Abstract
The Mycobacterium avium complex is a source of disseminated infections in patients with advanced AIDS. This group of mycobacteria is distinguished by the presence of highly antigenic, surface-exposed glycopeptidolipids, and these glycolipids possess variant oligosaccharide structures that are the chemical basis of the 28 distinct serovars of the M. avium complex. We previously described the ser2 gene cluster, encoding the synthesis of the haptenic oligosaccharide (2,3-dimethylfucose-rhamnose-6-deoxytalose-) of the serovar 2-specific glycopeptidolipid, and revealed a locus (ser2A) encoding a putative rhamnosyltransferase. Sequencing of the ser2A locus demonstrated the presence of three open reading frames, two of which yielded significant homology to several glycosyltransferases, and the deduced amino acid sequences of these two putative glycosyltransferases had 63% identity. These two genes were expressed in Mycobacterium smegmatis, and the resulting recombinant glycopeptidolipids were characterized by thin-layer chromatography and gas chromatography-mass spectrometry. These analyses demonstrated that only one of these genes, termed rtfA, encoded the rhamnosyltransferase responsible for the transfer of rhamnose to 6-deoxytalose. The identification of rtfA will permit further evaluation of glycopeptidolipid biosynthesis and the construction of isogenic mutants of multiple M. avium complex serovars. Moreover, such mutants will help define the role of glycopeptidolipids in the intracellular survival of these bacteria.
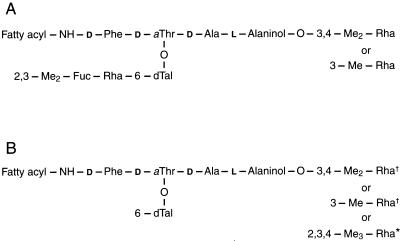
The mycobacterial cellular envelope contains a unique array of glycolipids, with individual Mycobacterium spp. exhibiting distinct glycolipid profiles (2, 7). Mycobacterium avium-M. intracellulare, the M. avium complex (MAC), are distinguished from all other Mycobacterium spp. by the presence of highly antigenic cell surface molecules termed glycopeptidolipids (GPLs), which are subdivided into nonspecific and serovar-specific forms (7). All GPLs have in common an N-acylated lipopeptide core that is glycosylated at the terminal alaninol residue with either 3,4-di-O-Me- or 3-O-Me-rhamnose (Rha) (Fig. 1). The nonspecific GPLs (nsGPLs) are present in all serovars of MAC and possess a single O-linked 6-deoxytalose (6-dTal) residue attached to d-allo-threonine (Thr) (Fig. 1). In addition, individual serovars of MAC contain serovar-specific GPLs (ssGPLs) that present extended oligosaccharides attached to the d-allo-Thr residue, and these haptenic oligosaccharides are responsible for serological specificity (Fig. 1) (2, 8). In all, 28 ssGPLs of MAC have been identified, and the structures of their respective oligosaccharides have been elucidated (2). Interestingly, the reducing termini of all of these oligosaccharides, except those of ssGPLs 5, 10, and 11, are comprised of a disaccharide of Rha-6-dTal (2). In the case of the three nonconforming GPLs, the Rha residue is replaced by a 3-O-Me-Rha (2).
FIG. 1.
General structures of the GPLs of MAC and M. smegmatis. (A) Structure of the ssGPL of M. avium serovar 2. The disaccharide 2,3-di-O-methyl-α-l-fucopyranosyl- (1→3)-α-l-rhamnopyranosyl is specific for the serovar 2 GPL. (B) Structure of the nsGPLs of MAC and M. smegmatis. The daggers indicate the methylated sugars attached to the alaninol of the nsGPLs of MAC and M. smegmatis. The asterisk indicates the methylated sugar attached only to the alaninol of the nsGPL of M. smegmatis.
Based on this structural information, the glycosyltransferase that catalyzes the addition of Rha to 6-dTal is hypothesized to be highly conserved in most serovars of MAC and to be essential for the expression of mature ssGPLs. We previously isolated a gene cluster (ser2) that encodes the enzymatic machinery required for the synthesis of the serovar 2-specific oligosaccharide (5). Further, four functional loci (ser2A, ser2B, ser2C, and ser2D) within this gene cluster were identified by transposon mutagenesis, and their participation in GPL biosynthesis was evaluated (17). One of these loci, ser2A, was proposed to possess the gene that encodes the rhamnosyltransferase responsible for the transfer of Rha to 6-dTal. In the present study, sequencing of the ser2A locus revealed two open reading frames (ORFs) whose predicted products had high homology to one another and homology to other glycosyltransferases. The functional expression of these ORFs and analysis of the resulting recombinant GPLs (rGPLs) provide strong evidence that one of the ORFs (termed rtfA) encodes the rhamnosyltransferase that modifies 6-dTal. The results support a model for ssGPL biosynthesis in which the sugars of the serovar-specific oligosaccharide are added sequentially to the lipopeptide core. The elucidation of the rtfA gene also provides the essential means for generation of M. avium isogenic mutants that express only nsGPLs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Culturing of Mycobacterium smegmatis mc2155 for use in electroporation was performed as described previously (20). Recombinant clones of this strain were grown in Middlebrook 7H9 broth or on 7H11 agar containing kanamycin (25 μg/ml). Escherichia coli DH5α was propagated with Luria-Bertani medium. Kanamycin (25 μg/ml) or ampicillin (100 μg/ml) was added for the growth of recombinant clones of E. coli. All bacterial strains were cultured at 37°C.
