Abstract
The complete nucleotide sequence for the aot operon of Pseudomonas aeruginosa PAO1 was determined. This operon contains six open reading frames. The derived sequences for four of these, aotJ, aotQ, aotM, and aotP, show high similarity to those of components of the periplasmic binding protein-dependent ABC (ATP binding cassette) transporters of enteric bacteria. Transport studies with deletion derivatives established that these four genes function in arginine-inducible uptake of arginine and ornithine but not lysine. The aotO gene, which encodes a polypeptide with no significant similarity to any known proteins, is not essential for arginine and ornithine uptake. The sixth and terminal gene in the operon encodes ArgR, which has been recently shown to function in regulation of arginine metabolism. Studies with an aotJ::lacZ translational fusion showed that expression of the aot operon is strongly induced by arginine and that this effect is mediated by ArgR. S1 nuclease and primer extension experiments showed the presence of two promoters, P1 and P2. The downstream promoter, P2, is induced by arginine and appears to be subject to carbon catabolite repression. The upstream promoter, P1, is induced by glutamate. Footprinting experiments established the presence of a 44-bp ArgR binding site that overlaps the −35 region for P2, as was shown to be the case for the arginine-inducible aru promoter, and the −10 region for P1, as was shown to be the case for arginine-repressible operons in P. aeruginosa. Sequence alignment confirms the architecture and the consensus sequence of the ArgR binding sites, as was previously reported.
Arginine is an important nutrient for Pseudomonas aeruginosa in the environment, as reflected in its ability to serve as one of the strongest chemotactic attractants to this organism (6). P. aeruginosa can utilize arginine under aerobic conditions as a sole source of carbon, energy, and nitrogen via the arginine succinyltransferase pathway (9). Despite the preference for an aerobic environment, P. aeruginosa can utilize arginine under anaerobic conditions via the arginine deiminase pathway (9). The resulting metabolic energy permits motility and slow growth, thus aiding adaptation under conditions of oxygen limitation (9).
Itoh (12) has reported preliminary evidence for an operon, designated aot, which consists of six genes that function in the transport of arginine and ornithine. The aot operon is located upstream of the aru operon, which encodes enzymes of the arginine succinyltransferase pathway (13). Recent work from our laboratory (22, 23) has shown that the sixth and terminal gene in this operon encodes a regulatory protein, ArgR, that functions in control of expression of certain genes of arginine biosynthesis and catabolism.
The present paper reports the cloning and characterization of the aot operon, which encodes components of a periplasmic binding protein (PBP)-dependent transporter for arginine and ornithine. This paper also reports that the aot operon is transcribed from two promoters that are differentially controlled and that ArgR participates in the control of both promoters.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) medium (18) or nutrient yeast broth (10) was used for strain construction with the following supplements as required: ampicillin at 50 μg/ml (Escherichia coli), carbenicillin at 200 μg/ml (P. aeruginosa), and 5-bromo-3-indolyl-β-d-galactoside (X-Gal) at 0.03% (wt/vol). The minimal medium P described by Haas et al. (10) was used for the growth of P. aeruginosa with either l-glutamate or l-glutamate and l-arginine (20 mM) as the sources of carbon and nitrogen as indicated.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80dlac ΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK−) supE44 λ− thi-1 gyrA96 relA1 | Bethesda Research Laboratory |
| P. aeruginosa | ||
| PAO1 | Wild type | D. Haas |
| PAO1214 | aotJ::Tn5-751 insertion mutant of PAO1 | 22 |
| PAO501 | argR; gentamicin resistant | 22 |
| PAO4447 | leu; kanamycin sensitive | Y. Itoh |
| PAO4449 | aotO::aph mutant of PAO4447 | This study |
| PAO4450 | aot::aph mutant of PAO4447; kanamycin resistant | This study |
| Plasmids | ||
| pUC119 | bla lacZ′ cloning vector | 27 |
| pUC19 | bla lacZ′ cloning vector | 31 |
| pQF52 | bla lacZ translational fusion vector | 23 |
| pST500 | aotJ::lacZ translational fusion of pQF52 | This study |
| pST26 | 360-bp PCR fragment of the aot promoter region cloned into SmaI site of pUC19 | This study |
| pMO011644 | Cosmid clone carrying the aot-argR and aru operons of P. aeruginosa | 13 |
| pYJ61 | 10-kb insert in pMO011644 subcloned into HincII site of pUC119 | This study |
| pYJ62 | Similar to pYJ61, but with the opposite orientation | This study |
| pTnL1 | Clone of PAO1214 carrying the aotJ regulatory region and the aph gene of Tn5-751 | This study |
Cloning and nucleotide sequencing of the aot operon.
