Abstract
Background
The Shiga toxin (Stx)-producing Escherichia coli (STEC) have become important global public health concerns. This study investigated the prevalence, antimicrobial resistance profile, and extended-spectrum beta-lactamase-producing E. coli in sheep and goat faeces.
Methods and results
A total of 53 E. coli isolates were confirmed by PCR targeting the uidA [β-D glucuronidase] gene. The Shiga toxin genes stx1 and stx2, as well as bfpA, vir, eaeA, lt and aafII virulence genes, were detected in this study. Of the 53 isolates confirmed to be STEC, 100% were positive for stx2 and 47.2% for stx1. Three isolates possessed a combination of stx1 + stx2 + eaeA, while four isolates harboured stx1 + stx2 + vir virulence genes. The isolates displayed phenotypic antimicrobial resistance against erythromycin (66.04%), colistin sulphate (43.4%), chloramphenicol (9.4%) and ciprofloxacin (1.9%). A total of 28.8% of the strains were phenotypically considered ESBL producers and contained the beta-lactamase blaCTX-M-9 and blaCTX-M-25 gene groups. A larger proportion of the E. coli strains (86.8%) contained the antibiotic sulphonamide resistant (sulII) gene, while 62.3%, 62.3%, 52.8%, 43.4%, 41.5%, 20.8%, 18.9%, 11.3%, 11.3%, 9.4%, 9.4% and 5.7% possessed mcr-4, floR, mcr-1, tet(A), sulI, tet(O), tet(W), parC, mcr-2, ampC 5, qnrS and ermB genes, respectively. Thirteen isolates of the ESBL-producing E. coli were considered multi-drug resistant (MDR). One Shiga toxin (stx2) and two beta-lactamase genes (blaCTX-M-9 and blaCTX-M-25 groups) were present in 16 isolates. In conclusion, the E. coli isolates from the small stock in this study contained a large array of high antibiotic resistance and virulence profiles.
Conclusions
Our findings highlight the importance of sheep and goats as sources of virulence genes and MDR E. coli. From a public health and veterinary medicine perspective, the characterization of ESBL producers originating from small livestock (sheep and goats) is crucial due to their close contact with humans.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11033-023-08987-0.
Keywords: E. coli, Antimicrobial resistance, Extended-spectrum β-lactamases, Virulence factors, Shiga-toxins
Introduction
Escherichia coli is a rod-shaped, Gram-negative coliform bacteria that is widely distributed in nature. They are a large and diverse group of bacteria that infects both humans and animals [1]. The Shiga toxin (Stx)-producing Escherichia coli (STEC) is an important zoonotic foodborne pathogen causing gastrointestinal complications in humans [2]. The term STEC refers to a pathotype of E. coli that produces Stx1 and/or Stx2 [2, 3]. There are two major families of Shiga toxins (Stx), Stx1 and Stx2, with 70 percent similar amino acid sequences [2, 4]. It is less likely that Stx1 will produce diseases in humans, while Stx2 is more likely to cause diseases like hemorrhagic colitis (HC) and hemolytic-uremic syndrome (HUS) [5].
More than 200 serotypes of STEC have been identified [6]. Due to its ubiquitous nature and sharing the same niche as other enteric pathogens and being transmissible by the same route, it contributes to the dissemination of antimicrobial resistance (AMR) [7]. Globally, AMR is considered a major threat to public health [7, 8] and affects both clinical settings and the community at large [9]. In the sentinel organism E. coli, resistance proportions are often called “prevalence” [10]. Prevalence is used to measure the proportion of bacteria that are resistant to a particular antibiotic. This can be used to predict the spread of antibiotic resistance in a population. Prevalence is an important factor in understanding the impact of antibiotics on bacterial populations, particularly as farmers use antibiotics as animal fodder [11].
Antibiotics such as β-lactams can cause multidrug resistance in E. coli isolates, mostly due to the synthesis of extended-spectrum β-Lactamases (ESBL) and/or plasmid-mediated ampC β-lactamases [12, 13]. The genes that encode ESBL enzymes are found on plasmids or on single chromosomes of bacteria, which also contain genes for resistance to other antimicrobial agents, including quinolones, aminoglycosides, trimethoprim, sulphonamides, tetracyclines and chloramphenicol [14]. As plasmids can be transferred between bacterial species, the spread of ESBL enzymes is a major concern, as they are often accompanied by other genes that impart resistance to other antimicrobial agents, making them more difficult to treat.
Beta-lactam resistance is caused by the production of beta-lactamases such as ESBLs, metallo-β-lactamases (MBLs) and sometimes ampC plasmid β-lactamases [15]. These enzymes confer resistance to a broad range of β-lactam antibiotics, including penicillin and cephalosporins. As a result, treatment of infections caused by β-lactamase-producing bacteria is difficult. Multiple antibiotic resistance genes are a trait of Gram-negative bacteria, which has effects on treatment and the environment. Gram-negative bacilli such as Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, E. coli and Enterobacter spp. all possess the ESBL genes blaCTXM-1, blaTEM and blaCTXM-8 as well as the carbapenemase genes blaNDM, blaIMP, blaOXA and blaVIM [16, 17]. From the “One Health” perspective, the World Health Organization (WHO) has identified ESBL-producing E. coli as an indicator of antimicrobial resistance [9, 18]. This means that monitoring the spread of ESBL-producing E. coli can provide a snapshot of the prevalence of antimicrobial resistance in a given area. It also helps inform public health interventions that can help reduce the spread of antimicrobial resistance.
Due to the widespread use of antibiotics, ESBL-producing E. coli with virulence and resistance profiles have been identified from livestock more frequently, which has facilitated the establishment of infections that are multidrug resistant. There are not many reports of E. coli isolates causing AMR in sheep and goats in South Africa. The current study therefore sought to ascertain the frequency of E. coli generating extended-spectrum beta-lactamases (ESBL) and the Shiga toxin in sheep and goats in the Matlwang community in Potchefstroom, North-West Province, South Africa.
Materials and methods
Ethical statement
The study was approved by the Faculty of Natural and Agricultural Sciences Ethics Committee (NWU-00776-21-A9) of North‒West University.
Study area and sample collection
The sample collection in this study was carried out in the small farming community of Matlwang in Potchefstroom, North West Province, South Africa. Samples were collected between February 2022 and April 2022. Fecal samples were collected from four different sheep and goat flocks, which were selected randomly. A total of 57 fecal samples (37 from sheep and 20 from goats) were collected from the study site and were isolated for the presence of E. coli. Immediately after collection, samples were packaged in clear polyethylene bags, kept in a cooler box and transported to the laboratory. Samples were processed within three to five hours after collection.
Microbiological techniques and analysis
Five grams of each animal fecal sample was added to 45 ml of peptone enrichment broth for Enterobacteriaceae and incubated at 37 °C for 24 h. The selective media used was sorbitol MacConkey agar (SMA). The enriched broth was inoculated on SMA using the spread plate method. Bacterial growth was enumerated and identified after 24 h of incubation at 37 °C on SMA. The E. coli appears yellowish on MacConkey agar because it does not ferment sorbitol or ferments it at a very slow rate. The isolates were confirmed as presumptive pathogenic E. coli isolates, after phenotypic identification. Pure isolates were preserved in 20% glycerol (Merck, SA) and stored at − 80 °C for further analysis.