Cloning and restriction endonuclease mapping.
A 14.5-kb HindIII DNA fragment of the ser2 gene cluster of M. avium TMC 724 was obtained from the recombinant cosmid pJTB71 (5) and ligated into the HindIII site of pBluescript II SK(−) (Stratagene, La Jolla, Calif.) to produce plasmid pJTB71-1 (Fig. 2). Restriction endonuclease mapping of this plasmid was performed by using the enzymes BamHI, ClaI, KpnI, and PstI. Plasmid pJTB71-1-1 was constructed by isolating the 4.1-kb PstI-KpnI fragment from pJTB71-1 and ligating it into pBluescript II SK(−) digested with the same restriction endonucleases. Restriction endonuclease mapping of the 4.1-kb insert was accomplished with BamHI, BglII, EcoRI, EcoRV, and SmaI. Subclones of the 4.1-kb insert were constructed by digestion of pJTB71-1-1 with BamHI, EcoRI, EcoRV, KpnI, SacI, SmaI, or PvuII, and combinations of these restriction endonucleases, and subsequent ligation of the restriction fragments into pBluescript II SK(−). These subclones were used as double-stranded DNA templates for DNA sequencing. Restriction endonucleases were obtained from Boehringer (Mannheim, Germany) or from Gibco BRL (Gaitherburg, Md.). DNA ligation was performed with T4 ligase (Gibco BRL) as described previously (25). All recombinant plasmids were amplified in E. coli DH5α and isolated by the standard alkaline sodium dodecyl sulfate method (25).
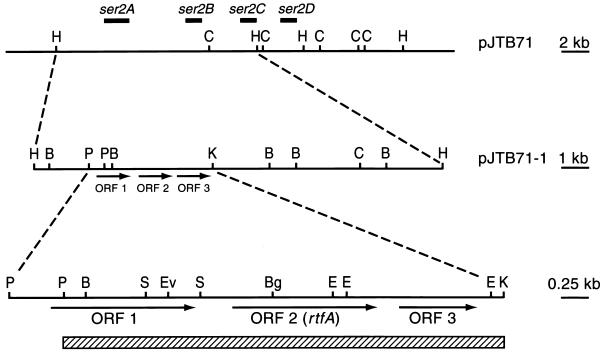
FIG. 2.
Restriction endonuclease maps of the 35-kb M. avium insert of cosmid pJTB71, the 14.8-kb HindIII insert of pJTB71-1, and the 4.6-kb region of the pJTB71-1 insert that was sequenced. The solid boxes indicate the locations of the ser2 loci within the ser2 gene cluster, and the hatched box represents the insert of pJTB71-1-1. B, BamHI, Bg, BglII; C, ClaI, E, EcoRI; Ev, EcoRV; H, HindIII; K, KpnI; P, PstI; and S, SmaI.
Plasmids for expression of ORF 1 and ORF 2 were constructed by using the mycobacterial expression vector pMV261 (28). Specifically, PCR products corresponding to ORF 1 and ORF 2 were obtained by using pJTB71-1 as the template and Vent polymerase (New England Biolabs, Beverly, Mass.). The primers used for amplification of ORF 1 were 5′GATGTGTGCTGGCCAGCTACG 3′ (forward) and 5′TGTAAGCTTACCTGCGGGGTAGCAATCCTG 3′ (reverse), and those for ORF 2 were 5′GATTCGCTGTGGCAAGTTATG 3′ (forward) and 5′CGTAAGCTTATCGGTCACGGCCAAAACTTG 3′ (reverse); underlined sequences indicate the HindIII site that was introduced. The 5′ nucleotide of each forward primer corresponds to nucleotide 5 of ORF 1 and ORF 2 and has been changed from A to G. The PCR products were digested with HindIII and ligated into the BalI-HindIII site of pMV261. Digestion of pMV261 with BalI permitted the use of the hsp60 promoter and the start codon (ATG) of the hsp60 gene fragment of pMV261 for the generation of inframe fusion products with single amino acid changes of lysine to glycine at position 2. Plasmids pTME1 and pTME2 were selected for expression of ORF 1 and ORF 2, respectively. The inserts of these two plasmids were sequenced to confirm the fidelity of the PCR.
DNA sequencing.