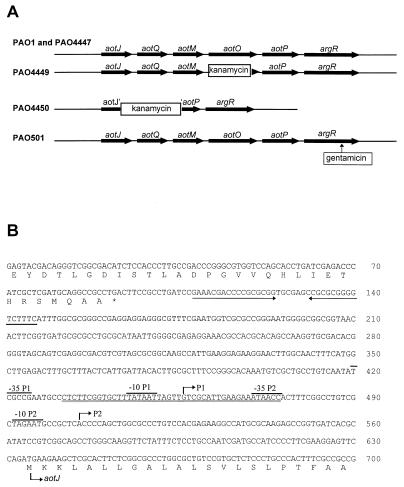
The 10-kb EcoRI fragment (Fig. 1A) carrying all of the aot structural genes was obtained from cosmid pMO011644 (13), its ends were filled by the Klenow fragment, and it was cloned into the HincII site of pUC119. The resulting plasmids, containing the aot genes in opposite orientations to each other, were designated pYJ61 and pYJ62 (Fig. 1A). The single EcoRI site on transposon Tn5-751 in the aotJ gene of PAO1214 chromosome (22) was used to clone the 5′ end of the aotJ gene and its upstream regulatory region into pUC19, giving rise to pTnL1. The DNA sequence of the entire aot operon and its regulatory region was determined with plasmid templates as described previously (13, 22).
FIG. 1.
(A) Schematic drawing of the genetic organization of the aot-arg locus on the chromosomes of P. aeruginosa PAO1 and its derivatives. The insertion sites of antibiotic cassettes and transposon Tn5-751 are indicated. (B) Nucleotide sequence of the promoters and flanking regions for the aotJQMOP-argR operon. The −10 and −35 regions of each promoter are indicated accordingly, and the transcriptional initiation sites are indicated by arrows above the nucleotides and labeled P1 and P2, respectively. The hypersymmetric sequence resembling that of a rho-independent terminator is defined by converging arrows. The ArgR binding site as determined in this study is double underlined.
Transport assays.
Cultures were grown in minimal medium P containing 20 mM arginine (inducible conditions) or 20 mM glutamate (noninducible conditions) as the sole source of carbon and nitrogen. Cells were harvested during logarithmic growth, washed three times with minimal medium P, and suspended in minimal medium P at a concentration of ca. 4 × 108 cells/ml, and then chloramphenicol was added at a concentration of 250 μg/ml. After incubation of the cell suspension for 5 min at 37°C, a 0.05-ml solution of 3H-labeled amino acid was added at a final concentration of 1 mM (74 Mbq/mmol), and samples (0.1 ml) were withdrawn at time intervals. Cells were collected on a cellulose acetate membrane filter (0.22-μm pore size) and washed with 10 ml of a solution containing 10 mM Tris-HCl (pH 7.3), 0.15 M NaCl, and 5 mM MgCl2. Incorporated radioactivity was counted in a liquid scintillation spectrometer. In competition experiments, cold amino acid was added, at the indicated concentrations, simultaneously with the radioactive amino acid.
Construction of aotJ::lacZ translational fusion.
A DNA fragment bracketed by nucleotides (nt) 142 to 649 (Fig. 1B) containing the 5′ terminus of aotJ and its upstream flanking region was amplified from pTnL1 by PCR employing two 27-mer oligonucleotide primers designed to generate HindIII and SmaI ends: oligo-1 (5′-AGCAAGCTTTCATTTGGCGCGGGCCGA-3′) and oligo-2 (5′-ATACCCGGGTGCGAGCTTCTTCATCTG-3′). The amplified fragment was digested with HindIII and SmaI and ligated into the translational fusion vector pQF52, previously cleaved with the same enzymes (Table 1). The resulting plasmid, pST500, was used to transform E. coli DH5α, and the transformants were selected on LB plates containing ampicillin and X-Gal. The orientation of the insert on the plasmid was confirmed by plasmid DNA sequencing.
Plasmid pST500 was used to transform E. coli and P. aeruginosa according to the method described by Chung et al. (5) for one-step preparation of competent cells.
β-Galactosidase assay.
Logarithmically growing cultures were harvested at an optical density of 0.5 at 600 nm. Cells were washed once and suspended in 20 mM potassium phosphate (pH 7.6) containing 1 mM EDTA. Cells were ruptured by passage through an Aminco French pressure cell, and the cell extract was centrifuged at 27,000 × g for 20 min at 4°C. β-Galactosidase activity was determined by the method of Miller (20). The relative levels of plasmid DNA under different growth conditions were determined by densitometric measurement of the stained and linearized DNA bands as described previously (15) and used to normalize β-galactosidase expression. Protein concentration was determined by the method of Bradford (3) with crystalline bovine serum albumin as a standard.
S1 nuclease mapping and primer extension experiments.
RNA extraction was carried out as previously described (23). S1 nuclease mapping was carried out basically as described by Greene and Struhl (8) with an end-labeled single-stranded probe. A clone containing the aotJ promoter (pST500) was used to prepare radioactively labeled single-stranded DNA. An 18-mer which can hybridize with nt 632 to 650 for aotJ (Fig. 1) was end labeled with [γ-32P]ATP and annealed to double-stranded DNA of HindIII-cleaved pST500. Extension of the labeled oligonucleotide for preparation of the single-stranded probe was carried out with Klenow fragment at 37°C, and the extended probe was eluted from a 6% polyacrylamide gel with a gel eluter (IBI). Experiments were performed quantitatively to permit comparison of levels of transcripts under different growth conditions. The relative levels of transcripts were determined by scanning autoradiographs with a Molecular Dynamics personal laser densitometer.
For primer extension experiments, the same 32P-labeled oligonucleotide described above was used for hybridization with 20 μg of total RNA, and the reaction was carried out as previously described (16).
Purification of ArgR.
The ArgR protein was purified to homogeneity from a recombinant strain of E. coli DH5α harboring pSM21 carrying argR from P. aeruginosa as previously described (23).
