ABSTRACT
DNA double-strand break (DSB) is the most dangerous type of DNA damage, which may lead to cell death or oncogenic mutations. Homologous recombination (HR) and nonhomologous end-joining (NHEJ) are two typical DSB repair mechanisms. Recently, many studies have revealed that liquid–liquid phase separation (LLPS) plays a pivotal role in DSB repair and response. Through LLPS, the crucial biomolecules are quickly recruited to damaged sites with a high concentration to ensure DNA repair is conducted quickly and efficiently, which facilitates DSB repair factors activating downstream proteins or transmitting signals. In addition, the dysregulation of the DSB repair factor’s phase separation has been reported to promote the development of a variety of diseases. This review not only provides a comprehensive overview of the emerging roles of LLPS in the repair of DSB but also sheds light on the regulatory patterns of phase separation in relation to the DNA damage response (DDR).
KEYWORDS: DNA damage response (DDR), DNA double-strand break (DSB), liquid-liquid phase separation (LLPS), condensates, homologous recombination (HR), nonhomologous end-joining (NHEJ)
Introduction
Recently, there has been increasing evidence of biomolecules interacting with each other through weak intermolecular bonds and undergoing phase separation during biological processes [1]. These biomolecules form concentrated condensates without membranes, such as the nucleolus, processing bodies, and stress granules [2,3]. The formation of these condensates can enhance intermolecular interactions and enzymatic reactions, ensuring the efficiency and accuracy of various biological activities [4]. This phenomenon is commonly observed in processes such as transcription, chromatin organization, X-chromosome inactivation, autophagy, DNA damage repair, anti-bacterial autophagy, innate immune signaling, and cell division [5–9].
Among different types of DNA damage, the most dangerous is DNA double-strand break (DSB), which can be caused by ionizing radiation, chemicals, or errors in DNA metabolic processes. Failure to promptly repair DSBs can lead to chromosomal translocations and aneuploidy, which are associated with oncogenic transformations and an increased risk of cancer [10]. In eukaryotic cells, two major pathways, homologous recombination (HR) and nonhomologous end-joining (NHEJ), are responsible for repairing DSBs. HR requires an intact sister chromatid and primarily occurs during the S/G2 phase of the cell cycle, while NHEJ directly ligates the DSBs throughout the entire cell cycle [11,12]. This review discusses the regulation of DNA repair factors in DSB repair through phase separation.
Liquid–liquid phase separation (LLPS)
Liquid–liquid phase separation (LLPS) plays an important role in various biochemical processes, particularly in the formation of membrane-less organelles [13]. The study of LLPS has provided valuable insights into the physiological processes of living organisms. LLPS occurs when proteins and nucleic acids, which are the constituents of condensates, form liquid droplets in response to changes in their concentration or environmental factors such as pressure, ionic strength, temperature, pH, and crowding [14–17]. The amino acid composition of proteins plays a crucial role in facilitating LLPS processes. Proteins that undergo LLPS tend to have an enrichment of polar and charged amino acids, which can engage in electrostatic interactions [18]. Additionally, hydrophobic interactions and other weak interactions, such as hydrogen bonds, also contribute to phase separation of proteins [19]. Intrinsically disordered regions (IDRs) are protein regions that lack a well-defined three-dimensional structure. These IDRs are highly flexible and dynamic, allowing proteins to undergo conformational changes and interact with other molecules. Post-translational modifications (PTMs) of proteins within IDRs can play pivotal roles in regulating LLPS and the formation of condensates. There are several types of PTMs that can occur within IDRs, including phosphorylation, acetylation, methylation, ubiquitination, and many others. These modifications can introduce changes to specific amino acid residues within the IDRs, altering their physicochemical properties and affecting protein–protein interactions [20,21]. The phenomenon of multivalence arises from the presence of diverse repetitive interaction sites within a polypeptide or nucleic acid molecule, which can be achieved through folded binding modules or IDRs. Multivalent interactions play crucial roles in various biological processes by enhancing binding affinity, specificity, and the formation of higher-order complexes [22]. Moreover, proteins recruit rapidly at specific regions by LLPS to active downstream responses [23]. The reversibility of high-concentration condensates provides a mechanism for cells to efficiently execute specific reactions while preventing unnecessary interactions. This characteristic contributes to the functional organization and regulation of cellular processes.
DNA double-strand break response
DNA damage is a common occurrence in living organisms due to various factors such as exposure to environmental agents, errors during DNA replication, and endogenous cellular processes. To counteract the detrimental effects of DNA damage, cells have evolved intricate mechanisms known as DNA damage responses (DDR). When DNA damage occurs, specialized proteins recognize and bind to the damaged sites, initiating a signaling cascade. This cascade activates various cellular pathways that halt the cell cycle, allowing time for DNA repair to take place. It also triggers the recruitment of repair enzymes and factors to the damaged sites. There are several types of DNA repair mechanisms including HR and NHEJ. HR primarily occurs during the S and G2 phases of the cell cycle, while NHEJ pathway is responsible for repairing DSB throughout the entire cell cycle [24]. By dynamically modifying the key players in HR and NHEJ, PTMs help control the competition between these repair pathways and ensure the appropriate choice for efficient and accurate DNA repair. The precise regulation of PTMs is crucial for maintaining genome stability and preventing the accumulation of DNA damage that can lead to various diseases, including cancer [25].