Bacterial genomic DNA extraction
Genomic DNA (gDNA) extraction was performed on the 55 presumptive E. coli isolates. Bacterial cells grown overnight were used to extract genomic DNA. A chromogenic DNA kit (ZYMO Research Quick-DNA Microbe Mini-prep commercial Kit) was used to extract and purify the genomic DNA by following the manufacturer’s protocol. A NanoDrop Lite 1000 spectrophotometer (Thermo-Fisher Scientific, USA) was used to determine the concentration and purity of DNA, which was ultimately stored at − 80 °C until further analysis.
Identification of diarrheagenic Escherichia coli using uidA PCR assay
A PCR assay was conducted to amplify the E. coli housekeeping uidA [β-D glucuronidase] gene as described by Omolajaiye et al. [19] using the following primers: UidA-F AAAACGGCAAGAAAAAGCAG and UidA-R ACGCGTGGTTACAGTCTTGCG, which produces a 147 bp amplicon size. The PCR consisted of 12.5 μL of AmpliTaq Gold® DNA Polymerase, 0.05 units/L Gold buffer, 930 mM Tris/HCl pH 8.05, 100 mM KCl, 0.4 mM of each dNTP, and 5 mM MgCl2] (New England Biolabs, USA), 8.5 μL of RNase-nuclease free PCR water, 1 μL of 10 μM each primer and 2 μL of template gDNA. The cycling consisted of an initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 40 s, 60 °C for 30 s and 72 °C for 40 s and a final extension of 72 °C for 7 min using the ProFlex PCR System (Applied Biosystems, USA). The negative control was a DNA-free template (nuclease-free water). To allow standardization, molecular weight markers of 1 kb and 100 bp DNA (PROMEGA, Madison, WI, USA) were used to determine the size of the PCR amplicons. For product size confirmation and yield estimation, 5 µL of the PCR products were loaded onto 1% agarose gels, subjected to electrophoresis for 45 min at 80 V and stained with ethidium bromide.
Identification of diarrheagenic E. coli using the 16S rRNA gene
The E. coli that was positive for the uidA PCR assay was further screened by 16S rRNA PCR using the following pairs of primers: 27F: AGA GTT TGA TCM TGG CTC AG and 1492R: GGT TAC CTT GTT ACG ACT T [20]. To set up PCR assays for virulent genes, a total of 25 µL reaction mixture consisted of 12.5 µL of the 2X [AmpliTaq Gold® DNA Polymerase, 0.05 units/L, Gold buffer, 930 mM Tris/HCl pH 8.05, 100 mM KCl, 0.4 mM of each dNTP, and 5 mM MgCl2], 10 µM of each primer, 2 µL of template DNA and 8.5 µL nuclease-free water. PCR conditions were optimized as follows: initial denaturation step at 96 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 30 s and extension at 72 °C for 1 min and finally a single and final extension step at 72 °C for 10 min. The representative PCR products were cleaned up using ExoSAP-IT (ThermoScientific, USA), subjected to cycle sequencing using the BigDye Terminator v3.1 kit (ThermoScientific, USA), and sequenced on the SeqStudio genetic analyzer at North‒West University, Potchefstroom, South Africa.
Detection of virulence genes
PCR was used to detect the presence of virulence genes, including the eaeA gene of enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC) lt, enteroinvasive E. coli (EIEC) vir, enteroaggregative E. coli (EAEC) aafII and Shiga toxin-producing E. coli (STEC) (stx1 and stx2). The reaction was carried out in a total volume of 25 μL consisting of 12.5 μL of 2X DreamTaq Green Master Mix (New England Biolabs, USA), 8.5 μL of RNase-nuclease free PCR water, 1 μL of each primer and 2 μL of template DNA. The PCR conditions were performed as described and optimized by Omolajaiye et al. [19] with slight modifications. The confirmation of virulence genes in the isolates are shown in Table 1. The detection of the stx1 and stx2 genes was performed using a multiplex PCR assay [21].
Table 1.
Primer pairs used in this study to determine the E. coli pathotypes of the isolates
| Pathotype | Target Gene | Primer | Sequence of primers (5–3′) | Amplification length (bp) | Annealing temp. (°C) | Reference |
|---|---|---|---|---|---|---|
| ETEC | lt | FW | GCACACGGAGCTCCTCAGTCTCC | 218 | 56 | [19] |
| RV | TTCATCCTTTCAATGGCTTT | |||||
| EAEC | aafII | FW | CACAGGCAACTGAAATAAGTCTGG | 378 | 56 | [19] |
| RV | ATTCCCATGATGTCAAGCACTTC | |||||
| EIEC | vir | FW | AGCTCAGGCAATGAAACTTTGAC | 822 | 61 | [19] |
| RV | TGGGCTTGATATTCCGATAAGTC | |||||
| EIEC | eaeA | EAE1 | TCAATGCAGTTCCGTTATCAGTT | 450 | 55 | [19] |
| EAE2 | GTAAAGTCCGTTACCCCAACCTG | |||||
| STEC | stx1 | EVT1 | CAACACTGGATGATCTCAG | 349 | 55 | [21] |
| EVT2 | CCCCCTCAACTGCTAATA | |||||
| STEC | stx2 | EVS1 | ATCAGTCGTCACTCACTGGT | 110 | 55 | [21] |
| EVS2 | CTGCTGTCACAGTGACAAA |
Antimicrobial susceptibility testing and ESBL detection
A Kirby-Bauer disc diffusion technique was used to determine the antimicrobial susceptibility profile of the isolates [22]. The antimicrobial agents selected for this study are some of the most commonly used prophylactics in small and large stocks in South Africa. Aminoglycosides [streptomycin (10 μg)], fluoroquinolones [ciprofloxacin (5 μg)], nalidixic acid (30 μg), macrolides [erythromycin (15 μg)], penicillin beta-lactam [ampicillin (10 μg)], polymyxins [colistin sulfate (300 μg)], and phenicols [chloramphenicol (30 μg)] were obtained from Mast Diagnostics, UK. The pure E. coli isolates were inoculated in nutrient broth and incubated at 37 °C for 24 h. The bacterial suspension was spread onto sterile Muller-Hinton agar (MHA) plates using a sterile spreader and allowed to dry for 10 min at room temperature. Then, the antibiotic discs were placed onto the agar plates and incubated at 37 °C for 24 h. After incubation, the diameter of the zone around the colony was measured and compared with the Clinical and Laboratory Standards Institute (CLSI) to categorize it into resistant (R), intermediate (I) and susceptible (S) [23]. A multidrug-resistant isolate was defined as any isolate showing resistance to more than two classes of antibiotics [24]. Escherichia coli ATCC 25922 was used as a reference for quality control in the antimicrobial susceptibility test. The ESBL-resistance phenotype was detected using the double disc synergy test (DDST). Using Muller-Hinton agar (Oxoid, UK), pairs of disks containing 30 mg of ceftaxime (CTX) and 30 mg of ceftazidime (CAZ) were inoculated with 20/10 mg of amoxicillin-clavulanic acid (AMC) through the same inoculated plate. A positive test result was defined as a 5 mm increase in the zone diameter compared to that of a disk without clavulanic acid [23]. It was classified as multidrug resistant (MDR) when it was resistant to at least three antimicrobial groups.