Gel retardation analysis.
A DNA fragment containing the aotJ regulatory region (nt 290 to 650 of Fig. 1) was generated by PCR and cloned into the SmaI site of pUC19. The insert of the resulting plasmid, pST26, was purified from a 1% (wt/vol) low-melting-point agarose gel after BamHI and EcoRI restriction digestions. The DNA probe was prepared by labeling with either [α-32P]dATP or dGTP with the Klenow fragment.
The binding reactions were carried out as described previously (16, 23). The radioactively labeled DNA probe (0.1 pM) was allowed to interact with different concentrations of ArgR in 20 μl of a mixture of 20 mM Tris-HCl (pH 7.6), 50 mM KCl, 1 mM EDTA, 5% (vol/vol) glycerol, and 50 μg of bovine serum albumin per ml. Reaction mixtures were allowed to equilibrate for 30 min at 25°C, the reactions were terminated by the addition of an excess of cold DNA probe (10 pM), and then the mixtures were applied to a 5% polyacrylamide gel while the gel was running. After autoradiography of the gel, the intensities of the bound and unbound DNA bands were measured with a Molecular Dynamics personal laser densitometer. The apparent equilibrium binding constant, defined as the protein concentration required for half-maximal binding, was subsequently determined from a plot of the percentage of DNA bound versus protein concentration.
DNA footprinting analysis.
A 360-bp DNA fragment containing the aot regulatory region in plasmid pST26 was prepared and radioactively labeled as described above. DNase I footprinting, premethylation interference, and missing-contact experiments were carried out as previously described (16, 23). The reaction mixtures contained 10 pM operator DNA, and the concentration of the ArgR protein used in the reactions was empirically determined as indicated in the figure legends. The final reaction mixtures were dissolved in 20 μl of formamide-dye mixture, and the reaction products were analyzed with an 8% denaturing polyacrylamide sequencing gel against a G+A sequencing ladder (19).
Nucleotide sequence accession number.
The sequence data for aot have been deposited in the GenBank database under accession no. AF012537.
RESULTS
Nucleotide sequence analysis of the aot operon.
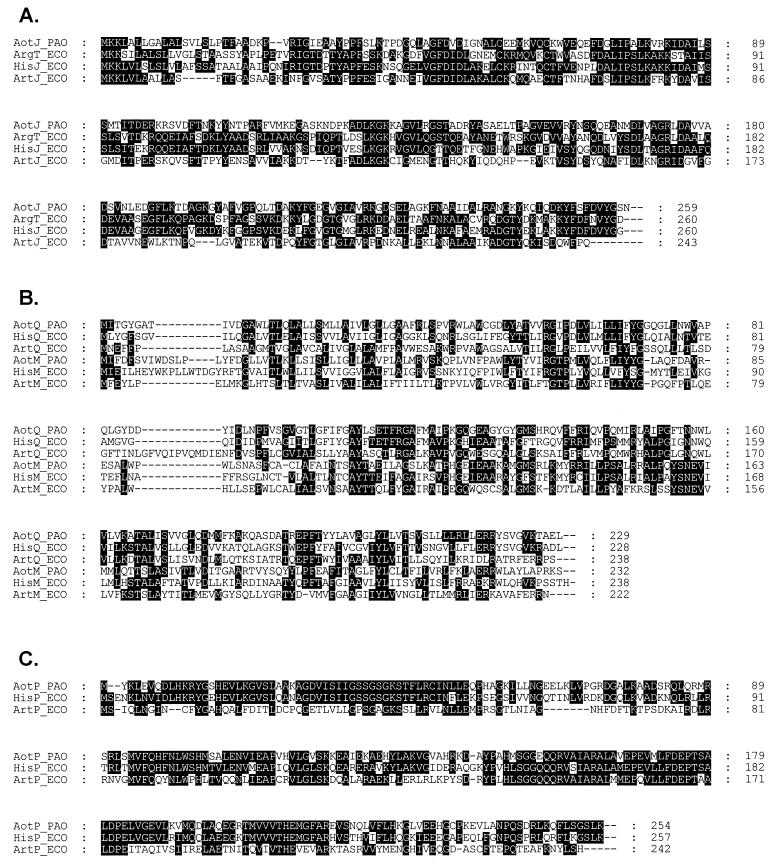
The nucleotide sequence of the aot operon and the flanking regions (GenBank accession no. AF012537) was determined from pYJ60, pYJ62, and pTnL1. Analysis of the resulting nucleotide sequence revealed the presence of six open reading frames (ORFs) (Fig. 1A). The first gene of the operon, designated aotJ, encodes a polypeptide of 259 amino acids with a predicted molecular mass of 28.0 kDa. The derived amino acid sequence shows high similarity to PBDs of other organisms (Fig. 2A). It exhibits the highest level of similarity to ArgT and HisJ of E. coli (46% identity), followed by ArtJ and ArtI of the same organism (40 and 37% identity, respectively). The second ORF, designated aotQ, encodes a polypeptide of 229 residues with a predicted molecular mass of 25.1 kDa. The third ORF, aotM, encodes a polypeptide of 232 residues with a predicted molecular mass of 26.1 kDa. AotQ and AotM exhibit similarity to membrane components of other PBP-dependent transporters (Fig. 2B), with the highest similarity to HisQ and HisM of E. coli (50 and 45% identity, respectively). The fourth ORF, designated aotO, has a coding capacity for 370 amino acids, yielding a polypeptide with a predicted molecular mass of 40.2 kDa. The derived amino acid sequence of AotO exhibits no significant similarity to any known protein. The fifth ORF, designated aotP, encodes a polypeptide of 254 amino acids with a predicted molecular mass of 28.0 kDa. The derived amino acid sequence of AotP shows 70% identity to the HisP protein of S. typhimurium (Fig. 2C), which is the ATP-binding protein of the histidine transport permease complex. The six and terminal gene in the operon corresponds to argR, which has been recently shown to function in regulation of arginine metabolism (22).