Sequencing of the subclones of pJTB71-1-1 was performed with the pBluescript T3 and M13-20 primers. Custom primers were synthesized as necessary to resolve sequence ambiguities and to sequence regions flanking the 4.1-kb PstI-KpnI fragment in pJTB71-1. Sequencing of DNA and synthesis of primers were performed by Macromolecular Resources Facility, Colorado State University. Contiguous DNA sequences, ORFs, and codon usage were determined with Sequencher 3.0 software (Gene Codes Corporation, Ann Arbor, Mich.). Putative transmembrane domains were predicted with the program TMpred (12) (http://ulrec3.unil.ch/software/TMPRED_form.html).
Extraction and TLC of glycopeptidolipids.
To assess the expression of recombinant glycolipids, cells from 150-ml broth cultures of M. smegmatis/pMV261, M. smegmatis/pTME1, and M. smegmatis/pTME2 were harvested by centrifugation and lyophilized. Lipids were extracted with CHCl3-CH3OH (2:1), and enrichment of the GPLs was achieved by mild alkaline hydrolysis (200 mM NaOH for 30 min at 40°C) (16). The resulting alkali-stable lipids were dissolved in CHCl3 (10 mg/ml) and applied to a silica gel Sep-Pak cartridge (Millipore Corp., Milford, Mass.) and eluted with increasing concentrations of CH3OH in CHCl3. The total GPL population was eluted with 30% CH3OH. Thin-layer chromatography (TLC) of the GPLs was performed in CHCl3-CH3OH-H2O (90:10:1) on aluminum-backed silica gel 60 plates (E. Merck, Darmstadt, Germany). Lipids were visualized by spraying with 10% H2SO4 in ethanol and heating at 120°C.
Analytic methods.
Glycosyl analysis of the GPLs from recombinant strains of M. smegmatis was performed by gas chromatography-mass spectrometry (GC-MS) as described previously (16). Briefly, lipids were hydrolyzed with 2 M CF3COOH, and the free sugars were reduced with NaBD4 and acetylated with acetic anhydride. The resulting alditol acetates were resolved on a BPX70 capillary column (SGE Analytic Products, Austin, Tex.) by using a Hewlett-Packard model 5890 gas chromatograph and analyzed with a Hewlett-Packard model 5970 mass detector.
The release of the carbohydrate moiety glycosidically linked to the d-allo-Thr of the GPL core was accomplished by beta-elimination with 500 mM NaOH in ethanol and 6.4 μM NaBD4 and heating at 60°C for 24 h (16). The aqueous soluble products were collected, desalted with AG 501-X8 (Bio-Rad Laboratories, Hercules, Calif.), and acetylated with pyridine-acetic anhydride (1:1) at 80°C for 2 h (11). The resulting peracetylated oligoglycosyl alditols were separated and analyzed by GC-MS.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the GenBank database under accession no. AF060183.
RESULTS
Restriction endonuclease mapping and sequencing of the ser2A locus.
Previous studies used transposon mutagenesis of cosmids containing the ser2 gene cluster and analysis of rGPL production in M. smegmatis to define a 2-kb genetic locus (ser2A) that apparently encoded a rhamnosyltransferase involved in the biosynthesis of the oligosaccharide of the serovar 2-specific GPL (17). To obtain a more detailed map of this region, the 14.5-kb HindIII fragment containing the ser2A locus was cloned into pBluescript II SK(−) to give pJTB71-1 (Fig. 2). Restriction endonuclease mapping of this fragment and comparison to the Tn5 insertion map of the ser2 gene cluster (17) indicated that the ser2A locus was within a 4.1-kb PstI-KpnI fragment (Fig. 2). Nucleotide sequencing of this 4.1-kb fragment along with 500 bp upstream revealed three putative ORFs (Fig. 2).
The deduced amino acid sequences of ORF 1 and ORF 2 (nucleotides 361 to 1635 and nucleotides 2042 to 3325, respectively) had 63% identity and 89% similarity to one another. Moreover, the deduced amino acid sequences of these two ORFs demonstrated 83 to 89% similarity to two putative glycosyltransferases of Mycobacterium tuberculosis (MTCY19G5_2 and MTCY19G5_4c) that were identified through the M. tuberculosis genome sequencing project (22) and to a putative glycosyltransferase of Mycobacterium leprae (U00023_2c) (24). Significant similarity (61.3%) to the rhlB gene product of Pseudomonas aeruginosa, a rhamnosyltransferase utilized in the synthesis of rhamnolipids (19), was also observed. The initiation codons of ORF 1 and ORF 2 were based on their proximity to possible Shine-Dalgarno sequences (nucleotides 343 to 348 and 2027 to 2032 for ORF 1 and ORF 2, respectively) and alignment of their deduced amino acid sequences with those of defined and putative glycosyltransferases. Additionally, ORFs 1 and 2 exhibited G+C contents of 69 and 65%, respectively, and codon usage was consistent with that of Mycobacterium spp. (1). Such nucleotide sequence analysis indicated that either ORF 1 or ORF 2 could encode the targeted rhamnosyltransferase.