Detection of antibiotic resistance genes by PCR
The gDNA extracted from E. coli isolates was screened for the presence of the antibiotic resistance genes tetracycline (tetA, tetO, tetX, tetP, tetW and tetK), erythromycin (ermB), colistin (mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5), chloramphenicol (catI, catII, catIII, catIV, and floR), sulfonamide (sulI, sulII and sulIII), quinolone (qnrA, qnrD, qnrS and parC), and aminoglycoside (strA, strB and aadA). To set up PCR assays for antibiotic resistance genes, a 25 µL reaction mixture consisted of 12.5 µL of the PCR Master Mix (AmpliTaq Gold® DNA Polymerase, 0.05 units/L, Gold buffer, 930 mM Tris/HCl pH 8.05, 100 mM KCl, 0.4 mM of each dNTP, and 5 mM MgCl2), 10 µM of each primer, 2 µL of template DNA, and 8.5 µL nuclease-free water. The amplification program consisted of the following steps: 94 °C for 5 min, followed by 35 cycles of amplification divided into 1 min of denaturation at 94 °C, 1 min of primer annealing at the temperature according to Supplementary Table S1, and 1 min of primer extension at 72 °C, followed by a 10 min final extension step at 72 °C. Furthermore, multiplex PCR screening for mcr-1 to 5 colistin-encoding genes was performed as described previously [25]. Electrophoresis of PCR amplicons was conducted as described above.
Molecular screening of β-lactamase-encoding genes
The E. coli isolates were screened for the β-lactam-encoding genes ampC, blaSHV, blaOXA, blaCARB, blaTEM, blaCTX-M, blaCTX-M-1 group, blaCTX-M-2 group, blaCTX-M-8 group, blaCTX-M-9 group, blaCTX-M-15 group and blaCTX-M-25 group using primers described previously [26–28]. To set up PCR assays for β-lactam-encoding genes, a total of 25 µL reaction mixture consisted of 12.5 µL of the PCR Master Mix (AmpliTaq Gold® DNA Polymerase, 0.05 units/L, Gold buffer, 930 mM Tris/HCl pH 8.05, 100 mM KCl, 0.4 mM of each dNTP, and 5 mM MgCl2), 10 µM of each primer, 2 µL of template DNA and 8.5 µL nuclease-free water. The expected PCR product sizes are listed in Supplementary Table S1.
Data analysis
Statistical analysis was carried out using Microsoft Excel 2016 (Microsoft Corporation, Redmond, DC, USA) and Statistical Package for the Social Sciences v. 26 (IBM Corporation, Armonk, NY, USA). The sequenced 16S rRNA gene of the representative isolates were aligned to nucleotide sequences available in GenBank and identified by comparing them with those available in the National Center for Biotechnology Information database (NCBI) using BLASTn (http://www.ncbi.nlm.nih.gov/BLAST/). Heatmap plots of the virulence and antibiotic resistance profiles were generated using ChipPlot (https://www.chiplot.online/#).
Results
Identification of d E. coli
In this study, nonduplicated (one isolate per sample) isolates of E. coli were recovered from 53 fecal samples. All 53 E. coli isolates were confirmed by the uidA gene PCR assay. However, the two isolates did not possess the uidA gene. A high (64.9%) incidence of E. coli was observed in sheep samples and a comparatively low occurrence was observed in goats (35.1%).
Nucleotide sequence identity
The 16S rRNA gene sequence analysis of the E. coli isolates (n = 3) revealed a high percentage of nucleotide similarity (96.4 to 97.5%) to the reference GenBank sequences of the E. coli strains. The BLASTn results of 16S rRNA gene E. coli detected in this study (GenBank accession number: OR123648, OR123649 and OR123650) confirmed that it matches with relevant E. coli species on the NCBI database (GenBank accession numbers: KY458548.1, CP041429.1 (STEC367) and CP063518.1).
Detection of virulence genes in E. coli
Six virulence genes were screened among the 53 E. coli isolates identified by PCR. Approximately 14 of the isolates were considered to belong to the E. coli O177 serogroup, as they contained the wzy gene. The Shiga toxin gene stx2 (100%) was the most commonly detected gene, followed by stx1 (47.2%), vir (18.9%) and (16.9%) eaeA, while lt and aafII were not detected. The majority (43.4%) of the isolates carried a combination of stx2 + stx1, 5.7% carried a combination of stx1 + stx2 + eaeA, 22.6% harbored a mix of stx1 + stx2 + eaeA genes and 7.5% of the isolates possessed a mix of stx1 + stx2 + vir genes.
Antibiotic susceptibility and resistance genes detected in E. coli isolates
Antimicrobial susceptibility tests revealed that all 53 E. coli isolates were resistant to nalidixic acid and ampicillin. Most of the isolates also showed resistance to erythromycin (66.04%; n = 37), colistin sulfate (43.4%; n = 23), chloramphenicol (9.4%; n = 5) and ciprofloxacin (1.9%; n = 1). In contrast, all isolates were susceptible to streptomycin. It was observed that 94.1% (n = 43/53) of the E. coli isolates were resistant to at least three classes of antibiotics and were all considered MDR.
The E. coli isolates (n = 53) from sheep and goat fecal samples harbored the antibiotic resistance genes sulII 46 (86.8%), mcr-4 33 (62.3%), floR 33 (62.3%), mcr-1 28 (52.8%), tet(A) 23 (43.4%), sulI 22 (41.5%), tet(O) 11 (20.8%), tet(W) 10 (18.9%), parC 6 (11.3%), mcr-2 6 (11.3%), ampC 5 (9.4%), qnrS 5 (9.4%), and ermB 3 (5.7%). Meawhile the tet(X), tet(P), ermB, mcr-3, mcr-5, catI, catII, catIII, catIV, sulIII, qnrA, while qnrD genes were not detected in all 53 isolates (Table 2). Most (28.3%) of the isolates carried a combination of mcr-1 + mcr-4 + sulII, while 20.8% and 3.8% possessed tet(A) + mcr-1 + mcr-4 + sulII and a combination of tet(W)9 + mcr-1 + floR + sulII resistance genes, respectively. In this study, correlations between phenotypic antibiotic resistance patterns and antibiotic resistance genes encoding four antibiotics, nalidixic acid, erythromycin, chloramphenicol, ampicillin, and colistin sulfate, were observed in the isolates (Fig. 1).
Table 2.