FIG. 2.
Amino acid sequence alignments of components for l-arginine uptake in P. aeruginosa PAO1 (PAO) and E. coli K-12 (ECO). Sequences were aligned with the Clustal W program (26). (A) Comparison of the PBPs. (B) Comparison of the membrane components. (C) Comparison of the components with an ATP-binding cassette.
The nucleotide sequence of the upstream region flanking aotJ shows a truncated ORF at the beginning of the determined sequence (Fig. 1B). The derived amino acid sequence of this truncated ORF shows a very high similarity to the corresponding C-terminal sequences of the acetyl coenzyme A synthetase of various microorganisms (50 and 60% identity to the E. coli and yeast counterparts, respectively). A stem-loop structure resembling the rho-independent terminator was found immediately downstream from the stop codon of this ORF (Fig. 1B). The stop codon of this ORF and the start codon of AotJ are separated by 570 bp with no apparent coding capacity, as determined by the codon bias profile of P. aeruginosa generated by the MacVector package (Oxford Molecular Group).
Arginine and ornithine transport in PAO.
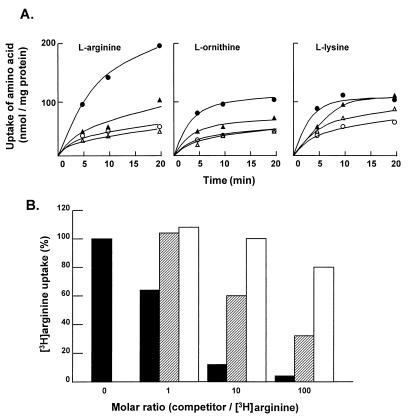
The similarity of the Aot proteins to periplasmic arginine-binding protein-dependent transport proteins of E. coli and Salmonella typhimurium and the presence of argR as the terminal gene in the encoding operon suggest that the aot operon functions in the uptake of l-arginine. In order to investigate this hypothesis, arginine uptake was examined in a pair of isogenic strains of P. aeruginosa, PAO4447 (wild type for aot) and PAO4450 (aotJQMOP deletion [Fig. 1A]). The deletion mutant strain retained the ability to use arginine as the sole source of carbon and nitrogen. Cultures of PAO4447 and PAO4450 cells were grown in minimal medium P with either l-arginine or l-glutamate as the sole source of carbon and nitrogen. The uptake of arginine, ornithine, and lysine in cell suspensions was examined. The results (Fig. 3A) show that uptake of arginine and ornithine is significantly induced by the presence of exogenous arginine in the growth medium of both the parent (PAO4447) and its aot deletion derivative (PAO4450). However, the levels of arginine-inducible uptake of both arginine and ornithine are significantly lower in the aot deletion derivative. While lysine uptake is also induced, although to a lesser extent by arginine, the deletion of the aot genes does not appear to affect lysine uptake. These results indicate that the aot genes function in the uptake of arginine and ornithine but not lysine.
FIG. 3.
(A) Effect of exogenous arginine on the uptake of arginine, ornithine, and lysine in a pair of isogenic strains of P. aeruginosa, PAO4447 (wild type for aot) (circles) and PAO4450 (aotJQMOP deletion) (triangles). Cultures grown in minimal medium P with either arginine (solid symbols) or glutamate (open symbols) as the sole source of carbon and nitrogen were harvested in the early log phase and used for l-arginine transport assays, as described in Materials and Methods. (B) Competition tests of arginine uptake in P. aeruginosa. The wild-type strain, PAO1, was grown in minimal medium P with arginine as the sole source of carbon and nitrogen, and arginine transport assays were done with different molar ratios of cold arginine (solid bars), ornithine (shaded bars), and lysine (open bars) to [3H]arginine as indicated.
Transport experiments with P. aeruginosa PAO4449, a derivative from which aotO was deleted, retained the arginine uptake pattern of the wild-type parent. Accordingly, the AotO protein, encoded by the fourth ORF in the operon, does not appear to function in arginine transport.
The arginine-inducible uptake pattern of PAO4447 was also observed in the wild-type strain, PAO1 (data not shown). The solute specificity of the arginine uptake system in PAO1 was analyzed by competition experiments. The results (Fig. 3B) show that uptake of l-[2, 3-3H]arginine is strongly inhibited by the addition of cold l-arginine, whereas cold l-ornithine exerts a moderate inhibition effect. In contrast, no significant inhibition was observed in the presence of cold l-lysine; only 20% inhibition was observed in the presence of a 100-times excess of l-lysine.
Regulation of an aotJ::lacZ translational fusion in P. aeruginosa.