HR primarily operates during the late S and G2 phases of the cell cycle when sister chromatid is available as template for repair. During HR, the MRE11-RAD50-NBS1 (MRN) complex plays a crucial role in recognizing and processing DNA double-strand breaks (DSBs) [26]. After MRE11 subunit of MRN complex cleaves the DNA near the DSB site, it generates short single-stranded DNA (ssDNA) overhangs [27]. These ssDNA overhangs are then bound by replication protein A (RPA) filaments. RAD51, with the assistance of BRCA1 and BRCA2, replaces RPA and forms nucleoprotein filaments on ssDNA overhangs [28]. These filaments help to find homology and initiate strand invasion, resulting in the extension of a displacement loop (D-loop) mediated by polη and facilitating the repair of DSB through homologous recombination. After the formation of D-loop, the repair process of DSB continues with DNA chain elongation [29]. This involves the synthesis of new DNA strands using intact homologous DNA molecule as a template. The elongation of DNA chains results in the formation of a structure called a double Holliday junction (DHJ). Various enzymes, such as resolvases, endonucleases, and helicases, are involved in the resolution of DHJ. These enzymes cleave and separate DNA strands at crossover points, allowing for the proper segregation of genetic material and the completion of repair processes [30].
NHEJ is the primary mechanism for DSB in mammalian somatic cells, it can occur throughout cell cycle and does not require the presence of a sister chromatid. When a DSB occurs, Ku70/Ku80 heterodimer, also known as Ku protein complex, recognizes and binds to broken DNA ends [31]. Following recognition and binding of Ku protein complex, a series of events is initiated to repair DSB. The phosphorylation of H2A.X by ATM kinase triggers a signaling cascade that leads to rapid aggregation of 53BP1 on chromatin near the site of DNA damage [32]. This aggregation results in the formation of irradiation-induced foci (IRIF). 53BP1 plays a crucial role in safeguarding DNA ends and promoting efficient repair. It can discriminate between nucleosomes that possess specific histone modifications and engages in interactions with proteins like RAP1-interacting factor 1 (RIF1) and shieldin complex to protect DNA ends. The C-terminus of Ku70/80 facilitates recruitment of DNA-dependent protein kinase catalytic subunits (DNA-PKcs) to form complex DNA-PK [33]. These complex recruit other NHEJ proteins and activates end-processing enzymes, polymerases, and ligases such as LIG4, XRCC4, and XLF. These enzymes are responsible for processing and joining broken DNA ends, ultimately completing repair process [34].
Phase separation in DNA double-strand break response
LLPS is involved in various biological processes, such as transcriptional regulation, chromatin organization, control of intracellular signaling, and DNA damage response [35]. In the context of DNA damage response, multiple proteins involved in this process have been found to undergo LLPS. These proteins form liquid-like droplets at sites of DNA damage, which facilitate the recruitment and assembly of other repair factors. This spatial organization allows for efficient and coordinated repair of damaged DNA [36,37].
MRNIP
Once a DSB occurs, MRN complex recognizes damaged DNA and initiates a signaling cascade to activate cellular response to repair the break [38]. MRN complex facilitates autophosphorylation of ataxia telangiectasia mutated (ATM) at serine 1981 (S1981) and autoacetylation of ATM at lysine 3106 (K3106) [39,40]. Additionally, ATM-mediated phosphorylation of H2A.X at S139 serves as a scaffold for the assembly of repair-related complexes, ensuring proper progression of DSB repair [41]. DNA end resection is dependent on the recruitment of MRN and CtBP-interacting protein (CtIP), followed by exonuclease 1 (Exo1) bind to it [42,43]. However, MRN cannot work reliably without MRNIP. MRNIP is the binding partner of MRN complex and is involved in promoting genomic stability and protecting replication forks [44,45]. When a DSB occurs, MRNIP droplets rapidly migrate to the sites of DNA damage. These droplets concentrate MRN complex into condensates. The condensates formation help activate downstream proteins involved in DNA repair or transmits signals to initiate repair processes (Figure 1a) [46]. MRNIP contains two IDRs, and IDR1 (amino acids:123–176) is essential for its LLPS. In a study by Wang et al., it was reported that colorectal cancer patients with high expression of MRNIP may have an increased likelihood of experiencing shorter survival time and exhibiting radio resistance. This suggests that MRNIP expression levels could potentially serve as a prognostic marker in colorectal cancer [44].
Figure 1.