In contrast to the high homology between ORF 1 and ORF 2, the hydrophobicity plots of the deduced amino acid sequences differed significantly. Specifically, ORF 1 lacked putative transmembrane domains, while ORF 2 was predicted to have three such regions (amino acid positions 95 to 114, 243 to 265, and 325 to 343). These data suggest that the gene products of ORF 1 and ORF 2, although homologous, have separate subcellular locations.
The deduced amino acid sequence of ORF 3 demonstrated significant homology to several putative methyltransferases. This result was unexpected, given that the two loci for methylation of the fucosyl residue of the serovar 2 oligosaccharides (ser2B and ser2D) were located 3.8 and 10.6 kb away from ser2A, respectively (17).
Recombinant expression of rhamnosyltransferase activity.
We previously demonstrated that M. smegmatis, with its singly glycosylated nsGPLs and the absence of the multiglycosylated ssGPLs, acted as a suitable host for expression of M. avium genes responsible for extended glycosylation of GPLs (5). Thus, the present work utilized the same system for recombinant expression and facile functional analysis of ORF 1 and ORF 2. The putative glycosyltransferase genes of the ser2A locus were amplified by PCR, and each PCR product was ligated into the BalI-HindIII site of the mycobacterial expression vector pMV261 (28), resulting in pTME1 (ORF 1) and pTME2 (ORF 2). This strategy allowed for recombinant gene expression from the mycobacterial hsp60 promoter and generation of recombinant gene products possessing a single amino acid change of lysine to glycine at position 2 (see Materials and Methods).
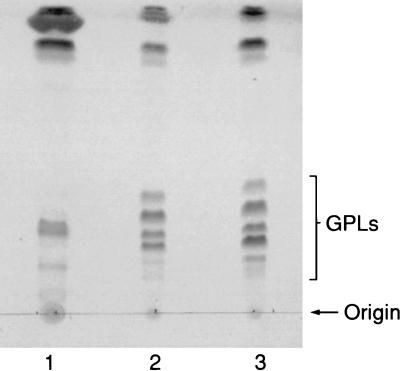
To assess the correlation between rhamnosyltransferase activity and the cloned ORFs, the alkali-stable lipid fractions of M. smegmatis/pTME1, M. smegmatis/pTME2, or M. smegmatis/pMV261 were analyzed by TLC and GC. The TLC profile of M. smegmatis/pTME2 revealed a dramatic shift in the rGPL population compared to that of the vector control, M. smegmatis/pMV261 (Fig. 3). Specifically, the four GPL spots observed for M. smegmatis/pMV261 were barely detectable in the alkali-stable lipid fraction of M. smegmatis/pTME2 and were replaced by two new GPL spots. Such a shift was not observed with the same lipid fraction of M. smegmatis/pTME1. This initial analysis indicated that ORF 2 but not ORF 1 encoded a transferase that allowed for further glycosylation of the nsGPLs of M. smegmatis.
FIG. 3.
TLC of alkali-stable lipids isolated from recombinant strains of M. smegmatis. Lanes: 1, M. smegmatis/pTME2; 2, M. smegmatis/pTME1; 3, M. smegmatis/pMV261 (vector control). The lipids were resolved on a silica gel TLC plate in CHCl3-CH3OH-H2O (90:10:1).
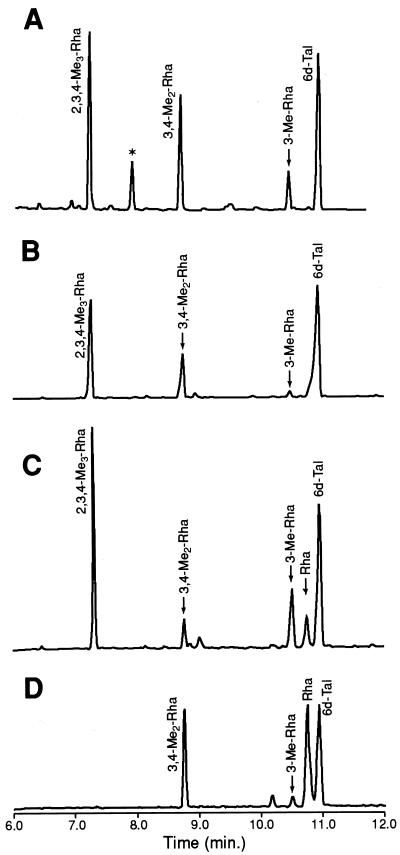
To confirm these results and to demonstrate the nature of the recombinant glycosylation, the partially purified GPLs from each recombinant strain were hydrolyzed, and their sugars were converted to alditol acetates for analysis by GC-MS. This exercise demonstrated that M. smegmatis/pTME2 possessed a significant amount of GPL-associated Rha (Fig. 4C). In contrast, the glycosylation pattern of the GPLs of M. smegmatis/pTME1 did not differ from that of the vector control (Fig. 4A and B). In all, these experiments provided strong evidence that ORF 2 of the ser2A locus encoded the rhamnosyltransferase required for formation of the serovar 2-specific oligosaccharide of M. avium. Thus, this ORF was designated rtfA. Carbohydrate analysis of the rGPLs also revealed that M. smegmatis/pTME2 yielded a significant increase in the amount of 3-O-Me-Rha. We previously observed this same manifestation in M. smegmatis expressing the entire ser2 gene cluster (5); however, a similar result with the expression of a single gene (rtfA) encoding a rhamnosyltransferase was unexpected. The increased content of 3-O-Me-Rha in these lipids was possibly a result of heterogeneous glycosyl substitution of the talosyl residue or a shift in the methylation pattern of the rhamnosyl residue covalently attached to alaninol of the lipopeptide core (Fig. 1).