Antibiotic resistance phenotypes, genotype and prevalence of β-lactamase-encoding genes in E. coli isolates
| No | Isolate codes | Resistance phenotype | Resistance genotype | β-lactamase-encoding genes |
|---|---|---|---|---|
| 1 | S1DEC* | AMP, ERY, CS, CHL, NA | tet(A), mcr-1, floR, sulI, sulII | blaSHV, CARB, TEM, CTX-M, CTX-M-2, CTX-M-9, CTX-M-25, CTX-M-15 |
| 2 | S2DEC | AMP, ERY, NA | tet(W), mcr-4, ampC, floR, sulII | CTX-M-15 |
| 3 | G1DEC | AMP, ERY, CS, NA | mcr-1, mcr-2, mcr-4, ermB, sulI, sulII, parC | TEM, CTX-M, CTX-M-2, CTX-M-9 CTX-M-25, CTX-M-15 |
| 4 | S3DEC | AMP, NA | tet(A), mcr-1, mcr-3, mcr-4, floR, sulI, sulII | SHV, CTX-M-25 |
| 5 | G2DEC* | AMP, ERY, NA | tet(A), mcr-1, mcr-4, ampC, sulII | TEM, CTX-M-2, CTX-M-9, CTX-M-25 |
| 6 | G3DEC | AMP, ERY, CS, NA | mcr-1, mcr-2, mcr-4, floR, sulI, sulII | – |
| 7 | S4DEC* | AMP, NA | floR, sulII, parC | SHV, CTX-M, CTX-M-9, CTX-M-25, CTX-M-15 |
| 8 | S5DEC* | AMP, CS, NA | tet(A), mcr-1, mcr-4, | CARB, CTX-M, CTX-M-2, CTX-M-9, CTX-M-25 |
| 9 | S6 DEC* | AMP, ERY, CS, NA | tet(A), mcr-1, ermB, floR, sulI, sulII | SHV, TEM, CTX-M-2, CTX-M-9, CTX-M-25, CTX-M-15 |
| 10 | G4DEC | AMP, ERY, NA | mcr-4, floR, sulII | – |
| 11 | S7DEC | AMP, ERY, NA | tet(A), mcr-2, mcr-4, sulI, sulII | TEM, TEM, CTX-M, CTX-M-25 |
| 12 | S8DEC | AMP, ERY, CHL, NA | mcr-1, mcr-4, floR, sulI, sulII | SHV, CARB, CTX-M-2, CTX-M-9, CTX-M-15 |
| 13 | S9DEC | AMP, NA | mcr-1, mcr-4, sulI, sulII, parC | TEM, CTX-M-9, CTX-M-25 |
| 14 | G5DEC | AMP, ERY, NA | mcr-1, mcr-4, ampC, floR, sulI, sulII parC | – |
| 15 | S11DEC* | AMP, ERY, NA | tet(A), mcr-1, sulII | SHV, TEM, CTX-M, CTX-M-2, CTX-M-9, CTX-M-25, CTX-M-15 |
| 16 | S12DEC | AMP, NA | tet(W), mcr-1, floR, sulII | – |
| 17 | S13DEC | AMP, ERY, | mcr-2, mcr-4, sulII | CARB, CTX-M, CTX-M-25 |
| 18 | S14DEC | AMP, CS, NA | tet(A), sulI, sulII | TEM, CTX-M-9, CTX-M-25, CTX-M-15 |
| 19 | S158DEC | AMP, NA | mcr-4, floR, sulII | CTX-M-9 |
| 20 | S15DEC* | AMP, ERY, CS, NA | tet(A), mcr-1, mcr-4, floR, sulII | CARB, TEM, CTX-M-9, CTX-M-25, CTX-M-15 |
| 21 | S16DEC | AMP, ERY, CS, NA | tet(A), mcr-4, floR, sulI, sulII | CTX-M-15 |
| 22 | S17DEC | AMP, ERY, NA | tet(A), tet(W), mcr-4, floR, | TEM, CTX-M, CTX-M-25 |
| 23 | G7DEC | AMP, ERY, NA | mcr-1, mcr-4, | CARB, CTX-M-25, CTX-M-15 |
| 24 | S18DEC | AMP, CS, NA | tet(W), mcr-1, mcr-4, floR, sulI, sulII | CTX-M, CTX-M-15 |
| 25 | S19DEC | AMP, NA | tet(W), mcr-4, floR, sulI, sulII | – |
| 26 | S20DEC | AMP, ERY, CS, NA | tet(A), mcr-1, floR, sulI, sulII | SHV, CTX-M-2, CTX-M-9, CTX-M-25 |
| 27 | S21DEC | AMP, CS, NA | tet(W), mcr-1, floR, sulI, sulII | CTX-M-25 |
| 28 | G8DEC | AMP, CS, NA | tet(A), mcr-1, mcr-4, floR, sulII | CARB, CTX-M |
| 29 | S22DEC | AMP, ERY, NA | tet(A), mcr-4, sulI, sulII | CTX-M-9, CTX-M-25 |
| 30 | S23DEC | AMP, ERY, CS, CHL, NA | tet(A), mcr-1, sulII | CTX-M-25 |
| 31 | S24DEC | AMP, ERY, CS, NA | tet(A), mcr-4, ampC, sulII | – |
| 32 | S25DEC* | AMP, NA | tet(A), floR, sulII | SHV, CARB, CTX-M-2, CTX-M-9, CTX-M-25 |
| 33 | S26DEC | AMP, ERY, CS, | mcr-4, floR, sulII | TEM, CTX-M-15 |
| 34 | S27DEC | AMP, CS, NA | CTX-M-9, CTX-M-15 | |
| 35 | S28DEC | AMP, ERY, CS, NA | floR, sulI, sulII | CTX-M-9, CTX-M-15 |
| 36 | S29DEC* | AMP, ERY, CS, NA | mcr-4, sulI, sulII | TEM, CTX-M-2, CTX-M-9, CTX-M-25 |
| 37 | S30DEC | AMP, CIP, ERY, NA | tet(A), tet(W), ermB, floR, sulII | – |
| 38 | S31DEC | AMP, ERY, NA | tet(A), floR, sulII, parC | CTX-M-9, CTX-M-15 |
| 39 | S32DEC | AMP, ERY, CS, NA | tet(A), mcr-1, mcr-4, sulI, sulII | – |
| 40 | S33DEC* | AMP, CS, NA | tet(W), mcr-1, mcr-4, sulII | CARB, CTX-M-2, CTX-M-9, CTX-M-25 |
| 41 | S34DEC | AMP, ERY, NA | mcr-4, sulII | TEM, CTX-M-15 |
| 42 | S35DEC | AMP, ERY, NA | tet(A), mcr-1, mcr-2, mcr-4, floR, sulI, sulII | SHV, CTX-M |
| 43 | G9DEC* | AMP, ERY, NA | sulII | CARB, CTX-M-2, CTX-M-9, CTX-M-25 |
| 44 | S36DEC | AMP, ERY, NA | mcr-1, floR, sulII | SHV |
| 45 | S37DEC* | AMP, ERY, NA | tet(A), mcr-4, floR, sulII | CTX-M-2, CTX-M-9, CTX-M-25 |
| 46 | S38DEC | AMP, NA | tet(W), floR, sulII | CTX-M-15 |
| 47 | S39DEC | AMP, ERY, NA | mcr-4, floR, sulII, parC | SHV, CARB, TEM, CTX-M, CTX-M-25 |
| 48 | G10DEC | AMP, NA | mcr-1, mcr-4, ampC, floR, sulI, sulII | – |
| 49 | S40DEC* | AMP, ERY, NA | mcr-1 sulII | SHV, CTX-M, CTX-M-2, CTX-M-9, CTX-M-25, CTX-M-15 |
| 50 | S41DEC | AMP, ERY, CHL, NA | mcr-1, mcr-2, sulII | TEM, CTX-M-9, CTX-M-15 |
| 51 | S42DEC | AMP, ERY, NA | mcr-4, floR, sulI, | CTX-M, CTX-M-2, CTX-M-9 |
| 52 | G11DEC* | AMP, ERY, CS, NA | tet(A) | CARB, CTX-M-9, CTX-M-25, CTX-M-15 |
| 53 | S43DEC* | AMP, ERY, CS, CHL, NA | tet(W) | SHV, TEM, CTX-M-2, CTX-M-25 |
S streptomycin, CIP ciprofloxacin, NA nalidixic acid, E erythromycin, AMP ampicillin, CS colistin sulfate, CHL chloramphenicol
*ESBL-producing E. coli
Fig. 1.