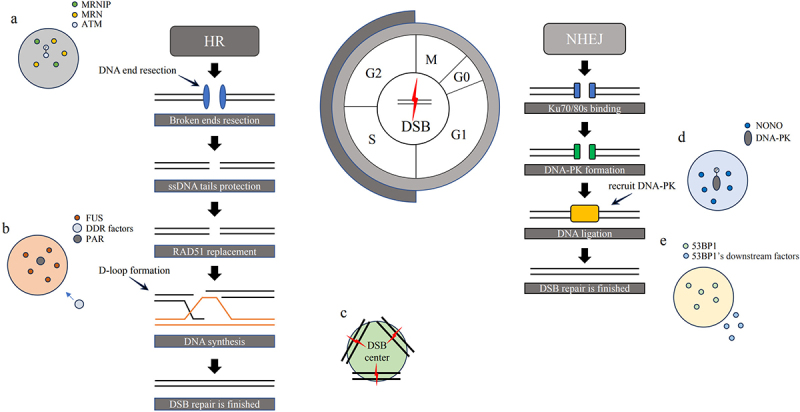
DNA repair factors LLPS involved in HR and NHEJ processes. HR occurs in cell S and G2 phases. (a) MRNIP droplets rapidly migrate to the sites of DNA damage, leading to the activation of downstream proteins and facilitating the autophosphorylation of ATM. MRE11-mediated resection leads to the formation of single-stranded DNA (ssDNA) tails. RPAs bind to the tails to protect them. BRCA2 assists RAD51 in displacing RPA. RAD51-ssDNA invades the homologous sequence, leading to synthesis of the DNA strands. (b) In this process, the assistance of PAR facilitates the essential role of FUS phase separation in the efficient assembly of DNA repair complexes and is necessary for recruiting DDR factors to DNA damage sites. (c) RAD52 droplets collaborate with various types of nuclear filaments to facilitate the formation of DNA repair centers. NHEJ is active throughout the cell cycle. Ku70/80s first quickly recognize DSB sites and bind them to promote DNA-PK formation. (d) NONO enhances DNA-PK phosphorylation at T2609 in response to DNA damage by generating high-concentration droplets. (e) 53BP1 has the ability to form condensates and subsequently recruit downstream factors such as RIF1, PTIP, and the shieldin complex.
PAR
PARP1-mediated synthesis of poly (ADP-ribose) (PAR) is an initial response that happens promptly following the occurrence of DSBs [47]. PARP1 is classified as one of the numerous abundant Poly (ADP-ribose) polymerases (PARPs) that possess polymerase activity. It catalyzes the transfer of ADP ribose from nicotinamide adenine dinucleotide (NAD+) to serine, glutamate and aspartate residues, thereby connecting the molecules sequentially to generate extensive branches of ADP ribose chains [48,49]. PARP1, as a highly efficient detector of DNA damage, facilitate Mre11 and NBS1 recruit at sites of DSBs through intramolecular folding and dimerization mechanisms [50,51]. PAR, as a nucleic acid-like polymer, serves as a transient signal for facilitating DNA repair. Additionally, it plays a crucial role in promoting LLPS of FET (FUS, EWSR1, and TAF15) proteins. For example, PAR assists in the formation of FUS compartment [52]. Each PAR unit contains two groups of ribose that are connected with a phosphorylated adenosine. This structure results in an increased negative charge and additional spatial capacity. As a result, PAR can recruit additional positively charged molecules through electrostatic action [53]. The structural diversity and adaptability of PAR enable it to interact with various molecules and form condensates. The abundance of ADP units in PAR facilitates the recruitment of multiple proteins, allowing for the formation of protein complexes [54,55]. For compartmentalization, FUS interacts with PAR through a positively charged area arginine-glycine-glycine (RGG). Moreover, the recruitment of non-POU domain-containing octamer-binding protein (NONO) is contingent upon its interaction with PAR through a specific RNA recognition motif 1 (RRM1) during the process of DSB repair, specifically to promote NHEJ [56].
Paraspeckles
Paraspeckles are subnuclear bodies that are formed by long nonprotein-coding RNA known as nuclear paraspeckle assembly transcript 1 (NEAT1) [57]. It has been demonstrated to be a target for p53, which is a significant contributor to DNA damage repair in both direct and indirect ways [58]. As major constituents of paraspeckles, FUS and NONO have been implicated in LLPS.
FUS is a protein integral to the processes of DNA and RNA metabolism. The majority of FUS is primarily localized within nucleus and has potential to forming condensates [59]. FUS contains several functional domains, including N-terminal low-complexity prion-like domain (FUS-LC), three RGG repeat motifs, a zinc finger, an RNA recognition motif, and a C-terminal nuclear localization signal [60]. Due to the arrangement of its amino acid residues FUS could undergo phase separation by hydrophobic interactions and its LLPS can precisely regulate the response in a specific location at a specific time [61]. FUS phase separation plays a crucial role in the efficient assembly of DNA repair complexes and is required for the recruitment of DDR factors at DNA damage sites (Figure 1b) [62].