FIG. 4.
Gas chromatograms of alditol acetate derivatives of sugars released from GPL-enriched fractions by hydrolysis with CF3COOH. (A) GPL-enriched lipids of M. smegmatis/pMV261; (B) GPL-enriched lipids of M. smegmatis/pTME1; (C) GPL-enriched lipids of M. smegmatis/pTME2; (D) purified ssGPLs of M. avium serovar 1. *, MS analysis of this product indicated that it was not carbohydrate.
Analysis of the d-allo-Thr-linked disaccharide of the recombinant GPL.
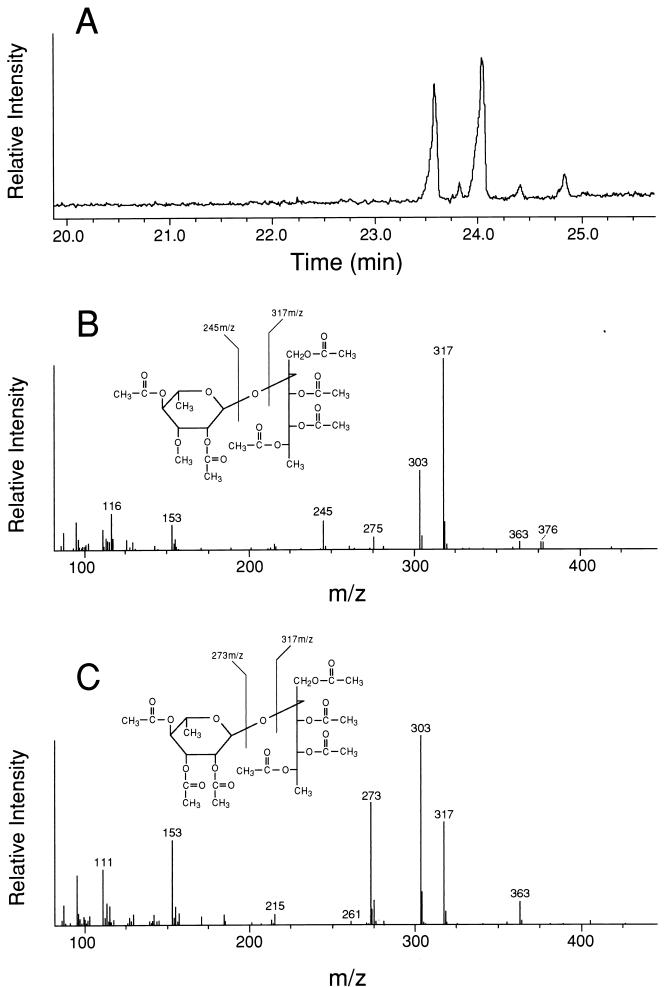
To confirm that the rGPLs of M. smegmatis/pTME2 possessed a d-allo-Thr-linked disaccharide of Rha-6-dTal-, and to determine the structural location of the 3-O-Me-Rha, purified rGPLs from M. smegmatis/pTME2 were subjected to beta-elimination, and the released oligoglycosyl alditols were peracetylated and analyzed by GC-MS. This resulted in the resolution of two peaks, with retention times of 23.6 and 24.0 min (Fig. 5A). Fragmentation ions of m/z 245 and 317 were detected for the 23.6-min peak (Fig. 5B). These ions are diagnostic for a peracetylated alditol of 6-deoxyhexose linked to a monomethylated peracetylated 6-deoxyhexose. The fragmentation ions detected for the 24.0-min peak (m/z 273 and 317) demonstrated the presence of a peracetylated alditol of 6-deoxyhexose linked to peracetylated 6-deoxyhexose (Fig. 5C). These data coupled with the analysis of alditol acetates derived from the rGPLs of M. smegmatis/pTME2 demonstrated that the d-allo-Thr residue of the lipopeptide core was glycosylated with either Rha-6-dTal- or 3-O-Me-Rha-6-dTal-.
FIG. 5.
GC-MS of alditol derivatives of the oligoglycosyl moieties released from rGPLs of M. smegmatis/pTME2 by beta-elimination. (A) Gas chromatogram of oligoglycosyl alditols; (B) mass spectrum and proposed structure of the oligoglycosyl alditol corresponding to the GC peak with a retention time of 23.6 min; (C) mass spectrum and proposed structure of the oligoglycosyl alditol corresponding to the GC peak with a retention time of 24.0 min. The temperature program used for the resolution of oligoglycosyl alditols was 100°C for 1 min, 30°C/min to 200°C, 4°C/min to 260°C, and 260°C for 7 min.