Heatmap showing the antibiotic resistance profiles (phenotype and genotype) from the E. coli isolated from fecal samples of sheep and goats. Black indicates the presence of an antibiotic resistance phenotype, while brown represents the presence of antibiotic resistance genes. https://www.chiplot.online/
Prevalence of ESBL-producing E. coli isolates
The phenotypes of ESBL-producing E. coli were recovered from 15 samples, of which 3/15 (20%) were from goats and 12/15 (80%) were from sheep. Therefore, 38/52 (71.7%) E. coli isolates were excluded from further screening for ESBL. Of the 15 ESBL-positive E. coli isolates, 15/15 (100%) harbored both the blaCTX-M-9 and blaCTX-M-25 genes. Meanwhile, 13/15 (81.3%) harbored the blaCTX-M-2 gene, 10/15 (62.5%) blaSHV, 7/15 (43.8%) blaCARB, 7/15 (43.8%) blaTEM, 7/15 (13.2%) blaCTX-M-15 and 5/15 (31.3%) blaCTX-M. All the isolates were negative for the blaOXA, blaCTX-M-1, and blaCTX-M-8 groups (Table 2). Thirteen (81.3%) of the ESBL-producing E. coli isolates were considered MDR.
Coexistence of ESBL-producing E. coli and virulence genes
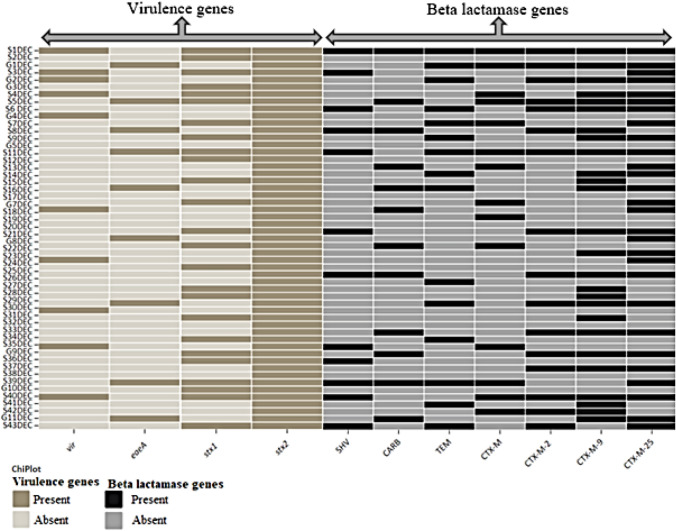
Only 16 isolates consisted of one virulence gene (stx2) and two bla genes (blaCTX-M-9 and blaCTX-M-25 groups), while six isolates harbored two virulence genes (stx1 and stx2) and two bla genes (blaCTX-M-9 and blaCTX-M-25 groups). One isolate (S1DEC) harbored all the bla genes screened in this study and three virulent genes (stx1, stx2 and vir). Another isolate (S4DEC) possessed four vir + eaeA + stx1 + stx2 virulence genes and three bla genes (blaCTX-M-9 + blaCTX-M-25 + blaCTX-M-15) (Fig. 2). The blaCTX-M-25 and blaCTX-M-9 groups, as well as stx1 and stx2, had the most genes detected in the E. coli strains isolated in this study.
Fig. 2.
Heatmap showing the clustering of the beta-lactamase and virulence genes in E. coli strains isolated from fecal samples of sheep and goats. Dark and light gray colors indicate the presence and absence of the genes, respectively. https://www.chiplot.online/
Discussion
This study investigated the existence of STEC in sheep and goats as well as the isolates’ capacity to produce extended-spectrum beta-lactamase (ESBL) and the Shiga toxin. Cattle, goats and sheep serve as natural reservoirs for E. coli in their guts [29]. As a result of their virulence factors, diarrheagenic E. coli strains cause diarrhea in both animal and human hosts [30]. In the present study, the uidA gene was used to confirm the isolates as E. coli. All the E. coli strains obtained from this study were positive for at least one of the virulence genes.
In this study, the Shiga toxin gene stx2 (100%) was the most detected gene, followed by stx1 (47.2%), as well as enteroinvasive E. coli (vir) 18.9%, and enterohemorrhagic E. coli (eaeA) 16.9% virulence genes. This observation was different to the findings of the previous studies conducted in South Africa, where stx1 had the highest prevalence in human [31]. Different findings were reported by Dela et al. [32] in Ghana, whereby none of the E. coli isolates tested positive for stx1, stx2 and eaeA. The combination of stx2/stx1 genes was observed in (43.4%) of the isolates. This is the most relevant finding because it suggests that STEC strains are circulating among sheep and goats. Furthermore, strains harboring the stx2 gene are considered more virulent [33–35]. Additionally, the strain carrying stx1 may cause diarrhea in immunocompromised individuals [33]. Nine isolates of E. coli (16.9%) consisted of the eaeA gene. This is a gene of enteropathogenic E. coli (EPEC) that is necessary for intimate attachment to epithelial cells [36]. These findings are higher than those reported in China, where 9.5% of E. coli strains harbored this gene [37]. The eaeA gene encodes an outer membrane that mediates the adhesion of STEC/EPEC to the intestinal epithelium [34]. Enterotoxigenic E. coli (ETEC) strains expressing the eaeA gene are potentially capable of causing attaching and effacing lesions in humans [38]. In this study, the eaeA gene was detected in 9.4% of the E. coli isolates. The study conducted by Ercoli et al. [39] in Italy reported a high prevalence of this gene (50%) in swine fecal samples, in South Africa Ramatla et al. [40], reported a high prevalence of this gene (14%) in Rattus species fecal samples. Variations in detection and isolation methods of STEC strains may also contribute to differences in prevalence rates [39, 41]. The ETEC strains that contain the lt and aafII genes were not detected in this study. These results are in agreement with the reports of other studies from South Africa and Iran, where these genes were not detected in children with acute diarrhea samples and riding horse samples, respectively [19, 42]. However, the observations differ from the study conducted in Turkey, whereby the lt gene was detected in one isolate from clinical mastitis bovine milk [38].