NONO is a member of drosophila behavior human splicing (DBHS) family, which also includes splicing factor proline and glutamine (SFPQ) and paraspeckle component 1 (PSPC1) [63]. DBHS proteins share common structural features, including two RNA recognition motif (RRM) domains, a NonA/paraspeckle domain (NOPS), and disordered low complexity domains (LCDs). These structural elements facilitate the formation of obligate complexes that can bind to DNA or RNA, leading to the oligomerization of proteins into condensates [64]. For DNA damage repair processes, NONO impacts γ-H2A.X foci formation and promotes NHEJ in vivo. NONO contains a nuclear localization signal (NLS) and localizes in a specific subnuclear region, where is related to the sensitivity of cells to ionizing radiation. NONO facilitates the recruitment and interaction of nuclear epidermal growth factor receptor (EGFR) and DNA-PK by generating high-concentration droplets, thereby enhancing DNA-PK phosphorylation at T2609 in response to DNA damage (Figure 1d) [65]. Lacking of NONO and its functional homologs protein PAPC1 led to severe radiosensitivity and delayed resolution of DSB repair foci [66]. RNA-binding proteins harboring intrinsically disordered domains could be recruited to DNA damage sites by PAR and create liquid compartments [67].
RAD52
Radiation-sensitive protein 52 (RAD52) is a member of single-strand annealing protein (SSAP). It helps RAD51 assemble onto ssDNA and promotes ssDNA annealing [68]. RAD52 consists of two parts, its C-terminal region is essential for accumulation at DNA damage sites [69]. During the process of homologous directed recombination (HDR), the N terminus of RAD52 binds to ssDNA, while the disordered C terminus assists RAD51 in displacing RPA and forming droplets [70]. The physical characteristics of RAD52 in foci conform to LLPS model, and foci formed by SUMOylation of Rad52 are denser [71]. RAD52 foci can recruit more than one DSB and serve as the center of DSB repair [72]. Additionally, PTI-DIM also helps RAD52 phase separation, RAD52 droplets collaborate with different types of nuclear filaments facilitate forming DNA repair center (Figure 1c) [73].
53BP1
The competition between 53BP1 and BRCA1 plays a decisive role in determining the choice of DSB repair pathway [74]. The collaborative action of Pax2 transactivation domain interaction protein (PTIP), replication timing regulatory factor 1 (RIF1), and shieldin (SHLD1, SHLD2, SHLD3, and REV7) with 53BP1 prevents DNA end excision and ssDNA’s formation, promoting DSB repair through NHEJ pathway [75]. 53BP1 can form condensates at heterochromatin which can maintain the stability of mitosis and genome (Figure 1e) [76]. AHNAK restrains 53BP1 phase separation by interacting with its OD domain to control p53 gene network in DSB [77].
Many domains enable 53BP1 to perform crucial functions in DSB. Oligomerization domain (OD) promotes 53BP1 dimerization or polymerization; accumulation of 53BP1 on DNA break sites is attributable to LC8 domain; and 28 amino-terminal Ser/Thr-Gln (S/T-Q) polypeptides are phosphorylated by ATM to promote DNA damage response [78]. Among these, the structural features elucidated that OD and disordered sequence at C terminus are involved in LLPS of 53BP1 [79]. The study conducted by Kilic S et al. shows that 53BP1 undergoes phase separation in response to DNA damage, which is accompanied by frequent fusion and occasional fission events [80]. During DSB, RNA polymerase II (RNAP II) is recruited to generate damage-induced long non-coding RNA (dilncRNA). DlincRNA transforms into shorter double-stranded DNA damage response DDR RNA (DDRNA), which is crucial for LLPS of 53BP1. Impairment of 53BP1 condensates may occur as a result of the degradation of dilncRNA [81–83].
AHNAK controls LLPS of 53BP1, thereby influencing the interaction between 53BP1 and p53. 53BP1 condensate play a role in detecting DNA damage, increasing p53 activation, inducing the expression of DNA damage repair-related genes, and promoting protein modification [77,84]. Additionally, 53BP1 protects broken DNA ends from nucleolytic degradation and separates damaged sites from intact sites, facilitating the repair of damaged DNA. LLPS of 53BP1 is also essential for maintaining the structural integrity of heterochromatin [76].
Implications and perspectives
DSBs serve as a highly perilous form of DNA damage that can have detrimental effects on both genome integrity and cell viability. Upon DNA damage, multiple DNA repair-related proteins are recruited to damaged DNA sites sequentially, including ATM, ATR, PARP1, and 53BP1, inducing the formation of DNA repair foci. Many DNA repair factors, such as 53BP1, NONO, and DNA damage-induced paraspeckles, are capable of undergoing LLPS and thus promoting DNA repair.
Phase-separation-related DSB factors are found to efficiently recruit DSB repair factors and support enzymatic responses, such as promoting the recruitment of MRN complex to facilitate DNA damage repair. LLPS of FUS is essential for the assembly of SFPQ at DNA damage foci. Most of the previous studies have described the important role of phase separation in DDR, however, the role of LLPS is virtually unexplored. In addition, LLPS seems to play an important role in DSB response pathway; 53BP1 was found to promote p53 pathway for DSB activation. Advanced imaging techniques such as super-resolution and single-molecule imaging may provide technical support for the understanding of this issue [85].