DISCUSSION
Extensive chemical characterization and structural elucidation of the cellular envelopes of Mycobacterium spp. demonstrate an abundant use of glycosyl units in the modification of lipids and in the construction of complex molecules by these organisms (7). However, studies of the molecular mechanisms of glycosylation have not kept pace with the elaborate structural work. This statement is underscored by the fact that embAB, which participate in arabinogalactan biosynthesis, are the only mycobacterial genes to be functionally defined as glycosyltransferases (4). The only other glycosylation event that has been evaluated at a molecular level is the synthesis of the serovar 2-specific oligosaccharide of M. avium, for which a 22- to 27-kb gene cluster termed ser2 was identified (5). Further, Tn5 mutagenesis of this genomic region led to the identification of four functional loci that are proposed to possess genes encoding a rhamnosyltransferase (ser2A), methyltransferases (ser2B and -D), and fucose biosynthesis and/or a fucosyltransferase (ser2C) (17). The present work has extended these earlier findings and has provided functional definition of a gene termed rtfA, which encodes the rhamnosyltransferase essential for the synthesis of serovar-specific oligosaccharides.
Nucleotide sequencing of the ser2A locus revealed two putative glycosyltransferases. The recombinant expression of these two ORFs in M. smegmatis coupled with detailed chemical analysis demonstrated that only one of these (rtfA) allowed for the production of rGPLs possessing a disaccharide of Rha-6-dTal-. The rtfA gene product demonstrated limited but significant homology to a rhamnosyltransferase (RhlB) involved in the formation of rhamnolipids of P. aeruginosa (19), but no homology to other defined bacterial glycosyltransferases, such as those that participate in the synthesis of the O-antigens of gram-negative bacteria (15, 18, 27), was observed. One feature shared by RhlB and RtfA is the presence of putative transmembrane domains (19), a structural characteristic not associated with the rhamnosyltransferases involved in O-antigen biosynthesis (15, 18, 27). Thus, the limited homology to RhlB may be due to similarity in the subcellular location of catalytic activity. Alternatively, RtfA and RhlB may utilize similar donor molecules. In the case of lipopolysaccharide biosynthesis, it is well established that the repeating glycosyl units are formed from nucleotide diphosphosugars; however, it is not known whether TDP-Rha or a rhamnosylphosphorylpolyisoprenol is the donor involved in either GPL or rhamnolipid biosynthesis (19). The similarity between RtfA and RhlB cannot be attributed to the use of like acceptor molecules, since RtfA utilizes a deoxyhexose (6-dTal) covalently linked to the lipopeptide and RhlB donates a rhamnosyl unit to β-hydroxydecanoyl-β-hydroxydecanoate (5, 19).
An even greater degree of homology between RtfA and putative rhamnosyltransferases of M. leprae and M. tuberculosis was observed (22, 24). Although neither of these Mycobacterium spp. produces GPLs, both are noted for the synthesis of phenolic glycolipids substituted with rhamnose-containing oligosaccharides (2, 14, 30). Thus, the analysis of the rtfA gene of M. avium has led us to hypothesize that ORF MTCY19G5_2 or MTCY19G5_4c of M. tuberculosis and ORF U0023_2c of M. leprae encode rhamnosyltransferases involved in the production of the oligosaccharides associated with phenolic glycolipids. Rha is also present in the linker region of the arabinogalactan-peptidoglycan complex; however, the rhamnosyltransferase involved in the formation of this particular structure does not show homology to RtfA (16a).
The biochemical analysis of recombinant rtfA expression coupled with our previous observations (5, 17) provides additional insight into the biosynthesis of the glycosyl units of the ssGPLs. In particular, the observation that only rtfA was required for rGPL production in M. smegmatis supports our hypothesis that the sugars of the serovar-specific oligosaccharides are added sequentially to the lipopeptide core. An alternative pathway, using lipopolysaccharide biosynthesis as a paradigm (23), would be that the recombinant disaccharides are synthesized on a lipid carrier and then transferred to the lipopeptide core. Such a mechanism would require a gene encoding the transfer of the oligosaccharide in addition to rtfA. Presently available data indicate that an oligosaccharide transferase is not encoded by the ser2 gene cluster; however, it is possible that the genome of M. smegmatis possesses the corresponding gene and that in M. avium serovar 2 this gene is located outside the ser2 gene cluster. It is interesting that the ser2A locus possesses a second putative glycosyltransferase gene (ORF 1) that encodes a product with 89% homology to RtfA and that recombinant expression of this ORF failed to provide the targeted rhamnosyltransferase activity. Although the function of ORF 1 is undefined, the proximity and homology of this gene to rtfA suggest that it participates in GPL biosynthesis, possibly encoding the talosyltransferase or the transferase responsible for the addition of Rha to the alaninol residue. Moreover, the absence of putative transmembrane domains within the gene product of ORF 1 suggests a subcellular location separate from that of RtfA and that specific steps of GPL biosynthesis are compartmentalized in different cellular locations. The physical separation of the enzymes responsible for nsGPL production from those that result in extended glycosylation (ssGPL production) provides one explanation for the presence of the large quantities of both nsGPLs and ssGPLs in M. avium. However, the exact function of ORF 1, as well as that of ORF 3 (encoding a predicted methyltransferase), and the proof of enzymatic segregation await further genetic and biochemical evaluation.
