD, et al. Hypoxia-activated evofosfamide for treatment of recurrent bevacizumab-refractory glioblastoma: a phase I surgical study. Neuro Oncol. 2018;20(9):1231–1239. doi: 10.1093/neuonc/noy015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Brenner AJ, Floyd J, Fichtel L, et al. Phase 2 trial of hypoxia activated evofosfamide (TH302) for treatment of recurrent bevacizumab-refractory glioblastoma. Sci Rep. 2021;11(1):2306. doi: 10.1038/s41598-021-81841-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rey S, Schito L, Wouters BG, et al. Targeting hypoxia-inducible factors for antiangiogenic cancer therapy. Trends Cancer. 2017;3(7):529–541. doi: 10.1016/j.trecan.2017.05.002 [DOI] [PubMed] [Google Scholar]
- [45].Méndez-Blanco C, Fondevila F, García-Palomo A, et al. Sorafenib resistance in hepatocarcinoma: role of hypoxia-inducible factors. Exp Mol Med. 2018;50(10):1–9. doi: 10.1038/s12276-018-0159-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. doi: 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- [47].Monkkonen T, Debnath J. Inflammatory signaling cascades and autophagy in cancer. Autophagy. 2018;14(2):190–198. doi: 10.1080/15548627.2017.1345412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- [50].Wu HB, Yang S, Weng HY, et al. Autophagy-induced KDR/VEGFR-2 activation promotes the formation of vasculogenic mimicry by glioma stem cells. Autophagy. 2017;13(9):1528–1542. doi: 10.1080/15548627.2017.1336277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sun L, Xiong H, Chen L, et al. Deacetylation of ATG4B promotes autophagy initiation under starvation. Sci Adv. 2022;8(31):eabo0412. doi: 10.1126/sciadv.abo0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Shi YH, Ding ZB, Zhou J, et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7(10):1159–1172. doi: 10.4161/auto.7.10.16818 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www. researchdata.org.cn).