LLPS of some DDR factors (such as 53BP1, NONO and FUS) also facilitate processes of cancer and neurodegenerative diseases [86,87]. In colon cancer, sentrin/SUMO-specific proteases 1 (SENP1) can promote DNA damage and drug resistance by reducing LLPS of RING finger protein 168 (RNF168) [88]. NONO drives the oncogenic transcriptional program by promoting Transcriptional coactivator with PDZ‐binding motif (TAZ) LLPS [89]. Amyotrophic lateral sclerosis (ALS) was also associated with LLPS of FUS. Through the study of phase separation, DSB repair process would be deeply understood, and strategies targeting LLPS of DNA repair factors may be an effective way to improve radiation therapy outcome, and study of phase separation of related molecules provides new insights for cancer treatment.
Funding Statement
The work was supported by the the National Science Fund for Distinguished Young Scholars [82225040]; Guangdong Natural Science Funds for Distinguished Young Scholars [2021B1515020022]; the National Science Fund for Excellent Young Scholars [82122057]; the National Natural Science Foundation of China [32000135].
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Author contributions
X.J.F. and X.B.W. conceived this review, H.L.L., H.N., W.W.Z., X.J.F. and X.B.W. wrote the manuscript. All authors read and approved the final manuscript.
Data availability statement
The data that support the findings of this study are openly available in figshare at http://doi.org/10.6084/m9.figshare.23671068.
References
- [1].Alberti S, Gladfelter A, Mittag T.. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019 Jan 24;176(3):419–10. doi: 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hofmann S, Kedersha N, Anderson P, et al. Molecular mechanisms of stress granule assembly and disassembly. Biochim Biophys Acta, Mol Cell Res. 2021. Jan;1868(1):118876. doi: 10.1016/j.bbamcr.2020.118876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ilık İA, Aktaş T. Nuclear speckles: dynamic hubs of gene expression regulation. FEBS J. 2022. Nov;289(22):7234–7245. doi: 10.1111/febs.16117 [DOI] [PubMed] [Google Scholar]
- [4].O’Flynn BG, Mittag T. The role of liquid-liquid phase separation in regulating enzyme activity. Curr Opin Cell Biol. 2021. Apr;69:70–79. doi: 10.1016/j.ceb.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pinheiro EDS, Preato AM, Petrucci TVB, et al. Phase-separation: a possible new layer for transcriptional regulation by glucocorticoid receptor. Front Endocrinol. 2023;14:1160238. doi: 10.3389/fendo.2023.1160238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Miao C, Zhang Y, Yu M, et al. HSPA8 regulates anti-bacterial autophagy through liquid-liquid phase separation. Autophagy. 2023. Jun;13:1–17 doi: 10.1080/15548627.2023.2223468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu D, Lum KK, Treen N, et al. IFI16 phase separation via multi-phosphorylation drives innate immune signaling. Nucleic Acids Res. 2023 Jun 7;51(13):6819–6840. doi: 10.1093/nar/gkad449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zheng H, Wen W. Protein phase separation: new insights into cell division. Acta Biochim Biophys Sin (Shanghai). 2023 May 30;55(7):1042–1051. doi: 10.3724/abbs.2023093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yasuhara T, Zou L. Impacts of chromatin dynamics and compartmentalization on DNA repair. DNA Repair. 2021. Sep;105:103162. doi: 10.1016/j.dnarep.2021.103162 [DOI] [PubMed] [Google Scholar]
- [10].Ui A, Chiba N, Yasui A. Relationship among DNA double-strand break (DSB), DSB repair, and transcription prevents genome instability and cancer. Cancer Sci. 2020. May;111(5):1443–1451. doi: 10.1111/cas.14404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Oh JM, Myung K. Crosstalk between different DNA repair pathways for DNA double strand break repairs. Mutat Res Genet Toxicol Environ Mutagen. 2022. Jan;873:503438. doi: 10.1016/j.mrgentox.2021.503438 [DOI] [PubMed] [Google Scholar]
- [12].Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016. Jan;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang K, Huang M, Li A, et al. DIAPH3 condensates formed by liquid-liquid phase separation act as a regulatory hub for stress-induced actin cytoskeleton remodeling. Cell Rep. 2023 Jan 31;42(1):111986. doi: 10.1016/j.celrep.2022.111986 [DOI] [PubMed] [Google Scholar]
- [14].Schvartzman C, Zhao H, Ibarboure E, et al. Control of enzyme reactivity in response to osmotic pressure modulation mimicking dynamic assembly of intracellular organelles. Adv Mater. 2023 May 7;35(33):e2301856. doi: 10.1002/adma.202301856 [DOI] [PubMed] [Google Scholar]