In order to investigate promoter activity in the upstream region flanking aotJ, an aotJ::lacZ translational fusion, pST500, was constructed. The chromosomal insert of pST500 contains the first 18 nt of aotJ and 493 nt of the upstream flanking sequence, with the 6th codon of aotJ fused in frame to lacZ on the broad-host vector pQF52.
Comparison of blue colorations on X-Gal plates showed that expression of pST500 is much weaker in E. coli than in P. aeruginosa.
P. aeruginosa PAO1 cells harboring pST500 were grown in minimal medium P supplemented with various carbon and nitrogen sources, and β-galactosidase expression from the fusion was determined with cell extracts. The results (Table 2) show that arginine strongly induces expression of aotJ when it serves as the sole source of carbon and nitrogen. The level of induction is reduced when succinate, a good carbon source for P. aeruginosa, is present in the growth medium. The use of ammonium as a nitrogen source does not significantly affect the expression of aotJ. The translational fusion, pST500, was also introduced into P. aeruginosa PAO501, in which argR was inactivated by gene replacement (Fig. 1A) (23). Measurements of β-galactosidase expression in the absence or presence of arginine show that aotJ expression in the fusion is no longer induced by arginine in PAO501 (Table 2).
TABLE 2.
Expression of β-galactosidase from an aotJ::lacZ fusion in PAO1 and PAO501
| Host strain (genotype)a | Growth conditionb | Sp act (nmol/mg/ml)c |
|---|---|---|
| PAO501 (argR::Gm) | S+N | 940 (47) |
| G | 1,030 (31) | |
| G+A | 1,038 (50) | |
| PAO1 (wild type) | S+N+A | 6,021 (310) |
| S+N | 1,654 (31) | |
| S+A | 9,586 (470) | |
| G | 3,662 (37) | |
| N+A | 42,368 (420) | |
| G+A | 23,928 (240) | |
| A | 44,436 (430) |
The aot::lacZ translational fusion, pST500, was constructed and transformed into PAO1 and PAO501 as described in Materials and Methods.
Cells were grown in minimal medium P supplemented with 20 mM succinate (S), ammonium (N), glutamate (G), and arginine (A) as indicated.
Specific activities represent the average of cultures under each growth condition, with standard errors being shown in parentheses. The values shown are normalized relative to plasmid DNA.
Mapping of transcriptional initiation.
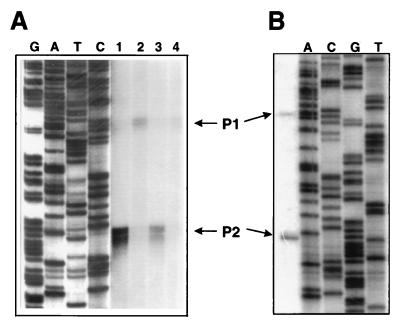
The 5′ terminus of aotJ mRNA was determined by S1 nuclease mapping with an end-labeled single-stranded DNA probe with a sequence complementary to nt 149 to 650 of Fig. 1. The results (Fig. 4A) show four consecutive bands corresponding to nt 503 to 506 of the sequence in Fig. 1B. Multiple bands are usually considered the result of nibbling by S1 nuclease. These bands are preceded by a good −10 sequence (five of six bases) and a weak −35 sequence (three of six bases) on the basis of homology to the consensus sequence of sigma-70 promoters of E. coli. Additional weak signals corresponding to nt 453 to 457 of the sequence in Fig. 1B are also evident. A perfect −10 sequence (six of six bases) and a weak −35 sequence are found preceding these signals. These two putative promoters of aotJ were designated as P1 and P2 for the upstream and downstream promoters, respectively.
FIG. 4.
(A) S1 nuclease mapping of the 5′ end of the aotJ transcripts in P. aeruginosa. A single-stranded DNA fragment covering the regulatory region of the aot operon was prepared and labeled at the 5′ end as described in Materials and Methods. After hybridization to cellular RNA, the hybridization mixture was treated with S1 nuclease and analyzed on a 6% sequencing gel. The dideoxy ladder (lanes G, A, T, and C) was derived with the same primer that was used to generate the probe for S1 nuclease mapping. Total RNA samples were purified from cultures grown with 20 mM arginine (lane 1), glutamate (lane 2), succinate and arginine (lane 3), or succinate and ammonium chloride (lane 4). (B) Primer extension. The reaction was carried out with RNA extracted from cultures grown with succinate and arginine. The 5′ termini of the two transcripts, designated as P1 and P2, correspond to nt 456 and 505, respectively, of Fig. 1B.
The presence of two transcriptional initiation sites for the aot operon as indicated above was also confirmed by primer extension experiments (Fig. 4B), and the most likely transcriptional initiation sites for P1 and P2 were determined to correspond to nt 456 and 505, respectively, of the sequence in Fig. 1B.
Quantitative S1 experiments were carried out to investigate the responses of P1 and P2 to arginine and succinate, which were shown above to affect expression of the aotJ::lacZ translation fusion. Densitometric measurement of the aotJ transcripts in the resulting autoradiographs (Fig. 4A) showed that the level of the P2 transcript is induced 10-fold relative to that obtained with glutamate (compare lanes 1 and 2) when arginine is the sole source of carbon and nitrogen in the growth medium. The presence of exogenous succinate in the growth medium reduces the level of P2 to fourfold (compare lane 1 and lane 3). In contrast, a higher level of the P1 transcript is induced when glutamate is the sole source of carbon and nitrogen in comparison to the two other growth conditions (compare lane 1 to lane 2 or 3).