Globally, diarrheagenic bacterial pathogens are becoming increasingly resistant to antimicrobials, especially in less developed areas [19, 43]. Resistance to a majority of antibiotics has been developing as a result of improper use and overuse of antimicrobials [44]. In this study, all E. coli strains were resistant to nalidixic acid and ampicillin. The majority of the isolates also showed resistance to erythromycin (66.04%), colistin sulfate (43.4%), chloramphenicol (9.4%) and ciprofloxacin (1.9%). Resistance to antibiotics, especially chloramphenicol, ampicillin and tetracycline [39] and fluoroquinolones [15], has been reported in previous studies on E. coli isolates. The correlation between phenotypic and antimicrobial resistance genes encoding four antibiotics, nalidixic acid, erythromycin, chloramphenicol, ampicillin and colistin sulfate, was observed in this study.
Antimicrobial resistance genes allow bacteria to survive and resist the effects of antibiotics [45]. These genes can be passed on to future generations of bacteria, making them more resistant to antibiotics and leading to the development of superbugs [46]. This is a serious health concern, as it reduces the effectiveness of antibiotics and can result in increased mortality rates. The majority of the isolates harbored multiple resistance genes and some contained up to six antibiotic resistance genes. These genes were associated with resistance to multiple classes of antibiotics, including erythromycin, colistin sulfate, chloramphenicol, and ciprofloxacin. In the current study, 94.1% (n = 43/53) of the E. coli isolates were resistant to multiple antibiotics. This suggests that antibiotic resistance is widespread in E. coli isolates. The MDR bacteria are resistant to multiple antibiotics, so infections caused by them can be difficult to treat. This can lead to prolonged hospital stays, increased medical costs and even death in some cases [47].
Antibiotic resistance genes such as sulII, mcr-4, floR, and mcr-1 were the most frequently detected in 46 (86.8%), 33 (62.3%), 33 (62.3%) and 28 (52.8%) E. coli isolates, respectively. The colistin mcr-1, mcr-2 and mcr-4 resistance genes were detected in this study. This is because colistin is utilized to stimulate animal growth [48] and it can be found in concentrations that are too high for humans to consume safely in some countries. Despite having only 6% of the world’s population, Brazil, China, India, Russia and South Africa together account for 13% of colistin use, according to a statistical analysis from 2000 to 2010 [49]. Due to its success in treating infections that are resistant to antibiotics, colistin use has increased dramatically in these nations in recent years.
Resistance to antibiotics belonging to the beta-lactam group has been increasing recently, probably due to the high prescription of these antibiotics [50]. There is also an increasing recognition of livestock carrying extended-spectrum beta-lactamase-producing Escherichia coli as a potential source of the spread of these microorganisms to humans, where livestock and humans share the same residence [51]. This increase in resistance is due to the overuse of these antibiotics. As a result, these antibiotics become less effective in treating bacterial infections. In our study, 15 (28.8%) ESBL-E. coli isolates were identified. As expected, of the 15 ESBL-E. coli isolates, blaCTX was the most prevalent gene in our ESBL-positive isolates. The blaCX-M-9 and blaCX-M-25 genes were found to be the most prevalent ESBL-encoding genes in E. coli isolates, and their prevalence was 100%. The blaCTX-M-15, which remains the most widely disseminated genotype worldwide, was detected in seven ESBL-producing E. coli isolates. There was a low frequency of E. coli carrying the blaCTX-M gene (5: 31.3%) among our isolates, which is similar to what has been reported by Nakhaei et al. [52] in Iran. Differences in animal husbandry practices and the use of antibiotics in food animals may also play a role in the varying rates. Thus, there is a need for increased surveillance and preventive measures to reduce the spread of antibiotic resistance.
Conclusion
This is the first study to report the prevalence of STEC, including the ESBL producing E. coli (STEC) in sheep and goats in South Africa. The E. coli strains from this study displayed high levels of resistance traits against the erythromycin and colistin sulphate antibiotic groups. This is especially concerning since E. coli is a common commensal in humans and is known to cause severe gastrointestinal infections. An interesting point to highlight from the results of this study is the presence of STEC isolates expressing a combination of the wzy/eaeA/stx1/stx2 virulent genes and three bla genes (CTX-M-9/CTX-M-25/CTX-M-15). The high proportion of the mcr and stx2 gene detected in E. coli represents a serious public-health threat. This highlights the need for increased vigilance and proactive measures to ensure the safety of the population from these potentially harmful pathogens.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank farmers at Matlwang for their cooperation.
Abbreviations
- STEC
Shiga toxin producing Escherichia coli
- EHEC
Enterohemorrhagic E. coli
- ETEC
Enterotoxigenic E. coli
- EIEC
Enteroinvasive E. coli
- EAEC
Enteroaggregative E. coli
- AMR
Antimicrobial resistance
- MDR
Multidrug resistant
- CLSI
Clinical and laboratory standards institute
- WHO
World Health Organization
- DNA
Deoxyribonucleic acid
- ESBL
Extended spectrum beta lactamase
- BLAST
Basic local alignment search tool
- SMA
Sorbitol MacConkey agar
- NCBI
National center for biotechnology information
- PCR
Polymerase chain reaction
Author contributions
TR, OT and KL, study conceptualization and design. TR, TM, MT and LdeW performed laboratory analysis. KL and OT funding acquisition. OT and KL supervised the study. PM, TR and KL, data analysis and curation. TR interpreted the data and drafted the manuscript. All authors reviewed, edited and approved the manuscript.
Funding
Open access funding provided by North-West University. This research was funded by NRF Thuthuka (GUN: 129525) made available to KT.
Data availability
The data and materials of the study will be available from the corresponding author on reasonable request. The sequences of the two strains analyzed were deposited in the National Library of Medicine, National Center for Biology Information (NCBI), GenBank nucleotide sequence database. The accession numbers assigned as OR123648 (https://www.ncbi.nlm.nih.gov/nuccore/OR123648), OR123649 (https://www.ncbi.nlm.nih.gov/nuccore/OR123649) and OR123650 (https://www.ncbi.nlm.nih.gov/nuccore/OR123650).