- [15].Sahin C, Motso A, Gu X, et al. Mass spectrometry of RNA-Binding proteins during liquid-liquid phase separation reveals distinct assembly mechanisms and droplet architectures. J Am Chem Soc. 2023 May 17;145(19):10659–10668. doi: 10.1021/jacs.3c00932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pintado-Grima C, Bárcenas O, Ventura S. In-silico analysis of pH-Dependent liquid-liquid phase separation in intrinsically disordered proteins. Biomolecules. 2022 Jul 12;12(7):974. doi: 10.3390/biom12070974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lu T, Nakashima KK, Spruijt E. Temperature-responsive peptide-nucleotide coacervates. J Phys Chem B. 2021 Apr 1;125(12):3080–3091. doi: 10.1021/acs.jpcb.0c10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ukmar-Godec T, Hutten S, Grieshop MP, et al. Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat Commun. 2019 Jul 2;10(1):2909. doi: 10.1038/s41467-019-10792-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin Y, Fichou Y, Longhini AP, et al. Liquid-liquid phase separation of tau driven by hydrophobic interaction facilitates fibrillization of tau. J Mol Biol. 2021 Jan 22;433(2):166731. doi: 10.1016/j.jmb.2020.166731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fang XD, Gao Q, Zang Y, et al. Host casein kinase 1-mediated phosphorylation modulates phase separation of a rhabdovirus phosphoprotein and virus infection. Elife. 2022 Feb 22;11:e74884 doi: 10.7554/eLife.74884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Saito M, Hess D, Eglinger J, et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat Chem Biol. 2019. Jan;15(1):51–61 doi: 10.1038/s41589-018-0180-7 [DOI] [PubMed] [Google Scholar]
- [22].Song X, Yang F, Yang T, et al. Phase separation of EB1 guides microtubule plus-end dynamics. Nat Cell Biol. 2023. Jan;25(1):79–91 doi: 10.1038/s41556-022-01033-4 [DOI] [PubMed] [Google Scholar]
- [23].Lin CC, Suen KM, Jeffrey PA, et al. Receptor tyrosine kinases regulate signal transduction through a liquid-liquid phase separated state. Mol Cell. 2022 Mar 17;82(6):1089–1106.e12. doi: 10.1016/j.molcel.2022.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Scully R, Panday A, Elango R, et al. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019. Nov;20(11):698–714 doi: 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Averbek S, Jakob B, Durante M, et al. O-GlcNAcylation affects the pathway choice of DNA double-strand break repair. Int J Mol Sci. 2021 May 27;22(11):5715. doi: 10.3390/ijms22115715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Syed A, Tainer JA. The MRE11–RAD50–NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu Rev Biochem. 2018 Jun 20;87(1):263–294. doi: 10.1146/annurev-biochem-062917-012415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cannavo E, Reginato G, Cejka P. Stepwise 5’ DNA end-specific resection of DNA breaks by the Mre11-Rad50-Xrs2 and Sae2 nuclease ensemble. Proc Natl Acad Sci U S A. 2019 Mar 19;116(12):5505–5513. doi: 10.1073/pnas.1820157116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Belan O, Barroso C, Kaczmarczyk A, et al. Single-molecule analysis reveals cooperative stimulation of Rad51 filament nucleation and growth by mediator proteins. Mol Cell. 2021 Mar 4;81(5):1058–1073.e7. doi: 10.1016/j.molcel.2020.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McIlwraith MJ, Vaisman A, Liu Y, et al. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005 Dec 9;20(5):783–792. doi: 10.1016/j.molcel.2005.10.001 [DOI] [PubMed] [Google Scholar]
- [30].Agmon N, Yovel M, Harari Y, et al. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011 Sep 1;39(16):7009–19. doi: 10.1093/nar/gkr277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhao B, Rothenberg E, Ramsden DA, et al. The molecular basis and disease relevance of non-homologous DNA end joining. Nat Rev Mol Cell Biol. 2020. Dec;21(12):765–781 doi: 10.1038/s41580-020-00297-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair. 2006 May 10;5(5):534–543. doi: 10.1016/j.dnarep.2006.01.012 [DOI] [PubMed] [Google Scholar]
- [33].Chen X, Xu X, Chen Y, et al. Structure of an activated DNA-PK and its implications for NHEJ. Mol Cell. 2021 Feb 18;81(4):801–810.e3. doi: 10.1016/j.molcel.2020.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Andres SN, Vergnes A, Ristic D, et al. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012. Feb;40(4):1868–1878 doi: 10.1093/nar/gks022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Peng PH, Hsu KW, Wu KJ. Liquid-liquid phase separation (LLPS) in cellular physiology and tumor biology. Am J Cancer Res. 2021;11(8):3766–3776. [PMC free article] [PubMed] [Google Scholar]
- [36].Shi J, Chen SY, Shen XT, et al. NOP53 undergoes liquid-liquid phase separation and promotes tumor radio-resistance. Cell Death Discovery. 2022;8(1):436 doi: 10.1038/s41420-022-01226-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ji Y, Li F, Qiao Y. Modulating liquid-liquid phase separation of FUS: mechanisms and strategies. J Mater Chem B. 2022 Nov 3;10(42):8616–8628. doi: 10.1039/D2TB01688E [DOI] [PubMed] [Google Scholar]