Binding of ArgR to the aotJ regulatory region.
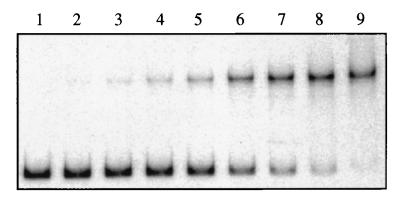
Gel retardation experiments were carried out with the purified ArgR protein of P. aeruginosa and a DNA fragment carrying the regulatory region of aotJ (Fig. 5). The relative amounts of free and bound DNA bands were estimated densitometrically. A plot of the percentage of bound DNA versus the concentration of ArgR yields an apparent dissociation constant of 21 pM. The gel retardation experiments shown in Fig. 5 were performed in the absence of arginine from all reagents. Inclusion of l-arginine (1 mM) in the reaction mixture did not significantly affect the dissociation constant for the aotJ operator (data not shown).
FIG. 5.
Gel retardation experiments. The radioactive 32P-labeled aotJ operator DNA fragment was incubated with various ArgR concentrations in the absence of l-arginine. The reactions were terminated by the addition of excess unlabeled operator DNA and applied to a 5% polyacrylamide gel while the gel was running. Lanes 1 to 9 contained ArgR concentrations of 0, 1, 3, 6, 12, 26, 50, 100, and 200 pM, respectively.
DNA footprinting analyses.
Three footprinting approaches, DNase I, premethylation, and depurination, were used to characterize the interactions of ArgR with the control region of the aot operon. DNase I footprinting analysis was used to define the target site for ArgR binding. The results (Fig. 6) show that binding of ArgR protects a 44-bp region against nuclease digestion. Specific base interactions that are crucial for ArgR binding were analyzed by premethylation interference and missing-contact probing experiments. As shown in Fig. 6a and b, methylation of G25, G28, and G36 residues on the sense strand and G16′ and G18′ residues on the antisense strand of the aot operator sequence strongly interferes with the binding of ArgR. Fig. 6a and b also show that depurination of these guanine residues as well as most of adenine residues in the region defined by DNase I footprinting inhibits the formation of the ArgR-aot operator complex. A schematic presentation of the combined data (Fig. 6c) clearly indicates that the ArgR binding site in the aot regulatory region completely overlaps the putative −35 region of the arginine-inducible P2 promoter and the −10 region of the upstream P1 promoter for this operon (Fig. 1B).
FIG. 6.
Footprinting analyses of ArgR with the aotJ control region. The DNA fragments used were labeled at the 3′ end of the sense strand (a) and antisense strand (b) of the aotJ operator. Lanes: 1, the corresponding G+A Maxam-Gilbert sequencing ladder; 2 to 6, DNase I footprinting with decreasing concentrations of ArgR: 9, 1.8, 0.36, 0.07, and 0 pM, respectively (the protected region is indicated by a bar); 7 and 8, premethylation footprinting sequences of the unbound and bound DNA, respectively; 9 and 10, depurination footprinting sequences of unbound and bound DNA, respectively. These two forms of DNA have been separated and modified as described in Materials and Methods. Also shown are several guanine residues, which are numbered according to the description of panel c. (c) Summary of the results of the three footprinting analyses. The nucleotide sequence of a region defined by DNase I footprinting is shown and numbered accordingly. Nucleotides detected by premethylation interference footprinting are indicated by solid squares, and those detected by depurination footprinting are indicated by solid triangles. Also shown is the −35 region of P1 (Fig. 1B).
DISCUSSION
The organization of the aot operon of P. aeruginosa is similar to those of operons encoding the binding protein-dependent (BPD) transport systems of enteric bacteria (for a review, see reference 2). In general, all of the protein components of a BPD transport system are encoded by a single operon, sometimes together with other proteins not directly involved in the transport system. This is also the case here, with aotO and argR being the terminal two genes in the operon. Furthermore, as is the case for most of the BPD operons, AotJ, the homolog of the enteric PBP, is encoded by the first gene of the operon. Interestingly, a sequence structure similar to that of the rho-independent terminator is located in the aotJ-aotQ intergenic region. Similar structural elements have also been found in other BPD transport operons (2), namely, glycerol-3-phosphate (ugp), phosphate (pst), histidine (his), and maltose (mal). By analogy to the proposed function in these systems, the terminator-like sequence could function here to generate a substantially higher level of AotJ than the other downstream components of the transport system and the ArgR protein. It has been suggested (2) that the presence of such differential expression levels is in accordance with the need for a higher level of the PBP for efficient uptake.
Recent work from our laboratory (22) has described the cloning and characterization of ArgR in P. aeruginosa, which has significant similarity to the AraC-XylS family of transcriptional regulators. These studies showed that ArgR induces the aru operon, which encodes enzymes of the arginine succinyltransferase pathway (13), which is considered the major pathway for aerobic utilization of arginine and ornithine in P. aeruginosa (9). In accordance with these findings, inactivation of argR abolished utilization of arginine or ornithine as the sole source of carbon and nitrogen but did not affect the utilization of other amino acids, including lysine (22).