Separately, analysis of the rGPL-associated carbohydrates demonstrated an increase in the presence of 3-O-Me-Rha. This methylated sugar is present in the native nsGPL of M. smegmatis and is linked to the alaninol of the lipopeptide core (3). However, isolation of the glycosyl moieties linked to the d-allo-Thr of the rGPL and subsequent analysis of this structure by GC-MS yielded a disaccharide of 3-O-Me-Rha-6-dTal- as well as the expected structure Rha-6-dTal-. Thus, the increased 3-Me-Rha content was a consequence of the ability of M. smegmatis to modify the Rha linked to 6-dTal and was not a result of a shift in the ratio of methylated rhamnosyl residues (3-O-Me-Rha/3,4-di-O-Me-Rha/2,3,4-tri-O-Me-Rha ratio) linked to the alaninol of the lipopeptide core. Given the strict specificity exhibited by glycosyltransferases (31), it is proposed that this methylation occurred after the formation of the Rha-6-dTal- disaccharide. This mechanism is further supported by the data of Mills et al. (17) that demonstrated that specific Tn5 insertions into the ser2 gene cluster disrupted the methylation of the fucosyl residue of the serovar 2-specific oligosaccharide but did not interfere with the incorporation of this carbohydrate residue. The ability of M. smegmatis to methylate the 3 position on the rhamnosyl residue of the recombinant disaccharide, along with the fact that the serovar 2 oligosaccharide possesses a dimethylated fucose with a 1→3 linkage to Rha, also explains the poor yield of fully glycosylated recombinant ssGPL2 when the complete ser2 gene cluster is expressed in M. smegmatis (5).
MAC is the most common opportunistic bacterial pathogen and a leading cause of mortality among patients in the advanced stage of AIDS (9). The use of in vitro assays has provided evidence that GPLs are capable of scavenging reactive oxygen intermediates, inducing secretion of tumor necrosis factor and prostaglandin E2, and reducing the lymphocyte response to mitogen-induced blastogenesis (10, 13, 29). Moreover, the oligosaccharides of the GPLs appeared to play an important part in some of these biological activities (21). However, the fact that some rough (presumably GPL-lacking) variants of M. avium are virulent (26) has led us to question the actual contribution of these structures to the pathogenesis of MAC infections (6). To resolve this issue, it is essential that a set of well-defined isogenic GPL mutants be developed for several MAC serovars. Thus, the functional characterization of the rtfA gene now provides a critical tool for the generation of isogenic ssGPL mutants of all MAC serovars that possess oligosaccharides with Rha-6-dTal- at their reducing termini.
ACKNOWLEDGMENTS
This work was supported by grants RO1 AI-18357 and RO1 AI-41925 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank Julia Inamine for her advice and the careful critique of the manuscript and Marilyn Hein for assistance in preparation of the manuscript.
REFERENCES
- 1.Andersson S G E, Sharp P M. Codon usage in the Mycobacterium tuberculosis complex. Microbiology. 1996;142:915–925. doi: 10.1099/00221287-142-4-915. [DOI] [PubMed] [Google Scholar]
- 2.Aspinall G O, Chatterjee D, Brennan P J. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv Carbohydr Chem Biochem. 1995;51:169–242. doi: 10.1016/s0065-2318(08)60194-8. [DOI] [PubMed] [Google Scholar]
- 3.Asselineau C, Asselineau J. Lipides specifiques des mycobacteries. Ann Microbiol (Inst Pasteur) 1987;129A:49–69. [PubMed] [Google Scholar]
- 4.Belanger A E, Besra G S, Ford M E, Mikusova K, Belisle J T, Brennan P J, Inamine J M. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci USA. 1996;93:11919–11924. doi: 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belisle J T, Pascopella L, Inamine J M, Brennan P J, Jacobs W R., Jr Isolation and expression of a gene cluster responsible of the glycopeptidolipid antigens of Mycobacterium avium. J Bacteriol. 1991;173:6991–6997. doi: 10.1128/jb.173.21.6991-6997.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belisle J T, Brennan P J. Molecular basis of colony morphology in Mycobacterium avium. Res Microbiol. 1994;145:237–242. doi: 10.1016/0923-2508(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 7.Brennan P J. Mycobacterium and other actinomycetes. In: Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. London, United Kingdom: Academic Press; 1988. pp. 203–298. [Google Scholar]
- 8.Brennan P J, Goren M B. Structural studies on the type-specific antigens and lipids of the Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum serocomplex. J Biol Chem. 1979;254:4205–4211. [PubMed] [Google Scholar]