- [38].Otahalova B, Volkova Z, Soukupova J, et al. Importance of germline and somatic Alterations in human MRE11, RAD50, and NBN Genes coding for MRN complex. Int J Mol Sci. 2023 Mar 15;24(6):5612. doi: 10.3390/ijms24065612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003 Jan 30;421(6922):499–506. doi: 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- [40].Kozlov SV, Graham ME, Peng C, et al. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006 Aug 9;25(15):3504–14. doi: 10.1038/sj.emboj.7601231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998 Mar 6;273(10):5858–68. doi: 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- [42].Zdravković A, Daley JM, Dutta A, et al. A conserved Ctp1/CtIP C-terminal peptide stimulates Mre11 endonuclease activity. Proc Natl Acad Sci USA. 2021 Mar 16;118(11):e2016287118. doi: 10.1073/pnas.2016287118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Eid W, Steger M, El-Shemerly M, et al. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010. Dec;11(12):962–968 doi: 10.1038/embor.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang YL, Zhao WW, Bai SM, et al. MRNIP condensates promote DNA double-strand break sensing and end resection. Nat Commun. 2022 May 12;13(1):2638. doi: 10.1038/s41467-022-30303-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bennett LG, Wilkie AM, Antonopoulou E, et al. MRNIP is a replication fork protection factor. Sci Adv. 2020. Jul;6(28):eaba5974 doi: 10.1126/sciadv.aba5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Staples CJ, Barone G, Myers KN, et al. MRNIP/C5orf45 interacts with the MRN complex and contributes to the DNA damage response. Cell Rep. 2016 Sep 6;16(10):2565–2575. doi: 10.1016/j.celrep.2016.07.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Caron MC, Sharma AK, O’Sullivan J, et al. Poly(adp-ribose) polymerase-1 antagonizes DNA resection at double-strand breaks. Nat Commun. 2019 Jul 4;10(1):2954. doi: 10.1038/s41467-019-10741-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nilov DK, Pushkarev SV, Gushchina, IV, et al. Modeling of the enzyme-substrate complexes of human Poly(ADP-Ribose) polymerase 1. Biochemistry (Mosc). 2020. Jan;85(1):99–107 doi: 10.1134/S0006297920010095 [DOI] [PubMed] [Google Scholar]
- [49].Altmeyer M, Neelsen KJ, Teloni F, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun. 2015 Aug 19;6(1):8088. doi: 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Matkarimov BT, Zharkov DO, Saparbaev MK. Mechanistic insight into the role of Poly(ADP-ribosyl)ation in DNA topology modulation and response to DNA damage. Mutagenesis. 2020 Feb 13;35(1):107–118. doi: 10.1093/mutage/gez045 [DOI] [PubMed] [Google Scholar]
- [51].Haince JF, McDonald D, Rodrigue A, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008 Jan 11;283(2):1197–208. doi: 10.1074/jbc.M706734200 [DOI] [PubMed] [Google Scholar]
- [52].Rhine K, Dasovich M, Yoniles J, et al. Poly(adp-ribose) drives condensation of FUS via a transient interaction. Mol Cell. 2022 Mar 3;82(5):969–985.e11. doi: 10.1016/j.molcel.2022.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miwa M, Ida C, Yamashita S, et al. Poly(adp-ribose): structure, physicochemical properties and quantification in Vivo, with special reference to Poly(ADP-ribose) binding protein modules. Curr Protein Pept Sci. 2016;17(7):683–692. doi: 10.2174/1389203717666160419145246 [DOI] [PubMed] [Google Scholar]
- [54].Kim JJ, Lee SY, Hwang Y, et al. USP39 promotes non-homologous end-joining repair by poly(ADP-ribose)-induced liquid demixing. Nucleic Acids Res. 2021 Nov 8;49(19):11083–11102. doi: 10.1093/nar/gkab892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kang HC, Lee YI, Shin JH, et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):14103–8. doi: 10.1073/pnas.1108799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Krietsch J, Caron MC, Gagné JP, et al. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012 Nov 1;40(20):10287–301. doi: 10.1093/nar/gks798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Taiana E, Ronchetti D, Todoerti K, et al. LncRNA NEAT1 in Paraspeckles: a structural scaffold for cellular DNA damage response systems? Noncoding RNA. 2020;6(3):26. doi: 10.3390/ncrna6030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Adriaens C, Standaert L, Barra J, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016. Aug;22(8):861–8 doi: 10.1038/nm.4135 [DOI] [PubMed] [Google Scholar]
- [59].Kang J, Lim L, Lu Y, et al. A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids. PLoS Biol. 2019. Jun;17(6):e3000327 doi: 10.1371/journal.pbio.3000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lee M, Ghosh U, Thurber KR, et al. Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS. Nat Commun. 2020 Nov 12;11(1):5735. doi: 10.1038/s41467-020-19512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Félix SS, Laurents DV, Oroz J, et al. Fused in sarcoma undergoes cold denaturation: Implications for phase separation. Protein Sci. 2023. Jan;32(1):e4521 doi: 10.1002/pro.4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Levone BR, Lenzken SC, Antonaci M, et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J Cell Bio. 2021 May 3;220(5):e202008030. doi: 10.1083/jcb.202008030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010. Jul;2(7):a000687 doi: 10.1101/cshperspect.a000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Passon DM, Lee M, Rackham O, et al. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci U S A. 2012 Mar 27;109(13):4846–50. doi: 10.1073/pnas.1120792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fan XJ, Wang YL, Zhao WW, et al. NONO phase separation enhances DNA damage repair by accelerating nuclear EGFR-induced DNA-PK activation. Am J Cancer Res. 2021;11(6):2838–2852. [PMC free article] [PubMed] [Google Scholar]