The results of amino acid uptake experiments (Fig. 3) show that exogenous arginine induces the uptake of l-arginine and l-ornithine and has no effect on the uptake of l-lysine. Deletion of the aotJQMOP genes greatly reduces arginine and ornithine uptake. The finding that the residual uptake of arginine or ornithine remains inducible by arginine indicates the presence of a second and yet uncharacterized arginine uptake system in P. aeruginosa. The results of competition experiments with PAO1 support the conclusion that the major arginine uptake system in P. aeruginosa is specific for arginine and ornithine but not lysine.
The presence of more than one system for uptake of arginine in P. aeruginosa reported here is similar to the situation in enteric bacteria, which possess multiple systems for uptake of this amino acid (1, 4, 29). One significant difference, however, is that while arginine uptake systems of P. aeruginosa are induced by the presence of exogenous arginine in the growth medium, the well-characterized uptake systems of enteric bacteria are either repressed or unaffected under these conditions. These differences likely reflect the different role of arginine in nutrition of these organisms: P. aeruginosa can utilize arginine efficiently as a sole source of carbon, energy, and nitrogen (9), whereas enteric bacteria can utilize this amino acid only as a source of nitrogen (25). Recently, cloning and characterization of the aru operon in P. aeruginosa have led to the identification of its equivalent ast operon of E. coli (13) and have confirmed the presence of the arginine succinyltransferase pathway in enteric bacteria (25).
We have previously shown that ArgR synthesis is induced by exogenous arginine (22). The S1 nuclease experiments reported here show the presence of two promoters for the aot-argR operon (Fig. 4). Quantitative S1 experiments and fusion studies show that the two promoters are controlled differently. The downstream promoter, P2, is clearly the site sensitive to arginine induction. Interestingly, arginine induction is dampened by the presence of exogenous succinate in the growth medium, indicating that this promoter is also subject to carbon catabolite repression. While the molecular mechanism of carbon catabolite repression has not been elucidated, cyclic AMP has been shown not to be involved (24), and tricarboxylic acid cycle intermediates such as succinate are known to exert a much stronger effect than glucose (30). The upstream promoter, P1, is induced when glutamate is the sole source of carbon and nitrogen. Additional studies are needed to investigate the regulation of this promoter.
Footprinting analyses (Fig. 6) identified an ArgR binding site in the aot regulatory region. Comparison of this site (Fig. 7) with the ArgR binding sites for the argF, car, and aru operons indicates a consensus sequence identical to that reported in our previous work (23). As previously noted (23), the ArgR binding site is composed of a consensus sequence of TGTCGCN8AAN5 in a direct repeat structure, and the first half-site of the ArgR binding site appears more degenerate than the second half-site.
FIG. 7.
Sequence alignment of ArgR binding sites. The first and second halves of the binding sites are depicted by arrows. The consensus sequence was deduced from the second half-sites, which are more conserved. Nucleotides identical to those of the consensus site are shaded.
The ArgR binding site in the aot regulatory region overlaps the −35 region of the arginine-inducible P2 promoter, as was found in our earlier work with the arginine-inducible aru operon (23). However, only one ArgR binding site is found in the aot regulatory region (Fig. 1B), in contrast to the two sites found in the regulatory region for the arginine-inducible aru operon (23). While these results indicate that a single ArgR binding site at the appropriate location is sufficient for promoter activation, the significance of the two contiguous sites in the aru operon remains to be elucidated. The single ArgR binding site of aot also overlaps the −10 region for P1 (Fig. 7 [and see Fig. 1B), as was found to be the case for the arginine-repressible car and argF operons (23). Thus, it appears that that ArgR functions as a repressor of P1. Consistent with this hypothesis, quantitative S1 nuclease experiments indeed indicate that the level of the P1 transcript is reduced in the presence of exogenous arginine.
The presence of l-arginine does not affect the in vitro binding of ArgR of P. aeruginosa to the aot operator, which was shown earlier (23) to be the case for the car, argF, and aru operators. The earlier work with ArgR (23) has shown that certain intermediates of arginine metabolism (ornithine, agmatine, and glutamate) also have no effect on the binding of ArgR to the argF operator. While these results do not preclude that another arginine metabolite might serve as the effector, it is possible that, as in the case of AraC, binding of the signal ligand induces the correct conformation of ArgR for transcriptional activation via its interactions with the RNA polymerase. Alternatively, another gene might be involved in induction of the aot operon. A candidate for such a locus was previously identified by mutations at 1 min of the PAO1 chromosome that result in a phenotype unable to utilize arginine or ornithine as the sole source of carbon and nitrogen (14). Recent work (20a) has shown that the derived amino acid sequence for this locus possesses features of a sensor-regulator two-component system.
This work and previous reports (22, 23) establish that ArgR plays an important role in regulation of arginine uptake and arginine metabolism in P. aeruginosa. We have also previously shown (22) that ArgR controls expression of the catabolic glutamate dehydrogenase. Interestingly, collaborative work with other laboratories indicates a wider regulatory role for ArgR of P. aeruginosa than originally envisioned. Collaborative work (21) with R. Hancock and coworkers (Vancouver, Canada) showed that ArgR participates in control of expression of the oprD operon (11), which encodes the outer membrane porin D2. Similarly, collaborative work (28) with D. Haas (Lausanne, Switzerland) indicated that ArgR participates in regulation of the arcDABC operon (7), which encodes enzymes for the arginine deiminase pathway. Furthermore, preliminary work from our laboratory (17) established a regulatory role for ArgR in the gltBD operon, which encodes glutamate synthase. Studies of the role of ArgR in regulation of these operons are in progress.