- 9.Brettle R P. Mycobacterium avium intracellulare infection in patients with HIV or AIDS. J Antimicrob Chemother. 1997;40:156–160. doi: 10.1093/jac/40.2.156. [DOI] [PubMed] [Google Scholar]
- 10.Brownback P E, Barrow W W. Modified lymphocyte response to mitogens after intraperitoneal injection of glycopeptidolipid antigens from Mycobacterium avium complex. Infect Immun. 1988;56:1044–1050. doi: 10.1128/iai.56.5.1044-1050.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee D, Khoo K-H, McNeil M R, Dell A, Morris H R, Brennan P J. Structural definition of the non-reducing termini of mannose-capped LAM from Mycobacterium tuberculosis through selective enzymatic degradation and fast atom bombardment-mass spectrometry. Glycobiology. 1993;3:497–506. doi: 10.1093/glycob/3.5.497. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann K, Stoffel W. Tmbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 13.Hooper L C, Barrow W W. Decreased mitogenic response of murine spleen cells following intraperitoneal injection of serovar-specific glycopeptidolipid antigens from the Mycobacterium avium complex. Adv Exp Med Biol. 1988;239:309–325. doi: 10.1007/978-1-4757-5421-6_31. [DOI] [PubMed] [Google Scholar]
- 14.Hunter S W, Brennan P J. Further specific extracellular phenolic glycolipid antigens and a related diacylphthiocerol from Mycobacterium leprae. J Biol Chem. 1983;258:7556–7562. [PubMed] [Google Scholar]
- 15.Liu D, Haase A M, Lindquist L, Lindberg A A, Reeves P R. Glycosyl transferases of O-antigen biosynthesis in Salmonella enterica: identification and characterization of transferase genes of group B, C2, and E1. J Bacteriol. 1993;175:3408–3413. doi: 10.1128/jb.175.11.3408-3413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNeil M, Chatterjee D, Hunter S W, Brennan P J. Mycobacterial glycolipids: isolation, structures, antigenicity, and synthesis of neoantigens. Methods Enzymol. 1989;179:215–242. doi: 10.1016/0076-6879(89)79123-0. [DOI] [PubMed] [Google Scholar]
- 16a.Mills, J. A., and M. McNeil. Personal communication.
- 17.Mills J A, McNeil M R, Belisle J T, Jacobs W R, Jr, Brennan P J. Loci of Mycobacterium avium ser2 gene cluster and their functions. J Bacteriol. 1994;176:4803–4808. doi: 10.1128/jb.176.16.4803-4808.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchison M, Bullach D M, Vinh T, Rajakumar K, Faine S, Adler B. Identification and characterization of the dTDP-rhamnose biosynthesis and transfer genes of the lipopolysaccharide-related rfb locus in Leptospira interrogans serovar Copenhageni. J Bacteriol. 1997;179:1262–1267. doi: 10.1128/jb.179.4.1262-1267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsner U A, Fiechter A, Reiser J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem. 1994;269:19787–19795. [PubMed] [Google Scholar]
- 20.Parish T, Stoker N G. Electroporation of mycobacteria. Methods Mol Biol. 1995;47:237–252. doi: 10.1385/0-89603-310-4:237. [DOI] [PubMed] [Google Scholar]
- 21.Pedrosa J, Florido M, Kunze Z M, Castro A G, Portaels F, McFadden J, Silva M T, Appelberg R. Characterization of the virulence of Mycobacterium avium complex (MAC) isolates in mice. Clin Exp Immunol. 1994;98:210–216. doi: 10.1111/j.1365-2249.1994.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillip W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raetz C R H. Bacterial LPS: a remarkable family of bioactive macromolecules. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B Jr, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1035–1063. [Google Scholar]
- 24.Robinson, K. 1993. Direct submission to GenBank.
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schaefer W B, Davis C L, Cohn M L. Pathogenicity of transparent, opaque and rough variants of Mycobacterium avium in chickens and mice. Am Rev Respir Dis. 1970;120:499–506. doi: 10.1164/arrd.1970.102.4.499. [DOI] [PubMed] [Google Scholar]
- 27.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F. New use of BCG for recombinant vaccines. Nature (London) 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 29.Tassell S K, Pourshafie M, Wright E L, Richmond M G, Barrow W W. Modified lymphocyte response to mitogens induced by the lipopeptide fragment derived from Mycobacterium avium serovar-specific glycopeptidolipids. Infect Immun. 1992;60:706–711. doi: 10.1128/iai.60.2.706-711.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe M, Yamada Y, Iguchi K, Minnikin D E. Structural elucidation of the new phenolic glycolipids from Mycobacterium tuberculosis. Biochem Biophys Acta. 1994;1210:174–180. doi: 10.1016/0005-2760(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 31.Watkins W M. Glycosyltransferases. Early history, development and future prospects. Carbohydr Res. 1986;149:1–12. doi: 10.1016/s0008-6215(00)90364-1. [DOI] [PubMed] [Google Scholar]