- [66].Li S, Li Z, Shu FJ, et al. Double-strand break repair deficiency in NONO knockout murine embryonic fibroblasts and compensation by spontaneous upregulation of the PSPC1 paralog. Nucleic Acids Res. 2014. Sep;42(15):9771–80 doi: 10.1093/nar/gku650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kai M. Roles of RNA-Binding Proteins in DNA Damage Response. Int. J. Mol. Sci. 2016, 17, 310. Int J Mol Sci. 2016;17(4):604–604. doi: 10.3390/ijms17040604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3077–82. doi: 10.1073/pnas.0813247106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Koike M, Yutoku Y, Koike A. The C-terminal region of Rad52 is essential for Rad52 nuclear and nucleolar localization, and accumulation at DNA damage sites immediately after irradiation. Biochem Biophys Res Commun. 2013 May 31;435(2):260–6. doi: 10.1016/j.bbrc.2013.04.067 [DOI] [PubMed] [Google Scholar]
- [70].Kinoshita C, Takizawa Y, Saotome M, et al. The cryo-EM structure of full-length RAD52 protein contains an undecameric ring. FEBS Open Bio. 2023 Jan 27;13(3):408–418. doi: 10.1002/2211-5463.13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Miné-Hattab J, Heltberg M, Villemeur M, et al. Single molecule microscopy reveals key physical features of repair foci in living cells. Elife. 2021 Feb 5;10:e60577. doi: 10.7554/eLife.60577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003. Jun;5(6):572–7. doi: 10.1038/ncb997 [DOI] [PubMed] [Google Scholar]
- [73].Oshidari R, Huang R, Medghalchi M, et al. DNA repair by Rad52 liquid droplets. Nat Commun. 2020 Feb 4;11(1):695. doi: 10.1038/s41467-020-14546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013 Mar 7;49(5):872–83. doi: 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- [75].Chen BR, Sleckman BP. The regulation of DNA end resection by chromatin response to DNA double strand breaks. Front Cell Dev Biol. 2022;10:932633. doi: 10.3389/fcell.2022.932633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhang L, Geng X, Wang F, et al. 53BP1 regulates heterochromatin through liquid phase separation. Nat Commun. 2022 Jan 18;13(1):360. doi: 10.1038/s41467-022-28019-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ghodke I, Remisova M, Furst A, et al. AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation. Mol Cell. 2021 Jun 17;81(12):2596–2610.e7. doi: 10.1016/j.molcel.2021.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014. Jan;15(1):7–18. doi: 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- [79].Piccinno R, Minneker V, Roukos V. 53BP1-DNA repair enters a new liquid phase. EMBO J. 2019 Aug 15;38(16):e102871. doi: 10.15252/embj.2019102871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kilic S, Lezaja A, Gatti M, et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38(16):e101379. doi: 10.15252/embj.2018101379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tosolini D, Antoniali G, Dalla E, et al. Role of phase partitioning in coordinating DNA damage response: focus on the apurinic apyrimidinic endonuclease 1 interactome. Biomol Concepts. 2020 Dec 23;11(1):209–220. doi: 10.1515/bmc-2020-0019 [DOI] [PubMed] [Google Scholar]
- [82].Kieffer SR, Lowndes NF. Immediate-early, early, and late responses to DNA double stranded breaks. Front Genet. 2022;13:793884. doi: 10.3389/fgene.2022.793884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Pessina F, Giavazzi F, Yin Y, et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol. 2019. Oct;21(10):1286–1299 doi: 10.1038/s41556-019-0392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Oda T, Gotoh N, Kasamatsu T, et al. DNA damage-induced cellular senescence is regulated by 53BP1 accumulation in the nuclear foci and phase separation. Cell Prolif. 2023. Jun;56(6):e13398 doi: 10.1111/cpr.13398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhang H, Shao S, Sun Y. Characterization of liquid-liquid phase separation using super-resolution and single-molecule imaging. Biophys Rep. 2022 Feb 28;8(1):2–13. doi: 10.52601/bpr.2022.210043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mehta S, Zhang J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat Rev Cancer. 2022. Apr;22(4):239–252. doi: 10.1038/s41568-022-00444-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Peng Q, Tan S, Xia L, et al. Phase separation in cancer: from the impacts and mechanisms to treatment potentials. Int J Biol Sci. 2022;18(13):5103–5122. doi: 10.7150/ijbs.75410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wei M, Huang X, Liao L, et al. SENP1 decreases RNF168 phase separation to promote DNA damage repair and drug resistance in colon cancer. Cancer Res. 2023 Jun 23;83(17):2908–2923. doi: 10.1158/0008-5472.CAN-22-4017 [DOI] [PubMed] [Google Scholar]
- [89].Wei Y, Luo H, Yee PP, et al. Paraspeckle protein NONO promotes TAZ phase separation in the nucleus to drive the oncogenic transcriptional program. Adv Sci. 2021. Dec;8(24):e2102653 doi: 10.1002/advs.202102653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in figshare at http://doi.org/10.6084/m9.figshare.23671068.