ACKNOWLEDGMENT
This work was supported in part by a research grant (GM47926) from the National Institute of General Medical Sciences.
REFERENCES
- 1.Ames G F-L. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 2.Boos W, Lucht J M. Periplasmic binding protein-dependent ABC transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1175–1209. [Google Scholar]
- 3.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Celis T F R. Chain-terminating mutants affecting a periplasmic binding protein involved in the active transport of arginine and ornithine in Escherichia coli. J Biol Chem. 1981;256:773–779. [PubMed] [Google Scholar]
- 5.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craven R, Montie T C. Regulation of Pseudomonas aeruginosa chemotaxis by the nitrogen source. J Bacteriol. 1985;164:544–549. doi: 10.1128/jb.164.2.544-549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamper M, Zimmermann A, Haas D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J Bacteriol. 1991;173:4742–4750. doi: 10.1128/jb.173.15.4742-4750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene, J. M., and K. Struhl. S1 analysis of mRNA using M13 template, p. 4.6.2–4.6.13. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 9.Haas D, Galimand M, Gamper M, Zimmermann A. Arginine network of Pseudomonas aeruginosa: specific and global controls. In: Silver S, Chakrabarty A-M, Iglewski B, Kaplan S, editors. Pseudomonas. Biotransformations, pathogenesis, and evolving biotechnology. Washington, D.C: American Society for Microbiology; 1990. pp. 303–316. [Google Scholar]
- 10.Haas D, Holloway B W, Schambock A, Leisinger T. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet. 1977;154:7–22. doi: 10.1007/BF00265571. [DOI] [PubMed] [Google Scholar]
- 11.Huang H, Siehnel R J, Bellido F, Rawling E, Hancock R E. Analysis of two gene regions involved in the expression of the imipenem-specific, outer membrane porin protein OprD of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1992;76:267–273. doi: 10.1016/0378-1097(92)90347-q. [DOI] [PubMed] [Google Scholar]
- 12.Itoh Y. Abstracts of the XVth International Arginine-Pyrimidine Conference and Symposium. 1996. Arginine and ornithine transport permease system of Pseudomonas aeruginosa PAO: primary structure and possible functions of the transport components, abstr. 18. [Google Scholar]
- 13.Itoh Y. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7280–7290. doi: 10.1128/jb.179.23.7280-7290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh Y, Matsumoto H. Mutations affecting regulation of the anabolic argF and the catabolic aru genes in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1992;231:417–425. doi: 10.1007/BF00292711. [DOI] [PubMed] [Google Scholar]
- 15.Kwon D H, Lu C-D, Walthall D A, Brown T M, Houghton J E, Abdelal A T. Structure and regulation of the carAB operon in Pseudomonas aeruginosa and Pseudomonas stutzeri: no untranslated region exists. J Bacteriol. 1994;176:2532–2542. doi: 10.1128/jb.176.9.2532-2542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C D, Houghton J E, Abdelal A T. Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. J Mol Biol. 1992;225:11–24. doi: 10.1016/0022-2836(92)91022-h. [DOI] [PubMed] [Google Scholar]
- 17.Lu C-D, Park S-M, Abdelal A T. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Role of ArgR in control of expression of glutamate synthase and NAD-dependent glutamate dehydrogenase in Pseudomonas aeruginosa, abstr. K-136; p. 348. [Google Scholar]
- 18.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20a.Nishijyo T, Park S-M, Itoh Y. Abstracts of the XVIth International Arginine-Pyrimidine Conference. 1998. The argSU two-component regulatory system involved in arginine and ornithine catabolism in Pseudomonas aeruginosa, abstr. 10. [Google Scholar]
- 21.Oche M M, Lu C-D, Abdelal A T, Hancock R E W. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Arginine-mediated regulation of the basic amino acid and imipenem-selective porin OprD of Pseudomonas aeruginosa, abstr. H-171; p. 305. [Google Scholar]
- 22.Park S-M, Lu C-D, Abdelal A T. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J Bacteriol. 1997;179:5300–5308. doi: 10.1128/jb.179.17.5300-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S-M, Lu C-D, Abdelal A T. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J Bacteriol. 1997;179:5309–5317. doi: 10.1128/jb.179.17.5309-5317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips A T, Mulfinger L M. Cyclic adenosine 3′,5′-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J Bacteriol. 1981;145:1286–1292. doi: 10.1128/jb.145.3.1286-1292.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitzer L J. Sources of nitrogen and their utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 380–390. [Google Scholar]
- 26.Thompson J D, Higgins D G, Gilson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4674–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 28.Winteler, H., C.-D. Lu, A. T. Abdelal, and D. Haas. Unpublished data.
- 29.Wissenbach U, Six S, Bongaerts J, Ternes D, Steinwachs S, Unden G. A third periplasmic transport system for L-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol Microbiol. 1995;17:675–686. doi: 10.1111/j.1365-2958.1995.mmi_17040675.x. [DOI] [PubMed] [Google Scholar]
- 30.Wolff J A, MacGregor C H, Eisenberg R C, Phibbs P V., Jr Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J Bacteriol. 1991;173:4700–4706. doi: 10.1128/jb.173.15.4700-4706.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]