Declarations
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Allocati N, Masulli M, Alexeyev MF, Di Ilio C (2013) Escherichia coli in Europe: an overview. Int J Environ Res Public Health 10(12):6235–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nada HG, El-Tahan AS, El-Didamony G, Askora A (2023) Detection of multidrug-resistant Shiga toxin-producing Escherichia coli in some food products and cattle faeces in Al-Sharkia, Egypt: one health menace. BMC Microbiol 23(1):127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etcheverria AI, Padola NL (2013) Shiga toxin-producing Escherichia coli: factors involved in virulence and cattle colonization. Virulence 4(5):366–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caprioli A, Tozzoli R, Morabito S, Strockbine NA (2012) Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50(9):2951–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melton-Celsa AR (2014) Shiga toxin (Stx) classification, structure and function. Microbiol Spectr. 10.1128/microbiolspec.EHEC-0024-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahzad A, Ullah F, Irshad H, Ahmed S, Shakeela Q, Mian AH (2021) Molecular detection of Shiga toxin-producing Escherichia coli (STEC) O157 in sheep, goats, cows and buffaloes. Mol Biol Rep 48(8):6113–6121 [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Vázquez AV, Vázquez-Villanueva J, Leyva-Zapata LM, Barrios-García H, Rivera G, Bocanegra-García V (2021) Multidrug resistance of Escherichia coli strains isolated from Bovine Feces and Carcasses in Northeast Mexico. Front Vet Sci 8:643802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandujano A, Cortés-Espinosa DV, Vásquez-Villanueva J, Guel P, Rivera G, Juárez-Rendón K, Cruz-Pulido WL, Aguilera-Arreola G, Guerrero A, Bocanegra-García V (2023) Extended-spectrum β-Lactamase-producing Escherichia coli isolated from food-producing animals in Tamaulipas. Mexico Antibiotics 12:1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa WF, Paranhos R, Mello MP, Pic˜ao RC, Laport MS (2023) Occurrence of extended-spectrum β-lactamases-producing Escherichia coli isolates over gradient pollution in an urban tropical estuary. Environ Microbiol 2023:1–8 [DOI] [PubMed] [Google Scholar]
- 10.Hesp A, Veldman K, van der Goot J, Mevius D, van Schaik G (2019) Monitoring antimicrobial resistance trends in commensal Escherichia coli from livestock, the Netherlands, 1998 to 2016. Euro Surveill 24(25):1800438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer RS, Finch R, Wegener HC, Bywater R, Walters J, Lipsitch M (2003) Antibiotic resistance—the interplay between antibiotic use in animals and human beings. Lancet Infect Dis 3(1):47–51 [DOI] [PubMed] [Google Scholar]
- 12.Laube H, Friese A, Von Salviati C, Guerra B, Käsbohrer A, Kreienbrock L, Roesler U (2013) Longitudinal monitoring of extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl Environ Microbiol 79(16):4815–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xexaki A, Papadopoulos DK, Alvanou MV, Giantsis IA, Papageorgiou KV, Delis GA, Economou V, Kritas SK, Sossidou EN, Petridou E (2023) Prevalence of antibiotic resistant E. coli strains isolated from farmed broilers and hens in Greece, based on phenotypic and molecular analyses. Sustainability 15(12):9421 [Google Scholar]
- 14.Haddadin RN, Collier PJ, Haddadin S (2023) Phenotypic ESBL and non-phenotypic ESBL isolates of Klebsiella pneumoniae exhibit differing responses to induced antimicrobials resistance and subsequent antibiotic cross-resistance. J Appl Microbiol 134(2):p.lxac082 [DOI] [PubMed] [Google Scholar]
- 15.Hawari AD, Al-Dabbas F (2008) Prevalence and distribution of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Jordan. Am J Anim Vet Sci 3:36–39 [Google Scholar]
- 16.Elbadawi HS, Elhag KM, Mahgoub E, Altayb HN, Ntoumi F, Elton L, McHugh TD, Tembo J, Ippolito G, Osman AY, Zumla A (2021) Detection and characterization of carbapenem resistant Gram-negative bacilli isolates recovered from hospitalized patients at Soba University Hospital. Sudan BMC Microbiol 21(1):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sajeev S, Hamza M, Rajan V, Vijayan A, Sivaraman GK, Shome BR, Holmes MA (2023) Resistance profiles and genotyping of extended-spectrum beta-lactamase (ESBL)-producing and non-ESBL-producing E. coli and Klebsiella from retail market fishes. Infect Genet Evol 2023:105446 [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization (WHO) (2021) WHO integrated global surveillance on ESBL-producing E. coli using a “One Health” approach: implementation and opportunities. World Health Organization, Geneva [Google Scholar]
- 19.Omolajaiye SA, Afolabi KO, Iweriebor BC (2020) Pathotyping and antibiotic resistance profiling of Escherichia coli isolates from children with acute diarrhea in amatole district municipality of Eastern Cape. South Africa Biomed Res Int 18(2020):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariam SH (2021) Isolation and characterization of gram-negative bacterial species from pasteurized dairy products: potential risk to consumer health. J Food Qual 2021:1 [Google Scholar]
- 21.Wolde A, Deneke Y, Sisay T, Mathewos M (2022) Molecular characterization and antimicrobial resistance of pathogenic Escherichia coli strains in children from Wolaita Sodo, Southern Ethiopia. J Trop Med 2022:9166209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496 [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute (CLSI) (2018) Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne [Google Scholar]
- 24.Ramatla T, Taioe MO, Thekisoe OM, Syakalima M (2019) Confirmation of antimicrobial resistance by using resistance genes of isolated Salmonella spp. in chicken houses of North West, South Africa. World Vet J 9(3):158–165 [Google Scholar]
- 25.Jousset AB, Bernabeu S, Bonnin RA, Creton E, Cotellon G, Sauvadet A, Naas T, Dortet L (2019) Development and validation of a multiplex polymerase chain reaction assay for detection of the five families of plasmid-encoded colistin resistance. Int J Antimicrob Agents 53(3):302–309 [DOI] [PubMed] [Google Scholar]
- 26.Liu G, Ding L, Han B, Piepers S, Naqvi SA, Barkema HW, Ali T, De Vliegher S, Xu S, Gao J (2018) Characteristics of Escherichia coli isolated from bovine mastitis exposed to subminimum inhibitory concentrations of cefalotin or ceftazidime. Biomed Res Int 2018:4301628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramatla T, Mileng K, Ndou R, Mphuti N, Syakalima M, Lekota KE, Thekisoe OM (2022) Molecular detection of integrons, colistin and β-lactamase resistant genes in Salmonella enterica serovars enteritidis and typhimurium isolated from chickens and rats inhabiting poultry farms. Microorganisms 10(2):313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gundran RS, Cardenio PA, Villanueva MA, Sison FB, Benigno CC, Kreausukon K, Pichpol D, Punyapornwithaya V (2019) Prevalence and distribution of bla CTX-M, bla SHV, bla TEM genes in extended-spectrum β-lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet Res 15:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persad AK, Lejeune JT (2015) Animal reservoirs of Shiga toxin-producing Escherichia coli. In: Hovde CJ, Sperandio V (eds) Enterohemorrhagic Escherichia coli and other Shiga toxin-producing E. coli. ASM Press, Washington, pp 211–230 [Google Scholar]
- 30.Ali DA, Tesema TS, Belachew YD (2021) Retracted article: molecular detection of pathogenic Escherichia coli strains and their antibiogram associated with risk factors from diarrheic calves in Jimma Ethiopia. Sci Rep 11(1):1–15 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Karama M, Mainga AO, Cenci-Goga BT, Malahlela M, El-Ashram S, Kalake A (2019) Molecular profiling and antimicrobial resistance of Shiga toxin-producing Escherichia coli O26, O45, O103, O121, O145 and O157 isolates from cattle on cow-calf operations in South Africa. Sci Rep 9(1):1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dela H, Egyir B, Majekodunmi AO, Behene E, Yeboah C, Ackah D, Bongo RN, Bonfoh B, Zinsstag J, Bimi L, Addo KK (2022) Diarrhoeagenic E. coli occurrence and antimicrobial resistance of extended spectrum beta-lactamases isolated from diarrhoea patients attending health facilities in Accra, Ghana. PLoS ONE 17(5):e0268991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrokh C, Jordan K, Auvray F, Glass K, Oppegaard H, Raynaud S, Thevenot D, Condron R, De Reu K, Govaris A, Heggum K (2013) Review of Shiga-toxin-producing Escherichia coli (STEC) and their significance in dairy production. Int J Food Microbiol 162(2):190–212 [DOI] [PubMed] [Google Scholar]
- 34.Montso PK, Mlambo V, Ateba CN (2019) The first isolation and molecular characterization of Shiga toxin-producing virulent multi-drug resistant atypical enteropathogenic Escherichia coli O177 serogroup from South African Cattle. Front Cell Infect Microbiol 9:333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karam M, Mainga AO, Cenci-Goga BT, Malahlela M, El-Ashram S, Kalake A (2019) Molecular profiling and antimicrobial resistance of Shiga toxin-producing Escherichia coli O26, O45, O103, O121, O145 and O157 isolates from cattle on cow-calf operations in South Africa. Sci Rep 9(1):11930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, Kaper JB, Levine MM (1993) Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Investig 92(3):1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Bai X, Zhang J, Sun H, Fu S, Fan R, He X, Scheutz F, Matussek A, Xiong Y (2020) Escherichia coli strains producing a novel Shiga toxin 2 subtype circulate in China. Int J Med Microbiol 310(1):151377 [DOI] [PubMed] [Google Scholar]
- 38.Yarar M, Turkyilmaz S (2019) Investigation of antibiotic resistance and important virulence genes of Escherichia coli isolated from clinical mastitic bovine milk. Isr J Vet Med 74(2):74–81 [Google Scholar]
- 39.Ercoli L, Farneti S, Zicavo A, Mencaroni G, Blasi G, Striano G, Scuota S (2016) Prevalence and characteristics of verotoxigenic Escherichia coli strains isolated from pigs and pork products in Umbria and Marche regions of Italy. Int J Food Microbiol 232:7–14 [DOI] [PubMed] [Google Scholar]
- 40.Ramatla TA, Mphuthi N, Ramaili T, Taioe M, Thekisoe O, Syakalima M (2022) Molecular detection of zoonotic pathogens causing gastroenteritis in humans: Salmonella spp., Shigella spp. and Escherichia coli isolated from Rattus species inhabiting chicken farms in North West Province South Africa. S Afr Vet Assoc 93(2):63–69 [DOI] [PubMed] [Google Scholar]
- 41.Tseng M, Fratamico PM, Manning SD, Funk JA (2014) Shiga toxin-producing Escherichia coli in swine: the public health perspective. Anim Health Res Rev 15(1):63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reshadi P, Heydari F, Ghanbarpour R, Bagheri M, Jajarmi M, Amiri M, Alizade H, Badouei MA, Sahraei S, Adib N (2021) Molecular characterization and antimicrobial resistance of potentially human-pathogenic Escherichia coli strains isolated from riding horses. BMC Vet Res 17(1):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heidary M, Momtaz H, Madani M (2014) Characterization of diarrheagenic antimicrobial resistant Escherichia coli isolated from pediatric patients in Tehran. Iran Iran Red Crescent Med J 16(4):e12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rather IA, Kim BC, Bajpai VK, Park YH (2017) Self-medication and antibiotic resistance: crisis, current challenges and prevention. Saudi J Biol Sci 24(4):808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haenni M, Dagot C, Chesneau O, Bibbal D, Labanowski J, Vialette M, Bouchard D, Martin-Laurent F, Calsat L, Nazaret S, Petit F (2022) Environmental contamination in a high-income country (France) by antibiotics, antibiotic-resistant bacteria and antibiotic resistance genes: status and possible causes. Environ Int 159:107047 [DOI] [PubMed] [Google Scholar]
- 46.García J, García-Galán MJ, Day JW, Boopathy R, White JR, Wallace S, Hunter RG (2020) A review of emerging organic contaminants (EOCs), antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in the environment: Increasing removal with wetlands and reducing environmental impacts. Bioresour Technol 307:123228 [DOI] [PubMed] [Google Scholar]
- 47.Van Duin D, Paterson DL (2016) Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin 30(2):377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar H, Chen BH, Kuca K, Nepovimova E, Kaushal A, Nagraik R, Bhatia SK, Dhanjal DS, Kumar V, Kumar A, Upadhyay NK (2020) Understanding of colistin usage in food animals and available detection techniques: a review. Animals 10:1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laxminarayan R, Matsoso P, Pant S, Brower C, Røttingen JA, Klugman K, Davies S (2016) Access to effective antimicrobials: a worldwide challenge. Lancet 387(10014):168–175 [DOI] [PubMed] [Google Scholar]
- 50.Atta HI, Idris SM, Gulumbe BH, Awoniyi OJ (2022) Detection of extended spectrum beta-lactamase genes in strains of Escherichia coli and Klebsiella pneumoniae isolated from recreational water and tertiary hospital waste water in Zaria, Nigeria. Int J Environ Health Res 32(9):2074–2082 [DOI] [PubMed] [Google Scholar]
- 51.Devi LS, Broor S, Chakravarti A, Chattopadhya D (2020) Livestock manure as potential reservoir of CTX-M type extended-spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae associated with carbapenemase production. J Pure Appl Microbiol 14(1):171–181 [Google Scholar]
- 52.Nakhaei MM, Hashemi BM, Amel JS, Ghahraman M (2014) Genetic properties of blaCTX-M and blaPER β-lactamase genes in clinical isolates of Enterobacteriaceae by polymerase chain reaction. Iran J Basic Med Sci 17(5):378–383 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials of the study will be available from the corresponding author on reasonable request. The sequences of the two strains analyzed were deposited in the National Library of Medicine, National Center for Biology Information (NCBI), GenBank nucleotide sequence database. The accession numbers assigned as OR123648 (https://www.ncbi.nlm.nih.gov/nuccore/OR123648), OR123649 (https://www.ncbi.nlm.nih.gov/nuccore/OR123649) and OR123650 (https://www.ncbi.nlm.nih.gov/nuccore/OR123650).